Abstract
Suppressor of fused (Sufu) is an essential negative regulator of the sonic hedgehog (Shh) pathway, but little is known about how Sufu itself is normally regulated. Here, we report that Sufu is phosphorylated at Ser-342 and Ser-346 by GSK3β and cAMP-dependent protein kinase A (PKA), respectively, and phosphorylation at this dual site stabilizes Sufu against Shh signaling-induced degradation. We further show that localization of Sufu in the primary cilium is induced by Shh signaling and is required for the turnover of both phosphorylated and total Sufu. Perturbing Sufu phosphorylation with PKA inhibitors or replacing Ser-346 with alanine reduced the stay and replacing Ser-342 and Ser-346 with aspartic acid prolonged the stay of Sufu in the cilia. Finally, ciliary localization of Gli2/3 also required Smo and was similarly influenced by perturbations of PKA activity or mutations at the dual Sufu phosphorylation site. Thus, Shh likely induced trafficking of phospho-Sufu into the primary cilium in a complex with Gli2/3, and dephosphorylation triggered a retrograde export, allowing Sufu to be degraded by the ubiquitin-proteasome system.
Keywords: Cell-Cell Interaction, Endosomes, Glycogen Synthase Kinase 3, Protein Degradation, Signal Transduction, Hedgehog, Primary Cilia, Sufu, Intraflagellar Transport, Phosphorylation
Introduction
The Shh pathway controls tissue patterning during vertebrate development and maintains stem cell fate in tissue regeneration and repair in the adult (1, 2). Inappropriate Shh signaling either due to pathway mutations or ligand misexpression is associated with a myriad of human congenital anomalies and cancers (3, 4); however, how Shh precisely elicit its signaling responses remains to be determined (5).
In mammals, the wiring logic of the Shh pathway is characterized by a series of negative regulatory steps (1, 6, 7), which begin with inhibition of a key membrane-bound activator, Smoothened (Smo), by the Shh receptor, Patched (Ptch). Transcription of Shh target genes is controlled by three Gli transcription factors of the Kruppel zinc finger DNA-binding protein family (8), among which Gli2 and Gli3 exist in two forms as follows: the full-length transcription activators and the truncated N-terminal fragments generated by a partial proteolysis of the C termini (5, 9, 10). Nascent Gli proteins are labile, requiring the binding of Sufu for stable expression (11–13). Sufu is a key downstream negative regulator of Shh signaling, but its mechanism of action remains controversial. Past and recent studies suggest that Sufu could act by sequestering Gli activators in the cytoplasm (14), promoting the production of the truncated repressor (12), or recruiting nuclear co-repressor complex to clamp down Gli transcriptional activity (15, 16).
Progress in the past decade has made it clear that much of vertebrate Shh signaling occurs at the primary cilium (17–19), a nonmotile flagellum-like protrusion present in interphase cells (20). Assembly and maintenance of cilium structure depend on intraflagellar transport (IFT)3 of large protein complexes driven by kinesin motors for anterograde movement from the base bodies to the tips of cilia along microtubule-based axonemes (21). Retrograde IFT movement is driven by dynein motors. Two types of IFT complexes, IFT-A and IFT-B, carry out coordinated but distinct ciliary functions. Mutations in IFT88 and IFT172 of the IFT-B complex and the motor proteins Kif3a and Dync2h1 all prevent cilium assembly and attenuate Shh signaling (17, 22), but mutation in IFT122 of the IFT-A complex results in altered cilium structure and elevates Shh signaling (23). A paradigm of Shh signaling through the primary cilium has emerged from a large body of experimental data (24, 25). In this model, Ptch is localized in the primary cilium in the absence of an Shh signal, thereby excluding the transport of Smo. Inhibition of Smo by Ptch is catalytic in nature rather than through physical contact. Binding of Shh ligand to Ptch induces its ciliary export, which alleviates the inhibition on Smo, allowing it to be rapidly incorporated into the primary cilium. The downstream pathway components Gli and Sufu are localized at ciliary tips, and intraflagellar transport through cilia is absolutely required for generating truncated Gli2 and Gli3 transcription repressors (26).
Mammalian Sufu is absolutely required for embryonic development (27, 28). Removal of Sufu by gene target inactivation or RNAi-mediated gene silencing is sufficient to fully activate Shh signaling (27, 29), but it is not completely understood how Sufu repression is alleviated by Shh. We previously demonstrated that Shh signaling could normally bypass Sufu inhibition by promoting its degradation in the proteasomes to activate downstream target genes (30). Here, we explore how this regulation is realized in the context of primary cilia.
EXPERIMENTAL PROCEDURES
Plasmids and Cell Lines
Human Sufu-Myc expression construct in pRK5 vector was described previously (16). Point mutations of Myc-tagged Sufu were generated by the QuickChange site-directed mutagenesis kit (Stratagene). HEK293 and NIH3T3 cells were purchased from ATCC. Gli-Luc 3T3 cells were purchased from StemRD. Primary MEFs from SmoFlox/Flox or wild type littermate embryos were isolated at 13.5 days post-coitum as described previously (30). The cells used in the experiments were in passage numbers 2–4. Ad-Cre adenoviral infection (Vector Laboratories) was used at 500 multiplicities of infection in serum-free DMEM for 12 h for genomic ablation of the Smo allele in SmoFlox/Flox MEFs, and loss of the Smo expression was confirmed by RT-PCR with the following primers in exons 9 and 11: 5′-TGGACCAAGGCCACCCTGCT-3′ and 5′-TGGCTCCTCTCCGAGCCACC-3′.
Antibodies
Two custom rabbit polyclonal antibodies were generated by Signalway Antibody (Nanjing, China) using the following synthetic phosphopeptides as immunogens: CRAPS(Pho)RKDSLESDS for Ab342P and CRAPSRKDS(Pho)LESDS for Ab346P. The antisera were affinity-purified against the phosphorylated peptide and then cross-absorbed against the nonphosphorylated peptide. The sources for other antibodies are as follows: anti-Myc (9E10) and anti-Sufu antibodies (Santa Cruz Biotechnology); acetylated α-tubulin antibodies (Sigma); and anti-GAPDH antibodies (Kangchen, China).
Small Molecular Inhibitors
The sources and working concentrations for small molecular inhibitors are as follows: purmorphamine (20 μm, Calbiochem), KAAD-cyclopamine (1 μm, Calbiochem), forskolin (10 μm, Tocris), isobutylmethylxanthine (40 μm, Sigma), SB216763 (10 μm, Enzo Life Sciences), MG132 (20 μm, Calbiochem), H89 (2.5 μg/ml, Sigma), KT5720 (2 μm, Santa Cruz Biotechnology), and (Rp)-cAMP (25 μm, Santa Cruz Biotechnology). DMSO was used as the solvent for these inhibitors and the vehicle control.
