Abstract
The anti-inflammatory effects of globular adiponectin (gAcrp) are mediated by IL-10/heme oxygenase 1 (HO-1)-dependent pathways. Although full-length (flAcrp) adiponectin also suppresses LPS-induced pro-inflammatory signaling, its signaling mechanisms are not yet understood. The aim of this study was to examine the differential mechanisms by which gAcrp and flAcrp suppress pro-inflammatory signaling in macrophages. Chronic ethanol feeding increased LPS-stimulated TNF-α expression by Kupffer cells, associated with a shift to an M1 macrophage polarization. Both gAcrp and flAcrp suppressed TNF-α expression in Kupffer cells; however, only the effect of gAcrp was dependent on IL-10. Similarly, inhibition of HO-1 activity or siRNA knockdown of HO-1 in RAW264.7 macrophages only partially attenuated the suppressive effects of flAcrp on MyD88-dependent and -independent cytokine signatures. Instead, flAcrp, acting via the adiponectin R2 receptor, potently shifted the polarization of Kupffer cells and RAW264.7 macrophages to an M2 phenotype. gAcrp, acting via the adiponectin R1 receptor, was much less effective at eliciting an M2 pattern of gene expression. M2 polarization was also partially dependent on AMP-activated kinase. flAcrp polarized RAW264.7 macrophages to an M2 phenotype in an IL-4/STAT6-dependent mechanism. flAcrp also increased the expression of genes involved in oxidative phosphorylation in RAW264.7 macrophages, similar to the effect of flAcrp on hepatocytes. In summary, these data demonstrate that gAcrp and flAcrp utilize differential signaling strategies to decrease the sensitivity of macrophages to activation by TLR4 ligands, with flAcrp utilizing an IL-4/STAT6-dependent mechanism to shift macrophage polarization to the M2/anti-inflammatory phenotype.
Keywords: Alcohol, AMP-activated Kinase (AMPK), Chemokines, Gene Expression, Gene Regulation, Innate Immunity, Interferon, Liver Injury, Macrophage, mRNA
Introduction
Resident tissue macrophages exhibit a tremendous plasticity in their response to external stimuli. In particular, the local metabolic and immune microenvironment contributes to this plasticity (1). In response to different stimuli, macrophages express distinct patterns of surface receptors and metabolic enzymes that ultimately generate the diversity of macrophage functions. Although the precise characterization of macrophage diversity is continuing to evolve, two distinct polarization states of macrophages, M1 and M2, have been characterized (1, 2). LPS and IFNγ can promote macrophage differentiation to a “classical” or M1 phenotype (1). The M1 activation pattern is associated with tissue destruction and inflammation and is responsible for up-regulating pro-inflammatory cytokines and increasing production of reactive nitrogen species and reactive oxygen species (1). In contrast, the “alternative” or M2 activation phenotype of macrophages is induced in response to IL-4 and IL-13. M2-polarized macrophages dampen the inflammatory process by producing anti-inflammatory factors, such as IL-10 and TGFβ. M2 macrophages also up-regulate mannose receptors, such as the macrophage mannose receptor (MMR)2 and mannose receptor, C type 2 (Mrc2c), IL-1 receptor antagonist (IL-1RA), and scavenger receptors, like cluster of differentiation 36 (CD36), as well as increased expression of arginase-1 (Arg-1). Arg-1 metabolizes arginine to ornithine and polyamines and thereby diminishes nitric oxide production (1). The M2 phenotype is thought to promote tissue repair after inflammation and/or injury (1).
Macrophages play a central role in resistance against pathogens, tissue remodeling, repair, and resolution of inflammation. However, inappropriate regulation of macrophage function contributes to a variety of chronic inflammatory diseases, such as alcoholic and nonalcoholic liver disease (3). Alcoholic liver disease is a complex process, progressing through several stages of tissue damage and liver dysfunction that are marked by pathological changes in the liver ranging from steatosis and hepatitis to cirrhosis. Chronic ethanol-induced inflammation in the liver is associated with increased sensitivity of Kupffer cells, the resident macrophage in the liver, to activation by TLR4 ligands or phagocytosis, leading to dysregulated expression of pro-inflammatory mediators (4, 5). Activation of TLR4 by LPS and signaling via MyD88-dependent and -independent pathways contribute to chronic ethanol-induced liver injury (6–8). Activation of Kupffer cells in the liver during chronic ethanol exposure is likely due to increased exposure to endotoxins (7), as well as in response to hepatocellular injury (9). Although it is clear that increased sensitivity of Kupffer cells to activation is critical to the progression of ethanol-induced liver injury (10, 11), the mechanisms for sustained inflammatory responses in the liver during chronic ethanol exposure are not well understood but may involve both increased sensitivity to pro-inflammatory signals, as well as an impaired capacity to resolve the inflammatory response.
The resolution of inflammation is a very active, highly coordinated response that is essential to sustain growth and survival in the face of infection and tissue damage (12). Recent evidence points to adipose tissue as a critical nexus in the regulation of immune responses (13). Adipose tissue not only provides an essential store of energetic substrates and inflammatory mediators to support an immune response but also promotes the termination of inflammation (13). Adiponectin (adiponectin is the generic name for all molecular forms of adiponectin), an abundant adipokine, has potent anti-inflammatory properties. Adiponectin reduces macrophage differentiation and migration (14) and chemokine production (15) and suppresses T-cell migration (15). Adiponectin potently desensitizes macrophages to TLR4-dependent MyD88-dependent and -independent signaling (16–20).
Adiponectin is present in the circulation as full-length adiponectin (flAcrp); flAcrp forms a number of oligomeric forms termed low molecular weight, middle molecular weight, and high molecular weight. Full-length adiponectin can be proteolytically cleaved to form globular adiponectin (gAcrp). Here, we will use the general term adiponectin to indicate all these different molecular forms and specify either the flAcrp or gAcrp forms when appropriate.
Although many of these anti-inflammatory responses of adiponectin are mediated by both gAcrp and flAcrp (see for example Refs. 17, 18, 21), it is not entirely understood if there are differential signaling pathways critical to the responses to gAcrp and flAcrp. Recent studies have identified an IL-10/heme oxygenase-1 (HO-1)-dependent pathway that mediates the anti-inflammatory effects of gAcrp in murine macrophages (17, 18), whereas the anti-inflammatory effects of flAcrp are independent of IL-10 in human macrophages (16).
Adiponectin also promotes macrophage polarization toward an anti-inflammatory M2 phenotype both in vivo and in cultured macrophages (22, 23). Adiponectin can either act directly on macrophages to shift polarization (23) or indirectly by priming macrophages to IL-4-mediated M2 polarization (24). Here, we have interrogated the mechanisms for adiponectin-mediated polarization of macrophages, and we report that flAcrp is a very potent inducer of M2 macrophage polarization, both in RAW264.7 macrophages and in primary cultures of rat Kupffer cells. In contrast, gAcrp was relatively ineffective at shifting macrophages to the M2 phenotype. flAcrp required adiponectin receptor 2 (AdipoR2) to effect macrophage polarization; this process was dependent on IL-4 and phosphorylation of STAT6.
