Abstract
Arrest of cell differentiation is one of the leading causes of leukemia and other cancers. Induction of cell differentiation using pharmaceutical agents has been clinically attempted for the treatment of these cancers. Epigenetic regulation may be one of the underlying molecular mechanisms controlling cell proliferation or differentiation. Here, we report on the use of proteomics-based differential protein expression analysis in conjunction with quantification of histone modifications to decipher the interconnections among epigenetic modifications, their modifying enzymes or mediators, and changes in the associated pathways/networks that occur during cell differentiation. During phorbol-12-myristate 13-acetate-induced differentiation of U937 cells, fatty acid synthesis and its metabolic processing, the clathrin-coated pit endocytosis pathway, and the ubiquitin/26 S proteasome degradation pathways were up-regulated. In addition, global histone H3/H4 acetylation and H2B ubiquitination were down-regulated concomitantly with impaired chromatin remodeling machinery, RNA polymerase II complexes, and DNA replication. Differential protein expression analysis established the networks linking histone hypoacetylation to the down-regulated expression/activity of p300 and linking histone H2B ubiquitination to the RNA polymerase II-associated FACT-RTF1-PAF1 complex. Collectively, our approach has provided an unprecedentedly systemic set of insights into the role of epigenetic regulation in leukemia cell differentiation.
Keywords: Chromatin Histone Modification, Chromatin Remodeling, DNA Methylation, Histone Modification, Mass Spectrometry (MS), Proteomics
Introduction
Both histone acetylation and H2B ubiquitination are associated with gene activation as defined by the “histone code” and “cross-talk” theories (1). As demonstrated previously in Berk's laboratory (2), small interfering RNA (siRNA)-mediated dual knockdown of CBP5 and p300 resulted in specific histone H3 Lys-18 hypoacetylation in vivo in both IMR90 and HeLa cells, indicating that p300/CBP, possibly together with their closely associated protein PCAF, are the major histone acetyltransferases required for maintaining global H3 Lys-18 acetylation. On the other hand, the putative histone H2B ubiquitination enzyme, UBCH6 (also known as UBE2E1), physically interacts with RNF20/40 (homologs of Bre1, the E3 ligase in yeast) and PAF1 to form a trimeric complex, which colocalizes with RNA polymerase II at transcriptionally active genes (3). RTF1, which has a function similar to Rad6 (mediates ubiquitination at lysine 123 of H2B in yeast) and is a component of the PAF1/RNA polymerase II complex, cooperatively interacts with RNF20/40, and FACT (facilitates chromatin transcription) to regulate H2B ubiquitination (4–8). Clearly, a link between histone modifications, histone modification enzymes, and transcription machinery has been established (9). However, few reports have focused on illustrating protein pathways and networks involved in the facilitation of this linkage prior to the emergence of functional proteomics.
Both loci and global changes in epigenetic modifications are associated with a variety of biological processes, including cell differentiation and proliferation (10). A few lines of evidence have recently emerged to support the perspective that “epigenetic switches determine cell fate” (11). For instance, a decrease in histone H4 acetylation was found prior to and during neural stem cell differentiation (12), and dysregulation of monocyte differentiation into dendritic cells due to oxidized phospholipids produced during an inflammatory reaction was accompanied by a reduction in histone H3 Ser-10 phosphorylation (13). In our previous work, we have also demonstrated that histone H3 propionylation was significantly reduced in U937 cells treated with the differentiation-inducing agent phorbol-12-myristate 13-acetate (PMA) (14).
PMA has been demonstrated as a model inflammatory stimulus that modulates cell differentiation (15–18). During the process of cell differentiation, several genes responsive to inflammation or the induction of cell differentiation, including TNF-α, IL-2, c-FMS, and MMA-9, that were once silent or expressed proteins at a low level in undifferentiated cells, can be reactivated by PMA treatment, accompanied by an elevated histone H3/H4 acetylation (15–18). However, few efforts have been made to examine genome-wide gene expression and global chromatin modifications in cell differentiation. Following our previous work (14), we reported, in this study, to use mass spectrometric approaches to differentially quantify protein expression levels in U937 cells and PMA-induced differentiated U937 cells in order to elucidate protein pathways and networks that regulate epigenetic modifications. Our results demonstrated that hypoacetylation of H3/H4 and hypoubiquitination of H2B, which is regulated by an array of pathways, including fatty acid synthesis and metabolism, protein degradation, chromatin remodeling, and DNA replication, is associated with cell differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture and Protein Extraction
U937 cells were cultured in RPMI 1640 (Mediatech, Inc., Manassas, VA) supplemented with 10% fetal bovine serum (FBS; PAA Laboratories Inc.), 1 mm l- glutamine (Mediatech, Inc.), and 200 units/ml penicillin and 200 μg/ml streptomycin (Lonza, Walkersville, MD) at a density of 2 × 105 cells/ml in a humid incubator with 5% CO2 at 37 °C. After cells grew to a density of 5 × 106 cells/ml, cells were split into two halves, in one of which the medium was replaced with freshly added 1.0 μm PMA. Cells grew for 72 h without further changes of medium before harvest. Cells were first rinsed with PBS supplemented with protease inhibitors (Roche Applied Science) and 10 mm sodium bicarbonate, lysed with Nonidet P-40 lysis buffer, and then centrifuged (19). A BCA assay on the protein extracts (the supernatant of previous centrifugation) was performed to determine protein concentration. Proteins were precipitated by cold acetone (precooled to −20 °C).
For histone isolation and purification, the nuclei were isolated and lysed in 0.4 n H2SO4 by incubation at 4 °C overnight. Trichloroacetic acid (TCA) was added to the supernatant of the centrifugation to reach a final concentration of 33% (v/v) so that histones were precipitated out. The core histones were further fractionated into H2A, H2B, H3, and H4 by reversed-phase HPLC (14).
Sample Preparation and Labeling
Around 300 μg of protein sample in 100 mm Tris-HCl (pH 8.6) and 0.1% SDS buffer solution was reduced by incubation in 10 mm DTT at 50 °C for 1 h, followed by carboxymethylation with 25 mm iodoacetamide in the dark for 2 h. Proteins were precipitated using cold acetone at −20 °C overnight. The protein pellet was generated by centrifugation at 15,000 rpm for 10 min, and the supernatant was removed using a glass Pasteur pipette. The protein pellet was dissolved in 80 mm triethylammonium bicarbonate buffer. Proteins were subsequently digested using trypsin at a protein/enzyme ratio of 40:1 (by mass), and the samples were incubated at 37 °C overnight. The tandem mass tags isobaric mass tagging kit (Thermo Fisher Scientific) was used for labeling the samples following the manufacturer's recommended conditions. The U937 control protein digests were labeled with the TMT labels 128 and 130, whereas the PMA-treated protein digests were labeled with TMT labels 127 and 129. Equal amounts of the labeled control and PMA-treated protein digests were combined for MS analysis.