Protein Turnover Assay
Myc-tagged Sufu and other indicated constructs were co-transfected into NIH3T3 cells with FuGENE HD (Roche Applied Science). 48 h after transfection, the cells were treated with cycloheximide (10 μm, Sigma) to block protein synthesis as indicated. At the end of each time point, the cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× EDTA plus protease inhibitors, and 1× phosSTOP phosphatase inhibitors) for Western analysis. The protein concentration of each cell lysate sample was determined by BCA assay, and an equal amount of total protein was loaded in each lane. To measure protein turnover of Sufu mutants, Myc-tagged Sufu and mutants were individually transfected into Sufu−/− MEFs with Lipofectamine and Plus reagent (Invitrogen) 48 h prior to cycloheximide treatment. The transfected cells were collected for Western analysis at the end of each time point. To measure protein turnover of endogenous Sufu, confluent Kif3a−/− and Kif3a+/+ cells were starved in DMEM containing 0.5% FBS for 24 h prior to cycloheximide treatment. The cells were lysed in RIPA buffer for Western blot analysis at the end of each time point.
In Vitro Kinase Assay
In vitro kinase reactions were carried out in 20 μl of kinase reaction buffer containing 5 μCi of [γ-32P]ATP (3000 Ci/mmol) with 1 μl of catalytically active PKA (PKAc, 2500 units/μl), CK-I (1000 units/μl), CK-II (500 units/μl), or GSK-3β (500 units/μl) at 30 °C for 30 min. All kinases were purchased from New England Biolabs and were used according to the manufacturer's suggestion. An equal amount of 2× SDS loading buffer was added to each reaction, and the samples were heated at 95 °C for 5 min before being resolved in 10% SDS-PAGE and visualized by autoradiography. 1 μg of GST-Sufu or GST alone was used. The phosphorylation mutants of Sufu were synthesized in the quick-coupled in vitro transcription and translation system (Promega) and were used in the reaction after immunopurification.
Mass Spectrometry Analysis of Phosphorylation Sites
4 μg of FLAG-tagged Sufu and 4 μg of PKAc were co-transfected into 2 × 106 HEK293 cells with FuGENE HD (Roche Applied Science). 48 h after transfection, the cells were lysed in RIPA buffer, including protease and phosphatase inhibitors. The transfected Sufu was immunopurified from 2 mg of cell lysates with anti-FLAG M2-agarose beads (Sigma) before being resolved by 7.5% PAGE. After Coomassie Blue staining, the band corresponding to Sufu was excised. The LC/MS-MS analysis was carried out at the Proteomics Center of Children's Hospital, Boston.
Measuring Phosphorylated Sufu Level
Myc-tagged Sufu or its mutants were transfected into HEK293 cells with other indicated constructs with Lipofectamine 2000 (Invitrogen). 48 h after transfection, transfected Sufu was immunopurified with anti-Myc antibody coupled to protein G beads before being subjected to 10% SDS-PAGE and Ab342P, Ab346P, or anti-Sufu blotting. To detect the phosphorylated level of endogenous Sufu, MEFs treated with compounds for the time indicated or from different genotype backgrounds were collected for Western analysis with the antibodies against phosphorylated Sufu.
Luciferase Reporter Assay
The Gli-Luc 3T3 cells and Shh ligand were purchased from StemRD. Approximately 0.6 × 105 cells per well were seeded in a 12-well plate. The next day, the culture medium was replaced with a low serum (0.5% calf serum) assay medium together with 20 μm purmorphamine or 20 ng/ml ShhN ligand. The luciferase activities were assayed after 24 h using the dual reporter luciferase system on a GloMax-96 luminometer (Promega).
Fluorescent-activated Cell Sorting
Cells transfected with various Sufu constructs were dissociated into a single cell suspension using 0.25% trypsin/EDTA. Prior to sorting, cell aggregates were removed by centrifugation through a 35-μm nylon mesh secured in a test tube (352235, BD Biosciences). FACS was carried out on a FACSAriaTM IIu cell sorter (BD Biosciences), gated for high levels of GFP expression. GFP-positive cells were plated out on an 8-well Lab-TEK chambered coverglass.
Confocal Microscopy
Approximately 0.6 × 105 cells per well were seeded in Lab-TEK chambered slides and cultured for 24 h. For each treatment described, the cells were starved in DMEM containing 0.5% FBS for 24 h before addition of compounds as indicated. The cells were fixed with 4% paraformaldehyde for 10 min at room temperature, and standard procedures for immunostaining were followed. To detect Sufu or Gli2/3, a confocal microscopic field was first set to a primary cilium in the channel of anti-acetylated α-tubulin staining. Then an image was captured in the channel of anti-Sufu or anti-Gli2/3 staining, and the intensity of staining at the ciliary tip was calculated after subtracting that from a background area with the identical size. The primary antibodies used were mouse anti-acetylated tubulin (1:2000), rabbit anti-Gli2 and rabbit anti-Gli3 (1:500), Ab342P (1:100), and goat anti-Sufu (1:50). The secondary antibodies used were donkey anti-mouse AlexaFluor 488, donkey anti-goat AlexaFluor 633, goat anti-mouse AlexaFluor 488, goat anti-rabbit AlexaFluor 594, and goat anti-mouse AlexaFluor 594 (1:400), all purchased from Invitrogen. Images were acquired on a Carl Zeiss confocal microscope (LSM510) and quantified with the Image-Pro software.
Live Cell Imaging Analysis
Sufu or Sufu-S342D/S346D mutant fused to the photoactivatable mCherry protein was constructed in the PA-mCherry1-N1 vector (31), and a somatostatin receptor 3-GFP (SR3-GFP) fusion was used for marking cilia (32). These plasmids were co-transfected into Ptch−/− cells, and 24 h later, the cells were starved in 0.5% FBS/DMEM for another 24 h before they were mounted on LSM710 (Carl Zeiss) live cell confocal imaging workstation. The cilia were identified in a green channel based on SR3-GFP fluorescence, and an area immediately covering the primary cilium was photoactivated for three iterations at 405 nm. Time-lapse images were taken in the red channel at a 15-s interval for a consecutive 10 min. The fluorescence intensity of mCherry-Sufu fusion proteins at each time point was normalized as percentage to time 0 after photoactivation and was corrected for photo-bleaching based on a photo-bleaching curve of pa-mCherry, which was determined under identical experimental conditions in separate Ptch−/− cells that had been transfected with the empty PA-mCherry-N1 vector.
