Abstract
Macrolide-specific efflux pump MacAB-TolC has been identified in diverse Gram-negative bacteria including Escherichia coli. The inner membrane transporter MacB requires the outer membrane factor TolC and the periplasmic adaptor protein MacA to form a functional tripartite complex. In this study, we used a chimeric protein containing the tip region of the TolC α-barrel to investigate the role of the TolC α-barrel tip region with regard to its interaction with MacA. The chimeric protein formed a stable complex with MacA, and the complex formation was abolished by substitution at the functionally essential residues located at the MacA α-helical tip region. Electron microscopic study delineated that this complex was made by tip-to-tip interaction between the tip regions of the α-barrels of TolC and MacA, which correlated well with the TolC and MacA complex calculated by molecular dynamics. Taken together, our results demonstrate that the MacA hexamer interacts with TolC in a tip-to-tip manner, and implies the manner by which MacA induces opening of the TolC channel.
Keywords: Antibiotics, Bacteria, Electron Microscopy (EM), Membrane Proteins, Protein-Protein Interactions
Introduction
Drug resistance of microbial pathogens presents an increasing threat to public health (1). In Gram-negative pathogens, high levels of intrinsic or acquired drug resistance are conferred by three-component multidrug efflux pumps, which are composed of the inner membrane transporter, the outer membrane factor (OMF), and the periplasmic membrane fusion protein (MFP)4 (2–5). These tripartite complexes span the entire two-membrane envelope of Gram-negative bacteria and expel various molecules into the medium, utilizing a proton gradient or ATP hydrolysis. The inner membrane transporters belong to one of three structurally dissimilar superfamilies of proteins: resistance-nodulation-cell division (RND), ATP-binding cassette (ABC), or major facilitator. The inner membrane transporters expel the substrates through the central channel of the OMF, as exemplified by Escherichia coli TolC, which spans the outer membrane (6). The MFP, which connects the other two components in the periplasm, is also essential for the function of the efflux pump.
In E. coli, AcrAB-TolC acts as a major multidrug efflux pump (7–9), where AcrB is the RND-type inner membrane transporter and AcrA belongs to MFP. The homotrimeric TolC is embedded in the outer membrane and continues ∼100 Å into the periplasmic space as an α-barrel composed of six α-hairpins that form the wall of a 35-Å inner-diameter cylindrical channel (10). The TolC channel is closed at the aperture end and the channel opening is induced only in the presence of the other components, the mechanism of which remains to be determined at the molecular level.
The MacAB-TolC pump has been identified in E. coli; the inner membrane transporter MacB belongs to non-canonic ABC-type transporters (8, 9, 11, 12), and MFP MacA shares structural similarity with AcrA (sequence similarity 44%) (13). Overproduction of MacAB results in increased resistance to the macrolide antibiotics in macrolide-susceptible AcrAB-deficient E. coli (8, 9, 11).
The structures of E. coli MacA and AcrA and its Pseudomonas aeruginosa homologue, MexA, have been determined and revealed that MFPs are elongated, sickle-shaped molecules comprised of four linearly arranged domains (membrane proximal (MP), β-barrel, lipoyl, and α-hairpin domains) (14–16). It is worth noting that MacA was crystallized as a funnel-like hexameric assembly with an α-barrel composed of six α-hairpins from each protomer. Genetic and biochemical studies further revealed that this hexameric assembly of MacA and the tip region of the α-barrel are functionally important. Based on the structural complementarity between the six-α-hairpin-membered α-barrels of MacA and TolC, we speculated a tip-to-tip interaction between MacA and TolC (17, 18). However, it remains to be elucidated which region of TolC makes contact with α-barrels of MacA.
To investigate the assembly mechanism of MacA-TolC interaction, we made a complex of MacA and TolC using a chimeric protein containing the tip regions of the α-barrel of TolC. Subsequent study of the complex revealed the assembly of MacA-TolC and provided insight into how the TolC channel is opened in association with MacA binding.
EXPERIMENTAL PROCEDURES
Construction of Chimeric Proteins Containing Aa MacA and E. coli TolC α-Helical Tip Regions
To construct the expression vector for Actinobacillus actinomycetemcomitans (hereafter referred to as Aa) MacA-TolCαR1-hybrid, DNA fragments encoding Aa MacA residues 29–123, E. coli TolC residues 136–159, and Aa MacA residues 148–394 (with/without the stop codon) were joined using the overlapping PCR technique, and then ligated into the NcoI and XhoI sites of pPROEX-HTA (Invitrogen), generating the vector pPRO-AaMacA-TolCαR1 and pPRO-AaMacA-TolCαR1-nostop, respectively. To construct the expression vector for Aa MacA-TolCαR2-hybrid, DNA fragments encoding Aa MacA residues 29–123, E. coli TolC residues 354–377, and Aa MacA residues 148–394 (including stop codon), were joined by the method described above, resulting in the vector pPRO-AaMacA-TolCαR2. To construct the expression vector for MacA-TolCα-hybrid-dimer, the DNA fragments encoding Aa MacA-TolCαR2-hybrid were inserted into the pPRO-AaMacA-TolCαR1-nostop vector at the XhoI and HindIII sites, resulting in the vector pPRO-MacA-TolCα-hybrid-dimer.
Construction of Aa MacA-TdeAαR1-hybrid
To construct the expression vector for Aa MacA-TdeAαR1-hybrid, the DNA fragments encoding Aa MacA residues 29–123, Aa TdeA residues 192–215, Aa MacA residues 148–394 were joined using the overlapping PCR technique, and then ligated into the NcoI and XhoI sites of pPROEX-HTA (Invitrogen). The sequences of the primers used for the plasmids are described in supplemental Table S1.
