Abstract
All strains of Staphylococcus aureus encode a putative copper-sensitive operon repressor (CsoR) and one other CsoR-like protein of unknown function. We show here that NWMN_1991 encodes a bona fide Cu(I)-inducible CsoR of a genetically unlinked copA-copZ copper resistance operon in S. aureus strain Newman. In contrast, an unannotated open reading frame found between NWMN_0027 and NWMN_0026 (denoted NWMN_0026.5) encodes a CsoR-like regulator that represses expression of adjacent genes by binding specifically to a pair of canonical operator sites positioned in the NWMN_0027–0026.5 intergenic region. Inspection of these regulated genes suggests a role in assimilation of inorganic sulfur from thiosulfate and vectorial sulfur transfer, and we designate NWMN_0026.5 as CstR (CsoR-like sulfur transferase repressor). Expression analysis demonstrates that CsoR and CstR control their respective regulons in response to distinct stimuli with no overlap in vivo. Unlike CsoR, CstR does not form a stable complex with Cu(I); operator binding is instead inhibited by oxidation of the intersubunit cysteine pair to a mixture of disulfide and trisulfide linkages by a likely metabolite of thiosulfate assimilation, sulfite. CsoR is unreactive toward sulfite under the same conditions. We conclude that CsoR and CstR are paralogs in S. aureus that function in the same cytoplasm to control distinct physiological processes.
Keywords: Copper, Metalloproteins, Metals, Sulfur, Transcription Regulation, Metalloregulation
Introduction
The Gram-positive opportunistic human pathogen Staphylococcus aureus is the causative agent of a wide range of hospital and community-acquired infections that are associated with significant morbidity (1). With the incidence of methicillin-resistant strains increasing in previously low prevalence areas (2), new antibiotic therapies that target novel metabolic pathways are urgently needed. One approach is to target those processes that allow a pathogen to respond to environmental stresses that might change depending on the microenvironmental host niche in which the organism finds itself. Resistance to host-mediated copper killing of Escherichia coli (3), Salmonella enterica (4), and Mycobacterium tuberculosis (5, 6) and sulfur assimilation and cysteine biosynthesis in M. tuberculosis (7, 8) are two such processes. S. aureus is particularly sensitive to rapid killing when exposed to copper or copper alloy surfaces, justifying this therapeutic direction (9, 10).
M. tuberculosis CsoR6 (copper-sensitive operon repressor) is a founding member of large family of regulators now known collectively to respond to Cu(I), Ni(II), and perhaps other stressors, the structural basis of which is not fully understood (11, 12). All CsoR family proteins lack a known canonical DNA binding domain and are projected to adopt the flat disc-shaped dimer of dimers homotetrameric structure characteristic of Cu(I)-sensing CsoRs, with individual dimers consisting of an antiparallel four-helix bundle flanked by a C-terminal α3 helix (13, 14). Two cysteine residues on opposite subunits within a dimer make coordination bonds to the Cu(I) ion, with the third ligand a His from the α2 helix (Cys36′, His61, Cys65), thus completing a trigonal S2N coordination complex (13). Additionally, two conserved second coordination shell residues, Tyr35′ and Glu81, play critical roles in driving allosteric negative regulation of DNA binding by Cu(I) within the tetramer (15, 16).
Some bacteria encode more than one CsoR family member. For example, pathogenic mycobacterial species encode as many as five CsoR-like proteins (13) and all strains of S. aureus appear to encode at least two. Both M. tuberculosis and S. aureus also encode at least one CsoR-like protein that conserves only the two Cys that coordinate Cu(I) in CsoR but otherwise lacks all other requisite features of a Cu(I)-sensing CsoR (16). This subfamily CsoR-like protein is also found in other Gram-positive microorganisms, including Bacillus subtilis (YrkD) and Streptococcus pneumoniae (SPD_0073), where their functions are completely unknown. This wide distribution and involvement in multiple regulatory pathways highlights the importance of understanding the molecular processes governed by this family of proteins.
In this work, we characterize the regulation of two stress response pathways in S. aureus by paralogs of the CsoR family of DNA-binding proteins. These transcriptional regulators are the copper sensor CsoR and a novel regulator denoted CstR (CsoR-like sulfur transferase regulator), which respond to distinct stressors with no detectable regulatory cross-talk in the cell.
EXPERIMENTAL PROCEDURES
Construction of ΔcsoR and ΔcstR Deletion Strains
The ΔcsoR mutant was constructed using established methods (17). Briefly, a PCR amplicon beginning 45 bp upstream of the corrected putative csoR (NWMN_1991) ORF containing the ∼1000 bp upstream sequence was amplified using primers CCCGGGAAAACACAACGTCAACACAAAG and GGGGACAAGTTTGTACAAAAAAGCAGGCTTTTACCTAAGTACTCATCACC. Another amplicon containing ∼50 bp of the putative csoR ORF together with ∼1000 bp downstream of the putative csoR ORF was amplified using primers CCCGGGCAGGAAGAGGCAATGGAAG and GGGGACCACTTTGTACAAGAAAGCTGGGTCTTTATCGTTGGTTTCGTCAC.These PCR-generated fragments were ligated together and cloned into pCR2.1 (Invitrogen). Next, the combined fragments were amplified using primers specific for 5′ and 3′ flanking sequences and the resultant PCR product was recombined into pKOR1 and used for allelic replacement into S. aureus strain Newman as described (17). An exactly analogous strategy was used to create the ΔcstR deletion strain with the exception that primers GGGGACAAGTTTGTACAAAAAAGCAGGCTTTTCTTTTTCATTACGTAGCGC and CCCGGGGTCATACCTCCACTTTTAATTG, and CCCGGGATTGGTGAAAAGTAAGTAATGG and GGGGACCACTTTGTACAAGAAAGCTGGGTCACGTAAATTTTTAATAGCTTCG, were used to amplify the 5′ and 3′ fragments, respectively.
Quantitative PCR
To prepare samples for RNA extraction, 5-ml cultures were grown overnight in 15-ml conical tubes at 37 °C in supplemented Chelex-treated RPMI (NRPMI) for copA expression experiments or TAB for NWMN_0026-NWMN_0029 expression experiments. The next morning the cultures were back diluted 1/100 into 5 ml of NRPMI in a 15-ml conical tube with or without 1 mm MnCl2 or 1 mm CuSO4 or TSB. The cultures were grown for 4 h at 37 °C with shaking at 180 rpm. At the end of the incubation, an equal volume of 1:1 acetone/ethanol was added to the cultures and the samples were frozen at −80 °C. To harvest RNA, the samples were thawed on ice and centrifuged to pellet the bacteria. The supernatant was removed and the bacterial pellet was air dried. RNA was harvested as previously described using a combination of Tri-Reagent (Sigma) and RNeasy Minikit purification (Qiagen, Valencia, CA) (18) with the exception that after the addition of TRIzol the samples were transferred to bead beater tubes and processed at 6 m/s for 40 s in a bead beater to aid in cell lysis. Random hexamers and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) were used to generate cDNA. Quantitative PCR was performed using the iQ SYBR Green Supermix (Bio-Rad) and the primer pairs as indicated under supplemental Table S3 (18). Quantitation of 16 S ribosomal RNA was used to normalize each sample.
Bacterial Expression Plasmid Construction and CsoR Purification
The complete open reading frame annotated as locus tag NWMN_1991 in S. aureus strain Newman (nucleotides 2212576–2212914; locus AP009351) (19) was PCR amplified and subcloned into pET3d between the NcoI and BamHI restriction sites. The resultant recombinant protein showed very poor solubility and no DNA binding to a 39-bp DNA derived from the promoter region of the S. aureus copA gene (see Fig. 1A) (data not shown). Further inspection of the DNA sequence of NWMN_1991 revealed a second initiation codon positioned at nucleotide 2212869, resulting in ORF 15 codons shorter than that annotated as NWMN_1991 and a consensus ribosome binding site just upstream of this initiation codon. A multiple sequence alignment of bona fide Cu(I)-sensing CsoRs revealed that no others contained an extended N-terminal region (supplemental Fig. S1). Therefore, the region corresponding to nucleotides 2212576–2212869 was hypothesized to represent the actual ORF corresponding to locus tag NWMN_1991 and was therefore PCR amplified from genomic DNA and subcloned into pET3d between the NcoI and BamHI restriction sites. The second residue was changed to an alanine as a result (T2A) of the subcloning and is referred to as wild-type CsoR here. Amino acid substitutions were introduced into this expression plasmid by site-directed QuikChange mutagenesis, and the sequences of all resultant plasmids were verified by DNA sequencing.