RESULTS
Phosphorylation by PKA and GSK3β Stabilizes Sufu
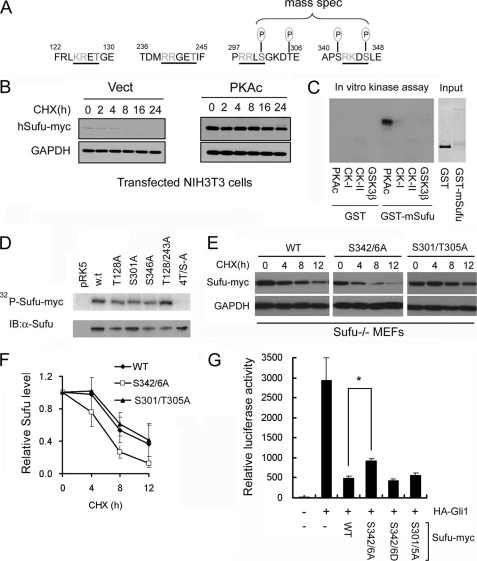
The primary sequence of mammalian Sufu contains four conserved cAMP-dependent protein kinase A (PKA) recognition sites (Fig. 1A). Forced expression of Sufu with the catalytic subunit of PKA (PKAc) allowed the exogenous Sufu to accumulate to a higher level compared with the vector control in Shh-responsive NIH3T3 (Fig. 1B) as well as HEK293 cells (supplemental Fig. 1). Purified GST-Sufu fusion protein can be phosphorylated directly by recombinant PKA (Fig. 1C) but not other kinases tested. We also generated alanine substitutions of each of the four PKA consensus sites and expressed the mutant Sufu as produced in the Quick-coupled transcription and translation system (PromegaTM). In the in vitro kinase assay, these substitution did not affect Sufu phosphorylation individually (Fig. 1D), probably due to target promiscuity under in vitro conditions, but if all four sites were mutated, Sufu phosphorylation was completely abolished (Fig. 1D). These results indicate that Sufu is a bona fide substrate of PKA. To identify the actual phosphorylation sites in vivo, we co-expressed the Myc-tagged Sufu with the catalytic subunit of PKA in HEK293 cells and isolated the phosphorylated Sufu for mass spectrometry analysis. The results showed two cluster sites at Ser-301 to Thr-305 and Ser-342 to Ser-346 (Fig. 1A); the latter is a classical dual phosphorylation site recognized by the priming PKA at the downstream Ser-346 and the lagging GSK3β at the upstream Ser-342 residue, respectively. Replacing these two serine residues with alanine destabilized the S342A/S346A mutant as tested in established Sufu null (Sufu−/−) MEFs, whereas the similar alteration at the Ser-301 to Thr-305 site did not show such an effect (Fig. 1, E and F). Co-expression of various Sufu mutants along with Gli1 and a Gli-responsive luciferase reporter construct in Sufu−/− cells showed that the S342A/S346A substitution dampened the repressor activity of Sufu on Gli1-mediated transcription (Fig. 1G), whereas S342D/S346D showed weak activating and S301A/T305A showed little effect. The stabilizing effect of PKA on Sufu is consistent with its negative regulation of Shh signaling (10, 33).
FIGURE 1.
PKA stabilizes Sufu through controlling phosphorylation of Ser-342 and Ser-346. A, Sufu sequences surrounding the four PKA consensus sites. Mass spectrometry (mass spec) analysis shows that Ser-301/Thr-305 and Ser-342/Ser-346 are the actual phosphorylation sites by PKA in vivo. B, Western analysis showing that co-expression of PKAc reduces Sufu turnover after cycloheximide (CHX) treatment. Vect, vector. C, autoradiogram of in vitro kinase assays reveals Sufu as a substrate of PKA but not casein kinase I, casein kinase II, or GSK3β. Input GST and GST-Sufu are shown in the right panel. D, upper panel, autoradiogram of in vitro PKA kinase assay as in C showing that replacing any one of the four PKA consensus sites with alanine is not sufficient to affect Sufu phosphorylation but replacing all four sites abolished it. Lower panel, Western analysis of Myc-Sufu and mutants expressed by in vitro translation. IB, immunoblot. Western blot analysis (E) and quantification (F) of the turnover rate of Sufu and its mutants in transfected Sufu−/− MEFs. The data presented in F were derived from three repeated experiments. G, luciferase reporter assay for various Sufu mutants co-transfected with Gli1 and the 8×GliBS construct in Sufu−/− cells with Renilla luciferase as an internal control. Each data point represents results from triplicate wells. Error bars are standard deviations. CHX, cycloheximide. *, p < 0.01.
Phosphorylation of Sufu Occurs in Vivo
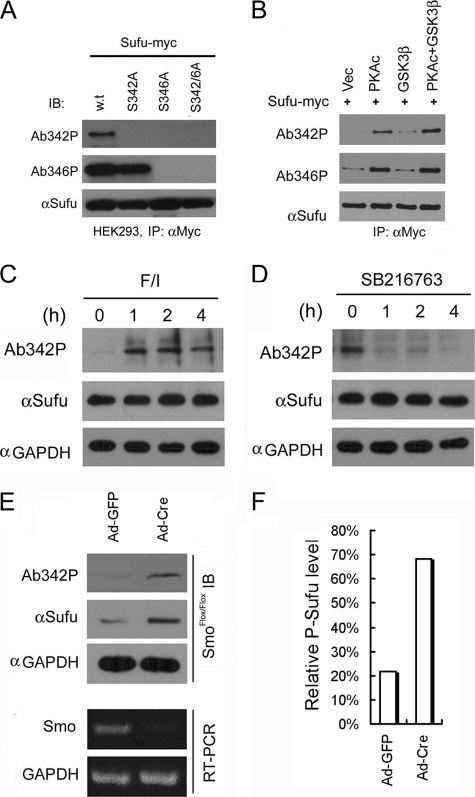
To determine whether the phosphorylation control of Sufu stability occurs in vivo, we generated two polyclonal antibodies, Ab-342p and Ab-346p, that specifically recognize the Ser-342- or Ser-346-phosphorylated form of Sufu (supplemental Fig. 2), respectively. In transfected Sufu−/− cells, Ab-346p recognized the exogenously expressed wild type and S342A mutant Sufu but not S346A or S342A/S346A double mutant, whereas Ab-342p only recognized the wild type Sufu (Fig. 2A). Co-expression of Sufu with the PKA catalytic subunit resulted in a robust phosphorylation at Ser-346 but a weaker one at Ser-342 (Fig. 2B). In contrast, co-expression with PKA and GSK3β, which by itself had little effect, caused strong phosphorylation of Sufu at both residues (Fig. 2B). These results suggested that phosphorylation of Sufu at Ser-346 serves as a priming event for GSK3β-mediated phosphorylation at Ser-342. Of the two phospho-specific antibodies, only Ab342P is capable of recognizing endogenous Sufu (supplemental Fig. 2). This allowed us to examine Sufu phosphorylation status at the lagging Ser-342 residue in primary MEFs following pharmacological perturbation to determine whether phosphorylation at this dual site occurs under physiological conditions. Our results indicated that activation of endogenous PKA activity by elevating the cAMP level with a combination of forskolin, an agonist of adenylyl cyclase, and isobutylmethylxanthine, an inhibitor of phosphodiesterase, strongly increased Sufu phosphorylation within 1 h of the treatment (Fig. 2C). Conversely, inhibition of GSK3β with SB216763 severely curtailed Sufu phosphorylation within the same time frame (Fig. 2D). Finally, we infected freshly isolated MEFs that are homozygous for a floxed Smo allele (SmoFlox/Flox) with a cre-expressing recombinant adenovirus (Ad-cre). This manipulation effectively abolished Smo expression as evident in RT-PCR detection of the Smo mRNA (Fig. 2E, lower panels). Under this condition, the level of total Sufu protein was markedly elevated in agreement with our earlier observation (30), but more importantly, the phosphorylated form of Sufu (p-Sufu) also increased to a level higher than that in the control SmoFlox/Flox MEFs infected with Ad-GFP (Fig. 2, E, upper panels, and F). These data revealed a sequential phosphorylation control of Sufu stability by PKA and GSK3β and suggested that blocking phosphorylation is likely an underlying mechanism by which Shh promotes Sufu turnover.