Measurement of Minimum Inhibitory Concentration (MIC)
Overnight grown cultures in LB medium containing appropriate antibiotics (100 μg of kanamycin, 100 μg of ampicillin, and/or 5 μg of tetracycline per ml) were 1 to 100 diluted in the same medium. At A600 = 0.1, the expression of proteins were induced by adding 0.1% arabinose and (/or) 1 mm IPTG to the cultures and incubated two more hours. Approximately 104 of the induced cells were then added to the same medium containing 0.1% arabinose and (/or) 1 mm IPTG and erythromycin at increasing concentrations. Cultures were grown for additional 16–20 h, and the lowest concentration of erythromycin that completely inhibited growth was designated as the MIC.
Size Exclusion Chromatography
To determine the molecular size of proteins or to learn whether the two proteins form a complex in solution, size-exclusion chromatography was performed at a flow rate of 0.5 ml/min on Superdex S-200 HR 10/30 (GE Healthcare) equilibrated with 20 mm Tris buffer (pH 8.0) containing 150 mm NaCl.
Isothermal Titration Calorimetry
ITC measurements were performed at 20 °C using a VP-ITC calorimeter (Microcal, Inc.). For E. coli MacA and Aa MacA-TolCα-hybrid-dimer, E. coli MacA and Aa MacA-TolCα-hybrid-dimer were concentrated to 318 and 12.5 μm, respectively, in a buffer containing that 20 mm Hepes (pH 7.0) and 150 mm NaCl. For Aa MacA and MacA-TdeAαR1-hybrid, the proteins were concentrated to 133 and 7 μm, respectively, in the same buffer. To titrate 1.4 ml of the MacA-TolCα-hybrid-dimer (or MacA-TdeAαR1-hybrid) solution, the E. coli MacA (or Aa MacA) solution was used as a titrant. The titration experiment was carried out by 20 10-μl injections. Blank titrations, which were carried out by injecting ligand into the buffer depending on the particular experiment, were subtracted from each data set.
Electron Microscopy and Image Processing
Data were acquired using a Tecnai F20 Twin transmission electron microscope operating at 120 kV, using a dose of ∼15 e−/Å2 and a nominal underfocus ranging from 1.0 to 2.0 μm. 1,292 images were automatically collected at a nominal magnification of 80,000× at a pixel size of 0.105 nm at the specimen level. All images were recorded with a Tietz F415 4k × 4k pixel CCD camera (15 μm pixel) utilizing the Leginon data collection software (19). Experimental data were processed by the Appion software package (20), which interfaces with the Leginon database infrastructure. 5,767 particles were automatically selected (21) from the micrographs and extracted at a box size of 448 pixels. Stacked particles were binned by a factor of 2 for the final reconstruction. The final stack contained 4,858 particles. The initial model was created by applying D6-symmetry (6-fold symmetry along the z axis, 2-fold rotational axis 90° from z) on manually selected two-dimensional averages, which were formed using the XMIPP reference-free maximum likelihood alignment procedure (22). The three-dimensional reconstruction was carried out using the IMAGIC-5 reconstruction package (23). Resolution was assessed by calculating the Fourier Shell Correlation (FSC) at a cutoff of 0.5, which provided a value of 25 Å resolution. Automatic Docking was performed using the Chimera software package (24).
In Silico Molecular Dynamics Simulation of the TolC-MacA Complex
We adopted a model for an open structure of TolC in which the structure of TolC α-barrel region was replaced by that of MacA hexamer. Having selected a putative cogwheel-to-cogwheel binding conformation for TolC-MacA complex consisting of 2,844 residues in total, we subjected it to the in silico molecular dynamics (MD) simulation to come up with an equilibrium complex-structure. The MD simulation of TolC-MacA complex in the explicit TIP3 water was performed with the atomistic force-field AMBER/v11.ff99SB under the SHAKE option. The TolC-MacA complex was immersed in a periodic rectangular three-dimensional box of TIP3 water molecules. The minimal distance of the TolC-MacA complex protein from the box boundary was 0.8 nm, containing about 108,067 water molecules. The cut-off distance for the electrostatic and van der Waals interaction was 0.8 nm, and the particle mesh Ewald method was used to include the long-range interactions. We first minimized the energy of the TolC-MacA complex by the steepest descent method for 1,000 steps followed by the conjugate gradient method for 1,000 steps. The water molecules were heated for 20 ps from 0 to 330 K. Then the MD simulation was performed with the NPT ensemble with a 2-fs integration time step. The temperature was regulated by Langevin dynamics, and the pressure was kept at 1 atm. Starting from an initial putative TolC-MacA complex, the MD simulation ran for 1.2 ns, from which the coordinates of all atoms were saved every 2 ps for the detailed analysis of not only the thermodynamic stability but also the structural characteristics of the TolC-MacA complex. The binding energy of the complex was calculated using the MM/GBSA algorithm in AMBER/v11. The solvent-accessible surface was calculated using AREAIMOL in the CCP4 Suite (25). The figures were generated with the program PYMOL (26).