FIGURE 1.
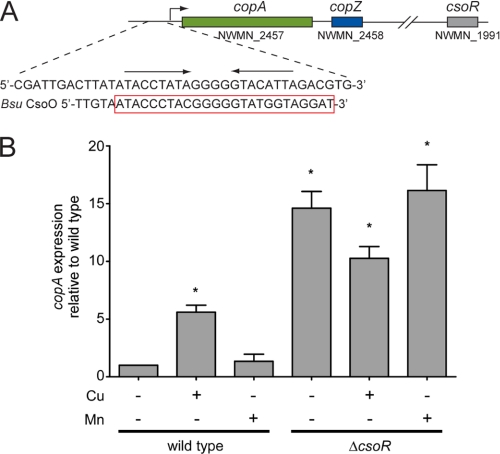
A, genomic location of copA, copZ, and the predicted csoR in S. aureus Newman. The DNA sequence in the copA promoter region is also shown compared with the Bsu copZA operator, which has been shown to be regulated and bound tightly by Bsu CsoR (15, 20). B, analysis of copA transcription by RT-PCR in WT and ΔcsoR grown in the absence or presence of 1.0 mm CuCl2 (Cu) or MnCl2 (Mn) as indicated. The results are the average of four independent experiments. *, p < 0.05 determined by Student's t test. Error bars = S.D.
Biochemical experiments confirmed the designation of the protein encoded by NWMN_1991 as CsoR and is therefore referred to as such (see below). Plasmids carrying wild-type or mutant Sau CsoRs were transformed into E. coli BL21(DE3)/pLysS to ampicillin resistance. A single colony from an LB agar plate containing 100 mg/liter of ampicillin was inoculated into 200 ml of LB medium containing 100 mg/liter of ampicillin and grown overnight in a 37 °C shaker. 20 ml of the overnight culture was then used to inoculate 1 liter of the same LB medium and grown at 37 °C until A600 reached 0.6–0.8. 0.4 mm Isopropyl 1-thio-β-d-galactopyranoside was then added and cells were grown for an additional 2 h prior to harvesting by low speed centrifugation. Cells were resuspended in 200 ml of Buffer E (25 mm Hepes, pH 7.0, 2 mm EDTA, 2 mm DTT) and lysed by sonication. After low speed centrifugation, CsoR was largely found in the lysis pellet, but was readily recovered in supernatant by stirring at 4 °C overnight in the same lysis buffer. 0.15% (v/v) of polyethyleneimine was added to the supernatant to precipitate the nucleic acids. Both wild-type and C41A Sau CsoRs were found in the polyethyleneimine pellet, which was then resuspended in Buffer E containing 0.5 m NaCl and reprecipitated, with the supernatant containing CsoR. In contrast, H66A Sau CsoR was found principally in the polyethyleneimine supernatant fraction. Each supernatant containing Sau CsoR was then subjected to ammonium sulfate precipitation and the resulting pellet resuspended in Buffer E and dialyzed exhaustively against Buffer E containing 0.05 m NaCl. The sample was then purified on a Q Fast Flow column with Buffer E using a salt gradient of 0.05–0.5 m NaCl. Fractions containing Sau CsoR were combined and concentrated to a final volume of ∼3 ml. 1 ml of the resultant protein was then loaded onto a Superdex 200 30/100GL size exclusion column (GE Healthcare) pre-equilibrated with Buffer E containing 0.3 m NaCl. The fractions containing Sau CsoR were combined and dialyzed against Buffer E containing 0.05 m NaCl and loaded onto a MonoQ column for further purification. Fractions from the MonoQ column containing Sau CsoR were then pooled and concentrated to a volume of ∼6 ml and dialyzed into Buffer S (10 mm Hepes, 0.2 m NaCl, pH 7.0) in an anaerobic glovebox. The purity of the final CsoRs was estimated by visualization of Coomassie-stained 18% Tris glycine SDS-PAGE gels to be ≥90% in each case. Protein concentration was determined by using a ϵ280 = 1615 m−1 cm−1. The free thiol content was determined by the 5,5′-dithiobis(nitrobenzoic acid) assay to be more than 90% of expected value in each case (2.0 expected) (13, 20). Less than 0.1% copper was detected by flame atomic absorption spectroscopy in all purified protein samples carried out as previously described (21).
CstR Purification
CstR is encoded by the complementary strand of nucleotides 37974–38234 in the S. aureus strain Newman genome. CstR was expressed in BL21(DE3)/pLysS cells under control of the lac repressor with coding sequences PCR-amplified and subcloned into pET3a between the NdeI and BamHI sites. Protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside when the cultures had reached an optical density (A600) ≈0.6 and allowed to grow for an additional 4–5 h at 37 °C at which time the cells were pelleted by centrifugation and stored at −20 °C overnight. The cell pellet was resuspended in 50 mm Hepes, 4 mm DTT, and 5 mm EDTA at pH 7.0 (Buffer A) with 1 m NaCl added to enhance the solubility of CstR. The solution was clarified by centrifugation. Standard 0.2% (v/v) polyethyleneimine precipitation removed most nucleotide contamination and the protein was then precipitated with 500 g/liter of (NH4)2SO4. The ammonium sulfate pellet was resuspended in Buffer A and extensively dialyzed against Buffer A plus 50 mm NaCl at 4 °C resulting in CstR precipitation. The dialysate was clarified by centrifugation and the pellet resuspended in degassed Buffer A with 1 m NaCl. Gel filtration chromatography using Superdex-200 in extensively degassed Buffer A (1 m NaCl) yielded pure CmtR (>95%) as visualized on an 18% acrylamide gel. A final anion exchange chromatography step was necessary to remove residual nucleotide contamination; in Buffer A (degassed), nucleotide-free CstR is present in the flow through at 300 mm NaCl. Dialysis into experimental buffer was carried out under an inert atmosphere (Vacuum Atmospheres glovebox). CstR was stored anaerobically at −80 °C.
Cu(I) X-ray Absorption Spectroscopy
Wild-type Sau CsoR was mixed with 0.8 mol eq of Cu(I) in 10 mm Hepes, 0.2 m NaCl, 30% (v/v) glycerol, pH 7.0, in an anaerobic environment and concentrated to a final protein concentration of ≈0.5–1.0 mm. Samples were loaded into standard XAS cuvettes or 5-well polycarbonate XAS cuvettes and immediately frozen in liquid N2. XAS data were collected at Stanford Synchrotron Radiation Lightsource (SSRL) on beamline 9-3. Extended x-ray absorption fine structure (EXAFS) data analysis was performed using EXAFSPAK software, using ab initio phase and amplitude functions computed with FEFF version 7.2, according to standard procedures as described (15, 16, 21).