EXPERIMENTAL PROCEDURES
Materials
Adult male Wistar rats weighing 140–150 g were purchased from Harlan Sprague-Dawley (Indianapolis, IN). Lieber-DeCarli ethanol diet (regular, number 710260) was purchased from Dyets (Bethlehem, PA). Recombinant human gAcrp expressed in Escherichia coli was purchased from PeproTech, Inc. (Rocky Hill, NJ). Recombinant human full-length Acrp expressed in HEK293 cells was purchased from Biovendor Research and Diagnostic Products (Candler, NC). Adiponectin preparations contained less than 0.2 ng of LPS/μg protein. Recombinant rat IL-10 and recombinant mouse IL-4 were purchased from R&D Systems, Inc. (Minneapolis, MN). Cell culture reagents were from Invitrogen. Antibodies were from the following sources: total ERK1/2 (Upstate Biotechnology; Lake Placid, NY); AMPK, phospho-STAT3, and phospho-STAT6 (Cell Signaling Technology, Inc., Danvers, MA); heat shock protein 70 (Hsc70) (Alpha Diagnostic, San Antonio, TX); STAT3 and STAT6 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-rat IL-4 (R&D Systems); and anti-mouse/human IL-4 (Millipore, Temecula, CA). Anti-rabbit and anti-mouse IgG-peroxidase antibodies were purchased from Roche Applied Science. LPS from E. coli serotype 026:B6 (tissue culture-tested, L-2654) was purchased from Sigma; all experiments were carried out with a single lot of LPS (lot number 064K4077). Mouse bone marrow macrophage and RAW264.7 cell nucleofection kit was purchased from Lonza (Cologne, Germany). Phycoerythrin-conjugated CD206, rat anti-mouse IgG1, and CD32/CD16 Fcγ receptor blocking antibodies were purchased from eBioscience (San Diego). Zinc protoporphyrin was purchased from Frontier Scientific, Inc. (Logan, UT).
Chronic Ethanol Feeding and Kupffer Cell Isolation
All procedures involving animals were approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic. Chronic ethanol feeding to rats, as well as the isolation and culture of Kupffer cells, were performed as described previously (25). Briefly, rats were allowed free access to the Lieber-Decarli high fat complete liquid diet for 2 days and then randomly assigned to pair-fed or ethanol-fed groups. Ethanol-fed rats were allowed free access to a liquid diet containing 17% of the calories (3.3% v/v) from ethanol for 2 days and then a liquid diet containing 35% of the calories (6.6% v/v) from ethanol for 4 weeks. Control rats were pair-fed a liquid diet in which maltose dextrins were substituted isocalorically for ethanol over the entire feeding period. Kupffer cells were isolated and cultured, as described previously (25). For experiments measuring IL-4 protein, Kupffer cells were isolated from chow-fed control rats. Briefly, isolated Kupffer cells were suspended in Connaught Media Research Laboratory (CMRL)-1066 media with 10% FBS, l-glutamine, and antibiotic/antimycotic (CMRL-FBS) at a concentration of 2 × 106 cells/ml. Cell suspensions were immediately plated onto 96-well (0.2 × 106 cells/well for analysis of mRNA) or 24-well (1.5 × 106 cells/well for flow cytometry and Western blot analysis) culture plates. One hour after plating the isolated Kupffer cells, nonadherent cells were removed by aspiration, and fresh media with or without gAcrp or flAcrp were added. After 18 h in culture, cells were treated with or without 100 ng/ml LPS, as indicated in the figure legends.
Nucleofection in Rat Kupffer Cells
Freshly isolated Kupffer cells were transfected using the Amaxa mouse macrophage Nucleofector kit according to the instructions of the manufacturer using the Y-001 program (Lonza, Cologne, Germany), except for the following modifications. Samples were processed individually, and the entire nucleofection procedure for each sample was completed in less than 5 min. For each nucleofection sample, 2 × 106 Kupffer cells were centrifuged for 10 min at 300 × g. The pellet was washed with 1 ml of PBS, collected at 300 × g for 5 min, and then resuspended in 105 μl of mouse macrophage nucleofector solution and transferred to a 1.5-ml Eppendorf tube for a final concentration of ∼2 × 106 cells/100 μl. Cells were then treated or not with 2.0 μg of specific or scrambled siRNA (siRNA sequences are provided in supplemental Table 2), transferred into the electroporation cuvette, and placed in the Nucleofector device. After nucleofection, cells were immediately removed from the cuvette and plated in a 96-well plate (150 μl/well) at 0.5 × 106 cells/well. After 4 h, the cell culture medium was replaced with fresh medium either with or without 45 nm gAcrp or 45 nm flAcrp for 18 h and then treated with or without LPS, as described in the figure legends.
Culture and Nucleofection of RAW264.7 Macrophages
The murine RAW264.7 macrophage-like cell line was routinely cultured in DMEM with 10% fetal bovine serum and penicillin/streptomycin at 37 °C and 5% CO2.
For siRNA knockdown experiments, RAW264.7 cells were transfected using the Amaxa Nucleofector apparatus (Lonza, Cologne, Germany). Briefly, 2 × 106 cells were resuspended in 105 μl of nucleofector solution and were nucleofected with 100 nm of specific or scrambled siRNA in the Nucleofector device using the D-032 program, according to the instructions of the manufacturer. After nucleofection, 500 μl of pre-warmed DMEM was added to the transfection mixture, and cells were transferred to 12-well plates containing 1.5 ml of pre-warmed DMEM per well. Transfected cells were seeded at 10.2 × 104/cm2 in 96-well plates and cultured for 24 h prior to experimental treatments. Validated Silencer Select siRNA pre-designed sequences were purchased from Ambion/Applied Biosystems. Efficiency of knockdown was determined by Western blot analysis and quantitative real time PCR (qRT-PCR) (supplemental Fig. 1).
RNA Isolation and qRT-PCR
Total RNA was isolated and reverse-transcribed, and qRT-PCR amplification was performed. The relative amount of target mRNA was determined using the comparative threshold (Ct) method by normalizing target mRNA Ct values to those of 18 S. RNA was isolated from Kupffer cells and RAW264.7 macrophages using the RNeasy micro kit (Qiagen), with on-column DNA digestion using the RNase-free DNase set (Qiagen) according to the manufacturer's instructions. Total RNA (200–300 ng) was reverse-transcribed using the RETROscript kit (Ambion/Applied Biosystems) with random decamers as primers. qRT-PCR amplification was performed in an Mx3000p (Stratagene, La Jolla, CA) using SYBR Green PCR core reagents (Applied Biosystems; Warrington, UK). All primers used for qRT-PCR analysis were synthesized by Integrated DNA Technologies (Coralville, IA). Statistical analysis of qRT-PCR data was performed using ΔCt values. Details of the primer sequences are provided in supplemental Table 1.