One- and Two-dimensional RPLC MS
Peptides were separated by online RPLC using an Eksigent nanoLC Ultra 1D Plus and nanoLC AS-2 Autosampler (Eksigent Technologies) coupled online with the Thermo Scientific LTQ Orbitrap Velos mass spectrometer. This hybrid instrument features a dual pressure ion trap configuration along with the OrbitrapTM detector and a higher energy collision induced dissociation (HCD) collision cell (20, 21). A 20-cm, 75-μm inner diameter, 5-μm particle size, Magic C18 analytical column (Michrom Bioresources) was used for the RPLC separations. Approximately 1 μg of protein sample was injected, and experiments were run in triplicate. For the two-dimensional separations, 10 μl of NH4Cl salt bumps were applied (10, 20, 30, 40, 60, 80, 120, and 400 mm), and around 1 μg of sample was loaded onto a 0.32 × 100-mm SCX column (5-μm particle size; Column Technology Inc.). A precolumn (DIONEX, Pep Map100 C18, 0.30 × 5 mm, 5-μm particle size) was brought inline with the analytical column, and a 3-h gradient (solvent A, 0.1% formic acid in water; solvent B, 0.1% formic acid in acetonitrile) from 5 to 30% solvent B was used for separating the peptides. The Orbitrap mass analyzer was set to acquire data at a 60,000 FWHM resolution for the parent full-scan mass spectrum, followed by data-dependent HCD MS/MS spectra for the top 10 most abundant ions acquired at a 7,500 FWHM resolution. Dynamic exclusion was applied where MS/MS was triggered twice within a repeat duration of 30 s with an exclusion list size of 500 and exclusion duration of 90 s.
Data Processing
Data were processed and searched using the Thermo Scientific Proteome Discoverer software suite 1.1 with MascotTM search engine and the Human IPI data base. 10 ppm precursor ion mass tolerance and 0.01 Da fragment ion tolerance and MUDPIT scoring was applied with a peptide cut-off score of 10 and peptide relevance score of 1. Peptides were filtered based on a false discovery rate cut-off of 1% (strict) and 5% (relaxed). Cysteine S-carbamidomethylation as fixed modification, methionine oxidation as variable modification, and up to two missing cleavages were considered during MASCOT searching. A total of 2,249 proteins were identified from the combined one- and two-dimensional experiments, and a total of 42,412 peptides, including 34,138 unique peptides, were detected within the selected filtering criteria. 98% of the total peptides detected were quantified. Protein ratios were reported as the mean values for the observed peptides. 85% of the proteins identified in the one-dimensional experiments and 72% of the proteins identified in the two-dimensional experiment were quantified with less than 20% variability in the protein quantitation ratios.
Quantification of Histone Modifications
Quantification of histone modifications was performed by using LC-MS/MS with multiple reaction monitoring (MRM) mode (22).
Western Blot Analysis
Cell lysates containing equal amount of total proteins were immunoprecipitated by anti-p300 or anti-CALM and analyzed by Western blot using a generic protocol, including PVDF membrane transferring, blockage of the epitope, and incubation with primary antibodies. Membranes were washed, incubated with fluoro-conjugated secondary antibodies, and imaged. For a separate aliquot of each cell lysate, a Western blot was run using anti-GAPDH as the loading control. GADPH was demonstrated not to change expression between undifferentiated and differentiated U937 cells on the basis of our proteomics-based differential protein expression analysis.
Histone Acetyltransferase Activity Assay
Whole cell extracts containing equal amounts of proteins (500 μg) from U937 and HL60 cells and their PMA-induced differentiated counterparts were incubated for 30 min with 3 μg of human recombinant histone H3 in the presence of 14C-acetyl-CoA. The proteins were separated by SDS-PAGE and analyzed by radiochromatography to determine the acetylation activity of the whole cell extracts. For p300-specific acetylation activity analysis, the assay was performed with a p300/CBP immunoprecipitation-HAT assay kit with slight modifications. Briefly, p300 proteins from the whole cells (containing 500 μg of total proteins) were immunoprecipitated with 10 μg of anti-p300. After immunoprecipitation, the protein G-agarose beads were incubated for 30 min with 50 μl of HAT assay mixture (10 μl of 5× HAT assay buffer, 20 μl of D.I. water, 10 μl of biotin-conjugated H4 peptides, and 10 μl of 100 μm acetyl-CoA containing 0.5 μCi/μl 14C-acetyl-CoA). After centrifugation, the supernatants were transferred to the P81 paper squares and equilibrated with the scintillation fluid overnight before the CMP was read by a scintillation counter.
Analysis of Global DNA Cytosine Methylation
Global cytosine methylation was measured by GC/MS as previously described (23).
RESULTS
PMA Treatment, Protein Identification, and Quantification
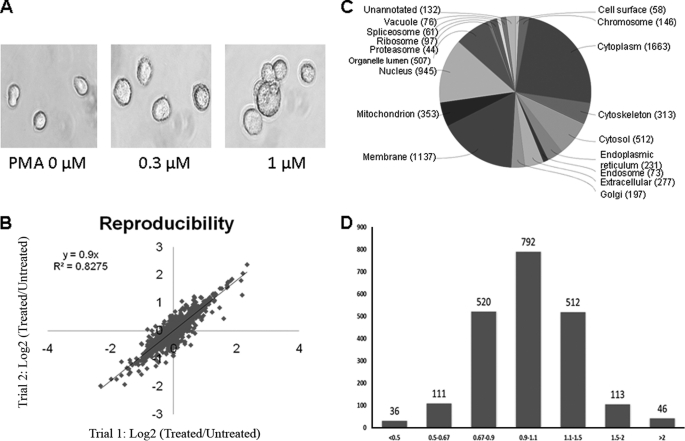
To understand the correlation between chromatin modifications and protein expression pathways, we employed TMT coupled with LC-MS/MS analysis to assess the alteration of protein expression in U937 cells when they were induced to differentiate by PMA. In order to perform cell differentiation and mass spectrometric experiments with the optimal dose of PMA, we initially treated cells with 0, 0.3, 1, and 3.0 μm PMA for 72 h and monitored cell morphology by using microscopy. As shown in Fig. 1A, the morphology of differentiated cells changed from the premonocytic cells into monocyte/macrophage-like cells that were larger and characterized by an increased cytoplasm/nucleus ratio, paler cytoplasm, more prominent granules, a greater degree of vacuolization in the cytoplasm, and ruffled plasma membranes. Occasionally, multinucleate giant cells were also observed (24). We found that cells were not completely differentiated upon treatment with 0.3 μm PMA. Although treatment with 3.0 μm PMA resulted in complete differentiation, ∼25% of cells were dead following such treatment, as revealed by a trypan blue exclusion assay. Cells were induced to full differentiation with 1.0 μm PMA, which resulted in less than 2% cell death. Therefore, 1 μm PMA was selected for subsequent cell culture experiments.