Cysteine Modifications and Analysis by Mass Spectrometry
100 μl of unmodified and fully reduced CsoR or CstR (20 μm protomer) were incubated with various concentrations of sodium thiosulfate (Na2S2O3), sodium sulfide (NaS8), methylmethanethiosulfonate (MMTS), or sodium sulfite (Na2SO3) in 25 mm Hepes, pH 7.0, 0.2 m NaCl, at 25 °C for 17 h. All chemicals were reagent grade quality and obtained from Sigma or AlfaAesar. Quantitation of reaction products obtained with intact proteins was carried out by LC-ESI-MS on an Agilent 1200 HPLC-6130 MSD Quadrupole instrument fitted with a C18 column using a 5–95% acetonitrile gradient in 0.1% formic acid. These data were processed with ProTrawler (BioAnalyte Software). To determine the nature of the cross-linked peptide in sulfite-treated CstR, 1 unit of proteomics grade trypsin, resuspended in degassed water, was incubated with 100 μl of 20 μm apo-CstR or 20 μm Na2SO3-treated CstR under rigorously anaerobic conditions overnight in 25 mm Hepes, pH 7.0, 0.2 m NaCl, at 25 °C, and ESI-MS data were recorded and analyzed in the same way.
Fluorescence Anisotropy Titrations
Fluorescein-labeled dsDNA constructs were prepared as previously described (16) and diluted to 5 or 10 nm as indicated under strictly anaerobic conditions. 1–5-μl aliquots of protein were anaerobically injected into 2 ml of dsDNA and allowed to equilibrate for 3–5 min. Fluorescence anisotropy, ri, was measured with an ISS PC1 spectrofluorometer using λex of 495 nm. The signal was normalized to starting (ro) and ending (rcomplex) anisotropies, the data were plotted versus total protein concentration (monomer) and fit to a two nondissociable tetramer binding model using DynaFit (22). The binding of CstR to unlabeled OP1 variants was conducted using a standard competition assay (23).
RESULTS
CopA Expression Is Induced by Copper Salts in a Manner That Requires the Gene Encoded by NWMN_1991
S. aureus strain Newman as well as all other sequenced staphylococci contain a homolog (NWMN_1991) of the Cu(I) sensor CsoR from M. tuberculosis (Mtb) (13). This homolog conserves all previously identified critical features of the Cu(I)-dependent metalloregulatory switch (16) (supplemental Fig. S1). We therefore designated NWMN_1991 as CsoR and predicted that that it would regulate the transcription of copA, a proposed copper exporter, in a Cu(I)-dependent manner (Fig. 1A). To test this, we constructed a ΔcsoR derivative and monitored the expression of copA by quantitative RT-PCR in wild-type (WT) and ΔcsoR in response to 1 mm CuCl2 or MnCl2 (Fig. 1B). Consistent with previous reports, copA expression is induced in the presence of copper salts by ≈6-fold (24, 25), but not by Mn(II) (Fig. 1B). The ΔcsoR strain has elevated copA expression that is nearly independent of copper (Fig. 1B) suggesting that CsoR is responsible for controlling the copper stress response in S. aureus. Consistent with this finding is the observation that ΔcsoR is detectably more resistant to copper toxicity versus the wild-type strain (supplemental Fig. S2).
Sau CsoR Binds 1 mol eq of Cu(I) per Monomer with High Affinity and Adopts a Three-coordinate S2N Complex
A direct anaerobic titration of CuCl into apo Sau CsoR results in intense ligand to metal charge transfer absorption at ≈240 nm with an ϵ ≈ 15,000 m−1 cm−1 (supplemental Fig. S3) and a 1:1 Cu(I):protomer stoichiometry (supplemental Fig. S3, inset). Similar features characterize Cu(I) binding to Mtb and Bsu CsoRs (13, 15, 16). The Cu(I) binding affinity (KCu) was further quantified using an anaerobic bathocuprione disulfonate competition assay with log KCu determined to be 18.1 ± 0.5 (supplemental Table S1). Substitution of Cys41 or His66 (equivalent to essential residues Cys36 and His61 in Mtb CsoR) (13) with alanine results in a significant decrease in KCu, with log KCu of 14.5 ± 0.1 and 15.3 ± 0.1 for C41A and H66A CsoRs, respectively (supplemental Table S1).
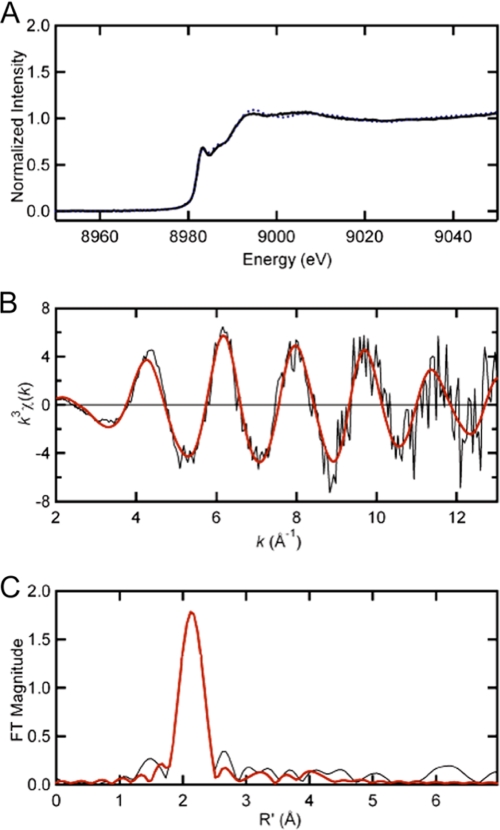
The Cu(I) coordination geometry was further investigated by x-ray absorption spectroscopy. As shown in Fig. 2A, the pre-edge peak of Cu(I)-CsoR at 8940 eV is consistent with a 1s → 4p excitation typical for 3-coordinate Cu(I) (13, 15, 26). The copper K-edge EXAFS spectrum as well as its Fourier transform for Sau CsoR with the best fit are shown in Fig. 2, B and C, respectively. The fitted parameters are compiled in supplemental Table S2. The fit suggests two Cu-S interactions at 2.20 Å and one Cu-N/O interaction at 2.01 Å. The significant outer shell scattering at 3–4 Å suggests that the third ligand is a histidine residue. Sau CsoR His66 corresponds to His61 in Mtb CsoR and His70 in Bsu CsoR (13, 15) and the Cu(I) binding affinity is significantly decreased in the H66A mutant (supplemental Table S1). These data reveal that His66 is a Cu(I) ligand that together with Cys41 and Cys70 complete the S2N coordination complex. DNA binding experiments (supplemental Fig. S4) show that Cu(I) binding to this site inhibits cop operator binding, whereas alanine substitutions of each of two Cu(I) ligands, Cys41 and His66, alters the DNA binding properties of S. aureus CsoR significantly (13, 16).
FIGURE 2.
X-ray absorption spectroscopy (XAS) of Cu(I)-bound Sau CsoR. A, copper K-edge x-ray absorption edge spectra of Cu(I)-bound WT Sau CsoR (solid black) and Mtb CsoR 1–106 (dashed blue) (16). The copper K-edge EXAFS spectrum and Fourier transforms (k3 weighted, k = 2–13 Å−1) for Cu(I)-bound WT Sau CsoR are shown in panels B and C, respectively. The solid red curves in panels B and C represent the best fits with parameters compiled under supplemental Table S2.