Western Blot Analysis
After experimental treatments, Kupffer cells or RAW264.7 macrophages were washed by cold PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer containing 100 μm sodium orthovanadate. Total cellular extracts (20 μg) were separated in a 10% SDS-polyacrylamide gel and transferred onto PVDF membranes. Membranes were first probed with either phospho-specific STAT3 or STAT6 antibody, followed by secondary antibody conjugated with HRP and then visualized with enhanced chemiluminescence detection reagents (Amersham Biosciences). The membranes were then stripped and reprobed with antibody against total STAT3, STAT6, or HSC70 as a loading control. Signal intensities were quantified by densitometry using ImageJ software (National Institutes of Health, Bethesda).
Flow Cytometry Analysis
After 18 h of culture with or without gAcrp, RAW264.7 cells were gently scraped and adjusted to 1 × 106 cells/ml with culture media. Cells were greater than 90% viable as determined by trypan blue exclusion. The cells were centrifuged at 100 × g for 10 min. The pellet was washed with PBS and resuspended in 100 μl of PBS with 0.1% sodium azide and then blocked with 1.0 μg of anti-mouse CD32/CD16 Fcγ receptor blocking antibody for 15 min at 4 °C. Cells were then stained with 0.5 μg of phycoerythrin-conjugated CD206 or isotype control (phycoerythrin-conjugated IgG1) diluted in PBS containing 0.1% sodium azide for 30 min. Cells were washed twice with PBS, resuspended in 0.5 ml of wash buffer (final concentration ∼106 cells in 0.5 ml), and kept on ice until flow cytometric measurements were performed on a FACScan flow cytometer (Immunocytometry Systems, Mountain View, CA). Data were acquired and processed using FlowJo software (BD Biosciences).
Statistical Analysis
Because of the limited number of Kupffer cells available from each animal, data from several feeding trials are presented in this study. All values are reported as means ± S.E. Data were analyzed by general linear models procedure (SAS; Carey, NC). Data were log transformed, if needed, to obtain a normal distribution. Follow-up comparisons were made by least square means testing.
RESULTS
Differential Role of IL-10 in Mediating the Effects of gAcrp and flAcrp in Kupffer Cells
Chronic ethanol exposure sensitizes Kupffer cells to stimulation with LPS (Fig. 1). Treatment of Kupffer cells from ethanol-fed rats with 45 nm gAcrp normalizes TLR4-dependent cytokine expression via both MyD88-dependent and -independent pathways via an IL-10/HO-1 dependent pathway (Fig. 1) (17, 18). Treatment of Kupffer cells with 45 nm flAcrp also reduced LPS-stimulated TNF-α expression (Fig. 1). This concentration of flAcrp (45 nm or 10 μg/ml) maximally suppresses LPS-stimulated TNF-α mRNA expression in Kupffer cells (25). However, this response was not attenuated by siRNA knockdown of IL-10 expression, compared with Kupffer cells that were not transfected or transfected with scrambled siRNA (Fig. 1).
FIGURE 1.
Anti-inflammatory effects of gAcrp and flAcrp are differentially dependent on IL-10 in Kupffer cells from ethanol- and pair-fed rats. Kupffer cells isolated from ethanol- and pair-fed rats were transfected or not with 100 nm IL-10 siRNA or scrambled siRNA and then cultured with or without 45 nm gAcrp or flAcrp for 18 h. Kupffer cells were then stimulated with 100 ng/ml LPS for 60 min, and TNF-α and 18 S mRNA were measured by qRT-PCR. n = 4. Values with different superscripts (a–d) within each treatment group are significantly different from each other (p < 0.05).
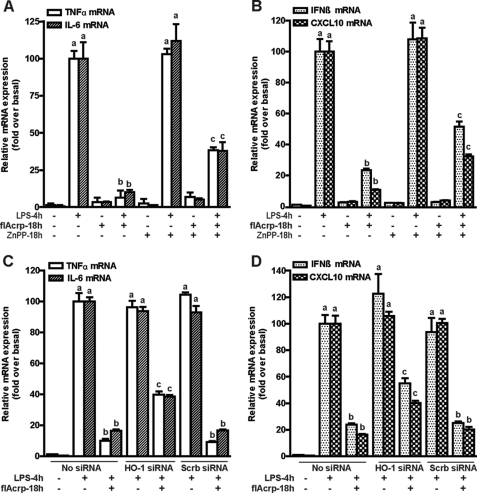
Decreased TLR4-mediated Cytokine Expression by flAcrp Is Only Partially Dependent on HO-1 in RAW264.7 Macrophages
To delineate more precisely the differential molecular mechanisms for the anti-inflammatory effects of gAcrp and flAcrp, studies were conducted in the macrophage-like cell line, RAW264.7. gAcrp suppresses both TLR4-MyD88-dependent and -independent signaling in RAW264.7 macrophages via an IL-10/HO-1 pathway (18). Culture of RAW264.7 macrophages with flAcrp for 18 h also decreased LPS-stimulated expression of MyD88-signature cytokines, TNF-α and IL-6 (Fig. 2, A and C), as well as MyD88-independent signature cytokines/chemokines, IFNβ and chemokine (CXC motif) ligand 10 (CXCL10) (Fig. 2, B and D). Both gAcrp and flAcrp increased expression of HO-1 mRNA and protein in RAW264.7 macrophages (supplemental Fig. 2). Although the anti-inflammatory effects of gAcrp are completely dependent on HO-1 expression (17, 18), inhibition of HO-1, either with the use of a zinc protoporphyrin (Fig. 2, A and B), a chemical inhibitor of HO-1 activity, or by siRNA knockdown of HO-1 expression (Fig. 2, C and D), only partially prevented the anti-inflammatory effects of flAcrp on expression of both MyD88-dependent and -independent cytokine signatures (Fig. 2). Taken together, these data suggest that, in contrast to gAcrp, the anti-inflammatory effects of flAcrp were independent of IL-10 and only partially dependent on HO-1 (Figs. 1 and 2).
FIGURE 2.
HO-1 partially mediates the inhibitory effects of flAcrp on MyD88-dependent and -independent cytokine signatures in RAW264.7 macrophages. A and B, RAW264.7 cells were cultured for 18 h in the absence or presence of 0.5 μm zinc protoporphyrin (ZnPP) in the presence or absence of flAcrp (45 nm). Cells were then stimulated with 100 ng/ml LPS for 4 h and expression of TNFα and IL-6 mRNA (A) and IFN-β and CXCL10 mRNA (B) were normalized to 18 S mRNA. C and D, RAW264.7 cells were transfected or not with 100 nm of HO-1 siRNA or scrambled siRNA and then cultured with or without 45 nm flAcrp for 18 h. Cells were then stimulated with LPS for 4 h. TNFα and IL-6 mRNA (C) and IFN-β and CXCL10 mRNA (D) were normalized to 18 S mRNA. n = 4. LPS-stimulated cells with different superscripts (a–c) are significantly different from each other (p < 0.05).