RESULTS
Construction of Chimeric Proteins Containing TolC-α-Barrel Tip
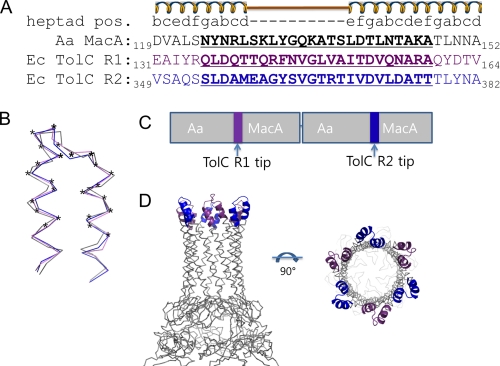
A previous study on the crystal packing of Aa MacA provided clues into how the MacA hexamer contacts the TolC trimer (18). The α-barrels from the neighboring Aa MacA hexameric units made a cogwheel-to-cogwheel interaction in a tip-to-tip manner in the crystal, suggesting that the α-barrel tip region of the upper Aa MacA hexamer might mimic the structure of the TolC aperture tip region in the MacA-TolC complex because of the resemblance between the α-barrels of the TolC trimer and the MacA hexamer. Even the inclinations of the α-hairpins in the α-barrels of the MacA and TolC are similar, although the central channel of the α-barrels of TolC becomes narrower at the end, while the diameter of the channel is virtually unchanged in MacA (18).
Given the interaction between the two MacA hexameric units in the crystal, albeit forced by the crystallization, and the similarities of MacA and TolC at the α-barrel tip, we hypothesized that the conformation of the tip region of the α-barrel of TolC might resemble that of MacA, and that only the α-barrel tip region of TolC is involved in the MacA binding. To test our hypothesis, we designed a chimeric form of Aa MacA in which the α-helical tip region (24 amino acids, residues 124–147) was substituted with the corresponding region of the TolC aperture tip region (24 amino acids, residues 136–159 or 354–377) (Fig. 1A). Because the TolC protomer comprises two structural repeats, two α-hairpins are found at the aperture tip region in a TolC protomer. To mimic the TolC protomer that contains two α-hairpins, we constructed the chimeric protein in two steps. Two chimeric Aa MacA proteins, whose α-helical tip regions were replaced with the first or second aperture tip region of TolC, were designated as Aa MacA-TolCαR1-hybrid and Aa MacA-TolCαR2-hybrid, respectively. Then the two chimeric proteins were linked in tandem to generate the Aa MacA-TolCα-hybrid-dimer construct, whose α-helical tip region was replaced with the first or second aperture tip region of TolC (Fig. 1). Because Aa MacA is present as a stable hexamer in solution unlike other MacA proteins, we chose Aa MacA as the scaffold for the construction of the chimeric protein to generate the hexameric assembly.
FIGURE 1.
The construction of chimera proteins of MacA and TolC α-barrel tip regions. A, sequence alignment of the α-hairpin end regions of Aa MacA (black) and the two repeats of E. coli TolC (violet and blue), based on the heptad rule of the α-helical coil. The secondary elements and the heptad positions are shown above the sequences. To make the chimeric protein, the MacA α-hairpin tip region (black, bold, and underlined) was replaced with the tip region of TolC repeat 1 (Ec TolC R1; violet, bold, and underlined) or TolC repeat 2 (Ec TolC R2; blue, bold, and underlined). B, superposition of the α-hairpin end regions from Aa MacA (gray), E. coli TolC repeat 1, and TolC repeat 2, whose sequences are shown in A. The residues marked with asterisks were subject to substitution during the construction of the chimeric protein. Superposition was done by the program SUPERPOSE in the CCP4 Suite (25). Although no notable sequence similarity was found between the tip regions of MacA and the TolC repeats, the structures were very similar (1.080 Å rmsd for MacA and TolC repeat 1 and 1.050 Å rmsd for MacA and TolC repeat 2). C, bar diagram for the construction of MacA-TolCα-hybrid-dimer. The first chimeric MacA harboring the substitution with TolC repeat 1 tip regions and the second chimeric MacA harboring the substitution with TolC repeat 2 tip regions are joined in tandem. D, expected structure of MacA-TolCα-hybrid-dimer. Aa MacA is known to form a funnel-like hexamer in solution (18). Because only the tip region, which is not involved in the hexamerization, is replaced with the TolC tip regions, the chimeric protein is highly likely to form the hexameric structure, as was confirmed by size exclusion chromatography (supplemental Fig. S2), dynamic light scattering (supplemental Fig. S1) and CD spectroscopy (supplemental Fig. S3).
The chimeric protein Aa MacA-TolCα-hybrid-dimer had the same molecular size in solution and the same circular dichroism spectrum as the hexameric Aa MacA protein (supplemental Figs. S1, S2, and S3), indicating that the chimeric protein exhibited a funnel-like hexameric structure with a similar fold, as expected. We used this chimeric protein as the putative structural imitators of the TolC α-barrel tip region in this study (Fig. 1D).