CstR (NWMN_0026.5) Is a Repressor of an Operon Predicted to Function in Sulfur Metabolism
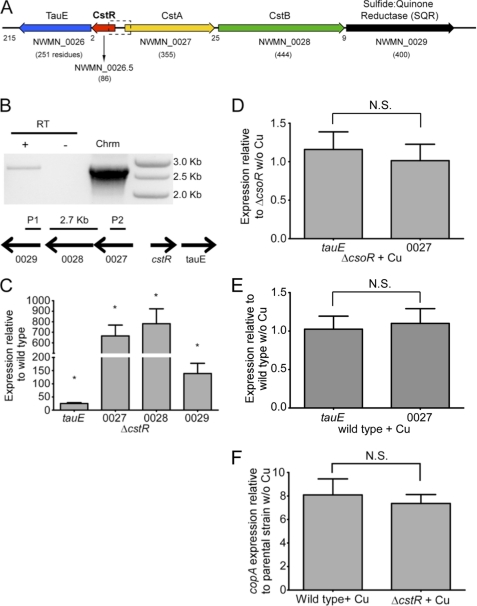
The experiments described above reveal that locus tag NWMN_1991 in S. aureus strain Newman encodes a bona fide Cu(I)-regulated CsoR. It was next of interest to determine the function of a previously unannotated open reading frame positioned between the NWMN_0026 and NWMN_0027 genes (Fig. 3A). This open reading frame, which we designate locus tag NWMN_0026.5, encodes a protein that is 35% identical and 65% similar to CsoR and is annotated in every other available S. aureus genome (supplemental Fig. S5). An analysis of the deduced amino acid sequence of NWMN_0026.5 reveals a CsoR/RcnR “WXYZ” four-amino acid fingerprint (11) of X-Cys-X-Cys (where X is any amino acid) that conserves the two Cu(I)-binding cysteines of CsoRs in the X and Z positions, but replaces the key Cu(I) coordinating His ligand (Y) with an Asn (supplemental Fig. S5). This suggests that the protein encoded by NWMN_0026.5 (CstR) cannot function as a Cu(I) sensor. To determine the genes regulated by CstR, we created a ΔcstR derivative and assessed expression of the immediately adjacent genes tauE, and NWMN_0027–0029 by quantitative RT-PCR. Loss of CstR gives rise to a massive increase in expression of all four genes (Fig. 3C). The genetic structure of this loci suggests that NWMN_0027 through NWMN_0029 forms an operon, which was confirmed by amplification of a PCR product spanning these genes from cDNA (Fig. 3B).
FIGURE 3.
CstR regulates the expression of immediately adjacent genes. A, schematic to scale representation of the genome encompassing locus tags NWMN_0026 to NWMN_0029 in S. aureus strain Newman in the immediate vicinity of NWMN_0026.5. Trivial names and the the number of amino acid residues of each encoded protein are shown, and the number of nucleotides that separate each ORF, for reference. TauE, putative sulfite/sulfonate effluxer; CstA and CstB, CsoR-like sulfur transferase-regulated genes A and B; SQR, putative sulfide:quinone reductase. SauSQR exhibits 57% identity to bona fide SQR from Bacillus stereothermophilus (Bst) (heavy metal tolerance-2 protein, HMT-2), which has been shown to catalyze sulfide-dependent menaquinone reduction (53). B, NWMN_0027 through NWMN_0029 form an operon. cDNA from ΔcstR was subjected to PCR using primers that anneal with the sqr and cstA genes and amplify a 2.7-kb product. No cDNA (RT) was used as a negative control, whereas chromosomal (Chrm) was used as a positive control. C–F, analysis of gene expression by quantitative RT-PCR. C, NWMN_0027 through 0029 and tauE levels in ΔcstR relative to wild type. NWMN_0027 and tauE levels in ΔcsoR (D) and wild-type (E) and in the presence and absence of copper. F, expression of copA in wild-type and ΔcstR strains. Results represent the average of three or more independent experiments. *, p < 0.05 determined by Student's t test. N.S., not significant. Error bars = S.D.
CstR Binds Specifically to Each of Two Operator Sites Situated between the cstR and tauE Genes
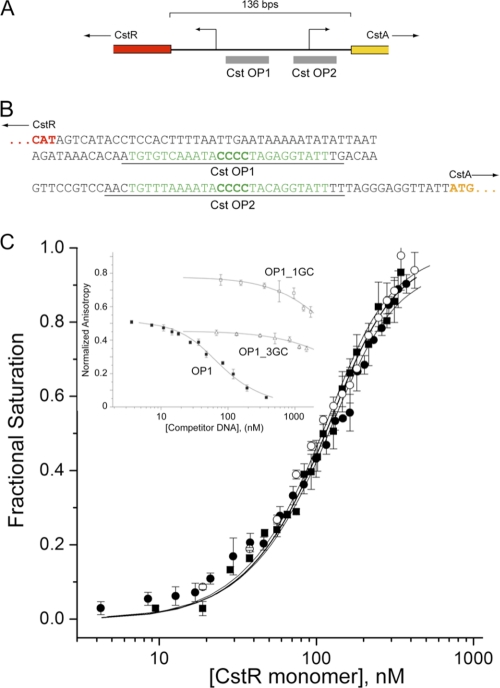
Two strong candidate tandemly repeated CstR operator sites, CstO OP1 and OP2, were found between the cstR and cstA genes. These sites were characterized by a run of four consecutive GC-base pairs and flanked by AT-rich regions (Fig. 4, A and B). We first purified recombinant, fully reduced Sau Newman CstR to ≥95% homogeneity from E. coli. CstR purified in this way migrates as a mixture of noncovalent dimers, tetramers, and octamers by gel filtration chromatography at 75 μm (monomer), with the primary species apparently octameric (supplemental Fig. S6). The binding of CstR to fluorescein-labeled OP1 and OP2 (see Fig. 4B, underlined sequences) was assessed by anaerobic titration measuring the fractional change in the anisotropy of the fluorescein fluorescence (Fig. 4C). CstR binds with high affinity to each cst operator site and the stoichiometry was determined using a model-independent method of analysis of two tetramers per operator, as previously determined for Mtb and Bsu CsoRs (15, 16) (supplemental Fig. S7). Fitting the binding data to a two-tetramer binding model gives an average tetramer-DNA association equilibrium constant, Ktet of ≈2.7 × 108 m−1, pH 7.0, 0.2 m NaCl, at 25 °C, with some evidence for positive cooperativity of stepwise binding of tetramers to the DNA (K2 > K1) (Table 1).
FIGURE 4.
Reduced CstR binds to cst operator (CstO) sites with high affinity in a manner dependent of the central run of four GC base pairs. A, schematic of the cstR-cstA intergenic region highlighting the positions of the two tandem candidate CstR operator sites. B, nucleotide sequence of the cstR-cstA intergenic region, highlighting the OP1 and OP2 operator sequences (green). Underlined bases correspond to the 5′-fluorescein-labeled duplex oligonucleotides used for DNA binding experiments. C, CstR binding isotherms for OP1 (solid squares, ■), OP2 (open circles, ○), and OP1_5GC (filled circles, ●) in which an additional GC base pair was inserted into the run of four GC base pairs. Inset, competition dissociation experiments with fluorescein-labeled apo-CstR-OP1 complexes with unlabeled wild-type OP1 (solid squares, ■), OP1-GC3 (open triangles, △), and OP1_1GC (open squares, □) duplexes. Fitted parameters derived from a two-tetramer binding model (15, 16) are compiled in Table 1. Conditions used were pH 7.0, 0.2 m NaCl, at 25.0 °C.
TABLE 1.
Equilibrium binding parameters for reduced S. aureus CstR or CsoR for fluorescein-labeled Cst OP1 or OP2 DNA duplexes
Conditions used were 10 mm Hepes, pH 7.0, 0.2 m NaCl, at 25.0 °C.
| Protein | DNA | ro | rcomplex | K1a | K2a | Kteta |
|---|---|---|---|---|---|---|
| × 107m−1 | ||||||
| CstR | OP1 | 0.129 | 0.226 | 0.7 (±0.3) | 55 (±5) | 27 (±5) |
| CstR | OP2 | 0.142 | 0.221 | 2.4 (±0.1) | 18 (±9) | 10 (±9) |
| CstR | OP1b | 6 (±5) | ||||
| CstR | OP1_5GC | 0.112 | 0.148 | 3 (±1) | 13 (±4) | 8 (±4) |
| CstR | OP1_3GCb | 0.5 (±0.1) | ||||
| CstR | OP1_1GCb | 0.09 (±0.01) | ||||
| CstR2S-S, CstR2S-S-S | OP1 | 0.129 | 0.226c | 0.31 (±0.03) | 0.15 (±0.02) | 0.23 (±0.03) |
| CstRS-SMe | OP1 | 0.129 | 0.226c | 0.15 (±0.04) | 0.0006 (±0.01) | 0.08 (±0.04) |
| Apo-CsoR | OP1 | 0.130 | 0.221 | 6 (±2) | 20 (±5) | 13 (±5) |
| Apo-CsoR | Cop | 0.092 | 0.101 | 0.2 (±0.2) | 5 (±6) | 3 (±6) |
| CsoRS-SMe | OP1 | 0.130 | 0.221d | 0.2 (±0.1) | 0.001 (±0.02) | 0.10 (±0.02) |
a Determined from a model that assumes two tetramers bind to each operator DNA with step-wise association constants of K1 and K2 and Ktet is the average macroscopic tetramer association constant (Ktet = sqrt(K1·K2).
b Determined by competitive dissociation experiments of fluorescently-labeled protein-OP1 complexes with unlabeled DNA competitors (23).
c Fixed to underivatized CstR.
d Fixed to underivatized CsoR.
