Abstract
Angiogenesis, or formation of new blood vessels, is crucial to cancer tumor growth. Tumor growth, progression, and metastasis are critically influenced by the production of the pro-angiogenic vascular endothelial growth factor (VEGF). Promising anti-angiogenic drugs are currently available; however, their susceptibilities to drug resistance and long term toxicity are serious impediments to their use, thus requiring the development of new therapeutic approaches for safe and effective angiogenic inhibitors. In this work, peptides were designed to mimic the VEGF-binding site to its receptor VEGFR-2. The VEGF conformational peptide mimic, VEGF-P3(CYC), included two artificial cysteine residues, which upon cyclization constrained the peptide in a loop native-like conformation to better mimic the anti-parallel structure of VEGF. The engineered cyclic VEGF mimic peptide demonstrated the highest affinity to VEGFR-2 by surface plasmon resonance assay. The VEGF peptide mimics were evaluated as inhibitors in several in vitro assays in which VEGF-dependent signaling pathways were observed. All VEGF mimics inhibited VEGFR-2 phosphorylation with VEGF-P3(CYC) showing the highest inhibitory effects when compared with unstructured peptides. Additionally, we show in several angiogenic in vitro assays that all the VEGF mimics inhibited endothelial cell proliferation, migration, and network formation with the conformational VEGF-P3 (CYC) being the best. The VEGF-P3(CYC) also caused a significant delay in tumor development in a transgenic model of VEGF+/−Neu2-5+/−. These results indicate that the structure-based design is important for the development of this peptidomimetic and for its anti-angiogenic effects.
Keywords: Antibodies, Anti-cancer Drug, Cancer Therapy, Peptide Chemical Synthesis, Peptide Conformation, Receptor Tyrosine Kinase, Angiogenesis, HER-2, Peptidomimetic, VEGF
Introduction
The formation of new blood vessels, called angiogenesis, is a process tightly regulated by a balance between pro- and anti-angiogenic factors, and physiologically, it is activated in wound healing, ovulation, and menstruation. However, it is also stimulated in pathological conditions such as cancer, macular degeneration in the eyes, psoriasis, and diabetes (1–5). Because most tumors cannot grow beyond a few millimeters in the absence of new blood vessel formation (6), angiogenesis inhibitors have been explored lately as a drug target to be used in combination with several cancer therapies (7–13). Several studies have explored the use of DNA vaccines, small tyrosine kinase inhibitors, siRNAs, ribozymes, antibodies, and receptor blocking agents aimed at better understanding the angiogenic mechanism and development of potential inhibitors (13–15). Specialized cancer treatments with anti-angiogenic agents approved by the Food and Drug Administration include monoclonal antibody, bevacizumab (Avastin) or small tyrosine kinase inhibitors, SU11248 (sunitinib), and BAY 43-9006 (sorafenib) (7, 16, 17). Although the clinical application of these drugs in cancer therapy is promising, drug resistance development and long term side effects like hypertension and endothelium dysfunction remain a concern (14).
The pro-angiogenic factor VEGF is the most studied growth factor in this field, due to its specificity and important role in the activation of all steps of angiogenesis in the endothelium vasculature (18–20). The splicing variant VEGF165 is the predominant form and has been shown to be up-regulated in tumor microenvironment by hypoxia or activation of oncogenes like HER-2 (21–26). VEGF is a glycoprotein and consists of an anti-parallel homodimer structure containing inter- and intra-disulfide bonds, and it has been shown to bind to three receptor types as follows: VEGFR-1 (flt-1), VEGFR-2 (flk-1 or KDR), and neurophilin-1 (NR-1). The VEGF-VEGFR-1 interaction exhibits high affinity, although the role of VEGFR-1 is not fully understood, but research suggests its function in activated pathways in macrophages or endothelial progenitor cells (27, 28). In the endothelial cells, the majority of angiogenesis signaling (proliferation, migration, and survival) proceeds via the interaction between VEGF and VEGFR-2 (18, 29–31).
The binding site of VEGF to its receptors has been characterized by crystal structure analysis as well as alanine scanning and reveals overlapping regions located at the poles in the homodimer (31–36). VEGF-VEGFR-2 interaction has been explored extensively using antibodies that bind VEGF as well as the extracellular domain of VEGFR-2 (13, 37–40), identifying VEGF epitopes in the binding region that inhibit VEGF-dependent pathways (33). The interaction between VEGF and VEGFR-2 has been identified and includes residues at a loop region formed by the anti-parallel β-sheets β5-β6 in the VEGF protein (31).
Blockade of receptor-ligand interaction offers a validated and proven approach in drug development because receptor-ligand interaction is usually confined to a defined portion of the ligand and the receptor, and recent technologies have allowed the accurate identification of these binding regions. Peptidomimetics is the approach of reproducing the biological activity or binding properties in a smaller molecule, like peptides or modified peptides that were designed to mimic the desired region. This approach demonstrated successful results in studies with melanocortin MC4, pallet GP IIb/IIIa, or CD28 costimulatory receptors (41–46). Peptides are excellent candidates for drug design because they demonstrate better target specificity and less susceptibility to drug resistance than small molecules. Additionally, peptides demonstrate several advantages like lower developing and manufacturing costs, improved organ or tumor penetration, and higher activity per mass when compared with antibodies or large proteins (47, 48). The main goal of this work was to create molecules that would retain the structural similarity with the binding site region and demonstrate bioactivity “in vivo.”
Our hypothesis to a peptide therapeutic approach is that key residues in the binding epitope, in particular side-chain functional groups responsible for a significant portion of the binding affinity to a given ligand, can be transferred to a much smaller fragment while maintaining the contributions to the binding largely intact. The goal was to direct critical amino acids into the same conformational space and orientation as in the bioactive surface yet retain sufficient flexibility to bind cooperatively and with complementarity to a given receptor. The therapeutic targeting of the tumor vasculature has multiple advantages over traditional cancer treatments (49) displaying minimal toxicity. Many new cancer therapies are being directed against VEGF and Flk-1 believed to be the most important for angiogenesis during tumor formation (50–52). Several anti-receptor therapeutic strategies have been pursued. Anti-VEGF strategies include the use of mAbs to VEGF (e.g. bevacizumab) or its receptors, use of ribozymes to decrease receptor expression, and small molecule inhibitors of VEGF receptor tyrosine kinase.
Here, we report that peptides corresponding to the natural VEGF amino acid sequence 102–122 (residues 76–96), which includes the important loop-binding residues of VEGF to its receptor, were successfully engineered to better mimic the conformational structure of this sequence in the protein. The conformational peptide mimics VEGF-P3(NC)2 and VEGF-P3 (CYC) sequences demonstrated the highest affinity to VEGFR-2 and were effective as inhibitors of VEGFR-2 phosphorylation and in several angiogenic in vitro assays such endothelial cell proliferation, migration, and network formation. The VEGF-P3(CYC) also caused a significant delay in tumor development in a transgenic model of VEGF+/−Neu2-5+/−. The observed results are consistent with the hypothesis that the structure-based design is important to the development of VEGF peptidomimetics and to its anti-angiogenic effectiveness.
MATERIALS AND METHODS
Peptide Synthesis
Peptides were synthesized on Milligen/Biosearch 9600 solid-phase peptide synthesizer (Bedford, MA) using Fmoc/t-butyl chemistry as described previously (53). In case of the synthesis of VEGF(102–122), preloaded Fmoc-Phe-CLEAR acid resin (0.41 mmol/g was used whereas VEGF-P3 and MVF-VEGF-P3 were synthesized using CLEAR amide resin (0.32 mmol/g)) (Peptides International, Louisville, KY). Peptide P3 was acetylated on resin, using acetyl imidazole reagent as reported earlier (53). The MVF-GPSL sequence was added on peptide resin of the H2N-VEGF-P3. All peptides were cleaved from the resin using the cleavage reagent B (trifluoroacetic acid/phenol/water/triisopropyl silane 90:4:4:2), and crude peptides were purified on preparative RP-HPLC using Vydac C-4 column and acetonitrile/water (0.1% TFA) gradient system. All fractions were analyzed on analytical RP-HPLC and characterized by MALDI (matrix-assisted laser desorption ionization mass spectroscopy) at Campus Chemical Instrumentation Center (Ohio State University, Columbus). RP-HPLC fractions showing same mass spectrum peak were pooled together and lyophilized. RP-HPLC pure peptides MVF-VEGF-P3 and VEGF-P3 containing two Cys residues in each peptide were cyclized using acetic acid/iodine method, further purified on RP-HPLC, and characterized by mass spectroscopy using established protocol as reported earlier (54). Amino acid sequences and molecular weight of all peptides and their molecular weights are shown in Table 1.
TABLE 1.
Amino acid sequences and molecular weight of VEGF mimic peptides
Cell Lines and Reagents
All culture media, FBS, and supplements were purchased from Invitrogen. HUVEC were purchased from GlycoTech and cultivated in F-12K Nutrient Mixture-Kaighn's Modification (F-12K) supplemented with 20% FBS, heparin (100 μg/ml), and endothelial (complement) cell growth factor supplement (ECGS) (50 μg/ml). 293/KDR cells (high expression of VEGFR2) were purchased from SibTech, Inc. (Brookfield, CT), and cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and puromycin (0.375 μg/ml). Cells were incubated in a 37 °C humidified 5% CO2 incubator.
Surface Plasmon Resonance
Binding assays were performed using a Biacore 3000 instrument (GE Healthcare). The experiments were performed at room temperature using HBS-EP buffer (25 mm HEPES, pH 7.4, 150 nm NaCl, 3.4 mm EDTA, and 0.005% surfactant P20). Peptides were coupled onto CM5 chip surfaces at 10 μl/min using a standard amine coupling protocol with N-ethyl-N′-[dimethylaminopropyl]carbodiimide/N-hydroxysuccinimide. KDR-Fc (R&D Systems) was used at several concentrations in HBS-EP buffer and was injected at a flow rate of 10 μl/min. Data analysis was performed with BIAsimulation software version 3.1 (GE Healthcare). For competition assay, KDR-Fc and rhVEGF (R&D Systems) were mixed in HBS-EP and incubated for 30 min at room temperature, and this mixture was injected over the chip where peptides were immobilized. To obtain measurement of anti-peptide binding affinities, a similar experiment was carried out, but rhVEGF was immobilized onto CM5 chip surface, and anti-peptides were injected at several concentrations.