Physical Interaction between the TolC Tip Region and the α-Hairpin Domains of MacA
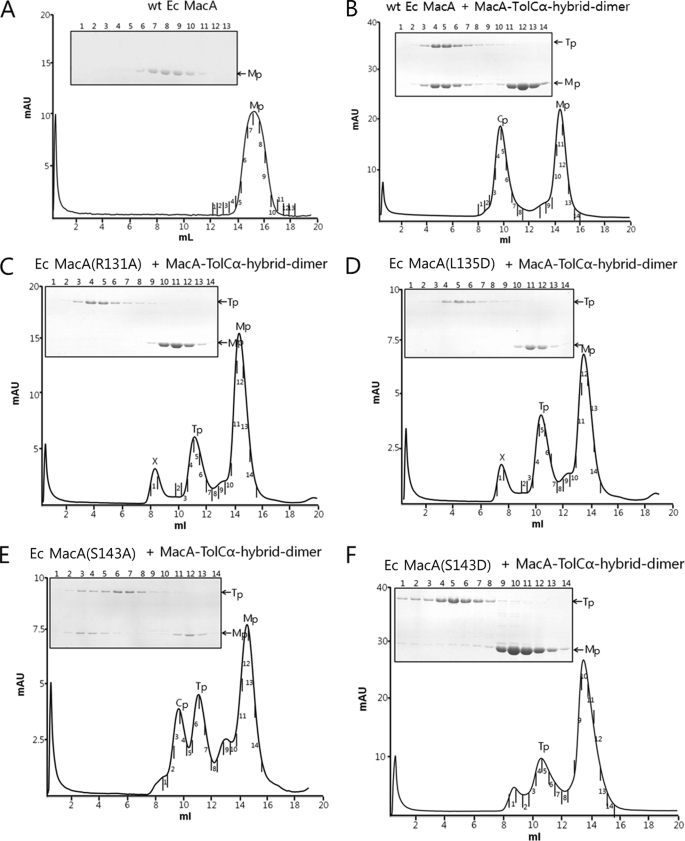
To probe the interaction between E. coli MacA and the TolC α-barrel tip region, size exclusion chromatography was applied using the purified proteins of E. coli MacA and the chimeric protein Aa MacA-TolCα-hybrid-dimer. The Aa MacA-TolCα-hybrid-dimer protein showed a strong interaction with the wild-type E. coli MacA protein (Fig. 2). The E. coli MacA was co-eluted with Aa MacA-TolCα-hybrid-dimer as a higher oligomer than the Aa MacA-TolCα-hybrid-dimer protein itself, while the E. coli MacA alone was eluted as a monomer (Fig. 2B). In contrast, the nonfunctional E. coli MacA variants containing a single amino acid substitution at the α-hairpin domains (R131A, L135D, or S143D) (17) did not interact with the Aa MacA-TolCα-hybrid-dimer, while E. coli MacA-S143A variant protein showed partial affinity for MacA-TolCα-hybrid-dimer, which is in good agreement with the functionality of MacA-S143A in vivo (Fig. 2E and Table 1). These results indicate that the α-helical tip region of MacA is important in the binding to the TolC α-barrel and that interactions between MacA and MacA-TolCα-hybrid-dimer reflect the functional binding between MacA and TolC (Fig. 2, C, D, and F). These results demonstrate that a pair of the TolC aperture consisting of 24 amino acids is sufficient to interact with the MacA α-helical tip region.
FIGURE 2.
Elution profiles of E. coli MacA (wild type and mutants) and MacA-TolCα-hybrid-dimer on size exclusion chromatographic column. The peak indicated by X does not contain protein, as revealed by SDS-PAGE. Structural integrities of the recombinant proteins used were confirmed by CD spectroscopy (data not shown). A, wild-type E. coli MacA. Mp stands for E. coli MacA, the elution volume of which is larger than that of MacA-TolCα-hybrid-dimer. B, mixture of Aa MacA-TolCα-hybrid-dimer (Tp) and wild-type E. coli MacA. Size exclusion chromatography clearly shows the complex formation of the two proteins. The complex peak (Cp) and the E. coli MacA peak (Mp) are shown. The SDS-polyacrylamide gel shows that the band intensity of Aa MacA-TolCα-hybrid-dimer is similar to that of E. coli MacA in the complex peak. C, elution profile of the mixture of E. coli MacA (R131A) and Aa MacA-TolCα-hybrid-dimer with SDS-PAGE analysis. Any binding was not observed. D, elution profile of the mixture of E. coli MacA (L135D) and Aa MacA-TolCα-hybrid-dimer with SDS-PAGE analysis. Any binding was not observed. E, elution profile of the mixture of E. coli MacA (S143A) and Aa MacA-TolCα-hybrid-dimer with SDS-PAGE analysis. Complex of E. coli MacA (S143A) and Aa MacA-TolCα-hybrid-dimer is observed. F, elution profile of the mixture of E. coli MacA (S143D) and Aa MacA-TolCα-hybrid-dimer with SDS-PAGE analysis. Binding was not observed.
TABLE 1.
The in vivo effect of a mutation at residue Ser-143 on E. coli MacA function
The effects of overexpressing wild-type MacB and wild-type or mutant MacA on the resistance of E. coli strain BW25113ΔacrAacrB to erythromycin were measured as MIC. These data have been previously reported (17).
| Background | BW25113ΔacrAB | |||
|---|---|---|---|---|
| Plasmid | pKAN6 | pMacAB1 | pMacAB1-S143D | pMacAB1-S143A |
| macA | - | Wt | S143D | S143A |
| macB | - | Wt | Wt | Wt |
| MIC (μg/ml) Erythromycina | 1.3 | 20.0 | 1.3a | 20.0 |
a Erythromycin concentrations used to measure MICs were 0, 1.3, 2.5, 5.0, 10.0, and 20.0 μg/ml.
One α-Helical End of TolC Is Sufficient for the Binding to MacA
Given that the periplasmic domain of the TolC protomer largely consists of two α-hairpin units, we tested whether one α-hairpin unit is sufficient for the binding to MacA.