To assess the DNA binding specificity of Cst OP1, we tested three mutant OP1 duplexes, one of which adds an additional GC base pair, OP1_5GC, whereas the others replace one (OP1_3GC) or three (OP1_1GC) GC base pairs with AT base pairs. The extended OP1_5GC duplex forms a complex with an affinity within 4-fold of OP1, whereas deletion of one or three GC pairs abolishes specific complex formation (Fig. 4C). These findings reveal that Sau CstR possesses sequence- and/or structure-specific DNA binding properties consistent with other CsoR/RcnR regulators (27).
The Cu(I) sensor CsoR binds to the non-cognate Cst OP1 with an affinity similar to that of CstR and ≈4-fold more tightly than to the cognate cop operator DNA (supplemental Fig. S8). This is not surprising given the similarity of the operator sequences, each of which is characterized by a run of 4–5 GC base pairs flanked by a 2–3 G-C base pairs (supplemental Fig. S9). This binding appears specific and is negatively regulated by Cu(I) (supplemental Fig. S10). However, the CstR regulon in wild-type or ΔcsoR strains is unaffected by copper added to the growth medium, indicating that CsoR has no role in CstR-regulated repression (Fig. 3, D–E). Likewise, Cu(I)-mediated depression of CsoR-regulated copA expression is unaffected by deletion of the cstR gene (Fig. 3F), further evidence that these two regulatory systems function independently of one another in the cell.
How Is cst Operator DNA Binding by CstR Regulated?
The cst regulon encodes a putative sulfite/sulfonate effluxer TauE (NWMN_0026) (28) and two of the four candidate rhodanese sulfur transferase domains in S. aureus (29) (CstA, NWMN_0027; CstB, NWMN_0028), a potential sulfur dioxygenase (CstB, NWMN_0027) (30), and a predicted sulfide quinone reductase (NWMN_0029) (see Fig. 3A) (31). These proteins may allow S. aureus to utilize thiosulfate (S2O32–) as a sulfur source for cysteine biosynthesis. To assimilate sulfur from thiosulfate, the S-S bond must ultimately be broken, perhaps catalyzed by a rhodanese domain, with the release of sulfite. S. aureus cannot metabolize sulfite, a potent oxidant or pro-oxidant that alters thiol-disulfide homeostasis, because it lacks both sulfite reductase and sulfite oxidase. We reasoned that sulfite may be toxic to S. aureus and could be effluxed by the CstR-inducible TauE. Growth curves on a chemically defined medium indeed reveal that S. aureus is capable of utilizing thiosulfate as a sole sulfur source for cysteine biosynthesis (32), with sulfide far less effective, and sulfite completely inhibitory to growth (supplemental Fig. S11). We therefore hypothesized that derivatization of CstR thiolates by thiosulfate, sulfite, or sulfide, rather than Cu(I) binding, might negatively regulate DNA binding.
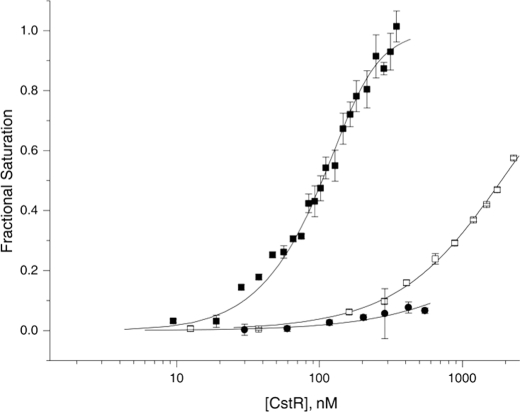
To test this, we performed anaerobic incubations of reduced CstR (20 μm protomer) with a 10–100-fold molar excess of sodium thiosulfate or sodium sulfide; these trials failed to yield any products other than the reduced CstR starting material as measured by ESI-MS of full-length proteins (Table 2). In striking contrast, anaerobic incubation with sodium sulfite at a 5–100-fold molar ratio over CstR cysteine resulted in three major oxidized products consistent with the following assignments: disulfide cross-linked dimer, CstR2S-S, and single and double trisulfide CstR2S-S—S cross-linked dimers where an additional sulfur atom bridges one or both cysteine pairs within the dimer (33) (Table 3). An ESI-MS spectrum of the interprotomer disulfide-bridged tryptic peptide obtained from a sulfite-treated CstR sample confirms the CstR2S-S assignment (Fig. 5), which is absent in untreated, reduced CstR starting material. This mixture of CstR2S-S and CstR2S-S—S binds weakly to the cst operator OP1, thus revealing that oxidation is strongly negatively regulatory (Fig. 6). In contrast, the Cu(I) sensor CsoR is unreactive toward sodium sulfite, even at a 100-fold molar excess of reagent (Table 2).
TABLE 2.
Molecular masses of reaction products obtained upon anaerobic incubation of reduced S. aureus CstR or CsoR with various sulfur-containing compounds
The conditions used are: 10 mm Hepes, pH 7.0, 0.2 m NaCl, at 25.0 °C. All 10-fold molar excess of Cys residues unless otherwise indicated, 20 μm protomer.
| Protein | Reactant | Molecular mass observed | Assignment | Expected modification | Molecular mass expecteda |
|---|---|---|---|---|---|
| Da | Da | ||||
| CstR | No addition | 9640.7 | 9641.2 | ||
| Na2S2O3 | 9640 | NRb | –SH (×2) | 9673.2 (9705.2) | |
| Na2S | 9640 | NR | –SH (×2) | 9673.2 (9705.2) | |
| Na2SO3 | 19280 | CstR2S-S | CstR2S-S | 19280.4 | |
| 19312 | CstR2S-S-S | CstR2S-S-S | 19312.4 | ||
| 19344 | CstR2(S-S-S)2c | CstR2(S-S-S)2c | 19344.4 | ||
| MMTS | 9732 | –SCH3 ×2 | –SCH3 (×2) | 9733.2 | |
| CsoR | No addition | 11035 | 11036.6 | ||
| Na2S2O3 | 11035 | NR | –SH (×2) | 11068.6 (11100.6) | |
| Na2S | 11035 | NR | –SH (×2) | 11068.6 (11100.6) | |
| Na2SO3d | 11035 | NR | CsoR2S-S | 22073.2 | |
| CsoR2S-S-S | 22105.2 | ||||
| MMTS | 11128 | –SCH3 ×2 | –SCH3 (×2) | 11130.6 |
a Expetced masses were calculated as average masses [M] with no modifictions at EXPASY.
b NR, no reaction.
c Both interprotomer cysteine linkages incorporate a sulfur atom in the dimer.
d 100-fold excess of reagent.
TABLE 3.