FIGURE 1.
PMA treatment and differential protein expression. A, cell morphology changes during cell differentiation induced by PMA. B, reproducibility of quantitative proteomics measurements. The abundance ratios in log2 scale (log ratio) of proteins measured from the two biological replicates were compared in order to benchmark the quantification reproducibility. C, protein annotation by gene ontology cellular localization. D, a gradient distribution of the expression ratio (treated/untreated) for the quantified proteins.
Protein expression analysis was performed by TMT-LC-MS/MS analysis on an LTQ-Orbitrap-Velos instrument as outlined in supplemental Fig. S1 and described in detail under “Experimental Procedures.” A total of 2,249 proteins were identified from the combined one- and two-dimensional experiments. A total of 42,412 peptides, including 34,138 unique peptides, were detected when the selected filtering criteria (as described under “Experimental Procedures”) were applied. 98% of all the identified peptides were quantified. The protein expression ratios are the mean of three LC-MS/MS runs and the average of two repeated samples that were labeled with different TMT labeling agents (supplemental Fig. S1) followed by normalization of the overall protein expression ratio (ratios from proteins with significant changes were excluded for calculation) to minimize systemic inconsistency between two samples. The quantification reproducibility between the two sample preparations of a direct comparison was measured with the Pearson correlation coefficient (r2) between the replicate protein abundance ratios in log2 scale (protein log ratio) (Fig. 1B). The excellent reproducibility suggested that we did indeed achieve a reliable protein quantification for subsequent molecular pathway analysis. From the log2 curve, 13 proteins were outliners (data points beyond the two paralleling lines ± 1 from the central line) and were not included in the final quantification results. To obtain an overview of the cellular distribution, the identified proteins were classified according to cellular components, molecular functions, and biological processes using the gene ontology annotation (Fig. 1C and supplemental Figs. S2 and S3). With regard to the “cellular compartment,” the top three components were cytoplasm (1,663 proteins), membrane (1,137 proteins), and nucleus (945 proteins). The full distribution map is illustrated in Fig. 1C. Several pathways, including DNA replication and repair, lysosome, and cell cycle, were overrepresented in KEGG pathways (supplemental Fig. S4). Several overrepresented domains, including MCM (replication licensing factors), PWWP (typical in methyltransferase), fn2 (plasminogen-related), and ubiquitin, were characterized by the “Pfam domains” and “InterPro domains” analysis (supplemental Fig. S4).
The distribution of the changes (treated/untreated) in protein expression levels induced by PMA is illustrated in Fig. 1D. Changes greater than 1.5-fold or less than 0.67-fold were considered significant. Based on this arbitrary criterion, the majority (86% of the total) of the 2,118 quantified proteins did not exhibit significant changes, whereas 306 (14% of the total) displayed significant changes upon PMA treatment, with 159 and 147 being up- and down-regulated, respectively. The quantification results for the proteins with significant changes are summarized in supplemental Table S1. Symbols of proteins, involved in the epigenetic network and used in the Ingenuity Pathways Analysis are specifically listed in a separate column and best kept consistent with nomenclatures frequently used in the literature. Protein markers associated with cell differentiation were positively confirmed by quantitative analysis of protein expressions (supplemental Table S2).
Cell Differentiation Induces Up-regulation of the Clathrin-coated Pit Endocytosis Pathway
The U937 cell line, derived from a diffuse histiocytic lymphoma, has a unique and reciprocal t(10;11)(p13;q14) translocation, leading to the fusion of AF10 at 10p13 to CALM at 11q14. The rearrangement of the CALM and AF10 transcripts at their breakpoints leads to the generation of a large fusion protein with 1,595 amino acids, CALM/AF10, by the joining of a nearly full-length CALM sequence (truncated of only the C-terminal four amino acids) with nearly the full-length AF10 protein, starting from amino acid 81. It has been postulated that the CALM-AF10 fusion protein is critical for the malignant phenotype of U937 (25). A well characterized functional domain in CALM is the clathrin-binding domain located next to the N terminus. CALM interacts with clathrin, particularly through its long chain (CLTC) and associates with adaptor proteins AP-1, AP-2, and phosphatidylinositol 4,5-bisphosphate (PIP2) to form an active clathrin-coated pit responsible for receptor-mediated endocytosis of molecules from the cytoplasm to the nucleus (26). The CALM-AF10 translocations might interfere with the expression and function of the wild-type CALM similarly to the knockdown of CALM expression by siRNA (27). A malfunctioning clathrin-coated pit has the potential to severely affect the efficient trafficking of signaling receptors for normal cell differentiation (28).