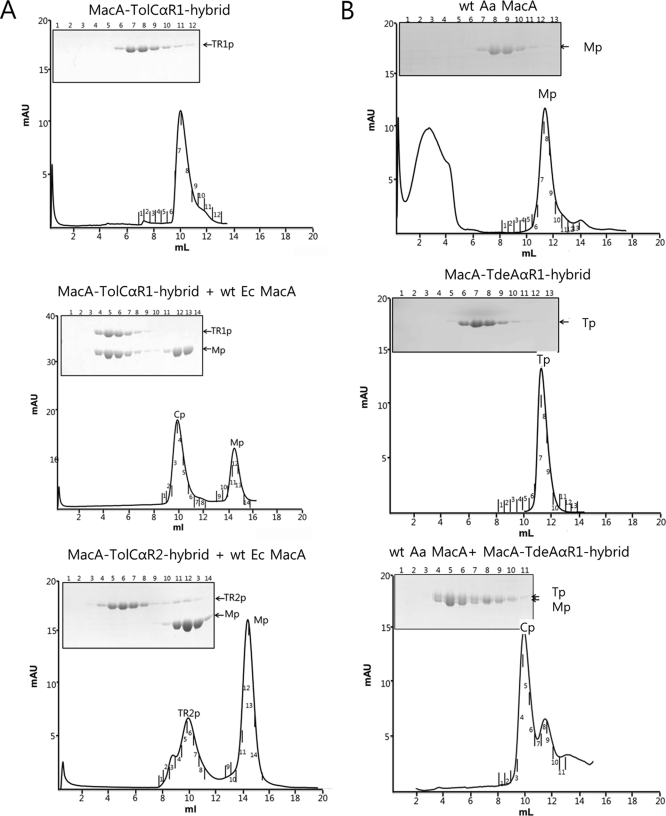
Interestingly, the molecular size of Aa MacA-TolCαR1-hybrid and Aa MacA-TolCαR2-hybrid were measured to be two times bigger in solution than that of Aa MacA, indicating that the chimeric proteins having only one repeat of TolC may form a dimer of the funnel-like hexamer (supplemental Fig. S1). The interactions between E. coli MacA and the hybrid proteins were also evaluated by size exclusion chromatography. The Aa MacA-TolCαR1-hybrid formed a complex with E. coli MacA protein as strong as the Aa MacA-TolCα-hybrid-dimer (Fig. 3A). This result further indicates that the dimer of the hexameric unit of Aa MacA-TolCαR1 is disrupted by the interaction with E. coli MacA.
FIGURE 3.
Repeat 1 region of TolC and it orthologue is able to bind to its cognate MacA in vitro. A, E. coli MacA binds to the α-helical tip barrel of TolC repeat 1. Top, a representative elution profiles for Aa MacA-TolCαR1-hybrid (TR1p). Middle, mixture of Aa MacA-TolCαR1-hybrid (TR1p) and wild-type E. coli MacA. Size exclusion chromatography clearly shows the complex formation of the two proteins. The complex peak (Cp) and the E. coli MacA peak (Mp) are shown. The SDS-polyacrylamide gel shows that the band intensity of MacA-TolCαR1-hybrid is similar to that of E. coli MacA in the complex peak. Mutant proteins of E. coli MacA used in Fig. 2 are also applied to the size exclusion chromatography with Aa MacA-TolCαR1-hybrid (TR1p), and the results are presented in supplemental Fig. S4. Bottom, mixture of Aa MacA-TolCαR2-hybrid (TR2p) and wild-type E. coli MacA. The result shows that the two proteins do not interact. B, Aa MacA binds to the α-helical tip barrel of Aa TdeA repeat 1. Top and middle, elution profile for wild-type Aa MacA (Mp) and MacA-TdeAαR1 (Tp), as indicated above the graphs. Bottom, an elution profile for the mixture of the two proteins. The SDS-polyacrylamide gels show the protein bands in the fractions indicated in the elution profiles. The protein bands in the complex peaks (Cp) indicate the proteins form a complex.
To examine the role of the E. coli MacA tip region, we evaluated the binding of E. coli MacA variants containing a mutation at the conserved Arg-131, Leu-135, or Ser-143, which are located at the tip region, with Aa MacA-TolCαR1. All the mutations partially or fully abolished to form a complex with Aa MacA-TolCαR1-hybrid on the size-exclusion chromatographic column, as was observed for Aa MacA-TolCα-hybrid-dimer (supplemental Fig. S4), indicating the importance of the α-barrel tip region of MacA in binding to the TolC α-barrel tip region. These results suggest that TolC repeat 1 may be responsible for the binding to MacA. In addition, Aa MacA-TolCαR2-hybrid, which has the α-hairpin end of TolC repeat 2, did not interact with E. coli MacA in vitro (Fig. 3A). Thus, these observations suggest that the six-bladed cogwheel structure composed of six α-hairpins of TolC or its homologues is important in interaction with MacA.
ITC Experiments Suggest the Strong Binding between MacA and the Chimeric Protein Containing the TolC (or TdeA) Tip Region
To measure the binding constant between MacA-TolCα-hybrid-dimer and E. coli MacA, ITC experiments were performed. We carried out experiments in which E. coli MacA in the injector syringe was titrated into MacA-TolCα-hybrid-dimer. The heat by the injection confirmed the interaction between the two proteins (supplemental Fig. S5). However, we failed to calculate the thermodynamic parameters from the titration curve. The titration curve becomes complicated due to the heat generated from the hexamerization of the monomeric E. coli MacA at higher concentrations.