Molecular masses of major species identified by LC-ESI-MS as a result of anaerobic incubation with sodium sulfite of fully reduced CstR
The conditions used were: 10 mm Hepes, pH 7.0, 0.2 m NaCl, at 25.0 °C. The indicated fold-excess is molar excess over Cys residues (20 μm protomer, 40 μm cysteine).
| Reaction condition | Molecular mass observed | Assignment | Molecular mass expected |
|---|---|---|---|
| Da | Da | ||
| No reaction | 9640.7 | CstRa | 9641.2 |
| 9662.8 | CstR + Na+ | 9663.2 | |
| +5X SO32- | 9639.8 | CstR | 9641.2 |
| 19280.0 | CstR2S-S | 19280.4 | |
| 19311.8 (+32)b | CstR2S-S-S | 19312.4 | |
| 19324.7 (+45) | CstR2S-S + 2 Na+ | 19324.4 | |
| 19343.7 (+64) | CstR2(S-S-S)×2 | 19344.4 | |
| +10X SO32- | 9640.4 | CstR | 9641.2 |
| 9661.2 | CstR + Na+ | 9663.2 | |
| 19280.7 | CstR2S-S | 19280.4 | |
| 19295.1 (+14) | CstR2S-S + Oc | 19296.4 | |
| 19312.4 (+32) | CstR2S-S-S | 19312.4 | |
| 19323.4 (+44) | CstR2S-S + 2 Na+ | 19324.4 | |
| 19334.0 (+53) | CstR2S-S-S + Na+ | 19334.4 | |
| 19357.6 (+77) | CstR2S-S-S + 2 Na+ | 19356.4 | |
| 19366.5 (+86) | CstR2(S-S-S)×2 + Na+ | 19366.4 | |
| +100X SO32- | 9640.1 | CstR | 9641.2 |
| 19280.0 | CstR2S-S | 19280.4 | |
| 19311.3 (+31) | CstR2S-S-S | 19312.4 | |
| 19332.7 (+43) | CstR2S-S-S + Na+ | 19334.4 | |
| 19343.0 (+63) | CstR2(S-S-S)×2 | 19344.4 |
a CstR, reduced CstR monomer (starting material).
b Mass increase relative to oxidized dimer (CstR2S-S).
c One disulfide linkage, with one sulfenic acid (–SOH) on the opposite side of the dimer; tentative assignment.
FIGURE 5.
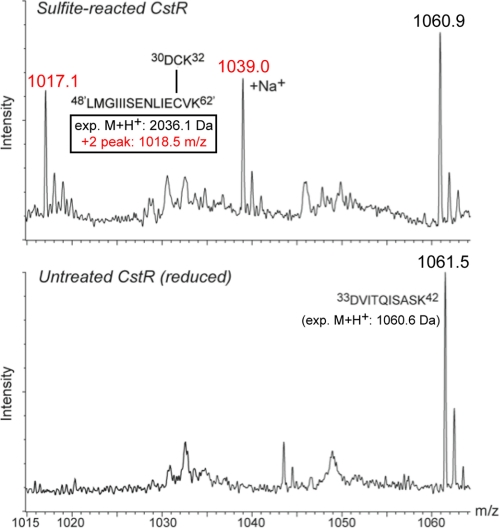
A portion of an ESI-MS of a tryptic digest of a sample of CstR reacted anaerobically with ×10 sulfite for 48 h versus one left untreated for 48 h. The peptide indicated is only found in the sulfite-reacted sample and has a molecular weight consistent with a disulfide cross-link between tryptic peptides corresponding to those containing the N-terminal Cys31 (30DCK32) and C-terminal Cys60, from opposite subunits within the dimer (expected monoisotopic M+H+ masses indicated).
FIGURE 6.
The DNA binding activity of CstR is negatively regulated by covalent cysteine disulfide and trisulfide derivatization by sodium sulfite. Unmodified fully reduced (solid squares, ■), sulfite-treated (open squares, □), and MMTS-reacted (solid circles, ●) CstRs were titrated into a solution of 10 nm Cst OP1 DNA and anisotropy (ri) was recorded in triplicate. Each data point represents the mean ± S.D. following each ith addition of protein, following normalization to the starting (ro) and ending (rcomplex) values of the anisotropy of the underivatized CstR complex. The solid line is a fit to a two-tetramer binding model, with the anisotropy of the 1:1 complex fixed to 0.5 × (rcomplex − ro). Fitted parameters are complied in Table 1. Conditions used were 10 mm Hepes, pH 7.0, 0.2 m NaCl, at 25.0 °C.
Anerobic incubation of reduced CstR with a 10-fold molar excess of the thiol modifying reagent MMTS quantitatively converts CstR to the doubly S-methylated derivative, denoted CstRSSMe (Table 2). The mixed disulfide RS-SCH3 is the methyl analog of a cysteine persulfide (RS-SH) as well as other mixed disulfides (RS-SR′) that could form with low molecular weight cellular thiols, and is thus of interest. CstRSSMe also binds weakly to the OP1 DNA, revealing strong inhibition of CstR binding upon S-methylation, like that observed for oxidation (Fig. 6). Treatment of the Cu(I) sensor CsoR under the same conditions yielded no reaction, although quantitative derivatization was possible at a 100-fold molar excess of MMTS (Table 2). CsoR derivatized in this way also results in weak non-cognate cst operator binding (supplemental Fig. S10).
DISCUSSION
In this work, we characterize two transcriptional regulatory responses controlled by paralogous regulators from the CsoR/RcnR family of proteins. One is a Cu(I)-stress response under control of a CsoR (13). The other encodes proteins that we predict will be required for sulfur assimilation from an inorganic sulfur source, thiosulfate, and is under control of a new regulator, termed CstR. CstR is the first characterized member of a subfamily of CsoR/RcnR regulators that do not appear to respond to transition metal stress, but are characterized by a high reactivity toward sulfite and the sulfur-modifying agent MMTS. Our data support the hypothesis that the interprotomer cysteine pair in CstR relative to CsoR readily forms a disulfide or trisulfide bond across this interface. This in turn drives a conformational change in the oligomer that reduces the affinity of CstR for the operator DNA, thus leading to transcriptional depression of the operon in vivo. CsoR, on the other hand, is not effectively derivatized by sulfite (Table 2).
Sulfur Assimilation in S. aureus
S. aureus is characterized by a unique thiol metabolism and is strongly restricted in its ability to obtain inorganic sulfur to make cysteine (32, 34). Sulfate is the preferred bacterial sulfur source but all sequenced S. aureus strains lack adenosine 5′-phosphosulfate reductase and thus cannot utilize sulfate as a sulfur source. Sulfite and alkylsulfonates, e.g. taurine, are likewise not assimilated and sulfite cannot be metabolized (35, 36). Organic sulfur sources, including glutathione and homocysteine, are preferred sources of cysteine, but both must be acquired by S. aureus from the host.
We establish here that S. aureus strain Newman is able to grow on thiosulfate as the sole sulfur source (36). Thiosulfate is a major excreted sulfur-containing compound in humans, and is thought to be generated by the oxidation of bacterially derived H2S by an essential mitochondrial sulfur dioxygenase in mucosal cells to protect against sulfide toxicity (30). Although the bioavailability of thiosulfate in S. aureus abscesses is not known, recent findings reveal that colonization of the gastrointestinal epithelium by Salmonella gives rise to an inflammatory burst that oxidizes thiosulfate to tetrathionate (S4O62–), which is used as an alternative terminal electron acceptor for respiration under anaerobic conditions (37). Although S. aureus lacks the tetrathionate respiratory complex, a major form of oxidized sulfur in inflamed tissues may well be tetrathionate. The physiological inducer of the cst regulon is as yet unknown.
One hypothesis is that the cst genes are induced by any condition that specifically alters thiol-disulfide homeostasis, including, but not limited to, sulfite stress. This would result in facile oxidation of the regulatory cysteines in CstR or formation of mixed disulfides with major low molecular weight thiols in S. aureus, e.g. bacillithiol (34, 38), coenzyme A, or cysteine, each of which is expected to drive transcriptional derepression on the basis of the observed weak DNA binding activity of S-methylated CstR (Fig. 6). General perturbation of thiol-disulfide homeostasis would signal to the organism to obtain sulfur for cysteine and/or bacillithiol biosynthesis from all available sources, including inorganic thiosulfate, particularly under conditions of low host-derived organic sulfur.