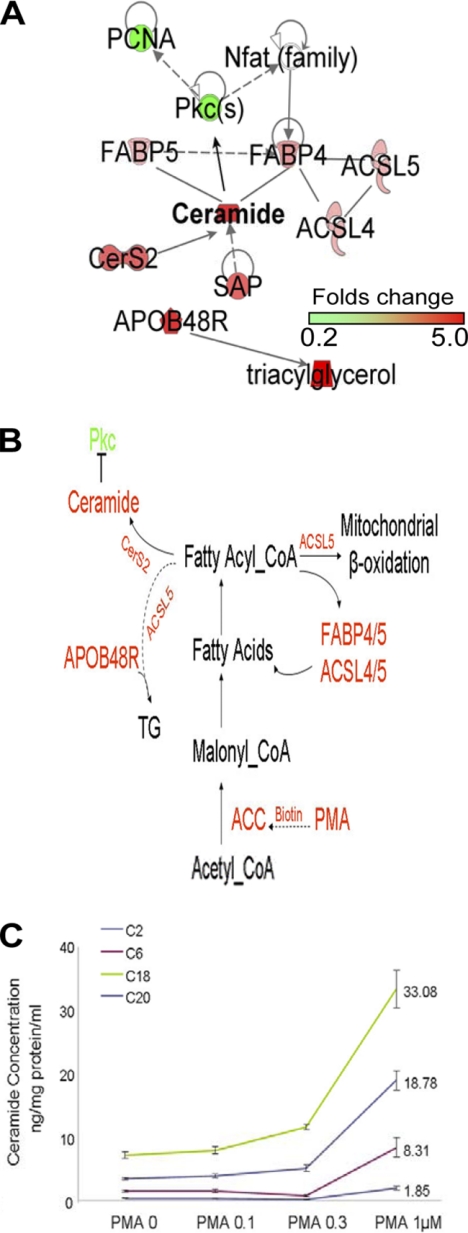
Our mass spectrometric quantification results revealed an overexpression of CALM (1.65-fold change; supplemental Table S3). Although only one peptide, ATTLSNAVSSLASTGLSLTK, was identified by LC-MS/MS (Fig. 2A) and quantified by the TMT reporter ions (Fig. 2B, top), the overexpression of CALM was confirmed by Western blot analysis with a CALM-specific antibody (Fig. 2B, bottom). Although the majority of the isoforms of the adapter proteins (AP-1, AP-2, and AP-3) associated with clathrin showed no significant changes in expression, AP-2β and AP-1γ exhibited changes in expression by 1.60- and 1.25-fold, respectively (supplemental Table S3). The PIP2 kinase responsible for PIP2 synthesis, PIP2-binding protein (GSN), proteins responsible for PIP2 transport (PITPNA and PITPNM1), and the PIP2-dependent ARF1 GTPase (PAP) (29) were overexpressed in the differentiated cells (supplemental Table S3), strongly suggesting a higher concentration of PIP2 in the PMA-treated cells versus untreated ones. The increase in PIP2 concentration might also emanate from the rise in ceramide concentration in the PMA-treated cells (Fig. 3C) because ceramide was found to regulate positively the PIP2 synthesis (30). In addition, the heavy chain of clathrin (CLTC) was overexpressed by 1.49-fold in differentiated cells. Collectively, our data from the differential protein expression analysis provided substantial evidence supporting the idea that the clathrin-coated pit endocytosis pathway is up-regulated during PMA-induced cell differentiation (Fig. 2C and supplemental Fig. S5).
FIGURE 2.
Up-regulation of CALM-associated clathrin-coated pit for signaling receptor endocytosis. A, the MS/MS spectrum of the precursor ion at m/z 794.47 (3+). The fragment ions, b and y, are differentiated by red and blue colors and labeled on the top of peaks, and their mass values are listed in the inset table, establishing the peptide sequence of CALM as shown at the top of the spectrum. B, a zoom-in spectrum of the reporter ions that were used for quantification (top) and the Western blot analysis (WB) of CALM expression using its specific antibody and GADPH as the control for equally loaded proteins (bottom). C, a presentation by IPA for the up-regulated components of clathrin-coated pit determined by mass spectrometry-based differential protein expression analysis.
FIGURE 3.
Regulated fatty acid synthesis and metabolism during cell differentiation. A, IPA links ceramide synthesis with the fatty acid metabolic pathway from differential protein expression analysis data. B, an outlined pathway of the fatty acid synthesis and metabolism induced by PMA. C, ceramide levels in U937 cells treated with a series of concentrations of PMA for 72 h. PCNA, proliferating cell nuclear antigen.
Cell Differentiation Induces Up-regulation of Fatty Acid Synthesis and Metabolic Pathways
Acetyl-CoA carboxylase is a biotin-dependent enzyme that catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA through its two catalytic activities, biotin carboxylase and carboxyltransferase (31). Probably through elevating cellular de novo biotin synthesis, PMA positively regulates acetyl-CoA synthesis in a time- and concentration-dependent manner, which accounts for the PMA-induced stimulation of de novo fatty acid synthesis. The induction of fatty acid synthesis is a key requirement for phagocytic differentiation of human monocytes (32). Fatty acids channel through the transporter system to undertake their cellular missions by binding to the lipid chaperones (i.e. fatty acid-binding proteins (FABP4 and FABP5)) or being directly converted by long chain acyl-CoA synthetases (ACSL3, ACSL4, and ACSL5) into transportable acyl-CoA esters (33–35). As shown in supplemental Table S1, FABP4/5 (FABP5 is not listed in supplemental Table S1 because the ratio is 1.4) and ACSL4/5 are overexpressed in U937 cells treated with PMA for 72 h, demonstrating an active metabolic process of fatty acids that contributes to an elevated synthesis of ceramide. The latter is further supported by the observation of a concomitant overexpression, by 2.10-fold, of CerS2 (ceramide synthetase 2) (36). The prosaposin protein (SAP1), overexpressed by 1.97-fold in differentiated U937 cells, is also involved in ceramide biosynthesis and metabolism (37). To confirm the predicted PMA-induced up-regulation of ceramide, we measured ceramide concentrations in treated and untreated cells (Fig. 3C). These results showed that the cellular ceramide content increased with the rise in the concentration of PMA used for treatment. In particular, treatment with 1.0 μm PMA induced a ∼4-fold increase in ceramide concentrations after 72 h of incubation. Furthermore, IPA suggested that the up-regulation of ceramide synthesis upon PMA treatment was from fatty acid synthesis and its metabolic process (Fig. 3A). Fig. 3B illustrates the pathway whereby PMA induces the activation of acetyl-CoA carboxylase possibly through an up-regulated biotin synthesis or its intake, resulting in an elevated synthesis of fatty acids, an enhanced production of fatty acid-related metabolites, and an accumulation of triacylglycerol.