FIGURE 2.
Shh signaling regulates Sufu phosphorylation in vivo. A, sequential recognition of Sufu at Ser-346 and Ser-342 by PKA and GSK3β, respectively. Myc-tagged wild type and mutant Sufu were individually force-expressed in HEK293 cells. The proteins were isolated by immunoprecipitation (IP) with anti-Myc antibodies and analyzed by Western blot with Ab342P and Ab346P for phosphorylated and anti-Sufu for total Sufu. IB, immunoblot. B, synergistic phosphorylation of Sufu by co-expression of PKAc and GSK3β. The loading in each lane was adjusted according to total Sufu to reveal changes in the level of phospho-Sufu and the Western analysis was carried out as in A. Vec, vector. C, activation of PKA with forskolin and isobutylmethylxanthine treatment dramatically increased, whereas in D, blocking GSK3β with SB216763 abolished phosphorylation of the endogenous Sufu. C and D, freshly isolated normal MEFs were treated with above compounds for the time as indicated, and the whole cell lysates were used in Western analysis. Note the exposure level of the Ab342P blot in C was intentionally set lower than that in D to avoid saturation of the film. E, inactivation of the Smo allele increased the endogenous levels of the total and phosphorylated Sufu in SmoFlox/Flox MEFs. Ad-Cre infection was carried out for 12 h before the proteins in the whole cell lysates were analyzed. This experiment was repeated, but only one representative Western blot was quantified in F.
Ciliary Localization of Sufu Is Induced by Shh and Depends on Smo Function
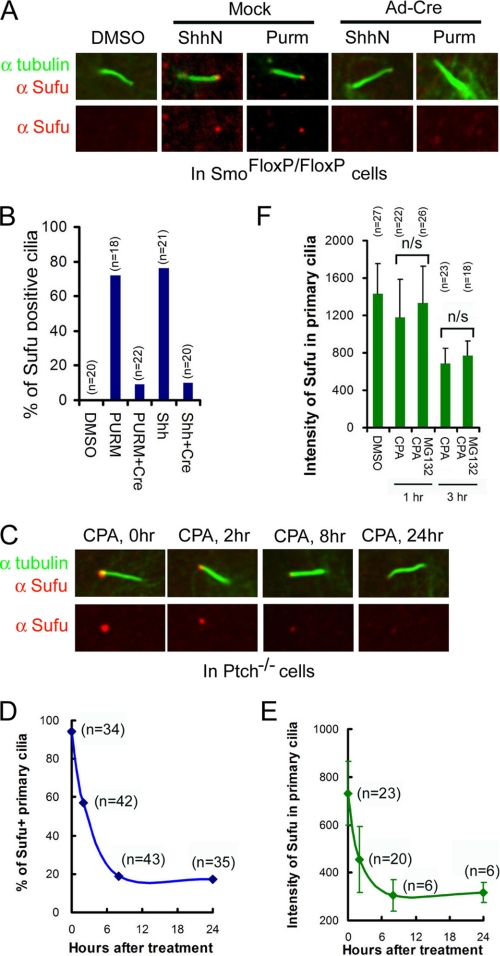
Recently, a number of laboratories reported the constitutive presence of Sufu at the tip of the primary cilium (26, 34). In our hands, Sufu was not detected at cilium tips in freshly isolated SmoFlox/Flox MEFs until the cells were activated with either a purified N-terminal fragment of Shh (Shh-N) or purmorphamine (Fig. 3, A and B). Inducible ciliary localization of Sufu was also detected in a subclone of NIH3T3 cells harboring chromosomal insertions of GliBS-luciferase reporters (supplemental Fig. 3, A and B). However, inactivation of the Smo locus by Ad-cre infection abolished the ciliary localization of endogenous Sufu (Fig. 3, A and B) as well as transiently expressed Sufu-GFP in SmoFlox/Flox MEFs (supplemental Fig. 3, C and D). In Ptch−/− cells, in which the Shh pathway is constitutively active, Sufu was readily detected at cilium tips (Fig. 3C). However, treating Ptch−/− cells with cyclopamine led to quantitative clearance of Sufu; within 8 h of the treatment, about 80% of primary cilia lost Sufu completely (Fig. 3D), and in those primary cilia that persistently stained positive for Sufu, the intensity of Sufu immunostaining decreased to the basal level during the same time frame (Fig. 3E). Additional treatment with MG132 did not reverse the effect of cyclopamine (Fig. 3F), thus the disappearance of Sufu from the primary cilium could not be accounted for simply by degradation. Taken together, the above data indicate that the Shh-induced and Smo-dependent ciliary localization of Sufu is most likely the result of a dynamic balance between anterograde and retrograde intraflagellar transport along cilium axonemes.
FIGURE 3.