Circular Dichroism
Circular dichroism (CD) measurements were performed on an AVIV model 62A DS instrument. All spectral measurements were obtained at 25 °C under continuous nitrogen purging of the sample chamber, using a quartz cuvette of 0.1-cm path length. Spectral measurements of VEGF-P3(NC) and VEGF-P3(CYC) were obtained at a concentration of 100 μm in water. Molar ellipticity values were calculated using the formula [θ]M,λ = (θ × 100 × Mr)/(n × c × l), where θ is the recorded ellipticity (degree); Mr indicates the molecular weight of the peptide; n indicates the number of residues in the peptide; c indicates the peptide concentration (milligrams/ml); and l indicates the path length of the cuvette (55).
Proliferation Assay
HUVEC (1 × 104 cells/well) were plated in 96-well flat-bottom plates overnight. Growth medium was replaced with low sera (1% FCS) medium, and the cells were incubated overnight. Media were removed from the wells and replaced with low sera medium containing VEGF mimic peptides at concentrations ranging from 50 to 50,000 ng/ml with or without rhVEGF (10 ng/ml). In the case of antibodies, low sera medium containing purified anti-VEGF peptide mimic antibodies at concentrations ranging from 0.15 to 150 μg/ml with or without rhVEGF (10 ng/ml) was used. Plates were incubated for an additional 72 h at 37 °C before adding 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (5 mg/ml) to each well. Plates were incubated 4 h at 37 °C; medium was discarded, and 100 μl of extraction buffer (20% SDS, 50% dimethylformamide, pH 4.7) was added to each well. Plates were incubated overnight at 37 °C and read on an ELISA plate reader at 570 nm with 655 nm background subtraction. Inhibition percentage was calculated as 100% × (VEGF-only treated cells − peptide-treated cells)/(VEGF-only treated cells).
Network Formation Assay Using Matrigel
Matrigel (60 μl) (BD Biosciences) was added to a 96-well plate and incubated for 30 min at 37 °C. HUVEC were kept overnight in low sera medium before cells (20,000/well) were seeded with low sera medium F-12K supplemented with 1% FBS and 10 ng/ml VEGF (R&D Systems) with or without inhibitor. The cells were fixed in 4% formaldehyde after overnight incubation at 37 °C. Pictures from ×40 magnification from light microscopy were taken and the sprout points counted using the software ImageJ (National Institutes of Health). Two set of experiments were combined and averaged.
Scratch Wound Assay
HUVEC were cultured on 0.1% gelatin-coated 24-well plates. Confluent cells were incubated overnight with starving media, and then they were scraped using sterilized 200-μl pipette tips and stimulated with 50 ng/ml rhVEGF with or without VEGF mimic peptides for 16 h at 37 °C. Cells were fixed, and images were captured immediately at ×40 magnification from light microscopy, and cells that migrated to the scraped area were counted using ImageJ software.
Phosphorylation Assay
HUVEC (5 × 105 cells/well) were grown on 6-well plates in FK-12 endothelial cell growth medium supplemented with ECGS and heparin until 80% confluence. After overnight incubation in starving medium (0.5% FBS), cells were treated with inhibitor (100 μm) for 30 min and then stimulated with 10 ng/ml rhVEGF for 5 min. When using KDR-Fc as inhibitor, it was incubated with rhVEGF for 30 min and then added to the cells for 5 min. Cells were washed in cold PBS supplemented with 1 mm sodium orthovanadate, harvested into RIPA lysis buffer (Santa Cruz Biotechnology, Santa Cruz, CA), and incubated on ice for 30 min. Cell lysate was collected after centrifugation at 13,000 rpm for 10 min and kept at −80 °C. Total protein (30 μg) from the cell lysate was separated in SDS-PAGE and then transferred onto PVDF membrane (Hybond-P, Amersham Biosciences). The membrane was blocked in 5% nonfat dried milk in TBST (0.05 m Tris base, 0.9% NaCl, 0.05% Tween 20, pH 7.4) and washed three times (10 min) in TBST before incubation overnight with anti-Tyr(P) KDR (Santa Cruz Biotechnology) in 2.5% milk in TBST at 4 °C. Membranes were washed four times in TBST (15 min), incubated for 1 h at room temperature with anti-rabbit IgG (Fab) monoclonal antibody HRP-conjugated (Thermo Fisher Scientific Inc., Rockford, IL), and then washed six times (15 min). Proteins on the Western blots were detected using the enhanced chemiluminescent detection system (Thermo Fisher Scientific Inc.). Membranes were stripped and probed for detection of total KDR using anti-KDR HRP-conjugated (Santa Cruz Biotechnology). HUVEC lysates were also used for Western blotting following the same procedure but probed for anti-phosphor-p44 and -p42 MAPK (ERK1 and ERK2) and re-probed with anti-CD31 for loading control. 293/KDR cells (5 × 105 cells/well) were seeded on 6-well plates in DMEM supplemented with 10% FBS. After overnight incubation in starving medium (no FBS), cells were stimulated with rhVEGF for 5 min. Cells were washed in cold PBS supplemented with 1 mm sodium orthovanadate, harvested into RIPA lysis buffer (Santa Cruz Biotechnology), and incubated on ice for 30 min, and the cell lysate was collected after centrifugation at 13,000 rpm for 10 min. Cell lysate was kept at −80 °C until used for Western blotting detecting phosphor-KDR and total KDR as described above.
Peptide Treatment in Transgenic Mouse
The VEGF+/− Neu2-5+/− mice group (n = 6) was treated with 500 μg of peptides dissolved in PBS and injected intravenously in the tail weekly from week 4 to 10. Mice were euthanized at week 10 and tumors removed. Tumors in each of the 10 mammary glands were measured for tumor volume. VEGF+/−Neu2-5+/− results were reported as the average of the single largest tumor per mouse. Tumor growth over time was analyzed using Stata's cross-sectional generalized estimating equations model, which fits general linear models that allow you to specify within animal correlation structure in data involving repeated measurements.
Immunization of Rabbits
New Zealand White rabbits were immunized with 1 mg of peptide dissolved in double distilled H2O emulsified (1:1) in Montanide ISA720 vehicle (Sepic) with 100 μg of N-acetylglucosamine-3-yl-acetyl-l-alanyl-d-isoglutamine. Rabbits were boosted with the respective doses at 3-week intervals. Rabbit blood was collected via the central auricular artery, and sera were tested for antibody titers. Anti-peptide antibodies were purified by affinity chromatography using a protein A/G column (Pierce) from high titer antibody sera.
ELISA for Anti-VEGF Antibodies
Plates were coated overnight at 4 °C with 100 μl of 2 μg/ml rhVEGF (R&D Systems), washed four times with 0.1% Tween 20/PBS, and blocked with of 100 μl of 1% BSA/PBS for 2 h at room temperature. Plates were washed four times with 0.1% Tween 20/PBS. Anti-peptide sera were added at several dilutions and incubated 2 h at room temperature. Plates were washed four times with 0.1% Tween 20/PBS; a 1:500 dilution of goat anti-rabbit IgG HRP was added and incubated for 1 h. Detection was done using 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) substrate and absorbance reading at 415 nm. Antibody titers were determined as described previously (56) and defined as the reciprocal of the highest serum dilution with an absorbance of 0.2 or greater after subtracting background.
Direct Peptide-Cell Binding Assay
The peptide VEGF-P3-CYC was biotinylated at the N terminus during synthesis. Peptide binding to the VEGFR2 was evaluated using both HUVECS and 293-KDR cells. 1 × 106 cells were incubated with the biotinylated peptide in 100 μl of 2% FCS in PBS for 2 h at 4 °C. Unbound peptides were removed by washing three times with PBS, and the cells were incubated with Alexa Fluor 594/streptavidin (Molecular Probe) for 1 h. Cells were then washed with PBS three times and fixed with 1% formaldehyde before being analyzed by phase contrast, fluorescence, and confocal microscopy.
Flow Cytometry
Binding of the VEGF P3 (CYC) peptide antibodies to KDR cells was evaluated using flow cytometry. 1 × 106 KDR cells were incubated with 100 μl of PBS + 5% BSA + 0.02% sodium azide for 30 min at 4 °C to block nonspecific binding. 50 μg of anti-peptide antibodies purified from rabbits were then added and incubated for 1 h. The cells were then washed by adding 2 ml of PBS + 0.05% BSA + 0.02% sodium azide and centrifuged for 5 min at 12,000 rpm. Cells were then suspended in 100 μl of PBS + 0.05% BSA + 0.02% sodium azide before adding 1 μl of Alexa Fluor 488 goat anti-rabbit IgG antibody at 1:100 dilution, mixed, and incubated for 30 min at 4 °C. The cells were then washed again as before and then fixed with 2% formaldehyde before being analyzed by a Coulter ELITE flow cytometer. A total of 10,000 cells was analyzed by light scatter assessment before single parameter histograms were drawn. Controls included cells alone and cells plus secondary antibody alone.