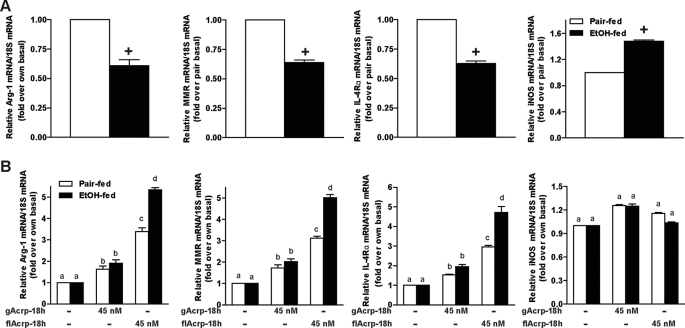
flAcrp Shifts Kupffer Cells to an M2 Phenotype
Macrophage polarization between M1 and M2 phenotypes is an important mechanism for the regulation of inflammatory responses. Kupffer cells isolated from ethanol-fed rats exhibited a more predominant M1 polarization, characterized by increased expression of inducible NOS, an M1 marker, and decreased expression of M2 markers, including Arg-1, IL-4Rα, and MMR compared with Kupffer cells from pair-fed controls (Fig. 3A). This polarization is consistent with increased production of inflammatory cytokines typically observed after chronic ethanol exposure (Fig. 1) (5). Because alternative macrophage polarization is dependent on IL-4- and/or IL-13-mediated signaling pathways (26), rather than IL-10, we hypothesized that flAcrp may act to desensitize Kupffer cells to LPS (Fig. 1) by differentially polarizing Kupffer cells to an M2 phenotype. Treatment of Kupffer cells with 45 nm flAcrp strongly increased the expression of markers of M2 polarization, including Arg-1, MMR, and IL-4Rα (Fig. 3B). This effect of flAcrp was more potent in Kupffer cells from ethanol-fed rats compared with pair-fed controls (Fig. 3B), consistent with previous data indicating that, after ethanol feeding, Kupffer cells are more sensitive to the anti-inflammatory effects of adiponectin compared with cells from pair-fed controls (25). In contrast, gAcrp had minimal effects on the expression of M2 markers (Fig. 3B). Neither gAcrp nor flAcrp decreased the expression of inducible NOS under these conditions (Fig. 3B).
FIGURE 3.
Chronic ethanol feeding shifts Kupffer cells to an M1 phenotype, and treatment of Kupffer cells with flAcrp promotes M2 macrophage polarization. Primary cultures of Kupffer cells isolated from pair-fed and EtOH-fed rats were cultured overnight in the absence or presence of 45 nm gAcrp or flAcrp for 18 h. Expression of Arg-1, MMR, IL-4Rα, and inducible NOS mRNA relative to 18 S mRNA was measured in Kupffer cells not treated with adiponectin (A) or after 18 h of culture with adiponectin (B). A, values are expressed relative to Kupffer cells from pair-fed rats not treated with gAcrp. n = 4. + indicates EtOH-fed compared with pair-fed (p < 0.05). B, values are expressed relative to Kupffer cells not treated with adiponectin. Values with different superscripts (a–d) are significantly different from each other (p < 0.05).
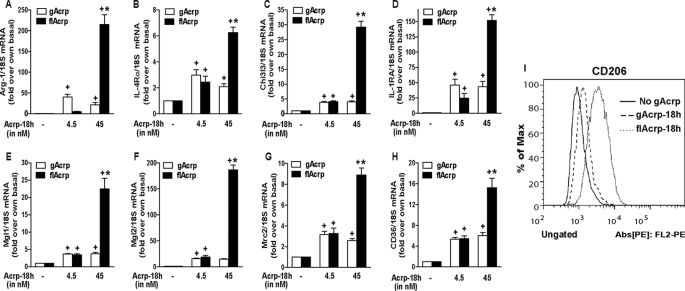
Dose-dependent Polarization of RAW264.7 Macrophages to the M2 Phenotype by flAcrp
RAW264.7 macrophages were cultured with 4.5 or 45 nm gAcrp and flAcrp. These concentrations were chosen based on the relative affinity of gAcrp for AdipoR1 and flAcrp for AdipoR2; gAcrp binds AdipoR1 with 1.14 nm affinity, and flAcrp binds AdipoR2 with 49 nm affinity (27). Although gAcrp had a modest effect on shifting the gene expression profiles of RAW264.7 macrophages to an M2 signature, flAcrp dose-dependently shifted RAW264.7 macrophages to an M2 phenotype (Fig. 4). flAcrp potently increased the expression of multiple markers of M2 macrophages, including Arg-1, scavenger receptors (CD36), surface lectins (macrophage galactose N-acetylgalactosamine-specific lectins 1 and 2 (Mgl1 and Mgl2)), secreted lectins (chitinase-3-like 3 (Chi3l3 or YM-1)), and cytokine receptors (IL-4 receptor α (IL-4Rα)) (Fig. 4). Surface expression of CD206, a marker for M2 macrophages, was increased to a greater extent by flAcrp compared with gAcrp (Fig. 4I).
FIGURE 4.
Differential effects of gAcrp and flAcrp on expression of M2 macrophage markers in RAW264.7 macrophages. A–I, RAW264.7 macrophages were treated either without or with 4.5–45 nm gAcrp or 4.5–45 nm flAcrp for 18 h. A–H, expression of markers of M2 polarization measured by qRT-PCR. mRNA of interest was normalized to 18 S mRNA. n = 4. + indicates p < 0.05 compared with cells not treated with adiponectin; * indicates p < 0.05 flAcrp-treated cells compared with gAcrp-treated cells. I, surface expression of CD206 was measured by flow cytometry. Histogram is representative of four independent experiments.