Cell Differentiation Induces Down-regulated Chromatin Remodeling-mediated Replication Licensing Factors
Chromatin remodeling complexes play important roles in facilitating the DNA replication process (38). DNA polymerase α is the only polymerase that initiates de novo synthesis of single-stranded DNA. After initiation of the first step of DNA synthesis (primer synthesis), DNA polymerases δ and ϵ replace DNA polymerase α for processive DNA synthesis. During the DNA elongation process, the ring-shaped DNA processivity factor, proliferating cell nuclear antigen, is loaded onto the DNA template by replication factor C. The cooperation of these molecules and the movement of histones to form new chromosomes are driven by ATP-dependent chromatin-remodeling complexes in which CAF1-ISWI/SNF2h is essential for efficient trafficking of the replication factors (DNA polymerases, proliferating cell nuclear antigen, and replication factor C) to the replication fork (39, 40). In order to facilitate the formation and elongation of the replication fork, the licensing factors and minichromosome maintenance protein complexes, are recruited together with Cdt1 at the replication origin (41). MCM proteins are composed of a hexamer of six related polypeptides (MCM2 to -7) that form a ring structure mediating DNA replication and growth arrest (42–45). After DNA replication, the DNA mismatch repair proteins (MSH) are in place instantly for the repair of postreplication errors and damage. The SMC (structural maintenance of chromosomes) proteins are subsequently incorporated into the process of replication and segregation for chromosome packaging and transmission. Differential protein expression analysis carried out in this study indicated that all of the above mentioned proteins except for Cdt1, which was not detected by mass spectrometry, possibly due to its low abundance, were down-regulated in PMA-treated U937 cells relative to untreated U937 cells (Fig. 4A and supplemental Table S5), suggesting that PMA inhibits chromatin remodeling-mediated DNA replication. Our data were also strengthened by a previous gene expression study in primitive hematopoietic cells of acute leukemia (42). In that study, the chromatin remodeling gene ISWI/SNF2h (homolog of SMARC5) was shown to be silenced during in vitro erythroid differentiation of MEL cells, whereas it was up-regulated in CD34+ hematopoietic progenitors of acute myeloid leukemia patients (46).
FIGURE 4.
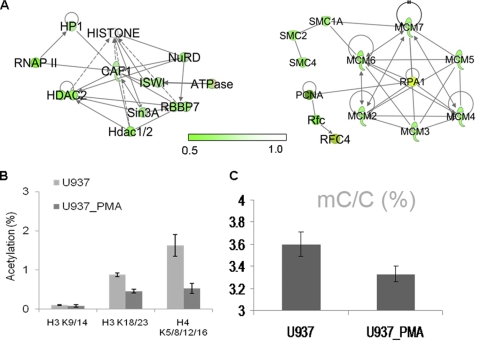
Down-regulation of DNA replication mediated by epigenetic factors. A, down-regulation of chromatin remodeling factors networked by IPA (left) and down-regulation of DNA replication factors (right). B, histone methylation measured by LC-MS/MS-MRM. C, DNA global methylation analyzed by GC/MS.
Chromatin replication involves the relocation of parental histones to newly synthesized DNA and the assembly of new nucleosomes by the chromatin remodeling complex. New histone H4 is diacetylated at Lys-5/12 by Hat1 (a HAT-B acetyltransferase) prior to its deposition on nascent DNA and subsequent incorporation into the new nucleosome, suggesting a coupling of histone synthesis and de novo histone acetylation with DNA replication (47). Fig. 4B and supplemental Fig. S6 show the quantification of histone acetylation and methylation level changes, indicating that the acetylation levels of H3 Lys-9/14, H3 Lys-18/23, and H4 Lys-5/8/12/16 dropped substantially after PMA treatment. Significantly decreased expression of Hat1, as demonstrated by quantitative mass spectrometric analysis (Table 1 and Fig. 5A), together with a decrease in global histone acetylation level, as determined by LC-MS/MS-MRM (Fig. 4B), underscored the direct correlation between DNA replication and histone acetylation. However, DNA global methylation decreased only 7.5% in differentiated cells (Fig. 4C), implicating that the DNA global methylation level does not change significantly when cells are replicated.
TABLE 1.
Differential expression of epigenetic regulators
| Change | ID | Symbol | Entrez gene name | Location | Type(s) |
|---|---|---|---|---|---|
| -fold | |||||
| DNA methylation | |||||
| 0.64 | IPI00220918.1 | DNMT1 | DNA (cytosine-5-)-methyltransferase 1 | Nucleus | Enzyme |
| 1.62 | IPI00220006.2 | DNMT3A | DNA (cytosine-5-)-methyltransferase 3 α | Nucleus | Enzyme |
| 3.50 | IPI00872623.1 | MECP2 | Methyl CpG binding protein 2 (Rett syndrome) | Nucleus | Transcription regulator |
| Histone acetylation/deacetylation | |||||
| 0.64 | IPI00619932.4 | CBP | CREB-binding protein | Nucleus | Transcription regulator |