RESULTS
Selection and Design of VEGF Peptide Mimics
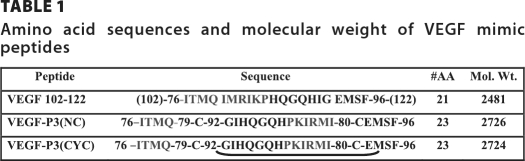
The crystal structure of the complex between VEGF and the Fab fragment of a humanized antibody (33) and analysis of the contact residues on both sides of the interface were published by Muller et al., (35, 57). Zilberberg et al. (58) also identified that the sequence 79–93 of VEGF is involved in the interaction with VEGF receptor-2. Although the VEGF residues critical for antibody binding are distinct from those important for high affinity receptor binding, they occupy a common region on VEGF demonstrating that the neutralizing effect of antibody binding results from steric blocking of VEGF-receptor interactions. Thus, it appears that only a small number of the residues buried in the VEGF-Fab interface are critical for high affinity binding and are concentrated in one continuous segment of the polypeptide loop between β5 and β6 (Fig. 1A). Several residues are important for VEGF receptor binding, including Met-81, Ile-83, Lys-84, Pro-85, Gln-89, and Gly-92 (31, 59). We have selected to use a peptide encompassing residues 102–122 (numbered as 76–96 in the crystal structure), which mimics the overlapping-VEGF binding sites to VEGFR-2 and Avastin.
FIGURE 1.
A, representation of the region selected for VEGF peptide mimic. The VEGF crystal structure (Protein Data Bank 2VPF) with the sequence 102–122 (residues 76–96) is shown in detail. VEGF fragment sequence 76–96 is a loop between anti-parallel β-sheets 5 and 6 and contains the residues involved in the VEGF-VEGFR-2 interaction. B, schematic representation of the VEGF peptide mimic design. VEGF sequence 102–122 (residues 76–96) from crystal structure (Protein Data Bank 2VPF) with labeled residues. Peptide mimic VEGF-P3(CYC) is shown. In black is shown the VEGF-P3(CYC) peptide sequence with labeled residues; arrows show anti-parallel β-sheet orientation based in the crystal structure as represented in A.
The strategy to create a conformational peptide consisting of an anti-parallel β-sheet is shown in Table 1, where the sequence was modified in a way that the ends were twisted to generate VEGF-P3(NC). For the peptide mimic, the peptide was synthesized starting with Ile-76 at the C-terminal end proceeding toward Phe-96 at the N terminus and going from left to right in that sequence. The strategy to create a conformational epitope consisting of an anti-parallel β-sheet as shown schematically in Fig. 1B requires two artificial cysteines to be introduced between Gln-79 and Gly-92 and between Ile-80 and Glu-93 for disulfide pairing to enable the formation of the twisted anti-parallel β-sheet. It also required two artificial cysteines to be introduced between Gln-79 and Gly-92 and between Ile-80 and Glu-93. After synthesis and purification of VEGF-P3 (NC) peptide, the disulfide bond was formed by oxidation reaction enabling the formation of the twisted anti-parallel β-sheet structure in the VEGF-P3 (CYC) (cyclized).
MS Analysis Peptides
All pure peptides showed uniform peaks on analytical HPLC (purity >95%) and were further characterized using MALDI mass spectroscopy analysis to confirm the calculated and observed (Cal/Obs) molecular weight. In brief, the following were used: VEGF(102–122) (M + H+) Cal/Obs 2482.24/2482.32; VEGF-P3(NC) (M + H+) Cal/Obs 2727.27/2727.61; VEGF-P3(CYC)(M + H+)Cal/Obs 2725.27/2725.43; MVF-VEGF-P3(NC)(M + H+)Cal/Obs 5023.67/5023.82; and MVF-VEGF-P3(CYC) (M + H+) Cal/Obs 5021.62/5021.19.
Structural Characterization
To verify the secondary structure of VEGF peptide mimics, circular dichroism experiments were carried out. CD analyses of VEGF peptide mimics demonstrated a shift in the minimum of the noncyclized peptide (197 nm) spectrum to minima in the cyclic peptide (203, 205, and 210 nm) (data not shown). The shift in the CD spectrum is characteristic of an assumed β-turn II conformational structure (60, 61), indicating that the cyclic peptide may adopt a configuration more similar of the anti-parallel β-sheet structure present in the loop of VEGF protein. This similarity in the binding region is expected to confer more binding ability to the receptor. We carried out surface plasmon resonance analysis with the purpose of evaluating the binding of VEGF peptide mimics to VEGFR-2.
Conformational VEGF Peptide Mimic Binds to VEGFR-2 with Higher Affinity
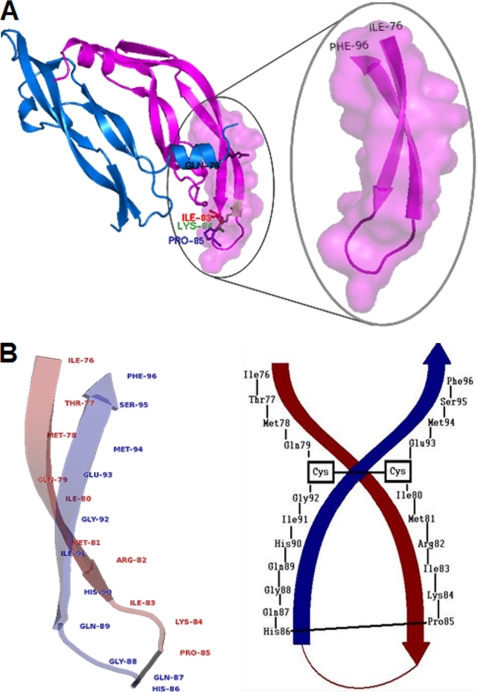
To determine the relative affinity of the VEGF peptide mimics to the receptor, we carried out binding assays using Biacore 3000 instrument. The VEGF natural sequence 102–122 (residues 76–96) and the engineered peptide mimics VEGF-P3(NC) and (CYC) were immobilized onto chip CM5, and the extracellular domain of VEGFR-2(KDR-Fc) was injected as the ligand. Sensograms in Fig. 2, A–C, show that VEGF receptor 2 demonstrated dose-dependent binding to the VEGF peptides VEGF(102–122), VEGF-P3(NC), and VEGF-P3(CYC). Global analysis was performed with data points fitting to a simple 1:1 (Langmuir) binding model. The values of association rate constant, Ka, and dissociation rate constant, Kd, are presented in Table 2. The equilibrium binding constant (KD) takes Ka and Kd values in consideration and represents the binding affinity. As can be seen in Table 2, the binding affinities for VEGF(102–122), VEGF-P3(NC), and VEGF-P3(CYC) are 45, 49, and 11 nm, respectively. The Ka for all three peptides demonstrated similar values, but the Kd value for VEGF-P3(CYC) is lower, resulting in a lower KD value that represents better affinity. These results confirm that the disulfide bond and cyclization confer a conformational structure in the designed VEGF mimic peptide that allows higher affinity to the receptor. Binding decrease of the extracellular domain of VEGFR-2 to VEGF mimic peptides was observed when rhVEGF was incubated with KDR-fc prior to injection (Fig. 2D) confirming that the extracellular domain of VEGFR-2-binding sites to VEGF mimic peptides and VEGF are located in the same region.
FIGURE 2.
Surface plasmon resonance assay. A–C, sensograms of VEGFR-2 binding to VEGF mimic peptides. VEGF(102–122) (A), VEGF-P3(NC) (B), and VEGF-P3(CYC) (C) were immobilized, and KDR-Fc at shown concentrations were injected at 10 μl/min. D, competition assay in which KDR-Fc at 500 nm was incubated with indicated rhVEGF concentrations for 30 min, prior to injection over peptide immobilized onto the chip. Percentage was calculated using the equation: 100% × (RUmax sample/RUmax control), where control was the KDR-Fc at 500 nm. RU, response units. Error bars represent means ± S.D.
TABLE 2.
Kinetic parameters of VEGF peptide mimics binding to VEGFR-2
Results were obtained from surface plasmon resonance assay where binding of peptide mimics to immobilized KDR was observed at different concentrations.
| Peptide | ka (1/ms) × 104 | kd (1/s) × 10−4 | KD |
|---|---|---|---|
| nm | |||
| VEGF(102–122) | 1.4 | 6.3 | 45 |
| VEGF-P3(NC) | 1.6 | 7.5 | 49 |
| VEGF-P3(CYC) | 1.9 | 2.0 | 11 |
Biotinylated Conformational VEGF Peptide Mimic Binds to Cells That Express VEGFR2
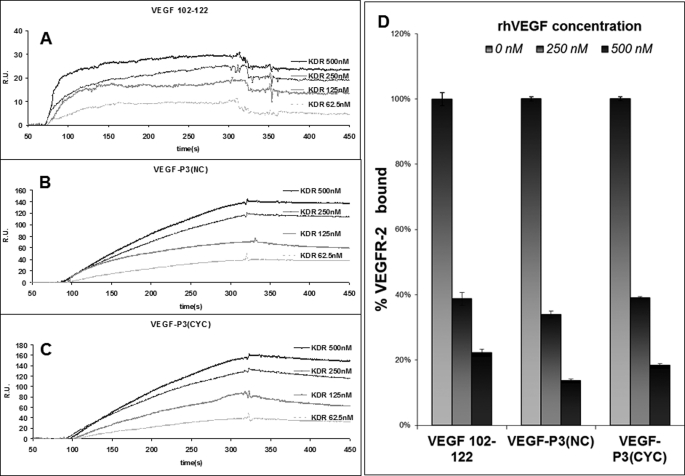
To determine whether the conformational peptide mimic VEGF P3(CYC) has the ability to recognize and bind cells that express VEGFR2, we biotinylated the peptide. The biotinylated VEGF-P3-CYC peptide was shown to specifically bind to VEGFR2-expressing cells as detected by streptavidin-Texas Red after incubation with the peptide (Fig. 3A). Results indicate binding and internalization in HUVEC and in a tumor cell line expressing only this receptor (293-KDR). Most interestingly, the binding of the peptide was seen unevenly distributed over HUVEC (Fig. 3, B and D), where the receptors are known to be expressed in clusters and reside in an endosomal population close to the plasma membrane (62). In the case of the 293-KDR cells, the binding could be seen all over the expressing cells (Fig. 3C) because expression is uniform throughout the cell surface. This explains the increased accumulation in some cells (293-KDR) and the clustering in others (HUVEC). These results clearly illustrate that the VEGF-P3-CYC peptide is specific to the VEGFR2 and recognize cells that are known to express the receptor, in a pattern that is consistent with its known distribution.