Smo dependence of Shh signaling-induced localization of Sufu in primary cilia. A, representative immunofluorescent staining of Sufu (red) at the tips of primary cilia or B, quantification thereof in SmoFlox/Flox MEFs infected with mock solution or Ad-cre viruses. Viral infection was for 12 h, and thereafter the cells were treated with either ShhN ligand or purmorphamine (Purm) for 24 h as indicated. “n” denotes the total number of primary cilia counted at each data point. Primary cilia were marked with anti-acetylated α-tubulin staining (green). C, representative immunofluorescent staining of Sufu at the tip of the primary cilium in Ptch−/− cells. Cyclopamine (CPA) treatment was carried out after the cell culture reached confluence. D, quantification of Sufu-positive primary cilia; E, intensity of Sufu in primary cilia affected by cyclopamine treatment. F, effect of MG132 and cyclopamine on the intensity of Sufu in primary cilia in Ptch−/− cells. n/s, not statistically significant (p > 0.1).
Primary Cilium Is Required for Sufu Degradation
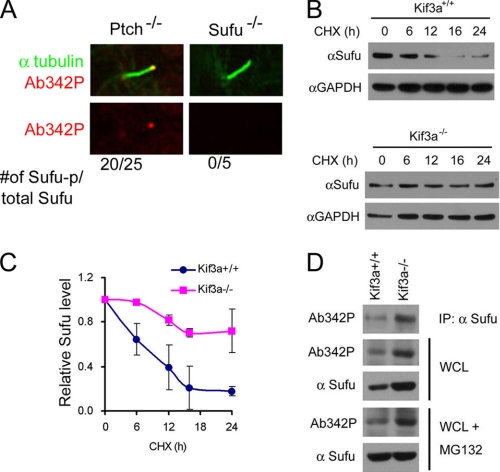
In Ptch−/− cells, we found that the tips of primary cilia could be decorated by immunostaining with Ab342-p phospho-specific antibody (Fig. 4A). Because the same staining was not detected in Sufu−/− cells, this indicates that phosphorylated Sufu is specifically localized at ciliary tips. In light of Shh signaling controls on phosphorylation and ciliary localization of Sufu, we asked if there is any link between these two types of regulation. We thus turned to Kif3a−/− cells, in which cilium formation and Shh signaling are both compromised (35). Western analyses showed that Sufu turned over more slowly in Kif3a−/− cells than in the isogenic control cells after protein synthesis was blocked by cycloheximide (Fig. 4, B and C). This result implies that in the absence of cilia, where Shh signaling is blocked (35), Sufu turnover is abrogated. Interestingly, p-Sufu also accumulated to a higher level in Kif3a−/− than in control cells (Fig. 4D). This was not a trivial correlation to the elevated level of total Sufu, because although blocking proteasomal degradation with MG132 brought total Sufu in control cells to a level comparable with that in Kif3a−/− cells, it failed to do so to p-Sufu (Fig. 4D), suggesting that p-Sufu continued to turn over into the nonphosphorylated form even without proteasome-mediated degradation. These results support that Sufu degradation is linked to translocation to primary cilia.
FIGURE 4.
Primary cilium is required for Sufu degradation. A, immunofluorescent staining of phospho-Sufu in primary cilia with Ab342P phospho-specific antibody. Sufu−/− cells were stained as negative controls. Western blot analysis (B) and quantification thereof (C) showing stabilization of Sufu in Kif3a−/− cells was carried out after blocking protein synthesis with cycloheximide (CHX) treatment as indicated. The data in the graph were derived from three repeated experiments. D, Western blot showing elevated phospho-Sufu in Kif3a−/− cells after anti-Sufu immunoprecipitation (IP) or in whole cell lysates (WCL). MG132 treatment for 6 h restored the level of total but not phosphorylated Sufu.
Phosphorylation Promotes Ciliary Retention of Sufu
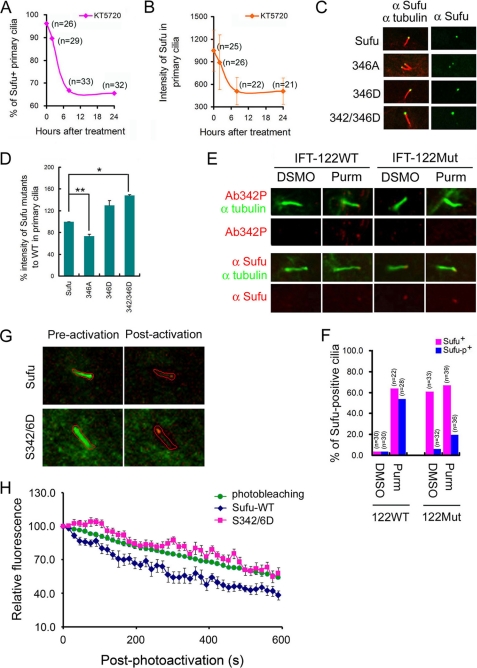
To further explore the relationship between phosphorylation and ciliary localization of Sufu, we treated Ptch−/− cells with three different PKA inhibitors, KT5720, H89, and (Rp)-cAMP, and we found that inhibition of endogenous PKA activity reduced the percentage of cilia that were stained positive for Sufu and the intensity of Sufu staining in those cilia that remained positive for Sufu staining (Fig. 5, A and B, and supplemental Fig. 4, A and B). The kinetics of Sufu clearance by all three compounds resembles that of cyclopamine treatment (comparing Fig. 5, A and B, to Fig. 3, D and E), albeit the amplitude of reduction was lower. These results of pharmacological intervention were corroborated by mutational studies in which we found that the nonphosphorylatable S346A mutant accumulated less, and the phospho-mimicking S346D and, especially, S342D/S346D mutants accumulated more at primary cilia than their wild type counterpart in transfected Sufu−/− cells under purmorphamine treatment (Fig. 5, C and D). Under these conditions, all Sufu mutants were expressed at the same level as that of wild type Sufu (supplemental Fig. 6). Because the nonphosphorylatable mutants were still detected at ciliary tips, it is unlikely that phosphorylation at Ser-342 and Ser-346 residues is required for Sufu to be transported into primary cilia. Rather, our results are consistent with phosphorylation being a ciliary retention signal of Sufu; for Sufu to exit cilia, it has to be dephosphorylated at the tip. This notion was supported by analyses of ciliary localization of Sufu in a mutant line of MEFs derived from IFT122 null embryos. IFT122 is part of the complex A IFT proteins, which together with Dync2h1 is required for retrograde transport from the ciliary tip to the base body (17, 22). Like other types of cells tested, purmorphamine treatment of matching wild type IFT122 control MEFs led to accumulation of Sufu at ciliary tips, but in IFT122 mutant MEFs, Sufu accumulated with or without purmorphamine treatment (Fig. 5, E and F). Interestingly, although Sufu accumulated in purmorphamine-treated cilia was the phosphorylated form, the one that accumulated as result of IFT122 mutation was unphosphorylated regardless of whether the cells were treated with purmorphamine (Fig. 5, E and F). Finally, to demonstrate that phosphorylation regulates retention of Sufu at ciliary tips, we established a live cell imaging system to directly measure the export rate of wild type Sufu and the phospho-mimicking S342D/S346D mutant. In this system, we fused Sufu or the S342D/S346D mutant to the PA-mCherry fluorescence protein and co-transfected the plasmids carrying the fusion cDNA in Ptch−/− cells together with a control encoding somatostatin receptor 3 fused to green fluorescence protein (SR3-GFP) for marking ciliary axonemes under the live culturing condition (32). After finding cilia based on fluorescence emanated from SR3-GFP (Fig. 5G), the mCherry-labeled Sufu or its mutant that had accumulated in the cilia was photoactivated with a laser. The decay of mCherry fluorescence at ciliary tips was then recorded by time-lapse confocal microscopy at a 15-s interval for 10 min (Fig. 2D). We estimated the photoextinction of the mCherry fluorescence protein in separate Ptch−/− cells transfected with the empty PA-mCherry vector (Fig. 5H). The export rate of mCherry-Sufu fusion proteins was calculated by dividing total fluorescence with that of mCherry photoextinction at each time point. When compared with wild type Sufu, the S342D/S346D mutant showed a markedly slower rate of export (Fig. 5H), suggesting that p-Sufu would have a tendency to stay longer than unphosphorylated Sufu at ciliary tips.