| 0.59 | IPI00020985.3 | p300 | E1A-binding protein p300 | Nucleus | Transcription regulator |
| 0.57 | IPI00024719.1 | HAT1 | Histone acetyltransferase type B catalitic subunit | Nucleus | Transcription regulator |
| 1.13 | IPI00300127.3 | NAT10 | N-Acetyltransferase 10 (GCN5-related) | Nucleus | Transcription regulator |
| 0.84 | IPI00013774.1 | HDAC1 | Histone deacetylase 1 | Nucleus | Transcription regulator |
| 0.51 | IPI00289601.1 | HDAC2 | Histone deacetylase 2 | Nucleus | Transcription regulator |
| Histone methylation/demethylation | |||||
| 0.61 | IPI00307783.9 | NSD3 | H3 Lys-4/27/36 methyltransferase (SET2 subfamily) | Nucleus | Transcription regulator |
| 0.82 | IPI00797257.1 | EHMT2/G9a | H3 Lys-9 methyltransferase | Nucleus | Transcription regulator |
| 0.92 | IPI00297579.4 | HP1γ | Chromobox homolog 3 (HP1 γ) | Nucleus | H3 Lys-9Me binding |
| 0.58 | IPI00024662.1 | HP1α | Chromobox homolog 5 (HP1 α) | Nucleus | H3 Lys-9Me binding |
| 0.55 | IPI00015568.1 | LRWD1 | Leucine-rich repeat and WD repeat-containing protein 1 | Nucleus | H3 Lys-9/27Me binding |
| 0.89 | IPI00171248.1 | EED | Embryonic ectoderm development, a PCG protein | Nucleus | H3 Lys-27Me3 binding |
| 0.99 | IPI00217540.7 | KDM1A/LASD1 | Lysine-specific demethylase 1A (H3 Lys-4 demethylase) | Nucleus | Enzyme |
| 1.47 | IPI00640875.2 | KDM5C/JARID1C | Lysine-specific demethylase 5C (H3 Lys-4 demethylase) | Nucleus | Enzyme |
| Histone H2B phosphorylation | |||||
| 1.44 | IPI00247439.3 | SLK | STE20-like kinase, histone H2B S14 phosphorylation | Nucleus | Kinase |
| Histone arginine methylation | |||||
| 0.88 | IPI00215734.3 | PRMT1 | Protein arginine methyltransferase 1, H4R3 methyltransferase | Nucleus | Enzyme |
| 0.71 | IPI00401321.1 | PRMT3 | Protein arginine methyltransferase 3, H4R3 methyltransferase | Nucleus | Enzyme |
| 1.01 | IPI00441473.3 | PRMT5 | Protein arginine methyltransferase 5, H4R3/H3R8 methyltransferase | Cytoplasm | Enzyme |
| H2B ubiquitination/deubiquitination | |||||
| 1.06 | IPI00251559.8 | RNF20 | Ring finger protein 20 | Nucleus | Other |
| 0.23 | IPI00303832.6 | RTF1 | Rtf1, Paf1/RNA polymerase II complex component | Nucleus | Other |
| 0.64 | IPI00300333.4 | PAF1 | Paf1, RNA polymerase II associated factor | unknown | Other |
| 0.52 | IPI00394926.1 | POLD3 | Polymerase (DNA-directed), δ 3, accessory subunit | Nucleus | Transcription regulator |
| 0.61 | IPI00031627.3 | POLR2A | Polymerase (RNA) II (DNA-directed) polypeptide A, 220 kDa | Nucleus | Enzyme |
| 0.81 | IPI00873948.1 | POLR2B | Polymerase (RNA) II (DNA-directed) polypeptide B, 140 kDa | Nucleus | Enzyme |
| 0.62 | IPI00026970.4 | FACT | Suppressor of Ty 16 homolog (Saccharomyces cerevisiae) | Nucleus | Transcription regulator |
| 0.86 | IPI00797418.1 | UBE2E1 | Ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast) | Cytoplasm | Enzyme |
| 0.42 | IPI00181504.2 | UBR7 | Ubiquitin protein ligase E3 component n-recognin 7 (putative) | unknown | Enzyme |
| 0.17 | IPI00066641.4 | UHRF1 | Ubiquitin-like with PHD and ring finger domains 1 | Nucleus | Transcription regulator |
| 1.26 | IPI00030915.1 | UBP8 | Ubiquitin-specific peptidase 8 | Cytoplasm | Enzyme |
| 1.04 | IPI00291946.8 | UBP10 | Ubiquitin-specific peptidase 10 | Cytoplasm | Enzyme |
| 1.02 | IPI00033130.3 | SAE1 | SUMO1-activating enzyme subunit 1 (histone sumoylation) | Cytoplasm | Enzyme |
| Chromatin remodeling | |||||
| 0.64 | IPI00645329.1 | CAF1 | Retinoblastoma-binding protein 4, CAF1 | Nucleus | Enzyme |
| 0.78 | IPI00395865.4 | RBBP7 | Retinoblastoma-binding protein 7 | Nucleus | Transcription regulator |
| 0.65 | IPI00297211.1 | ISWI/SNF2h | SWI/SNF-related | Nucleus | Transcription regulator |
| 0.62 | IPI00879096.1 | SNF5/INI1 | SWI/SNF-related | Nucleus | Transcription regulator |
| 0.63 | IPI00000846.1 | NuRD | Isoform 1 of chromodomain-helicase-DNA-binding protein 4 | Nucleus | Transcription regulator |
| 1.10 | IPI00041127.6 | ASF1B | ASF1 homolog B | Nucleus | Transcription regulator |
FIGURE 5.
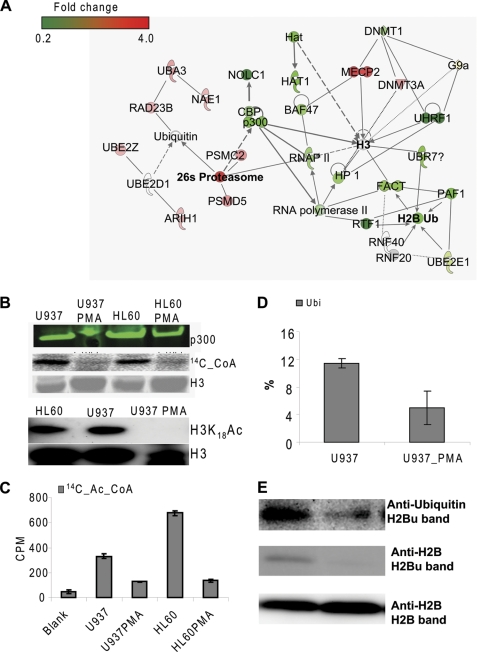
p300 degradation and histone hypoacetylation and H2B deubiquitination. A, network established by IPA from differential protein expression results determined by mass spectrometry. B, Western blot analysis of p300 expression (top), acetyltransferase activity of the whole cell lysates (middle), and Western blot analysis of H3 Lys-18 acetylation (bottom). C, acetyltransferase activity of immunoprecipitated p300. D, H2B Lys-120 ubiquitination analyzed by LC-MS/MS-MRM. E, Western blot analysis of H2B ubiquitination using anti-ubiquitin and anti-H2B.