FIGURE 3.
Binding of biotinylated VEGF-P3(CYC) peptide to cells expressing VEGFR2. A, HUVEC incubated with streptavidin-Texas Red reagent only (negative control). B, HUVEC incubated with peptide (100 μg) for 2 h, followed by streptavidin for 1 h. Accumulation of the fluorescent conjugate on plasma membrane patches and/or in endocytosis vesicles is apparent. C, staining of cells overexpressing the VEGFR2/KDR receptor. Note increased accumulation in some cells (arrows), although nonexpressors remained negative (arrowhead). D, optical sectioning of the peptide-labeled plasma membrane of a HUVEC (arrow). A and B, combined phase contrast and fluorescence microscopy. C and D, confocal microscopy. Blue, nuclear staining with DAPI.
Conformational VEGF Peptide Mimic Prevents VEGFR-2 Phosphorylation
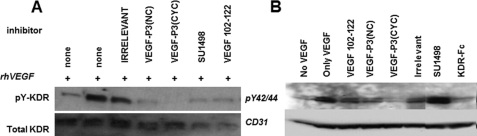
VEGFR-2 (also known as flt-1 or KDR) has been characterized as a tyrosine receptor type III. VEGFR-2 activation is promoted by dimerization upon VEGF binding. VEGFR-2 contains several tyrosine residues that can be phosphorylated, triggering several pathways such as proliferation, migration, and survival in the endothelial cells (18, 63, 64). Phosphorylation assay with HUVEC (cells that expresses physiological levels of VEGFR-2) was used to explore the ability of VEGF mimic peptides to block VEGF-VEGFR-2 interaction and consequently phosphorylation. All three VEGF mimic peptides were able to decrease the level of receptor phosphorylation (Fig. 4); however, the inhibitory effect was unable to be quantified, due to the limited detection of VEGFR-2 in the Western blotting. To gauge the effect of VEGF mimic peptides on VEGFR-2 phosphorylation, we used 293/KDR cells, which have been demonstrated to be an excellent model for VEGFR-2 phosphorylation because they overexpress VEGFR-2 (2.5 × 106 receptors per cell) (65). As seen in Fig. 5A, the degree of VEGFR-2 phosphorylation is notably increased in the presence of exogenous VEGF (10 ng/ml) and decreased when an exogenous receptor (KDR-Fc at 100 ng/ml) was used as a competitor. The level of inhibition in VEGFR-2 phosphorylation was similar with the VEGF natural sequence VEGF(102–122) or irrelevant control. When engineered peptides VEGF-P3(NC and CYC) were used as inhibitors, the level of VEGFR-2 phosphorylation was diminished, with the VEGF-P3(CYC) being the most potent inhibitor. These results were confirmed with the quantification of VEGFR-2 phosphorylation using the human phospho-VEGF R2/KDR DuoSet IC kit (Fig. 5B). The highest inhibition was observed with the VEGF-P3(CYC) (25%) followed by VEGF-P3(NC) (12%), although no inhibition was observed with irrelevant peptide (−2%) and a low level of inhibition with the natural sequence peptide VEGF(102–122) (4%). The percentage of inhibition was calculated assuming the phosphorylation level of control (only rhVEGF) was 100%, and the results are represented in Fig. 5B.
FIGURE 4.
Inhibition of phosphorylation in HUVEC. Cells were grown in 6-well plates and incubated with inhibitor as indicated for 30 min prior to stimulation with rhVEGF (10 ng/ml). Representative Western blots of cell lysates were dissolved in SDS-PAGE, transferred to PVDF membranes, and detected in phosphorylated KDR (A) and MAPK p44ERK1 and p42ERK2 (B) using specific anti-phospho- and anti-total KDR and anti-p44/42 MAPK and anti-CD31, respectively.
FIGURE 5.
Inhibition of KDR phosphorylation in 293/KDR cell line. Cells were grown in 6-well plates and incubated with inhibitor as indicated 30 min prior to stimulation with rhVEGF (10 ng/ml). A, representative Western blot using 293/KDR cell lysates that were solved in SDS-PAGE, transferred to PVDF membranes, and probed using specific anti-phospho- and anti-total KDR. B, percentage of VEGFR-2 phosphorylation calculated using human phospho-VEGF R2/KDR DuoSet IC kit (R&D Systems).
Activation of VEGFR-2 also triggers the MAPK pathway as one of the downstream signalings in the endothelial cells (66–68). The level of phosphorylation of MAPK p44ERK1 and p42ERK2 was observed using Western blotting and antibodies against phosphor-p44/42 (Fig. 4B). The decrease of phosphorylation level was greater when the VEGF-P3(CYC) was used as inhibitor followed by the noncyclic peptide VEGF-P3(NC) and VEGF(102–122). The small tyrosine kinase SU1498, used as one of positive controls, has been shown to accumulate phosphorylated MAPKs in endothelial cells because it interacts with other kinases such as ERK1/2, affecting other pathways (69). Tyrosine kinase inhibitors usually act by binding the kinase-active site blocking ATP binding; consequently, the phosphate is not transferred to the tyrosine residue. This mechanism of action has two major drawbacks as tyrosine kinase inhibitors as follows: low specificity and high susceptibility to resistance (enzymes often mutate themselves to recover activity) (70–72). VEGF peptide mimics demonstrated the same pattern of inhibition in VEGFR-2 and MAPK phosphorylation, indicating that downstream MAPK signaling of VEGFR-2 is being inhibited by decreased VEGFR-2 activation.
Conformational Peptide Inhibits HUVEC Proliferation
Endothelial cell proliferation is VEGF-dependent, and mostly activated by VEGFR-2 activation (30). Thus, angiogenesis inhibitors should inhibit HUVEC proliferation. This assay was carried out in the presence of several concentrations of VEGF mimic peptide to verify their ability to inhibit VEGF-dependent proliferation. Fig. 6A shows that all VEGF mimics can inhibit HUVEC proliferation in a dose-dependent way and that the conformational peptide VEGF-P3(CYC) demonstrated the highest inhibitory effect. The toxicity of the VEGF mimic peptides was verified using HUVEC proliferation assay in the absence of VEGF where no significant differences between peptide treated and untreated cells were observed (data not shown).
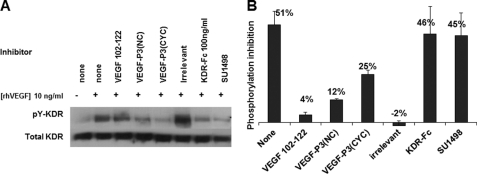
FIGURE 6.
A, inhibitory effects of peptides on HUVEC proliferation; B, network formation in Matrigel. The results represent an average from two set of experiments, each one done in triplicate, and are expressed as inhibition percent relative to the control (VEGF-stimulated cells without inhibitors). Pictures at ×40 magnification from light microscopy are shown for the Matrigel assay (B), and average sprout points counted using the software ImageJ (National Institutes of Health) are shown in C. Inhibitors were used at the indicated concentration. KDR-Fc is an extracellular domain from VEGFR2, and SU1498 (Calbiochem) is the small molecule tyrosine kinase inhibitor. ECGS represents a positive control where growth condition was used, i.e. medium supplemented with 20% FBS and ECGS. Irrelevant peptide represents a scrambled sequence with similar molecular weight. * represent p < 0.05 using Student's t test, indicating that only cells treated with irrelevant peptide were not significantly different from the control. Error bars represent mean ± S.E.
Conformational Peptide Decreases HUVEC Network Formation in Matrigel Assay
Activation of VEGFR-2 also triggers the MAPK pathway that leads to the formation of cell cords and tubes by the endothelial cells (73). The in vitro Matrigel assay is an appropriate model for assessing network formation as it takes advantage of the capacity of cell cord formation by HUVECs growing in an extracellular matrix (Matrigel).
Network formation is clearly VEGF-dependent as can be seen in Fig. 6B, where a cell network with several sprout points is more evident in the VEGF-treated HUVEC than the non-VEGF-treated HUVEC. A decrease in the network branching and tube formation was observed in VEGF-treated HUVEC in the presence of VEGF mimic peptides, and no significant effect was seen with the irrelevant control (Fig. 6C). The best inhibitory effect was demonstrated by engineered mimic peptides VEGF-P3 (NC and CYC). These results are in agreement with VEGFR-2 phosphorylation and HUVEC proliferation assay, indicating that VEGF mimic peptides can block VEGF and VEGFR-2 interaction.
VEGF Peptide Mimics Inhibit Cell Migration in a Scratch Wound Assay
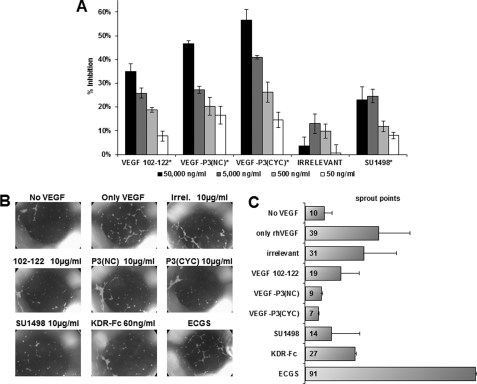
New blood vessel formation requires that the endothelial cells migrate toward the sources of growth factor. This process has similar characteristics with wound healing in which VEGF has been shown to play an important role throughout VEGFR-2 activation (74). We used the scratch wound assay with HUVEC to observe the ability of the VEGF mimic peptides in inhibiting endothelial cell migration. As can be seen in Fig. 7A, cells were able to migrate toward the scratched area in higher numbers when exogenous rhVEGF was added compared with the absence VEGF. Growth medium was supplemented with 20% FBS and endothelial cell growth supplements. Fig. 7B shows a slight increase in percentage of migrated cells, probably due to the complexity provided by the supplements. Irrelevant peptide control had a comparable number of migrated cells when compared with rhVEGF control, indicating no inhibition. All three VEGF mimic peptides demonstrated an ability to inhibit HUVEC migration at similar levels (∼50%) as the small VEGFR-2 tyrosine kinase inhibitor (SU1498) at a standard concentrations (Fig. 7B), indicating that VEGF mimic peptides are capable of blocking the VEGF-dependent migration in endothelial cells.