Another possible inducer of cst expression is the sulfonated β-amino acid taurine or an as yet unidentified taurine metabolite. S. aureus does not encode a taurine repressor TauR; thus, taurine might also be indirectly sensed by CstR as sulfite or another metabolite (39). Taurine is a highly abundant amino acid in mammals and a dedicated Na+-dependent uptake system for taurine is encoded by ssuBAC in S. aureus (35). However, S. aureus does not encode TauD or SsuDE and it is therefore unlikely that taurine is oxidized to aminoacetaldehyde with the release of sulfite. The other known alternative route of degradation of taurine is release of the NH4+ with the formation of negatively charged sulfoacetaldehyde (40). It is interesting to note that both routes of taurine degradation give rise to aldehydes, highly electrophilic carbon atoms that may well be quite reactive with CstR cysteines to form thioesters. Indeed, formaldehyde stress in E. coli has been proposed to be sensed by a CsoR family member FrmR, the mechanism of which is unknown (11, 41), and in B. subtilis, bacillithiol protects cells against electrophile stress (42).
DNA Binding and Cross-transcriptional Regulation of CsoR Versus CstR
The mechanism of operator recognition by these novel disc-shaped CsoR/RcnR family transcriptional repressors is not fully understood. Previous work proposed two general types of DNA operators, termed type 1 and type 2, which differ in the organization and number of GC base pair tracts (27) but seem to incorporate features of “shape selective” or “indirect readout”-based recognition of a pair of A-B helical junctions, and specific features of the flanking base pairs.
The type 2 operators possess two short stretches of GC base pairs (one block of 2–3 base pairs, another of 4–5 base pairs, often of opposite strand polarity) separated by 3–4 base pairs, and flanked by a nearly conserved palindromic ATA sequence sometimes in the context of a larger inverted repeat (supplemental Fig. S9). Both cop and cst operators in S. aureus as well as that of B. subtilis CsoR (15, 20) and all target sequences for M. tuberculosis RicR (6) conform to this consensus sequence. This places two predicted A-B helical junctions one helical turn (10–11 base pairs) apart (27), and this may facilitate the binding of two tetramers at these junctions, as validated for Bsu CsoR (15) and CstR here (supplemental Fig. S7). Such a model also explains why addition of a fifth consecutive GC base pair to Cst_OP1 is energetically nearly silent (Table 1); this simply increases the spacing between flanking palindromes from 10 to 11 base pairs, exactly as found in the Sau cop operator (supplemental Fig. S9). Reducing the number of consecutive GC base pairs to three (in CstOP1_3GC) may well change the structure of the operator, abolishing shape selective recognition by CstR.
The Sau cop operator sequence contains one consensus operator (Fig. 1), and the divergently transcribed cstR-cstA intergenic region contains two consensus operators, each of which binds two CstR tetramers (Fig. 4). As a result, tandem operators in the cstR-cstA intergenic region can therefore accommodate four tetramers (two octamers) in total, and thus can potentially adopt a higher order, strongly repressing a nucleoprotein complex that is unlikely to occur in the single operator cop sequence. Perhaps CsoR is not able to assembly such a higher order structure and thus does not strongly repress the cst operon in ΔcstR cells (Fig. 3) despite binding to a single consensus operator site with an affinity equal to or better that that of cognate CstR (Table 1).
S. aureus CsoR and Copper Toxicity
Control of copper homeostasis in bacteria is particularly important because nearly all prokaryotes, outside of photosynthetic bacteria, appear to lack an obligatory intracellular cytoplasmic copper requirement (43–45). Uncomplexed copper ions are extremely toxic, attributed to an autocatalytic Haber-Weiss reaction (46) or displacement of iron from solvent-exposed iron-sulfur clusters in proteins (47). As a result, cytoplasmic bioavailable copper levels must be strictly controlled and are proposed to be buffered at concentrations in the 10−18–10−21 m range despite a total concentration in the low micromolar range (48, 49).
Here, we establish that S. aureus CsoR is a Cu(I) sensing CsoR, binds Cu(I) with an affinity (KCu) of 1018 m−1, and possesses structural, allosteric and Cu(I) coordination features characteristic of Mtb and Bsu CsoRs (15, 16). A major point of departure from other bacterial systems is that the sensor is genetically unlinked from the expression of the copA and copZ resistance genes, thus making its identification and functional role more difficult to establish; furthermore, S. aureus CsoR is unique is that it oligomerizes beyond the stable tetramer assembly state under the conditions used here (supplemental Figs. S12 and S13). The significance of this finding is unknown.
Microarray experiments in S. aureus SH1000 reveal that the expression of CsoR is not strongly induced by copper stress, unlike the situation in M. tuberculosis (13, 50); as a result, constitutively expressed CsoR might play a role in buffering “free” Cu(I) in S. aureus in the ≈10−18 m range. On the other hand, the gene encoding the likely copper chaperone CopZ just downstream from copA (Fig. 1A) is induced by copper (24, 25), but may not be co-transcribed with copA (24). Although it is not yet known whether the transcription of copZ is also regulated by CsoR, inspection of the intergenic region between copA and copZ does not reveal an obvious candidate CsoR binding site like that found in the copA operator-promoter region, nor have we determined if copZ transcription is altered in the ΔcsoR strain.
Implications for Bacterial Virulence
It is interesting to note that the cst operon core (tauE-cstR-cstAB) and both CstR operator binding sites (Fig. 3) are duplicated in the genomes of major antibiotic-resistant S. aureus strains (N315, COL, and Mu50) (51). In contrast, the tauE gene is lost in at least one methicillin-sensitive Sau strain (MSSA) (51). These genomics data are consistent with the idea that this putative sulfur assimilation regulon provides a growth advantage to Sau during infection of the host. Recent findings suggest that alterations in sulfur metabolism influence the ability of Sau to form biofilms, an essential feature of its ability to thrive both inside and outside of the host (32). Microarray experiments carried out on mid-log liquid cultures of S. aureus SH1000 reveal that copper stress, sufficient to induce the CsoR-dependent expression of copA and perhaps other genes that function in oxidative stress resistance (25), negatively regulates biofilm formation via repression of the sae and agr regulons (52). Thus, CsoR and CstR-related stress response pathways may be physiologically linked by a connection to alteration in biofilm formation and oxidative stress and ultimately viability in the vertebrate host.
Acknowledgments
Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and the National Institutes of Health, National Center for Research Resources Biomedical Technology Program.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM042569 (to D. P. G.), U54 AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense (to E. P. S.), and R01 GM042025 (to R. A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods,” Tables S1–S3, Figs. S1–S13, and “Results.”
- CsoR
- copper-sensitive operon repressor
- MMTS
- methylmethanethiosulfonate
- EXAFS
- extended x-ray absorption fine structure
- ESI
- electrospray ionization
- OP
- operator-promoter.