To exclude this hexamerization effect of E. coli MacA, we chose Aa MacA that is present as a hexamer even at extremely low concentration (18). To make the cognate partner of Aa MacA, we generated a hybrid protein (MacA-TdeAαR1-hybrid) utilizing TdeA, an A. actinomycetemcomitans ortholog of TolC. The MacA-TdeAαR1-hybrid protein contains 24 amino acids of Aa TdeA corresponding to the α-helical tip of TolC repeat 1. We next confirmed that MacA-TdeAαR1-hybrid is able to form a complex with Aa MacA in the size exclusion chromatography, (Fig. 3B), as observed in MacA-TolCαR1-hybrid and E. coli MacA. Once again, the ITC experiments were carried out in which Aa MacA was titrated to MacA-TdeAαR1-hybrid. The results revealed that Aa MacA and Aa MacA-TdeAαR1-hybrid strongly interacted with an average KD of 0.69 nm, suggesting that strong interactions between tip regions of MacA and TolC homologs stem from their structural similarities (supplemental Fig. S6).
2:1 Binding Stoichiometry between MacA and TolC
Given the strong interaction between MacA and MacA-TolCα-hybrid-dimer, we analyzed the complex fractions on the size exclusion chromatographic column. As shown in supplemental Fig. S7, the intensities of the MacA-TolCα-hybrid-dimer protein band and the E. coli MacA protomer protein band on the SDS-polyacrylamide gel were measured. Similar intensities of the protein bands that were stained with Coomassie Blue indicate an equimolar binding of these proteins. Because the chimeric protein contains two units of MacA corresponding to one TolC protomer, our result thus suggests the 2:1 binding stoichiometry between a MacA protomer and an Aa MacA-TolCα-hybrid-dimer.
Electron Microscopy Reveals Tip-to-Tip Binding between TolC and MacA
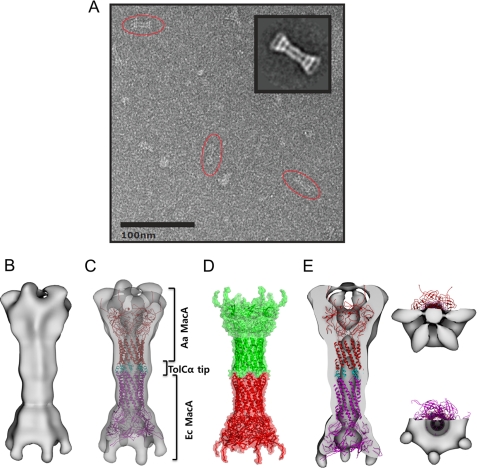
To visualize how MacA interacts with the TolC α-barrel tip region, a complex composed of E. coli MacA and Aa MacA-TolCα-hybrid-dimer was applied to electron microscopy. A projection matching approach using negatively stained single particles of the complex resulted in a three-dimensional density map at 26 Å resolution (FSC = 0.5) (Fig. 4, A and B). We constructed three starting models with different symmetries (C3, C6, and D6) by directly reconstructing a single selected side view class average using the applied symmetry factor. Additionally, a lowpass filtered density map of the generated pdb model (also used for docking, see below) was used for comparison. We refined the C3 and C6 initial models with their respective symmetries, the D6 starting model using C6-symmetry and the simulated density map using C1-symmetry. After several iterations all three-dimensional models converged to C6 symmetry. Despite the high similarity of the two protomers, an additional symmetry axis orthogonal to the 6-fold (D6-symmetry) could not be identified at this resolution.
FIGURE 4.
Electron microscopic images of the complex containing E. coli MacA and TolC α-barrel tip regions. A, representative electron microscopic image of E. coli MacA and Aa MacA-TolCα-hybrid-dimer, preserved in negative stain (uranyl-formate) and imaged at 80,000× magnification. Characteristic side views are encircled by red ellipses. A reference free class formed from an average of 119 side views is shown in the inset and can be compared with a projection of the reconstructed density map. B, surface representation of the reconstructed density map from the electron microscopy, displayed in side view. C, complex model of E. coli MacA and Aa MacA-TolCα-hybrid-dimer. E. coli MacA is shown in magenta, Aa MacA is in red, and the TolC α-barrel tip region (TolCα tip) is in cyan. D, two hexameric units of Aa MacA in the intermeshing cogwheel interaction in the crystal (18), shown in the ribbon and transparent surface representations. E, vertical section of the surface representation of the density map, displayed in the side and top views. The complex model of E. coli MacA and Aa MacA-TolCα-hybrid-dimer is docked to the density map.
The map comprises a volume of 985,200 Å and exhibits a characteristic dumb-bell-like (bridge symbol) side view with flared ends at both sides, a motif which can also easily be identified in the raw images (Fig. 4, A and B). A channel of varying diameter is observed through the center of the structure. The channel appears to be closed at the very end of one side but this is likely a low resolution artifact, as it disappears at a lower threshold, and is not evident in the crystal structure. We refrained from applying any masking during refinement to prevent bias in the reconstruction.
To dock into the density map, we generated a model for the complex in the intermeshing cogwheel fashion, where E. coli MacA (PDB code: 3FPP) is at the lower part and the chimeric Aa MacA containing the E. coli TolC α-barrel tip regions is at the upper part (Fig. 4C). The relative position between the lower part and the upper part was determined based on the two hexameric units of the Aa MacA structure that formed the intermeshing cogwheel interaction in the crystal structure (Fig. 4D), because we hypothesized that the overall structure of the two molecules was similar to Aa MacA and the packing interaction in the Aa MacA crystal may represent the binding mode between MacA and the TolC tip region (18).