FIGURE 7.
HUVEC migration in scratch wound assay using peptides as inhibitors. A, pictures taken at ×40 magnification in light microscopy. B, average percentage of migrated cells, counted using the software ImageJ (National Institutes of Health) assuming rhVEGF control as 100%. Inhibitors were used at indicated concentration. α-vegf represents a monoclonal antibody demonstrated to block VEGF-dependent pathways. Results shown represent an average of two different experiments each performed in duplicate, and error bars represent mean ± S.E. IRR, irrelevant.
VEGF Peptide Mimics as Treatment in a VEGF-overexpressing Mouse Model
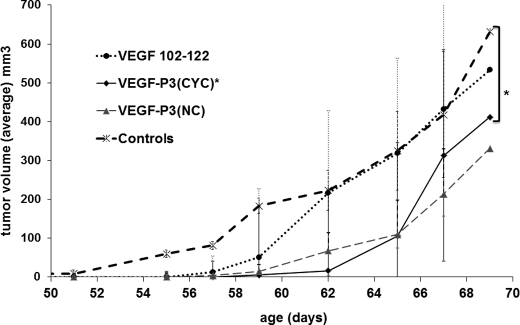
We have extensively studied conformational peptides of HER-2 as peptide-based immunotherapy. We used the double transgenic mice VEGF+/−Neu2-5+/− in our in vivo experiments taking into consideration that our ultimate goal was to combine HER-2 immunotherapy with anti-angiogenic therapy. The transgenic Neu2-5+/− mouse develops spontaneous mammary tumors at age 111 days due to a mutation in the neu gene (mouse homologue of HER-2). When they are crossed with the murine mammary tumor virus VEGF-164, which overexpress VEGF under an MMTV promoter, the offspring resulting is the double transgenic VEGF+/−Neu2-5+/− in which spontaneous tumors developed much earlier, around 51 days, due to excessive activation of angiogenesis by increased expression of VEGF. VEGF+/−Neu2-5+/− mice treated with VEGF peptide mimics developed tumors after 57 days, whereas nontreated mice had tumors around 51 days. However, using the statistical model cross-sectional generalized estimating equation, only the group treated with VEGF-P3(CYC) demonstrated statistical significance (p = 0.0074) in delaying tumor burden (Fig. 8). The tumor doubling time in the VEGF-P3(CYC) treated group was 1½ times longer than the control group (3.6 versus 2.3 days). The design of the constrained VEGF-P3(CYC) to mimic VEGF-binding sites seems to be important to produce the best inhibitor in vitro and in vivo.
FIGURE 8.
Passive treatment using peptides as inhibitors of mammary tumor development in mouse VEGF−/+/Neu2-5−/+. Groups of six 4–10-week-old mice were treated once a week with intravenous injection of 500 μg of peptide (in PBS). Control represents no treatment. The data are presented as the average tumor size per group and are reported as mm3. Average of largest tumor volume was calculated as follows: (long measurement × short measurement2)/2. Statistical analysis demonstrates significance in the growth tumor rate in mice treated with VEGF-P3(CYC) (*, p value = 0.0074), Error bars represent mean ± S.E.
Anti-VEGF Peptide Antibodies Validate Peptidomimetic Approach
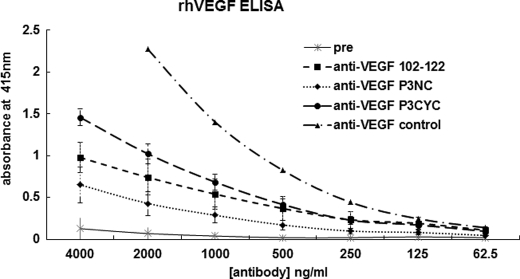
To develop therapeutic approaches to inhibit angiogenesis by using peptide mimics, we wanted to explore the ability of the VEGF peptide mimics to generate native-like antibodies. Such anti-VEGF antibodies could be used to further demonstrate the validity of the peptidomimetic approach and could themselves be used as inhibitors of angiogenesis. We have developed strategies for using epitope-based peptides to generate antibodies with better affinity and specificity. Similarly, we have successfully predicted oncogenic peptide epitopes to develop conformational peptides for cancer vaccine approaches (54, 56, 75–78). VEGF sequence 102–122 (residues 76–96) includes the region containing several residues important for antibody neutralization of VEGF. We hypothesized that antibodies elicited against the engineered peptide mimics VEGF-P3(NC and CYC) would retain or enhance the specificity for VEGF protein. A “promiscuous” T-cell epitope measles virus fusion protein 288–302, which has been demonstrated to enhance immune response (79), was incorporated into VEGF mimic peptides and used for raising antibodies in rabbits. All three constructs of VEGF peptide mimics demonstrated high immunogenicity (data not shown) and were able to recognize the rhVEGF in an ELISA (Fig. 9).
FIGURE 9.
Detection of anti-peptide antibodies by ELISA. Plate was coated with rhVEGF (1 μg/ml) overnight, and ELISA was done as usual. Purified anti-peptide antibodies were used as primary antibodies. The control represents a commercially available polyclonal antibody to detect VEGF (R&D Systems). Goat anti-rabbit antibody conjugated to HRP was used as secondary antibody at a 1:500 dilution factor. Pre represents blood drawn before immunization. Results shown represent an average of three different ELISAs each performed in duplicate, and error bars represent mean ± S.E.
Specificity of Anti-VEGF Peptide Mimic Antibodies
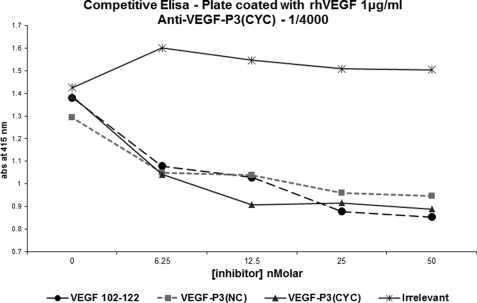
To confirm their specificity, we carried out competitive ELISA using rhVEGF on the plate with antibodies to the various mimics and VEGF mimic peptides as inhibitors. Fig. 10 shows the results for anti-MVF-VEGF-P3(CYC). Engineered VEGF-P3 peptides, in the cyclic form, were able to compete for the binding site in the anti-VEGF-P3(CYC) but not to the antibodies generated against the natural sequence. This indicates that the engineered peptides did not generate antibodies against the linear sequence of VEGF but most importantly that they mimic the conformational epitope in the VEGF protein.
FIGURE 10.
Competitive ELISA. Plates were coated with peptide as indicated. Competitive assay was carried out using constant amount of anti-peptide and several concentrations of peptides as competitors. Results represent average of two different experiments each done in duplicate.
Antibodies to Conformational VEGF Peptide Mimic VEGF P3(CYC) Have High Affinity for rhVEGF
Kinetic parameters of antibodies raised against VEGF mimic peptides were obtained by surface plasmon resonance using direct binding assay in BIAcore 3000. Anti-peptide antibodies were injected as ligands over rhVEGF immobilized onto CM5 chip. The binding affinity to the whole protein was higher for the antibody raised against the conformational epitope, anti-VEGF-P3(CYC) (KD = 146 nm), followed by the anti-VEGF-P3(NC) (KD = 251 nm), and the antibody raised against the natural sequence anti-VEGF(102–122) (KD = 552 nm). As can be seen in Table 3, the Ka value for the anti-VEGF mimic peptides demonstrated only a 10-fold decrease in Ka and a comparable Kd when compared with a commercially available monoclonal antibody against VEGF.
TABLE 3.
Kinetic parameters of anti-VEGF peptide antibodies binding to rhVEGF
Results were obtained from surface plasmon resonance assay where binding of anti-peptide antibodies to immobilized rhVEGF on the chip was observed at different concentrations.
| Antibody | ka (1/ms) × 103 | kd (1/s) × 10−3 | KD |
|---|---|---|---|
| nm | |||
| Anti-VEGF(102–122) | 6.2 | 3.6 | 552 |
| Anti-VEGF-P3(NC) | 6.7 | 1.4 | 251 |
| Anti-VEGF-P3(CYC) | 8.6 | 1.3 | 146 |
| Anti-mAb VEGF | 86.1 | 2.7 | 29 |
Reactivity of Peptide Antibodies to Conformational VEGF P3(CYC) with VEGFR2-expressing Cells (KDR)
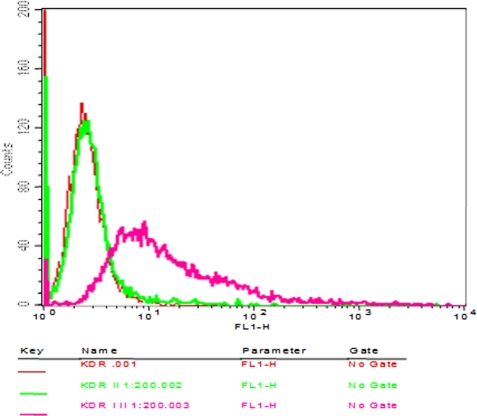
Binding of peptide antibodies to intact VEGFR2 cells was evaluated by immunofluorescence staining of single cell suspension of 293-KDR cells. The peptide antibodies were able to specifically bind the cells (Fig. 11). The binding was also dose-dependent with increased binding when higher concentrations of the antibodies were used (results not shown). These results demonstrate that the peptide vaccine was able to prevent VEGF binding to it receptor VEGFR2. The peptide when added before the anti-peptide antibodies was also able to prevent the binding of the antibodies illustrating its specificity for the VEGFR2 (results not shown).