REFERENCES
- 1. Conrady D. G., Brescia C. C., Horii K., Weiss A. A., Hassett D. J., Herr A. B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19456–19461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moen A. E., Storla D. G., Bukholm G. (2010) FEMS Immunol. Med. Microbiol. 58, 374–380 [DOI] [PubMed] [Google Scholar]
- 3. White C., Lee J., Kambe T., Fritsche K., Petris M. J. (2009) J. Biol. Chem. 284, 33949–33956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Osman D., Waldron K. J., Denton H., Taylor C. M., Grant A. J., Mastroeni P., Robinson N. J., Cavet J. S. (2010) J. Biol. Chem. 285, 25259–25268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ward S. K., Abomoelak B., Hoye E. A., Steinberg H., Talaat A. M. (2010) Mol. Microbiol. 77, 1096–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Festa R. A., Jones M. B., Butler-Wu S., Sinsimer D., Gerads R., Bishai W. R., Peterson S. N., Darwin K. H. (2011) Mol. Microbiol. 79, 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burns K. E., Baumgart S., Dorrestein P. C., Zhai H., McLafferty F. W., Begley T. P. (2005) J. Am. Chem. Soc. 127, 11602–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hong J. A., Bhave D. P., Carroll K. S. (2009) J. Med. Chem. 52, 5485–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santo C. E., Morais P. V., Grass G. (2010) Appl. Environ. Microbiol. 76, 1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weaver L., Noyce J. O., Michels H. T., Keevil C. W. (2010) J. Appl. Microbiol. 109, 2200–2205 [DOI] [PubMed] [Google Scholar]
- 11. Iwig J. S., Leitch S., Herbst R. W., Maroney M. J., Chivers P. T. (2008) J. Am. Chem. Soc. 130, 7592–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma Z., Jacobsen F. E., Giedroc D. P. (2009) Chem. Rev. 109, 4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu T., Ramesh A., Ma Z., Ward S. K., Zhang L., George G. N., Talaat A. M., Sacchettini J. C., Giedroc D. P. (2007) Nat. Chem. Biol. 3, 60–68 [DOI] [PubMed] [Google Scholar]
- 14. Sakamoto K., Agari Y., Agari K., Kuramitsu S., Shinkai A. (2010) Microbiology 156, 1993–2005 [DOI] [PubMed] [Google Scholar]
- 15. Ma Z., Cowart D. M., Scott R. A., Giedroc D. P. (2009) Biochemistry 48, 3325–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma Z., Cowart D. M., Ward B. P., Arnold R. J., DiMarchi R. D., Zhang L., George G. N., Scott R. A., Giedroc D. P. (2009) J. Am. Chem. Soc. 131, 18044–18045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bae T., Schneewind O. (2006) Plasmid 55, 58–63 [DOI] [PubMed] [Google Scholar]
- 18. Kehl-Fie T. E., Porsch E. A., Miller S. E., St. Geme J. W., 3rd (2009) J. Bacteriol. 191, 4976–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baba T., Bae T., Schneewind O., Takeuchi F., Hiramatsu K. (2008) J. Bacteriol. 190, 300–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smaldone G. T., Helmann J. D. (2007) Microbiology 153, 4123–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu T., Chen X., Ma Z., Shokes J., Hemmingsen L., Scott R. A., Giedroc D. P. (2008) Biochemistry 47, 10564–10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuzmic P. (1996) Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 23. Grossoehme N. E., Li L., Keane S. C., Liu P., Dann C. E., 3rd, Leibowitz J. L., Giedroc D. P. (2009) J. Mol. Biol. 394, 544–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sitthisak S., Knutsson L., Webb J. W., Jayaswal R. K. (2007) Microbiology 153, 4274–4283 [DOI] [PubMed] [Google Scholar]
- 25. Baker J., Sitthisak S., Sengupta M., Johnson M., Jayaswal R. K., Morrissey J. A. (2010) Appl. Environ. Microbiol. 76, 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kau L. S., Spira-Solomon D. J., Penner-Hahn J. E., Hodgson K. O., Solomon E. I. (1987) J. Am. Chem. Soc. 109, 6433–6442 [Google Scholar]
- 27. Iwig J. S., Chivers P. T. (2009) J. Mol. Biol. 393, 514–526 [DOI] [PubMed] [Google Scholar]
- 28. Weinitschke S., Denger K., Cook A. M., Smits T. H. (2007) Microbiology 153, 3055–3060 [DOI] [PubMed] [Google Scholar]
- 29. Cipollone R., Ascenzi P., Visca P. (2007) IUBMB Life 59, 51–59 [DOI] [PubMed] [Google Scholar]
- 30. Tiranti V., Viscomi C., Hildebrandt T., Di Meo I., Mineri R., Tiveron C., Levitt M. D., Prelle A., Fagiolari G., Rimoldi M., Zeviani M. (2009) Nat. Med. 15, 200–205 [DOI] [PubMed] [Google Scholar]
- 31. Marcia M., Ermler U., Peng G., Michel H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9625–9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soutourina O., Poupel O., Coppée J. Y., Danchin A., Msadek T., Martin-Verstraete I. (2009) Mol. Microbiol. 73, 194–211 [DOI] [PubMed] [Google Scholar]
- 33. Ball S., Milne J. (1995) Can. J. Chem. 73, 716–724 [Google Scholar]
- 34. Newton G. L., Rawat M., La Clair J. J., Jothivasan V. K., Budiarto T., Hamilton C. J., Claiborne A., Helmann J. D., Fahey R. C. (2009) Nat. Chem. Biol. 5, 625–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Giehl T. J., Qoronfleh M. W., Wilkinson B. J. (1987) J. Gen. Microbiol. 133, 849–856 [DOI] [PubMed] [Google Scholar]
- 36. Lithgow J. K., Hayhurst E. J., Cohen G., Aharonowitz Y., Foster S. J. (2004) J. Bacteriol. 186, 1579–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Winter S. E., Thiennimitr P., Winter M. G., Butler B. P., Huseby D. L., Crawford R. W., Russell J. M., Bevins C. L., Adams L. G., Tsolis R. M., Roth J. R., Bäumler A. J. (2010) Nature 467, 426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Helmann J. D. (2010) Antioxid. Redox Signal, in press [Google Scholar]
- 39. Wiethaus J., Schubert B., Pfänder Y., Narberhaus F., Masepohl B. (2008) J. Bacteriol. 190, 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krejcík Z., Denger K., Weinitschke S., Hollemeyer K., Paces V., Cook A. M., Smits T. H. (2008) Arch. Microbiol. 190, 159–168 [DOI] [PubMed] [Google Scholar]
- 41. Herring C. D., Blattner F. R. (2004) J. Bacteriol. 186, 6714–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gaballa A., Newton G. L., Antelmann H., Parsonage D., Upton H., Rawat M., Claiborne A., Fahey R. C., Helmann J. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6482–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tottey S., Harvie D. R., Robinson N. J. (2005) Acc. Chem. Res. 38, 775–783 [DOI] [PubMed] [Google Scholar]
- 44. Osman D., Cavet J. S. (2008) Adv. Appl. Microbiol. 65, 217–247 [DOI] [PubMed] [Google Scholar]
- 45. Waldron K. J., Robinson N. J. (2009) Nat. Rev. Microbiol. 7, 25–35 [DOI] [PubMed] [Google Scholar]
- 46. Solioz M., Stoyanov J. V. (2003) FEMS Microbiol. Rev. 27, 183–195 [DOI] [PubMed] [Google Scholar]
- 47. Macomber L., Imlay J. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., O'Halloran T. V. (1999) Science 284, 805–808 [DOI] [PubMed] [Google Scholar]
- 49. Changela A., Chen K., Xue Y., Holschen J., Outten C. E., O'Halloran T. V., Mondragón A. (2003) Science 301, 1383–1387 [DOI] [PubMed] [Google Scholar]
- 50. Ward S. K., Hoye E. A., Talaat A. M. (2008) J. Bacteriol. 190, 2939–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Highlander S. K., Hultén K. G., Qin X., Jiang H., Yerrapragada S., Mason E. O., Jr., Shang Y., Williams T. M., Fortunov R. M., Liu Y., Igboeli O., Petrosino J., Tirumalai M., Uzman A., Fox G. E., Cardenas A. M., Muzny D. M., Hemphill L., Ding Y., Dugan S., Blyth P. R., Buhay C. J., Dinh H. H., Hawes A. C., Holder M., Kovar C. L., Lee S. L., Liu W., Nazareth L. V., Wang Q., Zhou J., Kaplan S. L., Weinstock G. M. (2007) BMC Microbiol. 7, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Voyich J. M., Vuong C., DeWald M., Nygaard T. K., Kocianova S., Griffith S., Jones J., Iverson C., Sturdevant D. E., Braughton K. R., Whitney A. R., Otto M., DeLeo F. R. (2009) J. Infect. Dis. 199, 1698–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shibata H., Suzuki K., Kobayashi S. (2007) Can. J. Microbiol. 53, 1091–1100 [DOI] [PubMed] [Google Scholar]