Cell Differentiation Induces Down-regulation of CBP/p300
It has been reported previously that ceramides induced growth arrest in smooth muscle pericytes by inhibiting an upstream kinase in the ERK cascade and that ceramides rendered the human embryonic kidney 293 cells resistant to the mitogenic actions of growth factors and activators of protein kinase C (PKCδ and -ϵ) (48). These observations suggested a role for PKC in mediating ceramide inhibition of growth factor-induced activity and mitogenesis, and this possibility was further supported by an observation that exogenous ceramide inhibits both immunoprecipitated and recombinant PKCϵ (48). In a separate case, ceramide was reported to inhibit PKCα activity (49). In addition, it is known that inhibition of protein kinase C by chelerythrine and calphostin C resulted in an accumulation of ceramide in WEHI-231 cells (50). In our experiments, mass spectrometric analysis indicated that the PKCβ expression level was decreased by 2-fold, whereas concentrations of the five selected ceramides (C2, C6, C16, C18, and C20) were increased by ∼4-fold in the PMA-treated cells relative to untreated cells (Fig. 3C). These previous findings, combined with our current results, demonstrated that a mutually antagonistic relationship can occur between protein kinase C activity and ceramide in signaling events during cell differentiation and apoptosis. Down-regulation of PKC has been shown to inhibit the G1/S transition (51). Expression of PKCβ is associated with proliferation of cells that retain the capability to undergo further differentiation, as evidenced by overexpression of PKCβ in rat fibroblasts that decreased their sensitivity to growth inhibitory signals and increased their susceptibility to oncogenic transformation (52). PKCβ has also been implicated in epidermal hyperproliferation induced by deficiency in essential fatty acid (53). This is consistent with our observations because we noted that when U937 cells were treated with PMA to stop proliferation and induce differentiation, the fatty acid synthetic and metabolic pathways were up-regulated, whereas the expression of PKCβ was significantly reduced. This correlation was also underscored by a previous observation in which PKCβ was found to be down-regulated in differentiated F9 cells induced by ATRA (54). More recently, PKCβ has been shown to play a critical role in NF-κB-mediated CCL11 transcription by phosphorylation and activation of the p300/CBP and its associated factor (p/CAF), resulting in histone hyperacetylation (55). In contrast, a down-regulated PKCβ, as demonstrated in the PMA-induced differentiated U937 cells, would be correlated with a down-regulation and/or inactivation of p300/CBP.
In addition to the PKC deficiency-mediated down-regulation of CBP/p300, the ubiquitin-26 S proteasome degradation pathway may also be involved, resulting in an accumulative reduction of CBP/p300 expression. The quantitative mass spectrometric analysis indicated that polyubiquitin ligase E1, UBA1–6 and its associated protein NAE1, RAD23, ubiquitin ligase E2, ARIH1, and ubiquitin ligase E3-like protein UBE2E were all up-regulated in PMA-treated cells. Two components of proteasome complex, PSMC2 and PSMD5, were overexpressed by greater than 1.5-fold, and all other remaining quantifiable components showed slightly increased expression (supplemental Table S4), suggesting an up-regulated ubiquitin-26 S proteasome protein degradation pathway in the process of cell differentiation. Ubiquitinated p300 protein was first found in transcriptionally impaired cardiac cells (56) and then detected in RA-induced differentiated F9 cells (54). After ubiquitination, p300 is rapidly degraded (54, 56, 57). The same mechanism might be applicable to PMA-induced differentiated U937 cells because an up-regulated ubiquitin 26 S proteasome degradation pathway was noted. Therefore, a combination of down-regulation of PKCβ, mediated by fatty acid metabolism, and up-regulation of the ubiquitin/26 S proteasome degradation pathway, although it is not clear if these two pathways are mutually connected, could be cumulatively responsible for the decreased expression of CBP/p300 in differentiated cells and subsequent histone hypoacetylation.
As expected from the differential protein expression analysis by mass spectrometry, we observed a down-regulation of PKCβ, p300, CBP, and the CBP/p300-associated protein NOLC1 in PMA-treated U937 cells (Fig. 5A). The reduction of p300 expression in PMA-treated cells was further confirmed by Western blot analysis (Fig. 5B). The histone acetyltransferase activity of the whole cell lysate extracted from PMA-treated U937 cells was significantly lower than that of the whole cell lysate extracted from the untreated cells as measured by densitometry analysis of the SDS-polyacrylamide gel loaded with the incubation mixture in which the cell lysates were reacted with recombinant human histone H3 in the presence of 14C-acetyl-CoA (Fig. 5B). HL60 cells exhibited a similar pattern, suggesting that the reduction of acetyltransferase activity of the cell lysates was exclusively dependent on cell differentiation (Fig. 5B). Additionally, we observed a reduction of p300 acetyltransferase activity in PMA-treated cells, as determined by the p300-specific acetylation assay (Fig. 5C). The reduction of acetyltransferase activity of p300 might arise directly from the decrease in p300 expression. Moreover, we performed the comparative analysis of histone modifications at several selected sites by LC-MS/MS in the MRM mode. We found a significant reduction in both the acetylation of H3 and H4, and the dimethylation of H4 Lys-20 in PMA-treated U937 cells as compared with untreated counterparts (Fig. 4B and supplemental Fig. S6). In contrast, trimethylation of H3 Lys-9 and mono- and dimethylation of H3 Lys-79 were slightly increased in PMA-treated cells (supplemental Fig. S6). Consistent with the mass spectrometric data, Western blot analysis also demonstrated a substantial reduction of H3 Lys-18 acetylation in PMA-treated U937 cells (Fig. 5B, bottom). Taking into consideration that PKCβ and CBP/p300 are required for G1/S transition in the cell cycle (58), down-regulation of these proteins suggested a blockage of G1/S transition for PMA-treated U937 cells followed by an impaired DNA replication/synthesis, as previously demonstrated.
Cell Differentiation Induces Down-regulation of RNA Polymerase II-mediated H2B Ubiquitination
The RNAP II complex, RTF1-FACT-PAF1, has been demonstrated to be required for H2B ubiquitination (4–9). Our differential protein expression analysis by mass spectrometry showed a 3-fold decrease in RTF1 expression in PMA-treated U937 cells, being the most down-regulated component among the mediators/regulators required for the mono-ubiquitination of histone H2B. Although to a lesser extent than RTF1, FACT and PAF1 were also significantly down-regulated by 1.6- and 1.7-fold, respectively (Table 1 and Fig. 5A).
Despite the relatively small changes in protein expressions observed for the hypothetical ubiquitin E2 ligase UBE2E1 and RNF20, which was previously implicated in functioning together with FACT and PAF1 (7, 8), histone H2B ubiquitination was expected to be lower in differentiated cells due to the down-regulation of RTF1, FACT, and PAF1. Indeed, quantification by LC-MS/MS in MRM mode and Western blot analysis showed a marked reduction of histone H2B ubiquitination in differentiated cells, as shown in Fig. 5, D and E. UBP8 and UBP10 are the two known enzymes for H2B deubiquitination. No significant changes in protein expression were observed for these two proteins during cell differentiation (Table 1), suggesting that H2B deubiquitination might not contribute to the reduction of H2B ubiquitination in PMA-treated U937 cells. Meanwhile, we found a 1.5-fold decrease in protein expression level of the putative E3 ubiquitin ligase UBR7 in PMA-treated U937 cells based on the outcomes of quantitative protein analysis of the whole cell lysates. UBR7 belongs to the E3 family that has a characteristic 70-residue UBR box among which UBR1, UBR2, UBR4, and UBR5 can bind to and destabilize N-terminal residues of proteins according to the N-end rule (59), leading to their ubiquitination and subsequent degradation, whereas UBR7, UBR3, and UBR6 do not follow this rule, suggesting that UBR7 could alternatively function as a histone E3 monoubiquitin ligase.