Docking of the PDB structure into the experimental map using the automatic docking program Chimera (24) showed an excellent correlation between the map and the PDB structure. However, at this resolution we could not distinguish the two ends; only one of two possibilities is shown here (Fig. 4, C and E). The overall dimensions of the intermeshing α-barrels were exactly matched to the central long cylinder. In the funnel-mouth parts located on both sides, the lipoyl and the β-barrel domains were fitted into the density map adjacent to the long cylinder. However, the density at both ends was not docked by the model, which can be justified by the missing MP domain in the model. The overall size of the MP domain of P. aeruginosa MexA (27) correlates very well with the undocked electron density map from the electron microscopy (data not shown). Collectively, the electron density map is well matched to the generated model based on the intermeshing cogwheel interaction. Thus, our results demonstrate that TolC α-barrel tip regions make direct contact with the MacA α-barrel in an intermeshing cogwheel fashion. Our results further suggest that the α-barrel of the MacA hexamer may reflect the TolC channel open state when docked to MacA.
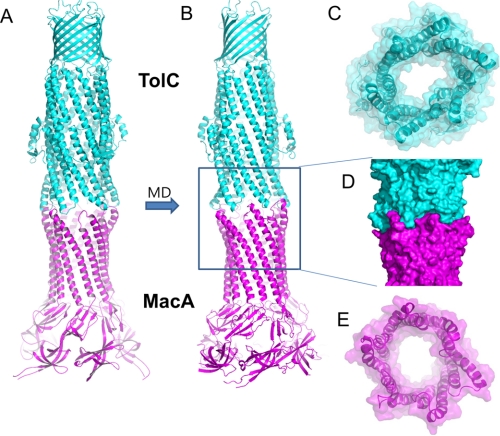
Molecular Dynamics Simulation of the TolC-MacA Interaction
To examine the feasibility of the cogwheel binding conformation between TolC and MacA and the stability of the tip-to-tip interaction between them, in silico molecular dynamics simulation was performed based on the crystal structures of TolC and MacA. We used a model of an open structure of TolC, in which the TolC α-barrel exhibits the conformation of the α-barrel structure of the MacA hexamer, as the starting model (Fig. 5A), because the TolC α-helical tip exhibiting the MacA α-barrel conformation showed a strong binding affinity to MacA in our experiment. We first generated putative cogwheel-to-cogwheel binding conformations by bringing the MacA hexamer close to TolC based on the electron microscopic tomography. The energy of a whole TolC-MacA complex, consisting of 2,844 residues, was minimized to remove the steric constraints (Fig. 5A). Subjecting this initial conformation to molecular dynamic simulation in the explicit TIP3 water using the atomistic force-field AMBER/v11.ff99SB produced an equilibrium structure of the TolC-MacA complex (Fig. 5B). Fig. 5D demonstrates how well the tip regions of the open form of the TolC α-barrel fit and complement those of the MacA α-barrel. The binding energy of this equilibrium cogwheel-to-cogwheel structure was stronger than that of the initial putative structure by 50%, demonstrating that the cogwheel binding conformation between TolC and MacA is possible and that the tip-to-tip interaction between them is more stable than that of initial putative structures. Formation of the docked complex buries 2,321 Å3 of solvent-exposed surface, which is comparable to the models based on in vivo chemical cross-linking experiments by Lobedanz et al. (28).
FIGURE 5.
TolC and MacA docking model produced by the in silico molecular dynamic simulation. A, an initial putative model of the TolC-MacA complex for MD simulation. The open conformation of TolC was generated by adopting the MacA α-barrel conformation to the TolC α-barrel end region. It was brought to the top of E. coli MacA hexamer to establish the intermeshing cogwheel interactions, and then energy-minimization was performed. B, an equilibrated conformation of the TolC-MacA complex after MD simulation. The cogwheel-to-cogwheel structure was stabilized via equilibration, thus maximizing the complementary tip-to-tip interactions between the TolC α-barrel region and MacA barrel region complementarily, which then resulted in the stabilized TolC-MacA complex. C, a bottom view of TolC in the docked complex displayed in B. The semi-transparent surface representation is shown with the ribbon representation. The central channel is wide open. D, a surface representation of the docked complex highlighting the intermeshing cogwheel interaction. E, a top view of the MacA hexamer in the docked complex. Structural resemblance to the TolC open structure is shown.
DISCUSSION
The crystal structures of MacA, suggested that MacA may function as a funnel-like hexamer (18). Subsequent studies revealed that its hexamerization is important for the function of the MacAB-TolC pump (18) and that the α-barrel tip region of the MacA hexamer plays a crucial role in binding to TolC (17). However, these studies did not show the nature of the interactions between the α-barrel tip region of the MacA hexamer and TolC, and which part of TolC is important in binding to MacA. We only speculated that the α-barrel tip region of MacA may make a tip-to-tip interaction with the TolC α-barrel although it is also possible that the α-barrel tip region of MacA wraps outside of the TolC α-barrel, as has been hypothesized for interaction between AcrA and TolC in the formation of AcrAB-TolC efflux pump (27–29). For this reason, in this study we explored the role of the α-barrel tip region of TolC in the binding to MacA using a chimeric protein containing the TolC α-barrel tip region, which indicated a tip-to-tip interaction between them. Electron microscopy studies further clearly showed that MacA is present as a homohexamer when binding to the TolC α-barrel tip region and that the TolC α-barrel tip and the MacA α-barrel tip interact in a tip-to-tip intermeshing fashion. Based on our results, we built an assembly model of TolC and MacA, where the two proteins interact in an intermeshing cogwheel-like manner. Molecular dynamics analysis showed that the cogwheel binding conformation is both possible and stable. Moreover, we present evidence that repeat 1 in TolC plays a more important role in the binding to MacA. Taken together, we conclude that MacA functions as a homohexamer in its functional state and forms an intermeshing cogwheel-type complex with TolC. Because the TolC α-barrel tip in the chimeric protein may represent the conformation of TolC in complex with MacA, our results also suggest that the conformation of TolC in the open state is the same as that of the chimer protein or the MacA hexamer, providing a clue how the opening of the TolC channel is induced. However, it is still unclear which residues are directly involved in this process. To reveal the assembly of this type of tripartite pump at the atomic level, crystal structures of the complex are needed.