FIGURE 11.
Cross-reactivity of antipeptide antibodies to VEGFR2. Flow cytometry was used to assess the binding capabilities of the peptide antibodies (I) to KDR cells. 50 μg of purified antibodies from immunized rabbits were used before adding secondary antibodies (KDR, I, II). The binding results was compared with cells alone (KDR) and cells with secondary antibodies alone (KDR, II). Antibody binding was detected by goat anti-rabbit Alexa Fluor 488 secondary antibodies (II). The x axis represents fluorescent intensity and the y axis represents relative cell number.
Anti-VEGF Peptide Antibodies Inhibits HUVEC Proliferation
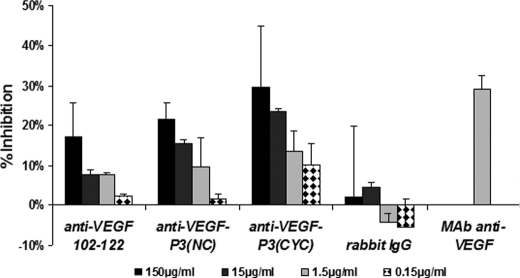
VEGF-neutralizing antibodies blocks the interaction of VEGF and VEGF receptors by binding to and occluding VEGF-binding sites. Because our anti-VEGF mimic peptide antibodies were able to bind VEGF, we tested their ability to inhibit the VEGF-dependent HUVEC proliferation assay. All three anti-VEGF peptide antibodies were able to inhibit HUVEC proliferation in a dose-dependent way when compared with the preimmune serum control. Anti-VEGF-P3(CYC) demonstrated the highest inhibition, and anti-VEGF-P3(NC) seems to be slightly more efficient than the natural sequence (Fig. 12). In this model the proliferation inhibition is believed to be due to blockage of interaction between VEGF and VEGFR-2, indicating that the engineered VEGF-P3(CYC), which contains twisted ends and the disulfide bond to mimic the binding region of VEGF, can generate antibodies against the conformational epitope that resulted in the highest neutralization effects.
FIGURE 12.
Proliferation assay using antibodies as inhibitors. Anti-VEGF(102–122), P3(NC), and P3(CYC) correspond to polyclonal antibodies against peptides MVF-VEGF(102–122), MVF-VEGF-P3(NC), and MVF-VEGF-P3(CYC) raised in our laboratory. Antibodies were purified from bleed 3Y+2 or 3Y+3. Rabbit IgG, rabbit total IgG; mAb anti-VEGF, monoclonal antibody against human VEGF protein (R&D Systems) shown to block proliferation. The results represent average from two set of experiments, each one done in triplicate and is expressed as inhibition percentage relative to the control (VEGF stimulated cells without inhibitors). Error bars represent mean ± S.E.
DISCUSSION
Protein-protein interactions trigger a wide variety of cellular pathways, representing a target for drug development. The active or passive binding sites of a protein are confined to a small set of amino acids; therefore, smaller sequence-like peptides can be designed to simulate these regions, potentially acting as an agonist or antagonist. Synthesis of peptides is easier and cheaper than proteins, and recent approaches have brought many new improvements to the delivery and stability of peptide in vivo. Recently, a 17-amino acid cyclic VEGF peptide (residues 79–93) has been shown to bind VEGF receptor-2 and block angiogenesis (58), and a peptide inhibitor of VEGF receptor KDR/Flk-1 was identified by phage display (80). Several peptides have been identified that block VEGF-VEGFR interactions and may be potent inhibitors of tumor angiogenesis and metastasis (40). Peptides that mimic the VEGFR-2-binding site of VEGF were designed to block VEGF-VEGFR-2 interaction, which has been characterized as the most important for angiogenesis activation.
Here, we report that peptides corresponding to the natural VEGF amino acid sequence 102–122 (residues 76–96) (Fig. 1A), which includes the loop region with the important binding residues of VEGF to its receptor, was engineered to better mimic the conformational structure of this sequence in the protein. The two conformational peptide mimics VEGF-P3(NC) and VEGF-P3 (CYC) sequence were specially designed such that the ends were twisted with cysteines artificially inserted to enable cyclization (Fig. 1B). CD analysis confirmed that the VEGF-P3(CYC) assumes characteristics of β-turn II, and surface plasmon resonance analysis demonstrates that VEGFR-2 had a higher binding affinity for this cyclic peptide VEGF-P3(CYC) than to the noncyclized version and/or the natural sequence (VEGF(102–122)), indicating the importance of the constrained structure for enhancing binding activity. Competition assay showed that VEGF peptide mimics and VEGF are binding to VEGFR-2 in the same region indicating the peptide mimics could act as antagonist to VEGF. The conformational peptide VEGF-P3(CYC) also be demonstrated to bind cells expressing VEGFR2. Antibodies raised in rabbits against VEGF mimic peptides were shown to be specific for each peptide and also to recognize the native protein rhVEGF. Anti-VEGF-MVF-P3(CYC) demonstrated better affinity for rhVEGF in surface plasmon resonance experiments, indicating that the conformational peptide construction is mimicking better the portion comprising the loop in VEGF.
Next, we evaluated whether inhibition of VEGFR-2 cascade signaling would be translated in inhibition of activation of endothelial cell network formation, migration, and proliferation. To determine these effects, we tested VEGF peptide mimics in several in vitro angiogenesis assays. Several in vitro assays have been established to explore VEGF-dependent angiogenesis (81, 82), and we carried them out to test whether the antagonist effect of VEGF peptide mimics could block VEGF action in these assays. Upon VEGF binding, VEGFR-2 dimerizes leading to phosphorylation of tyrosines in the kinase domain that triggers several pathways, including endothelial cell proliferation, migration, and survival. The inhibitory effects of VEGF peptide mimics on VEGFR-2 phosphorylation were evaluated indicating that they were able to inhibit VEGFR-2 phosphorylation in a cell line (HUVEC) physiologically expressing VEGFR-2, as well as in the overexpressing cell line (293/KDR). We also observed a decrease in p44/42 MAPK phosphorylation, which is one of the downstream signalings resulting from VEGFR-2 activation. The designed peptide VEGF-P3(CYC) displayed the best inhibitory effect on phosphorylation assay following the pattern observed with the surface plasmon resonance experiment, indicating that the design to better mimic conformational structure of the VEGF-binding site confers better inhibitory effects on VEGF-activated signaling. The biotinylated VEGF-P3-CYC peptide was also shown to specifically bind to cells that have different expression of the VEGFR2, and the pattern of binding was coherent with the receptor expression.
To confirm the effect of the VEGF peptide mimics as angiogenesis inhibitors, we used several in vitro angiogenic assays as follows: scratch wound (migration), Matrigel (network formation), and HUVEC proliferation assay (proliferation). All three VEGF peptide mimics were able to inhibit cell migration in the presence of exogenous rhVEGF in the wound assay, indicating that the peptides were effective inhibitors of VEGF-VEGFR-2 interaction. Our results of whether all three VEGF peptide mimics could inhibit the network formation in Matrigel show that the engineered peptide mimic VEGF-P3(CYC) had the largest inhibition compared with the natural sequence and/or the uncyclized peptide. Proliferation of endothelial cells is essential to formation of the new wall vessels, and inhibition of HUVEC proliferation was observed in a dose-dependent manner with VEGF-P3(CYC) as the most potent inhibitor. The conformational peptide VEGF-P3(CYC) demonstrated the best inhibitory effects and the highest binding affinity and is most likely due to the loop stabilization by the disulfide bond between the two cysteines. Our biochemical and in vitro experiment results were in agreement and established that VEGF-P3(CYC) had the best potential of inhibiting angiogenesis, further emphasizing that the receptor-ligand interaction is exquisitely dependent on the conformation of the peptide structure.
Peptides can be used as antigen to generate high affinity antibodies specific for an entire protein. These peptides must include the antigenic determinant residues that usually are hydrophilic and are exposed in the protein (83). These can be achieved by rational design of peptides that may include few modifications to obtain similar conformational structure of the protein. Our primary goal was to evaluate VEGF peptide mimic as angiogenesis inhibitors. However, the VEGF peptide mimic was designed to mimic the binding region of VEGF to VEGFR-2, which overlaps with a B-cell predicted epitope. We also tested if synthetic VEGF peptide mimics could be used to generated antibodies against native the VEGF protein. Because combining the B- and T-cell epitope have allowed us to increase the immunogenicity of peptides, we linked VEGF peptides to a promiscuous T-cell epitope from measles virus fusion protein. These peptides were highly immunogenic in outbred rabbits, and purified antibodies against all three VEGF peptide mimics recognized rhVEGF. We quantified the binding affinity of these antibodies by using the surface plasmon resonance experiments. Among anti-VEGF peptide mimics antibodies, anti-MVF-VEGF-P3(CYC) has the highest binding affinity, suggesting that the structural arrangements of VEGF-P3(CYC) were able to generate antibody that can bind tighter to the VEGF. Competitive ELISA results showed that the epitope recognized in VEGF by anti-VEGF peptide mimics are not the same, indicating that the anti-MVF-VEGF-P3 (NC) and (CYC) bind to VEGF by recognition of conformational instead of the linear epitopes.
VEGF-neutralizing monoclonal antibodies, such as Avastin®, binds to VEGF preventing VEGF-VEGFR-2 interaction and as consequence inhibits angiogenesis. Anti-peptide generated against VEGF peptide mimics were able to specifically recognize the native protein, and anti-MVF-VEGF-P3(CYC) demonstrated better affinity to rhVEGF. We further evaluated if these anti-peptide antibodies would block VEGF-VEGFR-2 interaction, and as expected, the inhibitory effect on HUVEC proliferation of anti-MVF-VEGF-P3(CYC) was slightly better than the other anti-VEGF peptide mimic antibodies.