FIGURE 5.
Phosphorylation promotes ciliary retention of Sufu. In Ptch−/− cells, immunofluorescent staining indicated that KT5720 treatment led to a rapid decrease of the percentage of Sufu-positive primary cilia (A) and the intensity of Sufu in primary cilia (B). Sufu was typically found in ∼95% of cilia, but this number decreased to ∼65% after KT5720 treatment for 24 h. C, representative autofluorescence images of Sufu-GFP and Sufu mutants in transfected Sufu−/− cells. Replacing Ser-346 with alanine decreased while replacing Ser-346 or both Ser-342 and Ser-346 with aspartic acid ciliary localization. Cilia were visualized by staining with anti-acetylated α-tubulin (red). The number of cilia counted for each data point was between 17 and 24. D, quantification of C. *, p < 0.01; **, p < 0.001. E, representative immunofluorescent staining of phospho-specific and total Sufu at ciliary tips in wild type or IFT122 null MEFs. Purmorphamine (Purm) treatment induced immunofluorescent staining of p-Sufu and total Sufu at the tips of primary cilia. In IFT122 null MEFs, p-sufu was not detected whereas total Sufu accumulated at the tip of cilia with or without purmorphamine treatment. F, quantification of E. G, live cell imaging of photoactivatable Sufu and S342D/S346D mutant force-expressed from PA-mCherry1-N1-Sufu and PA-mCherry1-Sufu-S342D/S346D, respectively, in Ptch−/− cells. Somatostatin receptor-3-GFP was co-transfected to mark for cilia. A type area to be photoactivated was marked, and images of merged green and red channels taken pre- and post-photoactivation were shown. H, relative fluorescence of PA-mCherry-Sufu or PA-mCherry1-Sufu-S342D/S346D was calculated at each time point after photoactivation by correcting for photoextinction of mCherry fluorescence from the total intensity recorded in the red channel at ciliary tips.
Sufu and Gli2/3 Are Transported into the Primary Cilium as a Complex
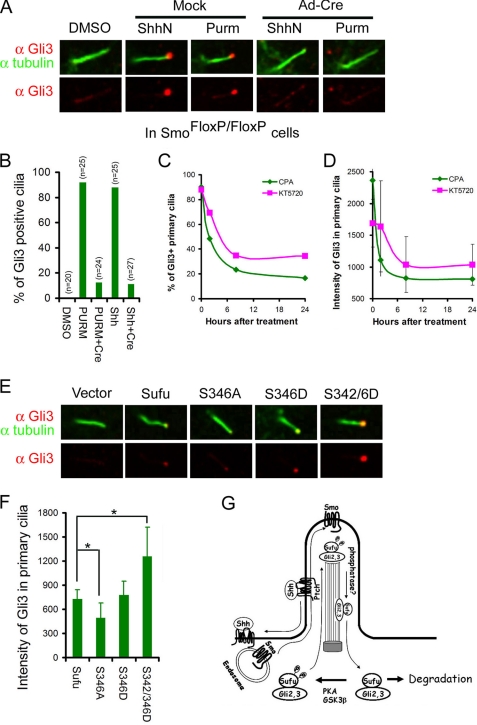
Despite its functional divergence in insects and mammals, Sufu was invariably identified as a binding partner of Gli transcription factors (14, 16, 36). In this regard, it would be reasonable to assume that Sufu and Glis are transported as a complex, making the ciliary localization of Glis also a subject of Shh signaling control. We confirmed the requirement of Smo for ciliary localization of Gli3 in SmoFlox/Flox MEFs, in which activation by either ShhN ligand or purmorphamine rendered a positive decoration of Gli3 immunofluorescence at the tip of the primary cilium and removal of Smo by Ad-cre infection abolished the Gli3 staining (Fig. 6, A and B). In Ptch−/− cells, Gli3 was constitutively present at ciliary tips (supplemental Fig. 5A), but treatment with cyclopamine led to the same decline of the percentage of Gli3-positive primary cilia as well as the intensity of Gli3 in primary cilia (supplemental Fig. 5A and Fig. 6, C and D) as that of Sufu. Likewise, we found that KT5720 treatment also led to the same decline in the above two measures (Fig. 6, C and D). However, because Gli3 is also phosphorylated by PKA, this latter result could not be used to distinguish if the effect of KT5720 treatment was due to blocking phosphorylation of Sufu or Gli3 directly. To circumvent this problem, we took advantage of a recent observation in Sufu−/− cells, in which the access of endogenous Gli3 to primary cilia was blocked unless the expression of Sufu was restored by transfection (34). Following the same experimental conditions, we found that the S346A mutant had a much weaker ability and the S342D/S346D mutants had a much stronger rescuing ability than the wild type Sufu in restoring the localization of Gli3 to the tip of the primary cilium (Fig. 6, E and F). Thus, the kinetic profiles of cyclopamine and KT5720 treatment and the influence of Sufu phosphorylation mutations strongly argue for a co-movement and, for that matter, a co-regulation between Sufu and Gli3. Because cyclopamine and KT5720 treatment also exhibited the same impact on the ciliary localization of Gli2 (supplemental Fig. 5, B–G), it is reasonable to assume that the above conclusion can be extended to Gli2.