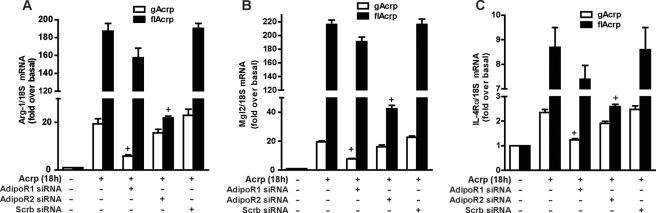
flAcrp Utilizes AdipoR2 to Shift Macrophage Polarization to the M2 Phenotype
To identify the adiponectin receptor required for M2 polarization by flAcrp, RAW264.7 macrophages were transfected or not with siRNA against AdipoR1, AdipoR2, or scrambled siRNA. Transfection with specific siRNAs effectively decreased expression of AdipoR1 and AdipoR2 (supplemental Fig. 1). In cells not transfected with siRNA or transfected with a scrambled siRNA, both gAcrp and flAcrp induced an mRNA expression profile characteristic of an M2 phenotype (Fig. 5). siRNA knockdown of AdipoR1 prevented the modest effects of gAcrp on expression of M2 markers but had no effect on the response to flAcrp (Fig. 5). In contrast, siRNA knockdown of AdipoR2 only prevented the response to flAcrp, whereas scrambled siRNA had no effect (Fig. 5). These data thus demonstrate that flAcrp acts primarily via AdipoR2 to polarize macrophages to an M2 phenotype, consistent with the higher affinity of flAcrp for AdipoR2 compared with AdipoR1 (27).
FIGURE 5.
Differential contribution of AdipoR1 and AdipoR2 in mediating the effects of gAcrp and flAcrp on expression of M2 macrophage markers. RAW264.7 cells were transfected or not with either 100 nm AdipoR1, AdipoR2 siRNA, or scrambled siRNA and then cultured with or without 45 nm gAcrp or flAcrp for 18 h. Arg-1, Mgl2, and IL-4Rα mRNA were measured by qRT-PCR and normalized to 18 S mRNA. n = 4. + indicates p < 0.05 compared with nontransfected cells treated with gAcrp or flAcrp.
flAcrp-mediated Macrophage M2 Polarization Is Dependent on IL-4 Expression
Alternative macrophage polarization is mediated via IL-4/IL-13-dependent pathways (1). flAcrp, but not gAcrp, induced IL-4 mRNA accumulation (Fig. 6A). Neither form of adiponectin significantly increased IL-13 mRNA accumulation (data not shown). Treatment with flAcrp increased IL-4 protein expression in RAW264.7 macrophages (Fig. 6A, inset). Similarly, primary Kupffer cells released IL-4 protein in response to flAcrp (Fig. 6A, inset).
FIGURE 6.
Differential contribution of IL-10, HO-1, and IL-4 to flAcrp-mediated M2 polarization in RAW 264.7 macrophages. A, RAW264.7 macrophages were treated with gAcrp or flAcrp for 18 h, and IL-4 mRNA accumulation was measured by qRT-PCR, and IL-4 protein expression was assessed by Western blot (inset). Kupffer cells isolated from chow-fed control rats were cultured with flAcrp for 18 h, and accumulation of IL-4 protein in the media was assessed by Western blot (inset). flAcrp increased IL-4 protein by 2.0 ± 0.4-fold (n = 5) in Kupffer cells and 1.6 ± 0.2-fold (n = 7) in RAW264.7 macrophages when compared with untreated cells. B–D, RAW264.7 cells were transfected or not with either 100 nm of IL-10, IL-4, HO-1. or scrambled siRNA and then cultured with or without 45 nm flAcrp for 18 h. Arg-1, Mgl2, and IL-4Rα mRNA were measured by qRT-PCR and normalized to 18 S mRNA. n = 4. Values from flAcrp-treated cells with different superscripts (a–c) are significantly different from each other, p < 0.05. E, Kupffer cells were treated with gAcrp or flAcrp for 60 min, and phosphorylation of STAT6 and STAT3 was assessed by Western blot. Expression of STAT6, STAT3, and Hsc70 was measured as loading controls. F, RAW264.7 macrophages were transfected with siRNA against IL-10 or IL-4 and then stimulated with gAcrp or flAcrp for 60 min. Phosphorylation of STAT6 and STAT3 was assessed by Western blot. Expression of Hsc70 was measured as a loading control. E and F, images are representative of at least four independent experiments.
To determine the mechanisms by which flAcrp polarizes macrophages to an M2 phenotype, RAW264.7 macrophages were transfected with siRNA to knock down expression of IL-10, HO-1, and IL-4. siRNA knockdown decreased expression of each of these target genes (supplemental Fig. 1). As expected from the differential role of IL-10/HO-1 in mediating the effects of gAcrp and flAcrp on LPS-stimulated cytokine expression (Figs. 1 and 2), siRNA knockdown of IL-10 had no effect on flAcrp-mediated M2 polarization, whereas siRNA knockdown of HO-1 only partially prevented M2 polarization in RAW264.7 macrophages (Fig. 6, B–D). In contrast, siRNA knockdown of IL-4 reduced flAcrp-mediated M2 polarization by more than 75% (Fig. 6, B–D).
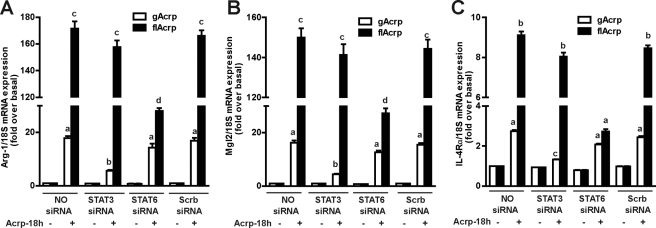
Stimulation of Kupffer cells (Fig. 6E) or RAW264.7 macrophages (Fig. 6F) with gAcrp differentially increased the phosphorylation of STAT3, whereas flAcrp increased phosphorylation of STAT6. siRNA knockdown of IL-10 prevented gAcrp-stimulated STAT3 phosphorylation but had no effect on flAcrp-stimulated STAT6 phosphorylation in RAW264.7 macrophages (Fig. 6F). In contrast, siRNA knockdown of IL-4 prevented flAcrp-stimulated STAT6 phosphorylation but not gAcrp-dependent STAT3 phosphorylation (Fig. 6F). If flAcrp acts via an IL-4-dependent, but IL-10-independent, mechanism to polarize macrophages, then siRNA knockdown of STAT6 should ameliorate the effects of flAcrp, whereas siRNA knockdown of STAT3 should have no effect. Knockdown of STAT6 potently inhibited flAcrp-mediated expression of M2 marker genes, including Arg-1, Mgl2, and IL-4Rα (Fig. 7). In contrast, knockdown of STAT3 reduced the modest impact of gAcrp to polarize RAW264.7 macrophages but had minimal effect on flAcrp-mediated expression of M2 markers (Fig. 7).
FIGURE 7.
gAcrp and flAcrp differentially modulate M2 polarization of RAW264.7 macrophages. RAW264.7 cells were transfected or not with 100 nm of STAT3, STAT6, or scrambled siRNA and then cultured with or without 45 nm gAcrp or flAcrp for 18 h. Arg-1, Mgl2, and IL-4Rα mRNA were measured by qRT-PCR and normalized to 18 S mRNA. n = 4. Values from adiponectin-treated cells with different superscripts (a–d) are significantly different from each other, p < 0.05.