The design of the peptide VEGF-P3(CYC) that would mimic a structural binding site of VEGF to its receptor was shown to be important in obtaining a better inhibitory molecule in several in vitro assays as well as in the transgenic mouse model of VEGF+/−Neu2-5+/−. These findings motivate the development and potential of using VEGF-P3(CYC) as an alternative of peptide therapeutic drugs to inhibit angiogenesis. Still, future analysis involving animal models of angiogenesis-dependent tumor formation will give insight into the efficacy of these peptides in inhibiting angiogenesis given the complexity of the tumor microenvironment. In the tumor vicinity, stromal cells are involved in angiogenesis, and they also can activate other processes like neovascularization, in which endothelial progenitor cells can initiate the formation of completely new blood vessels (84).
It also will be interesting to observe whether this peptide would have an effect on other important aspects of VEGF signaling via VEGFR-1 in other cells like macrophage or endothelial progenitor cells (85, 86). VEGF-P3(CYC) is not expected to interact with VEGFR-1 once it does not include the VEGF residues responsible for binding to VEGFR-1. However, VEGF-P3(CYC) may interfere with signaling activated by the heterodimer VEGFR-1/VEGFR-2, which can also activate angiogenesis (87, 88). VEGF-P3(CYC) may also be relevant in inhibiting autocrine activation in cancer cells once cells lines derived from breast cancer have been shown to overexpress VEGFR-2 that can be activated in an autocrine loop via up-regulation of VEGF (89).
In conclusion, we showed that VEGF receptor-specific peptides can interfere with the interaction between VEGF and VEGFR-2 inhibiting several VEGF-dependent pathways and indicating that VEGF mimic peptides have a clear potential as candidates in preclinical studies using animal models as alternatives to the development of new anti-angiogenesis therapeutic approaches.
Acknowledgments
We are grateful to Dr. Mirela Anghelina for skillful technical assistance and Dr. Nikanor Moldovan for useful discussions.
Footnotes
- NC
- noncyclized
- HUVEC
- human umbilical vein endothelial cell
- rh
- recombinant human
- Cal/Obs
- calculated and observed
- ECGS
- endothelial cell growth supplement
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- RP
- reversed phase.
REFERENCES
- 1. Benjamin L. E., Keshet E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8761–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bergers G., Benjamin L. E. (2003) Nat. Rev. Cancer 3, 401–410 [DOI] [PubMed] [Google Scholar]
- 3. Ng I. O., Poon R. T., Lee J. M., Fan S. T., Ng M., Tso W. K. (2001) Am. J. Clin. Pathol. 116, 838–845 [DOI] [PubMed] [Google Scholar]
- 4. Takahashi H., Shibuya M. (2005) Clin. Sci. 109, 227–241 [DOI] [PubMed] [Google Scholar]
- 5. Carmeliet P. (2005) Oncology 69, Suppl. 3, 4–10 [DOI] [PubMed] [Google Scholar]
- 6. Folkman J. (1992) Semin. Cancer Biol. 3, 65–71 [PubMed] [Google Scholar]
- 7. Ferrara N., Hillan K. J., Novotny W. (2005) Biochem. Biophys. Res. Commun. 333, 328–335 [DOI] [PubMed] [Google Scholar]
- 8. Drevs J., Hofmann I., Hugenschmidt H., Wittig C., Madjar H., Müller M., Wood J., Martiny-Baron G., Unger C., Marmé D. (2000) Cancer Res. 60, 4819–4824 [PubMed] [Google Scholar]
- 9. Ahmed S. I., Thomas A. L., Steward W. P. (2004) J. Chemother. 16, Suppl. 4, 59–63 [DOI] [PubMed] [Google Scholar]
- 10. Witte L., Hicklin D. J., Zhu Z., Pytowski B., Kotanides H., Rockwell P., Böhlen P. (1998) Cancer Metastasis Rev. 17, 155–161 [DOI] [PubMed] [Google Scholar]
- 11. Wood J. M. (2000) Medicina 60, Suppl. 2, 41–47 [PubMed] [Google Scholar]
- 12. Milkiewicz M., Ispanovic E., Doyle J. L., Haas T. L. (2006) Int. J. Biochem. Cell Biol. 38, 333–357 [DOI] [PubMed] [Google Scholar]
- 13. Verhoef C., de Wilt J. H., Verheul H. M. (2006) Curr. Pharm. Des. 12, 2623–2630 [DOI] [PubMed] [Google Scholar]
- 14. Morabito A., De Maio E., Di Maio M., Normanno N., Perrone F. (2006) Oncologist 11, 753–764 [DOI] [PubMed] [Google Scholar]
- 15. Bergers G., Hanahan D. (2001) Nat. Biotechnol. 19, 20–21 [DOI] [PubMed] [Google Scholar]
- 16. Sebolt-Leopold J. S., English J. M. (2006) Nature 441, 457–462 [DOI] [PubMed] [Google Scholar]
- 17. Dancey J. E., Chen H. X. (2006) Nat. Rev. Drug Discov. 5, 649–659 [DOI] [PubMed] [Google Scholar]
- 18. Ferrara N., Gerber H. P., LeCouter J. (2003) Nat. Med. 9, 669–676 [DOI] [PubMed] [Google Scholar]
- 19. Ferrara N. (2000) Curr. Opin. Biotechnol. 11, 617–624 [DOI] [PubMed] [Google Scholar]
- 20. Lee J., Gray A., Yuan J., Luoh S. M., Avraham H., Wood W. I. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1988–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. White F. C., Carroll S. M., Kamps M. P. (1995) Growth Factors 12, 289–301 [DOI] [PubMed] [Google Scholar]
- 22. Yoo P. S., Mulkeen A. L., Cha C. H. (2006) World J. Gastroenterol. 12, 4937–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Konecny G. E., Meng Y. G., Untch M., Wang H. J., Bauerfeind I., Epstein M., Stieber P., Vernes J. M., Gutierrez J., Hong K., Beryt M., Hepp H., Slamon D. J., Pegram M. D. (2004) Clin. Cancer Res. 10, 1706–1716 [DOI] [PubMed] [Google Scholar]
- 24. Wen X. F., Yang G., Mao W., Thornton A., Liu J., Bast R. C., Jr., Le X. F. (2006) Oncogene 25, 6986–6996 [DOI] [PubMed] [Google Scholar]
- 25. Kostopoulos I., Arapantoni-Dadioti P., Gogas H., Papadopoulos S., Malamou-Mitsi V., Scopa C. D., Markaki S., Karagianni E., Kyriakou V., Margariti A., Kyrkou E., Pavlakis K., Zaramboukas T., Skordalaki A., Bourli A., Markopoulos C., Pectasides D., Dimopoulos M. A., Skarlos D., Fountzilas G. (2006) Breast Cancer Res. Treat. 96, 251–261 [DOI] [PubMed] [Google Scholar]
- 26. Petit A. M., Rak J., Hung M. C., Rockwell P., Goldstein N., Fendly B., Kerbel R. S. (1997) Am. J. Pathol. 151, 1523–1530 [PMC free article] [PubMed] [Google Scholar]
- 27. Anghelina M., Krishnan P., Moldovan L., Moldovan N. I. (2004) Stem Cells Dev. 13, 665–676 [DOI] [PubMed] [Google Scholar]
- 28. Anghelina M., Krishnan P., Moldovan L., Moldovan N. I. (2006) Am. J. Pathol. 168, 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herley M. T., Yu Y., Whitney R. G., Sato J. D. (1999) Biochem. Biophys. Res. Commun. 262, 731–738 [DOI] [PubMed] [Google Scholar]
- 30. Kanno S., Oda N., Abe M., Terai Y., Ito M., Shitara K., Tabayashi K., Shibuya M., Sato Y. (2000) Oncogene 19, 2138–2146 [DOI] [PubMed] [Google Scholar]
- 31. Keyt B. A., Nguyen H. V., Berleau L. T., Duarte C. M., Park J., Chen H., Ferrara N. (1996) J. Biol. Chem. 271, 5638–5646 [DOI] [PubMed] [Google Scholar]
- 32. Fairbrother W. J., Champe M. A., Christinger H. W., Keyt B. A., Starovasnik M. A. (1998) Structure 6, 637–648 [DOI] [PubMed] [Google Scholar]
- 33. Fuh G., Wu P., Liang W. C., Ultsch M., Lee C. V., Moffat B., Wiesmann C. (2006) J. Biol. Chem. 281, 6625–6631 [DOI] [PubMed] [Google Scholar]
- 34. Fuh G., Li B., Crowley C., Cunningham B., Wells J. A. (1998) J. Biol. Chem. 273, 11197–11204 [DOI] [PubMed] [Google Scholar]
- 35. Muller Y. A., Li B., Christinger H. W., Wells J. A., Cunningham B. C., de Vos A. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7192–7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiesmann C., Christinger H. W., Cochran A. G., Cunningham B. C., Fairbrother W. J., Keenan C. J., Meng G., de Vos A. M. (1998) Biochemistry 37, 17765–17772 [DOI] [PubMed] [Google Scholar]
- 37. Zhang W., Ran S., Sambade M., Huang X., Thorpe P. E. (2002) Angiogenesis 5, 35–44 [DOI] [PubMed] [Google Scholar]