Acknowledgment
We thank Erica Jacovetty for assistance with EM data collection. EM imaging was performed at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health through the National Center for Research Resources P41 program (RR17573).
This work was supported in part by grants from the Korea Research Foundation (NRF-2010-0015298) (to N.-C. H.), and the 21C Frontier Microbial Genomics and Application Center Program (to K. L.). This work was also supported by the Joint Center for Innovation in Membrane Protein Production for Structure Determination (Grant RFA-RM-08-019) (to A. M.), and the National Creative Research Initiatives (Center for Proteome Biophysics, Grant No. 2008-0061984) (to I. C.) from the Ministry of Education, Science & Technology, Republic of Korea.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table S1.
- MFP
- periplasmic membrane fusion protein
- MIC
- minimum inhibitory concentration
- MD
- molecular dynamics.
REFERENCES
- 1. Levy S. B., Marshall B. (2004) Nat. Med. 10, S122–129 [DOI] [PubMed] [Google Scholar]
- 2. Koronakis V., Eswaran J., Hughes C. (2004) Annu. Rev. Biochem. 73, 467–489 [DOI] [PubMed] [Google Scholar]
- 3. Lewis K. (2000) Curr. Biol. 10, R678–R681 [DOI] [PubMed] [Google Scholar]
- 4. Zgurskaya H. I., Nikaido H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7190–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thanabalu T., Koronakis E., Hughes C., Koronakis V. (1998) EMBO J. 17, 6487–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fralick J. A. (1996) J. Bacteriol. 178, 5803–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hirakata Y., Srikumar R., Poole K., Gotoh N., Suematsu T., Kohno S., Kamihira S., Hancock R. E., Speert D. P. (2002) J. Exp. Med. 196, 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tikhonova E. B., Devroy V. K., Lau S. Y., Zgurskaya H. I. (2007) Mol. Microbiol. 63, 895–910 [DOI] [PubMed] [Google Scholar]
- 9. Kobayashi N., Nishino K., Hirata T., Yamaguchi A. (2003) FEBS Lett. 546, 241–246 [DOI] [PubMed] [Google Scholar]
- 10. Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. (2000) Nature 405, 914–919 [DOI] [PubMed] [Google Scholar]
- 11. Kobayashi N., Nishino K., Yamaguchi A. (2001) J. Bacteriol. 183, 5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin H. T., Bavro V. N., Barrera N. P., Frankish H. M., Velamakanni S., van Veen H. W., Robinson C. V., Borges-Walmsley M. I., Walmsley A. R. (2009) J. Biol. Chem. 284, 1145–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zgurskaya H. I., Yamada Y., Tikhonova E. B., Ge Q., Krishnamoorthy G. (2009) Biochim. Biophys. Acta 1794, 794–807 [DOI] [PubMed] [Google Scholar]
- 14. Mikolosko J., Bobyk K., Zgurskaya H. I., Ghosh P. (2006) Structure 14, 577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins M. K., Bokma E., Koronakis E., Hughes C., Koronakis V. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9994–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akama H., Matsuura T., Kashiwagi S., Yoneyama H., Narita S., Tsukihara T., Nakagawa A., Nakae T. (2004) J. Biol. Chem. 279, 25939–25942 [DOI] [PubMed] [Google Scholar]
- 17. Xu Y., Sim S. H., Song S., Piao S., Kim H. M., Jin X. L., Lee K., Ha N. C. (2010) Biochem. Biophys. Res. Commun. 394, 962–965 [DOI] [PubMed] [Google Scholar]
- 18. Yum S., Xu Y., Piao S., Sim S. H., Kim H. M., Jo W. S., Kim K. J., Kweon H. S., Jeong M. H., Jeon H., Lee K., Ha N. C. (2009) J. Mol. Biol. 387, 1286–1297 [DOI] [PubMed] [Google Scholar]
- 19. Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C. S., Carragher B. (2005) J. Struct. Biol. 151, 41–60 [DOI] [PubMed] [Google Scholar]
- 20. Lander G. C., Stagg S. M., Voss N. R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P. W., Lyumkis D., Potter C. S., Carragher B. (2009) J. Struct. Biol. 166, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roseman A. M. (2003) Ultramicroscopy 94, 225–236 [DOI] [PubMed] [Google Scholar]
- 22. Scheres S. H., Valle M., Nuñez R., Sorzano C. O., Marabini R., Herman G. T., Carazo J. M. (2005) J. Mol. Biol. 348, 139–149 [DOI] [PubMed] [Google Scholar]
- 23. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 24. Goddard T. D., Huang C. C., Ferrin T. E. (2007) J. Struct. Biol. 157, 281–287 [DOI] [PubMed] [Google Scholar]
- 25. CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 26. DeLano W. (2002) The PyMOL User's Manual, DeLano Scientific, Palo Alto [Google Scholar]
- 27. Symmons M. F., Bokma E., Koronakis E., Hughes C., Koronakis V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7173–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lobedanz S., Bokma E., Symmons M. F., Koronakis E., Hughes C., Koronakis V. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4612–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bavro V. N., Pietras Z., Furnham N., Pérez-Cano L., Fernández-Recio J., Pei X. Y., Misra R., Luisi B. (2008) Mol. Cell 30, 114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]