M2 Macrophage Polarization by flAcrp Is Dependent on AMPK in Kupffer Cells from Ethanol- and Pair-fed Rats
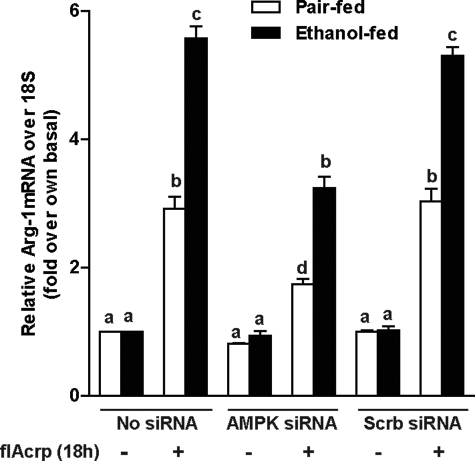
AMPK activation is associated with adiponectin receptor signaling (28), as well as M2 polarization in macrophages (29). When AMPK was knocked down in Kupffer cells from ethanol- and pair-fed rats, flAcrp-mediated increases in Arg-1 expression were partially attenuated, compared with cells transfected or not with scrambled siRNA (Fig. 8).
FIGURE 8.
M2 macrophage polarization by flAcrp is dependent on AMPK in Kupffer cells from ethanol- and pair-fed rats. Kupffer cells isolated from ethanol- and pair-fed rats were transfected or not with 100 nm of AMPK siRNA or scrambled siRNA and then cultured with or without 45 nm flAcrp for 18 h. Arg-1 and 18 S mRNA measured by qRT-PCR. n = 4, values with different superscripts (a–d) within each treatment group are significantly different from each other (p < 0.05).
M2 Polarization by flAcrp Involved Changes in Metabolic Activity of Macrophages
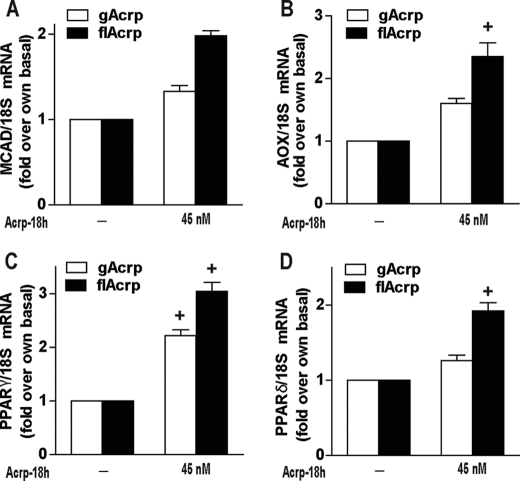
One of the most critical metabolic functions of flAcrp, acting via AdipoR2, is to increase oxidative metabolism in a number of target cells, most importantly hepatocytes (30). Increased oxidative metabolism leads to an increase in fatty acid oxidation and can prevent hepatic steatosis in both alcoholic and nonalcoholic liver disease (31). Interestingly, Chawla and co-workers (32) have discovered that M2 polarization of macrophages also involves a shift in macrophage metabolism to favor oxidative phosphorylation. Therefore, we next investigated whether M2 polarization by flAcrp also involved a shift in expression of genes involved in oxidative phosphorylation and lipid oxidation. flAcrp induced expression of mRNA for peroxisome proliferator-activated receptors (PPARδ and PPARγ) as well as medium chain acyl-CoA dehydrogenase and acyl-CoA oxidase (Fig. 9).
FIGURE 9.
Differential effects of gAcrp and flAcrp on expression of PPARs and oxidative phosphorylation genes in RAW264.7 macrophages. RAW264.7 macrophages were treated either without or with 45 nm gAcrp or flAcrp for 18 h, and expression of PPARδ, PPARγ, medium chain acyl-CoA dehydrogenase (MCAD), and acyl-CoA oxidase (AOX) was measured by qRT-PCR. mRNA of interest was normalized to 18 S mRNA. n = 4. + indicates p < 0.05 compared with cells not treated with adiponectin.
DISCUSSION
Activation of macrophages to secrete pro-inflammatory mediators is an essential response for defense against pathogens, as well as in response to tissue injury. However, the termination of pro-inflammatory signals is just as critical, both to prevent excessive damage during a response to pathogens and to promote a wound healing response. Decreased inflammatory responses by macrophages can be associated with decreased expression of TLR4 and/or via a shift to an alternative/M2 polarization (1). Adiponectin is an adipokine with potent anti-inflammatory properties (30). Adiponectin has complex effects on TLR4-dependent signal transduction signaling (16–20) and can also shift macrophages to an M2 polarization both in vivo and in isolated cells (22, 23). However, our understanding of the molecular mechanisms for adiponectin-mediated responses is very limited. We recently identified an AdipoR1 → IL-10 → HO-1-dependent pathway utilized by gAcrp to decrease TLR4 expression and dampen inflammatory cytokine expression in macrophages (17, 18). Here, we have characterized an additional anti-inflammatory pathway of adiponectin action that is differentially mediated by flAcrp and leads to a shift in macrophages to an M2 polarization. This M2 polarization in response to flAcrp is mediated via an AdipoR2 → IL-4 → STAT6-dependent signaling pathway.
Adiponectin is an abundant serum protein, with concentrations on the order of 10–40 μg/ml, circulating in its high molecular weight form. gAcrp is not typically detected in the circulation; instead, it is likely that secreted proteases act to generate gAcrp in localized environments (33). Therefore, it is likely that the local concentration of gAcrp is a critical factor for some of the anti-inflammatory effects of adiponectin on macrophages.
Adiponectin acts via interactions with AdipoR1 and AdipoR2. AdipoR2 has ∼67% homology at the amino acid level with AdipoR1 (27). AdipoR1 is a high affinity receptor for gAcrp and a low affinity receptor for flAcrp, whereas AdipoR2 binds both with flAcrp and gAcrp (27). In macrophages, the anti-inflammatory effects of gAcrp are dependent on AdipoR1 (17), whereas AdipoR2 is required for flAcrp-mediated M2 polarization (Fig. 5). The signaling modules activated by adiponectin receptors are not well characterized, but several studies suggest that there is an important role for adaptor protein containing the pleckstrin homology domain (34) in response activation of both AdipoR1 and AdipoR2. In hepatocytes and myocytes, adiponectin activation of AdipoR1 is associated with the activation of AMPK, whereas activation of AdipoR2 activates PPARα (35). Here, we find that AMPK is a partial mediator of flAcrp-dependent M2 polarization in Kupffer cells. Furthermore, flAcrp also increased expression of PPARγ and PPARδ (Fig. 9), with minimal effects on PPARα (data not shown) in RAW264.7 macrophages. Further research will be required to adequately understand the differential signaling mechanisms activated by each adiponectin receptor in the different cell types expressing these receptors.