- 38. Lu D., Shen J., Vil M. D., Zhang H., Jimenez X., Bohlen P., Witte L., Zhu Z. (2003) J. Biol. Chem. 278, 43496–43507 [DOI] [PubMed] [Google Scholar]
- 39. Böldicke T., Tesar M., Griesel C., Rohde M., Gröne H. J., Waltenberger J., Kollet O., Lapidot T., Yayon A., Weich H. (2001) Stem Cells 19, 24–36 [DOI] [PubMed] [Google Scholar]
- 40. Binétruy-Tournaire R., Demangel C., Malavaud B., Vassy R., Rouyre S., Kraemer M., Plouët J., Derbin C., Perret G., Mazié J. C. (2000) EMBO J. 19, 1525–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Allen S. D., Rawale S. V., Whitacre C. C., Kaumaya P. T. (2005) J. Pept. Res. 65, 591–604 [DOI] [PubMed] [Google Scholar]
- 42. Ercil N. E., Galici R., Kesterson R. A. (2005) Psychopharmacology 180, 279–285 [DOI] [PubMed] [Google Scholar]
- 43. Joppa M. A., Ling N., Chen C., Gogas K. R., Foster A. C., Markison S. (2005) Peptides 26, 2294–2301 [DOI] [PubMed] [Google Scholar]
- 44. Lender A., Yao W., Sprengeler P. A., Spanevello R. A., Furst G. T., Hirschmann R., Smith A. B., 3rd (1993) Int. J. Pept. Protein Res. 42, 509–517 [DOI] [PubMed] [Google Scholar]
- 45. Butenas S., Orfeo T., Kalafatis M., Mann K. G. (2006) J. Thromb. Haemost. 4, 2411–2416 [DOI] [PubMed] [Google Scholar]
- 46. Fear G., Komarnytsky S., Raskin I. (2007) Pharmacol. Ther. 113, 354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dass C. R., Choong P. F. (2006) Peptides 27, 3479–3488 [DOI] [PubMed] [Google Scholar]
- 48. Ladner R. C., Sato A. K., Gorzelany J., de Souza M. (2004) Drug Discov. Today 9, 525–529 [DOI] [PubMed] [Google Scholar]
- 49. Kerbel R., Folkman J. (2002) Nat. Rev. Cancer 2, 727–739 [DOI] [PubMed] [Google Scholar]
- 50. Carmeliet P., Jain R. K. (2000) Nature 407, 249–257 [DOI] [PubMed] [Google Scholar]
- 51. Kremer C., Breier G., Risau W., Plate K. H. (1997) Cancer Res. 57, 3852–3859 [PubMed] [Google Scholar]
- 52. Jain R. K. (2002) Semin. Oncol. 29, Suppl. 16, 3–9 [DOI] [PubMed] [Google Scholar]
- 53. Sundaram R., Lynch M. P., Rawale S. V., Sun Y., Kazanji M., Kaumaya P. T. (2004) J. Biol. Chem. 279, 24141–24151 [DOI] [PubMed] [Google Scholar]
- 54. Dakappagari N. K., Lute K. D., Rawale S., Steele J. T., Allen S. D., Phillips G., Reilly R. T., Kaumaya P. T. (2005) J. Biol. Chem. 280, 54–63 [DOI] [PubMed] [Google Scholar]
- 55. Gong Y., Yan K., Lin F., Anderson K., Sotiriou C., Andre F., Holmes F. A., Valero V., Booser D., Pippen J. E., Jr., Vukelja S., Gomez H., Mejia J., Barajas L. J., Hess K. R., Sneige N., Hortobagyi G. N., Pusztai L., Symmans W. F. (2007) Lancet Oncol. 8, 203–211 [DOI] [PubMed] [Google Scholar]
- 56. Dakappagari N. K., Pyles J., Parihar R., Carson W. E., Young D. C., Kaumaya P. T. (2003) J. Immunol. 170, 4242–4253 [DOI] [PubMed] [Google Scholar]
- 57. Muller Y. A., Chen Y., Christinger H. W., Li B., Cunningham B. C., Lowman H. B., de Vos A. M. (1998) Structure 6, 1153–1167 [DOI] [PubMed] [Google Scholar]
- 58. Zilberberg L., Shinkaruk S., Lequin O., Rousseau B., Hagedorn M., Costa F., Caronzolo D., Balke M., Canron X., Convert O., Laïn G., Gionnet K., Goncalvès M., Bayle M., Bello L., Chassaing G., Deleris G., Bikfalvi A. (2003) J. Biol. Chem. 278, 35564–35573 [DOI] [PubMed] [Google Scholar]
- 59. Muller Y. A., Christinger H. W., Keyt B. A., de Vos A. M. (1997) Structure 5, 1325–1338 [DOI] [PubMed] [Google Scholar]
- 60. Bush C. A., Sarkar S. K., Kopple K. D. (1978) Biochemistry 17, 4951–4954 [DOI] [PubMed] [Google Scholar]
- 61. Perczel A., Hollósi M., Sándor P., Fasman G. D. (1993) Int. J. Pept. Protein Res. 41, 223–236 [DOI] [PubMed] [Google Scholar]
- 62. Gampel A., Moss L., Jones M. C., Brunton V., Norman J. C., Mellor H. (2006) Blood 108, 2624–2631 [DOI] [PubMed] [Google Scholar]
- 63. Schramm R., Yamauchi J., Vollmar B., Vajkoczy P., Menger M. D. (2003) Transplantation 75, 239–242 [DOI] [PubMed] [Google Scholar]
- 64. Yang S., Toy K., Ingle G., Zlot C., Williams P. M., Fuh G., Li B., de Vos A., Gerritsen M. E. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 1797–1803 [DOI] [PubMed] [Google Scholar]
- 65. Backer M. V., Budker V. G., Backer J. M. (2001) J. Control Release 74, 349–355 [DOI] [PubMed] [Google Scholar]
- 66. Wu L. W., Mayo L. D., Dunbar J. D., Kessler K. M., Baerwald M. R., Jaffe E. A., Wang D., Warren R. S., Donner D. B. (2000) J. Biol. Chem. 275, 5096–5103 [DOI] [PubMed] [Google Scholar]
- 67. Shibuya M. (2006) J. Biochem. Mol. Biol. 39, 469–478 [DOI] [PubMed] [Google Scholar]
- 68. Shibuya M., Claesson-Welsh L. (2006) Exp. Cell Res. 312, 549–560 [DOI] [PubMed] [Google Scholar]
- 69. Boguslawski G., McGlynn P. W., Harvey K. A., Kovala A. T. (2004) J. Biol. Chem. 279, 5716–5724 [DOI] [PubMed] [Google Scholar]
- 70. Shor A. C., Agresta S. V., D'Amato G. Z., Sondak V. K. (2008) Cancer Control 15, 47–54 [DOI] [PubMed] [Google Scholar]
- 71. Deininger M. (2005) J. Natl. Compr. Cancer Netw. 3, 757–768 [DOI] [PubMed] [Google Scholar]
- 72. Noble M. E., Endicott J. A., Johnson L. N. (2004) Science 303, 1800–1805 [DOI] [PubMed] [Google Scholar]
- 73. Grove A. D., Prabhu V. V., Young B. L., Lee F. C., Kulpa V., Munson P. J., Kohn E. C. (2002) Clin. Cancer Res. 8, 3019–3026 [PubMed] [Google Scholar]
- 74. Perona R. (2006) Clin. Transl. Oncol. 8, 77–82 [DOI] [PubMed] [Google Scholar]
- 75. Sundaram R., Dakappagari N. K., Kaumaya P. T. (2002) Biopolymers 66, 200–216 [DOI] [PubMed] [Google Scholar]
- 76. Dakappagari N. K., Douglas D. B., Triozzi P. L., Stevens V. C., Kaumaya P. T. (2000) Cancer Res. 60, 3782–3789 [PubMed] [Google Scholar]
- 77. Allen S. D., Garrett J. T., Rawale S. V., Jones A. L., Phillips G., Forni G., Morris J. C., Oshima R. G., Kaumaya P. T. (2007) J. Immunol. 179, 472–482 [DOI] [PubMed] [Google Scholar]
- 78. Garrett J. T., Rawale S., Allen S. D., Phillips G., Forni G., Morris J. C., Kaumaya P. T. (2007) J. Immunol. 178, 7120–7131 [DOI] [PubMed] [Google Scholar]
- 79. Kaumaya P. T., Kobs-Conrad S., Seo Y. H., Lee H., VanBuskirk A. M., Feng N., Sheridan J. F., Stevens V. (1993) J. Mol. Recognit. 6, 81–94 [DOI] [PubMed] [Google Scholar]
- 80. Hetian L., Ping A., Shumei S., Xiaoying L., Luowen H., Jian W., Lin M., Meisheng L., Junshan Y., Chengchao S. (2002) J. Biol. Chem. 277, 43137–43142 [DOI] [PubMed] [Google Scholar]
- 81. Sepp-Lorenzino L., Rands E., Mao X., Connolly B., Shipman J., Antanavage J., Hill S., Davis L., Beck S., Rickert K., Coll K., Ciecko P., Fraley M., Hoffman W., Hartman G., Heimbrook D., Gibbs J., Kohl N., Thomas K. (2004) Cancer Res. 64, 751–756 [DOI] [PubMed] [Google Scholar]
- 82. Brown K. J., Maynes S. F., Bezos A., Maguire D. J., Ford M. D., Parish C. R. (1996) Lab. Invest. 75, 539–555 [PubMed] [Google Scholar]
- 83. Kaumaya P. T., VanBuskirk A. M., Goldberg E., Pierce S. K. (1992) J. Biol. Chem. 267, 6338–6346 [PubMed] [Google Scholar]
- 84. Li B., Sharpe E. E., Maupin A. B., Teleron A. A., Pyle A. L., Carmeliet P., Young P. P. (2006) FASEB J. 20, 1495–1497 [DOI] [PubMed] [Google Scholar]
- 85. Moldovan L., Moldovan N. I. (2005) Exs 127–146 [DOI] [PubMed] [Google Scholar]
- 86. Hiratsuka S., Maru Y., Okada A., Seiki M., Noda T., Shibuya M. (2001) Cancer Res. 61, 1207–1213 [PubMed] [Google Scholar]
- 87. Mac Gabhann F., Popel A. S. (2007) Biophys. Chem. 128, 125–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Neagoe P. E., Lemieux C., Sirois M. G. (2005) J. Biol. Chem. 280, 9904–9912 [DOI] [PubMed] [Google Scholar]
- 89. Weigand M., Hantel P., Kreienberg R., Waltenberger J. (2005) Angiogenesis 8, 197–204 [DOI] [PubMed] [Google Scholar]