FIGURE 6.
Phosphorylation promotes Shh signaling-induced co-localization of Sufu and Gli3 in primary cilia. Representative immunofluorescent staining of Gli3 (A) and quantification thereof (B) show Shh signaling-induced localization at the tip of the primary cilia in SmoFlox/Flox MEFs and curtailment by Ad-cre infection as in Fig. 3, A and B. C and D, effects of cyclopamine and KT5720 treatment on the percentage of Gli3 positive primary cilia and the intensity of Gli3, respectively. E, representative immunofluorescent staining and quantification of Gli3 (F) restored by Sufu and its phosphorylation site mutants that were force-expressed in Sufu−/− cells. *, p < 0.01. G, model for the regulation of Shh-induced co-localization of Sufu and Gli2/3 in primary cilia by phosphorylation. Purm, purmorphamine.
DISCUSSION
Sufu was once considered an enigmatic negative regulator of the Hedgehog pathway, partly because the absence of any recognizable domain in its protein coding sequence made it difficult to even postulate its function (13). However, mammalian Sufu is the most conserved member of the pathway, and it is absolutely required for embryonic development (14, 16, 27, 28, 36). Recent investigations have shown that mammalian Sufu plays an important function in stabilizing the full-length Gli2 and Gli3, allowing them to be processed into the truncated transcription repressors (11, 12, 34). Our current study casts new insight into the regulation of Sufu's inhibitory function, which must be overcome to turn on Shh target genes.
Drosophila Sufu was shown to be phosphorylated by Fu both in genetically altered imaginal discs as well as in cultured S2 cells (37, 38), but this phosphorylation is induced by Hedgehog signaling and the site of phosphorylation has not been determined. In contrast, the phosphorylation of mammalian Sufu that we reported here is not induced by Shh; rather, activation of Shh signaling leads to a reduction of the phospho-Sufu level in vivo (Fig. 2D). The difference between these two types of phosphorylation is another example of design shift of the mammalian Hedgehog pathway during evolution and, perhaps, a reflection of the functional divergence of the mammalian Fu kinase, which is involved in the ciliogenesis rather than directly participating in Shh signal transduction (39). The four consensus PKA recognition sites are not conserved in the sequence of Drosophila Sufu.
In transfected HEK293 and NIH3T3 cells, we observed limited but significant reduction in the stability of exogenous Sufu in response to treatments by the Shh ligand or agonists (30). However, the changes in the stability of endogenous Sufu were readily detectable by Western analysis in freshly isolated MEFs following pharmacological treatment (Fig. 2, C and D) or genetic inactivation of the Smo allele (Fig. 2, E and F). It is possible that the cellular capacity for processing Sufu turnover is limited and saturable by an overwhelming amount of Sufu expressed from the transfected plasmids. We postulate that only a small portion of total Sufu undergoes the Shh ligand-induced turnover such that it takes either complete inactivation (as in Smo−/− MEFs, Fig. 2E) or activation (as in Ptch−/− embryonic tissues, see Fig. 2D in Ref. 30)+ of the pathway to allow for visible changes in the total amount of endogenous Sufu in either direction.
The mechanism of stabilizing Sufu by phosphorylation is not clear nor is the identity of the E3 ligase required for Sufu ubiquitination. The dual phosphorylation site that we identified through mass spectrometry analysis resembles the recognition sequence of the F-box/WD domain containing E3 ubiquitin ligases, which requires phosphorylation for binding. However, this cannot be the site of action of a degrading ubiquitin E3 ligase because phosphorylation of Sufu at Ser-346 and Ser-342 by sequential actions of PKA and GSK3β leads to stabilization of Sufu. Nevertheless, there were reports in the literature showing a carboxyl region of Sufu binds and recruits GSK3β for efficient processing of Gli3 (40). We can speculate that sequential phosphorylation of Sufu by PKA and GSK3β stabilizes Sufu in a complex with Gli2/3, allowing the latter to be transported into the primary cilium for further modification that leads to proteolytic processing into truncated transcription repressors.
Our data of pharmacological treatment with cyclopamine and PKA inhibitors in Ptch−/− cells indicate that the ciliary localization of Sufu is a dynamic balance between anterograde and retrograde IFT, as opposed to Sufu being a residential protein at the ciliary tip (26). Because we demonstrated a dependence on Smo for the ciliary localization, it is tempting to speculate that the movement of Sufu into the primary cilium is likely driven by the same mechanism that powers Smo translocation, which is activated by Shh signaling (25). The same argument can be made for Gli2 and Gli3, because the clearance of the latter two from cilia showed the same kinetic characteristics under the above pharmacological treatment (Fig. 6, A–D, and supplemental Fig. 5). There are data reported in the literature showing that the membrane-bound Smo follows the endocytic route in the vesicular transport into the primary cilium (41) or, alternatively, moves laterally from the cell surface (42). It is possible that Shh signaling induces Sufu and Gli2/3 to hitch on the Smo-transporting vesicle in their movement into the primary cilium; however, Sufu and Gli2/3 must follow a different off-load control mechanism that renders them to be concentrated at ciliary tips as opposed to Smo being evenly distributed along the entire length of ciliary membrane. Therefore, we propose that under the influence of Shh signaling, phosphorylated Sufu traverses as a complex with Gli2/3 along the axoneme to the tip of the primary cilium, where a Smo-dependent mechanism likely dephosphorylates Sufu to mark a modification/processing event and sets forth the retrograde export (Fig. 6G). The system resets when Sufu is exported and degraded outside the primary cilium.
Acknowledgments
We thank Dr. Jonathan Eggenschwiler for carefully reading the manuscript and providing the IFT122 null cells; Drs. Jeremy Reiter for Kif3a null cells; Rune Toftgard for Sufu and Ptch null cells; Baolin Wang, Paotien Chuang, and Suzie Scales for Gli2 and Gli3 antibodies; and Joseph Brzostowski for assistance in confocal imaging.
This work was supported by Chinese National Science Foundation Grant 30771079 (to S. Y. C.), 973 State Key Research Project of Chinese Ministry of Science and Technology Grant 2009CB918403 (to S. Y. C.), and University Research Grant 08NMUM010 (to S. Y.). This work was also supported in part by National Institutes of Health NIAID intramural fund (to T. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- IFT
- intraflagellar transport
- MEF
- mouse embryo fibroblast
- PA
- photoactivatable.