Despite the lack of knowledge regarding the upstream signaling mechanisms for AdipoR1 and AdipoR2 in macrophages, it is clear that the anti-inflammatory responses to gAcrp and flAcrp are mediated via different downstream signaling pathways. Although the anti-inflammatory effects of gAcrp are mediated by an IL-10/HO-1-dependent pathway (17, 18), Libby and co-workers reported that flAcrp inhibits pro-inflammatory signaling in human macrophages via an IL-10-independent pathway (16). Here, we report that flAcrp-mediated M2 polarization in macrophages is independent of IL-10 but dependent on IL-4 expression (Fig. 6). Furthermore, although the anti-inflammatory effects of gAcrp are completely dependent on HO-1 (17, 18), here we find that the anti-inflammatory effects of flAcrp are only partially dependent on HO-1 expression. This partial role for HO-1 is likely due to induction of HO-1 by IL-4 (36).
Alternative macrophage polarization in vivo is prototypically driven by the Th2 cytokines IL-4 and IL-13; these cytokines then activate STAT6 in macrophages to evoke an alternative phenotype. IL-4 and IL-13 are expressed by a number of cells in the innate immune system, including macrophages (37). Here, we find that both RAW264.7 macrophages and primary Kupffer cells produce IL-4 protein in response to flAcrp (Fig. 6A). In RAW264.7 macrophages, knockdown of endogenously synthesized IL-4 prevented phosphorylation of STAT6 and M2 polarization by flAcrp (Fig. 6). Furthermore, flAcrp increased IL-4 mRNA and phosphorylation of STAT6, a signal typical of IL-4 receptor activation, but not IL-13 mRNA or phosphorylation of STAT3, a signal observed in response to IL-13 (1). Taken together, these data suggest an essential role for IL-4 in the regulation of macrophage polarization by flAcrp.
After chronic ethanol exposure, Kupffer cells are sensitized to TLR4-mediated activation of MyD88-dependent and MyD88-independent signaling (18). Here, we report that chronic ethanol feeding also promotes the polarization of Kupffer cells to an M1 phenotype. This polarization to an M1/inflammatory phenotype is consistent with numerous reports of enhanced TLR4-stimulated pro-inflammatory cytokine expression, via both MyD88-dependent and -independent pathways, by Kupffer cells after chronic ethanol exposure (5, 11, 18). Treatment with flAcrp increased expression of M2 markers in Kupffer cells from both ethanol- and pair-fed rats, associated with a suppression of LPS-stimulated TNF-α expression in primary cultures of Kupffer cells.
Circulating adiponectin concentrations are decreased in some murine models of chronic ethanol exposure (38), although the association in patients with alcoholic liver disease likely varies with stage of disease (39). In animal models of genetic or diet-induced obesity, adiponectin is also decreased and is associated with increased inflammatory responses in both adipose tissue and liver, as well as macrophage polarization to the M1 phenotype in adipose, muscle, and liver (32, 40). Treatment of obese mice with PPARγ or PPARδ agonists restores adiponectin concentrations (40) and induces M2 polarization of tissue macrophages (41). Although PPARγ and PPARδ agonists each can protect mice from chronic ethanol-induced liver injury (42, 43), the impact of these agonists on macrophage polarization during chronic ethanol exposure is not known. Evidence also indicates that changes in the M1/M2 phenotypic balance can impact diverse disease conditions, including tumorigenesis, pulmonary disease, as well as bacterial and viral infections (1). However, much less is known about the role of adiponectin in these nonmetabolic diseases.
Comprehensive gene analysis studies of macrophage differentiation and polarization suggest that genes regulating metabolism, in particular lipid metabolism, are very actively up-regulated during macrophage polarization (44). These changes likely support changes in metabolic demand in the M2 macrophages. Odegaard et al. (32, 41) hypothesize that PPARγ and PPARδ work coordinately to achieve maximal M2 polarization, with PPARγ increasing oxidative phosphorylation, thus shifting the metabolic programming of the macrophages, whereas PPARδ mediates expression of pattern recognition receptors and co-stimulatory molecules. In RAW264.7 macrophages, flAcrp increased expression of both PPARγ and PPARδ, as well as genes regulating oxidative phosphorylation. These data suggest that flAcrp is effectively changing gene expression patterns regulating both the metabolic capacity and immune activity of M2 macrophages. Interestingly, AdipoR2 activation by flAcrp is classically associated with increased fatty acid oxidation in hepatocytes and myocytes (28, 30), suggesting that the actions of flAcrp on primarily metabolic tissues, such as hepatocytes and muscle, and immune cells, such as macrophages, are likely mediated via regulation of some of the same metabolic pathways.
Acknowledgments
We thank Drs. A. Stavitsky, M. T. Pritchard, and Sanjoy Roychowdhury for useful discussions during the course of this study as well as their critical reading of the manuscript. We also thank Emmanuelle Ogier for technical expertise and additional support during the course of these studies.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1AA011975 and R37AA011876 (to L. E. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. 1 and 2.
- MMR
- macrophage mannose receptor
- AMPK
- AMP-activated kinase
- flAcrp
- full-length adiponectin
- gAcrp
- globular adiponectin
- qRT
- quantitative real time PCR
- PPAR
- peroxisome proliferator-activated receptor
- Arg-1
- arginase-1.