REFERENCES
- 1. Jiang J., Hui C. C. (2008) Dev. Cell 15, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ingham P. W., McMahon A. P. (2001) Genes Dev. 15, 3059–3087 [DOI] [PubMed] [Google Scholar]
- 3. Ruiz i Altaba A. (2008) Cancer Cell 14, 281–283 [DOI] [PubMed] [Google Scholar]
- 4. Beachy P. A., Karhadkar S. S., Berman D. M. (2004) Nature 432, 324–331 [DOI] [PubMed] [Google Scholar]
- 5. Wilson C. W., Chuang P. T. (2010) Development 137, 2079–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jacob L., Lum L. (2007) Sci. STKE 2007, cm6. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y., McMahon A. P., Allen B. L. (2007) Curr. Opin. Cell Biol. 19, 159–165 [DOI] [PubMed] [Google Scholar]
- 8. Ruiz i Altaba A., Mas C., Stecca B. (2007) Trends Cell Biol. 17, 438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang B., Fallon J. F., Beachy P. A. (2000) Cell 100, 423–434 [DOI] [PubMed] [Google Scholar]
- 10. Tempé D., Casas M., Karaz S., Blanchet-Tournier M. F., Concordet J. P. (2006) Mol. Cell. Biol. 26, 4316–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang C., Pan Y., Wang B. (2010) Development 137, 2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Humke E. W., Dorn K. V., Milenkovic L., Scott M. P., Rohatgi R. (2010) Genes Dev. 24, 670–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng S. Y., Yue S. (2008) Adv. Cancer Res. 101, 29–43 [DOI] [PubMed] [Google Scholar]
- 14. Kogerman P., Grimm T., Kogerman L., Krause D., Undén A. B., Sandstedt B., Toftgård R., Zaphiropoulos P. G. (1999) Nat. Cell Biol. 1, 312–319 [DOI] [PubMed] [Google Scholar]
- 15. Paces-Fessy M., Boucher D., Petit E., Paute-Briand S., Blanchet-Tournier M. F. (2004) Biochem. J. 378, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng S. Y., Bishop J. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5442–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. (2003) Nature 426, 83–87 [DOI] [PubMed] [Google Scholar]
- 18. Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. (2005) Nature 437, 1018–1021 [DOI] [PubMed] [Google Scholar]
- 19. Goetz S. C., Anderson K. V. (2010) Nat. Rev. Genet. 11, 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rosenbaum J. L., Witman G. B. (2002) Nat. Rev. Mol. Cell Biol. 3, 813–825 [DOI] [PubMed] [Google Scholar]
- 21. Silverman M. A., Leroux M. R. (2009) Trends Cell Biol. 19, 306–316 [DOI] [PubMed] [Google Scholar]
- 22. May S. R., Ashique A. M., Karlen M., Wang B., Shen Y., Zarbalis K., Reiter J., Ericson J., Peterson A. S. (2005) Dev. Biol. 287, 378–389 [DOI] [PubMed] [Google Scholar]
- 23. Cortellino S., Wang C., Wang B., Bassi M. R., Caretti E., Champeval D., Calmont A., Jarnik M., Burch J., Zaret K. S., Larue L., Bellacosa A. (2009) Dev. Biol. 325, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rohatgi R., Scott M. P. (2007) Nat. Cell Biol. 9, 1005–1009 [DOI] [PubMed] [Google Scholar]
- 25. Rohatgi R., Milenkovic L., Scott M. P. (2007) Science 317, 372–376 [DOI] [PubMed] [Google Scholar]
- 26. Haycraft C. J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E. J., Yoder B. K. (2005) PLoS Genet. 1, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Svärd J., Heby-Henricson K., Henricson K. H., Persson-Lek M., Rozell B., Lauth M., Bergström A., Ericson J., Toftgård R., Teglund S. (2006) Dev. Cell 10, 187–197 [DOI] [PubMed] [Google Scholar]
- 28. Cooper A. F., Yu K. P., Brueckner M., Brailey L. L., Johnson L., McGrath J. M., Bale A. E. (2005) Development 132, 4407–4417 [DOI] [PubMed] [Google Scholar]
- 29. Varjosalo M., Li S. P., Taipale J. (2006) Dev. Cell 10, 177–186 [DOI] [PubMed] [Google Scholar]
- 30. Yue S., Chen Y., Cheng S. Y. (2009) Oncogene 28, 492–499 [DOI] [PubMed] [Google Scholar]
- 31. Subach F. V., Patterson G. H., Manley S., Gillette J. M., Lippincott-Schwartz J., Verkhusha V. V. (2009) Nat. Methods 6, 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pazour G. J., Witman G. B. (2003) Curr. Opin. Cell Biol. 15, 105–110 [DOI] [PubMed] [Google Scholar]
- 33. Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., Ishii S. (1999) J. Biol. Chem. 274, 8143–8152 [DOI] [PubMed] [Google Scholar]
- 34. Chen M. H., Wilson C. W., Li Y. J., Law K. K., Lu C. S., Gacayan R., Zhang X., Hui C. C., Chuang P. T. (2009) Genes Dev. 23, 1910–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koyama E., Young B., Nagayama M., Shibukawa Y., Enomoto-Iwamoto M., Iwamoto M., Maeda Y., Lanske B., Song B., Serra R., Pacifici M. (2007) Development 134, 2159–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pearse R. V., 2nd, Collier L. S., Scott M. P., Tabin C. J. (1999) Dev. Biol. 212, 323–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ho K. S., Suyama K., Fish M., Scott M. P. (2005) Development 132, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 38. Lum L., Zhang C., Oh S., Mann R. K., von Kessler D. P., Taipale J., Weis-Garcia F., Gong R., Wang B., Beachy P. A. (2003) Mol. Cell 12, 1261–1274 [DOI] [PubMed] [Google Scholar]
- 39. Wilson C. W., Nguyen C. T., Chen M. H., Yang J. H., Gacayan R., Huang J., Chen J. N., Chuang P. T. (2009) Nature 459, 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kise Y., Morinaka A., Teglund S., Miki H. (2009) Biochem. Biophys. Res. Commun. 387, 569–574 [DOI] [PubMed] [Google Scholar]
- 41. Kovacs J. J., Whalen E. J., Liu R., Xiao K., Kim J., Chen M., Wang J., Chen W., Lefkowitz R. J. (2008) Science 320, 1777–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Milenkovic L., Scott M. P., Rohatgi R. (2009) J. Cell Biol. 187, 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]