REFERENCES
- 1. Gordon S., Martinez F. O. (2010) Immunity 32, 593–604 [DOI] [PubMed] [Google Scholar]
- 2. Stout R. D., Suttles J. (2004) J. Leukocyte Biol. 76, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tilg H., Diehl A. M. (2000) N. Engl. J. Med. 343, 1467–1476 [DOI] [PubMed] [Google Scholar]
- 4. McVicker B. L., Tuma D. J., Casey C. A. (2007) World J. Gastroenterol. 13, 4960–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagy L. E. (2003) Exp. Biol. Med. 228, 882–890 [DOI] [PubMed] [Google Scholar]
- 6. Hritz I., Mandrekar P., Velayudham A., Catalano D., Dolganiuc A., Kodys K., Kurt-Jones E., Szabo G. (2008) Hepatology 48, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rao R. (2009) Hepatology 50, 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao X. J., Dong Q., Bindas J., Piganelli J. D., Magill A., Reiser J., Kolls J. K. (2008) J. Immunol. 181, 3049–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cohen J. I., Roychowdhury S., McMullen M. R., Stavitsky A. B., Nagy L. E. (2010) Gastroenterology 139, 664–674.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thurman R. G. (1998) Am. J. Physiol. 275, G605–G611 [DOI] [PubMed] [Google Scholar]
- 11. Vidali M., Stewart S. F., Albano E. (2008) Trends Mol. Med. 14, 63–71 [DOI] [PubMed] [Google Scholar]
- 12. Serhan C. N., Savill J. (2005) Nat. Immunol. 6, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 13. Schäffler A., Schölmerich J. (2010) Trends Immunol. 31, 228–235 [DOI] [PubMed] [Google Scholar]
- 14. Yokota T., Oritani K., Takahashi I., Ishikawa J., Matsuyama A., Ouchi N., Kihara S., Funahashi T., Tenner A. J., Tomiyama Y., Matsuzawa Y. (2000) Blood 96, 1723–1732 [PubMed] [Google Scholar]
- 15. Okamoto Y., Folco E. J., Minami M., Wara A. K., Feinberg M. W., Sukhova G. K., Colvin R. A., Kihara S., Funahashi T., Luster A. D., Libby P. (2008) Circ. Res. 102, 218–225 [DOI] [PubMed] [Google Scholar]
- 16. Folco E. J., Rocha V. Z., López-Ilasaca M., Libby P. (2009) J. Biol. Chem. 284, 25569–25575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mandal P., Park P. H., McMullen M. R., Pratt B. T., Nagy L. E. (2010) Hepatology 51, 1420–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mandal P., Roychowdhury S., Park P. H., Pratt B. T., Roger T., Nagy L. E. (2010) J. Immunol. 185, 4928–4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wulster-Radcliffe M. C., Ajuwon K. M., Wang J., Christian J. A., Spurlock M. E. (2004) Biochem. Biophys. Res. Commun. 316, 924–929 [DOI] [PubMed] [Google Scholar]
- 20. Yamaguchi N., Argueta J. G., Masuhiro Y., Kagishita M., Nonaka K., Saito T., Hanazawa S., Yamashita Y. (2005) FEBS Lett. 579, 6821–6826 [DOI] [PubMed] [Google Scholar]
- 21. Park P. H., McMullen M. R., Huang H., Thakur V., Nagy L. E. (2007) J. Biol. Chem. 282, 21695–21703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fukushima J., Kamada Y., Matsumoto H., Yoshida Y., Ezaki H., Takemura T., Saji Y., Igura T., Tsutsui S., Kihara S., Funahashi T., Shimomura I., Tamura S., Kiso S., Hayashi N. (2009) Hepatol. Res. 39, 724–738 [DOI] [PubMed] [Google Scholar]
- 23. Ohashi K., Parker J. L., Ouchi N., Higuchi A., Vita J. A., Gokce N., Pedersen A. A., Kalthoff C., Tullin S., Sams A., Summer R., Walsh K. (2010) J. Biol. Chem. 285, 6153–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lovren F., Pan Y., Quan A., Szmitko P. E., Singh K. K., Shukla P. C., Gupta M., Chan L., Al-Omran M., Teoh H., Verma S. (2010) Am. J. Physiol. Heart Circ. Physiol. 299, H656–H663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thakur V., Pritchard M. T., McMullen M. R., Nagy L. E. (2006) Am. J. Physiol. Gastrointest Liver Physiol 290, G998–G1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gordon S., Taylor P. R. (2005) Nat. Rev. Immunol. 5, 953–964 [DOI] [PubMed] [Google Scholar]
- 27. Yamauchi T., Kamon J., Ito Y., Tsuchida A., Yokomizo T., Kita S., Sugiyama T., Miyagishi M., Hara K., Tsunoda M., Murakami K., Ohteki T., Uchida S., Takekawa S., Waki H., Tsuno N. H., Shibata Y., Terauchi Y., Froguel P., Tobe K., Koyasu S., Taira K., Kitamura T., Shimizu T., Nagai R., Kadowaki T. (2003) Nature 423, 762–769 [DOI] [PubMed] [Google Scholar]
- 28. Kadowaki T., Yamauchi T. (2005) Endocr. Rev. 26, 439–451 [DOI] [PubMed] [Google Scholar]
- 29. Sag D., Carling D., Stout R. D., Suttles J. (2008) J. Immunol. 181, 8633–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berg A. H., Combs T. P., Scherer P. E. (2002) Trends Endocrinol. Metab. 13, 84–89 [DOI] [PubMed] [Google Scholar]
- 31. Xu A., Wang Y., Keshaw H., Xu L. Y., Lam K. S., Cooper G. J. (2003) J. Clin. Invest. 112, 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Odegaard J. I., Ricardo-Gonzalez R. R., Goforth M. H., Morel C. R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A. W., Chawla A. (2007) Nature. 447, 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Waki H., Yamauchi T., Kamon J., Kita S., Ito Y., Hada Y., Uchida S., Tsuchida A., Takekawa S., Kadowaki T. (2005) Endocrinology 146, 790–796 [DOI] [PubMed] [Google Scholar]
- 34. Mao X., Kikani C. K., Riojas R. A., Langlais P., Wang L., Ramos F. J., Fang Q., Christ-Roberts C. Y., Hong J. Y., Kim R. Y., Liu F., Dong L. Q. (2006) Nat. Cell Biol. 8, 516–523 [DOI] [PubMed] [Google Scholar]
- 35. Steinberg G. R., Watt M. J., Febbraio M. A. (2009) Front. Biosci. 14, 1902–1916 [DOI] [PubMed] [Google Scholar]
- 36. Chaitidis P., O'Donnell V., Kuban R. J., Bermudez-Fajardo A., Ungethuem U., Kühn H. (2005) Cytokine 30, 366–377 [DOI] [PubMed] [Google Scholar]
- 37. Pouliot P., Turmel V., Gélinas E., Laviolette M., Bissonnette E. Y. (2005) Clin. Exp. Allergy 35, 804–810 [DOI] [PubMed] [Google Scholar]
- 38. Rogers C. Q., Ajmo J. M., You M. (2008) IUBMB Life 60, 790–797 [DOI] [PubMed] [Google Scholar]
- 39. Balmer M. L., Joneli J., Schoepfer A., Stickel F., Thormann W., Dufour J. F. (2010) Clin. Sci. 119, 431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lumeng C. N., Bodzin J. L., Saltiel A. R. (2007) J. Clin. Invest. 117, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Odegaard J. I., Ricardo-Gonzalez R. R., Red Eagle A., Vats D., Morel C. R., Goforth M. H., Subramanian V., Mukundan L., Ferrante A. W., Chawla A. (2008) Cell Metab. 7, 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pang M., de la Monte S. M., Longato L., Tong M., He J., Chaudhry R., Duan K., Ouh J., Wands J. R. (2009) J. Hepatol. 50, 1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shen Z., Liang X., Rogers C. Q., Rideout D., You M. (2010) Am. J. Physiol. Gastrointest Liver Physiol 298, G364–G374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martinez F. O., Gordon S., Locati M., Mantovani A. (2006) J. Immunol. 177, 7303–7311 [DOI] [PubMed] [Google Scholar]