In differentiated U937 cells, the expression of histone H3 Lys-9 methylation binding protein HP1α, LRWD1, UHRF1, and the major DNA maintenance methyltransferase DNMT1 is decreased by 1.7-, 1.8-, 5.9-, and 1.6-fold, respectively, whereas the methyl-CpG-binding protein MeCP2 is increased by 3.5-fold. The UHRF1 binds to hemimethylated CpG dinucleotides and recruits DNMT1, a major DNA maintenance methyltransferase, as well as histone deacetylase 1 (HDAC1) through its distinct SRA (SET- and RING-associated) domain (60). Down-regulation of components in this pathway could result in an activation of transcription. In contrast, MeCP2 and de novo DNA methyltransferase DNMT3a were up-regulated, indicating a repression of transcription (61). This discrepancy could be explained by the differential selection between the MeCP2 pathway and the UHRF1 pathway for specific gene locations or distinct usages for parental DNA sequences or de novo synthetic DNA sequences. An agonistic relationship between MeCP2 and PAF1 was also observed in F9 cells upon differentiation by ATRA (54).
DISCUSSION
Differential protein expression analysis by LC-MS/MS identified multiple pathways and their closely associated networks linked to epigenetic modifications and essential for regulating cell differentiation. Prominently, the clathrin-coated pit endocytosis pathway is up-regulated in differentiated cells. An active clathrin-coated pit endocytosis pathway may help ensure efficient delivery of signaling small molecules and proteins. Biotin or biotin-conjugated acyl-CoA carboxylases may be transported through this pathway to individual cellular locations to execute their functions. The biotin concentration is increased by 2-fold in both U937 cells and HL60 cells treated with 1.0 μm PMA for 72 h (data not shown), suggesting a surge in the supply of biotin-conjugated acetyl-CoA carboxylase that is required for fatty acid synthesis from the initial starter molecule, acetyl-CoA (Fig. 3B). Fatty acid synthesis and its metabolic process are facilitated by the overexpression of fatty acid-binding proteins FABP4/5 and long chain acyl-CoA synthetases (ACSL3, ACSL4, and ACSL5). As a result, ceramide is increased, possibly via CerS2, which is also up-regulated, in differentiated U937 cells. Ceramide inhibits protein kinase C (PKCβ) that phosphorylates and activates histone acetyltransferase p300/CBP and/or its associated protein PCAF. Biotin is also required to form a conjugate with propionyl-CoA carboxylase in order to convert cellular toxic propionyl-CoA to malonyl-CoA. A high concentration of biotin in differentiated cells accelerates the conversion of propionyl-CoA by biotin-conjugated propionyl-CoA carboxylase into malonyl-CoA, resulting in the depletion of propionyl-CoA and consequent reduction in H3 Lys-23 propionylation.
Differential protein expression analysis also demonstrated an up-regulated ubiquitin-26 S proteasome degradation pathway in differentiated U937 cells, which could result in the degradation of p300. A reduction of p300 might subsequently lead to the observed decrease in histone acetylation as verified by quantification with LC-MS/MS in MRM mode. The decrease in H3 Lys-18 acetylation is quite dramatic, as indicated by Western blot analysis with an H3 Lys-18 acetylation-specific antibody. Histone hypoacetylation at promoters and coding regions of genes correlates with transcriptional repression (62). Severe histone global hypoacetylation in differentiated U937 cells suggests that transcription of the majority of genes is repressed, as underscored by the down-regulation of the transcription regulation molecules, including RNA polymerase II, FACT, and the RTF1-PAF complex, which are also ubiquitin ligases or their mediators needed for histone H2B ubiquitination (4–8). Proteomics-based protein expression analysis also identified decreased expression of de novo histone H4 Lys-5/12 acetyltransferase Hat1 which is indispensable for DNA replication (63). DNA replication was impaired in differentiated cells, as evidenced by down-regulation of proteins around the DNA replication fork.
Distinct from histone modification, DNA methylation decreased slightly (∼7.5%) (Fig. 4C) in differentiated U937 cells as opposed to undifferentiated ones, demonstrating that cells manage to maintain a certain level of global DNA methylation through the regulation by two types of methyltransferases, DNMT1 and DNMT3A. Presumably, the down-regulation of DNMT1 and impaired DNA replication would result in a significant reduction in methylation of nascent DNA (23), whereas this loss of DNA methylation could be largely compensated for by the de novo methylation with DNMT3A, which was overexpressed in differentiated cells. As a result, the DNA cytosine methylation is maintained at a relatively stable level (64).
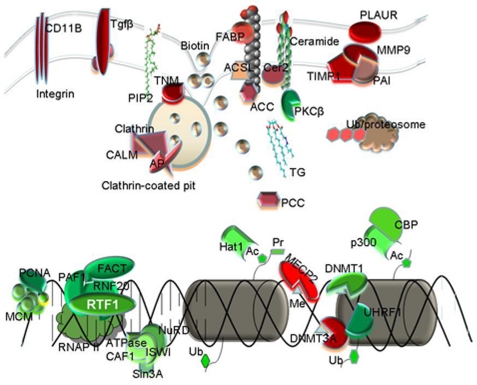
In conclusion, we found that the presented methodology of proteomics-based differential protein expression analysis, in combination with quantitative analysis of histone modifications by mass spectrometry and/or biochemical means, provided a rich set of information on how epigenetic modifications and their regulatory machinery functioned during the process of cell differentiation (Fig. 6). This approach may also provide a useful resource for epigenetic studies on the mechanism of leukemogenesis and the search for molecular targets for effective therapy.
FIGURE 6.
A summary of the regulation pathways/networks in U937 cell differentiation. Cell differentiation up-regulates the clathrin-mediated endocytosis pathway, resulting in overexpression of transforming growth factors as well as integrins that are required for cell morphology changes. Biotin uptake may also be elevated by endocytosis, resulting in increased biosynthesis of fatty acids and ceramides. Biotin is also required for the activation of propionyl-CoA carboxylase (PCC) to convert propionyl-CoA into methymalonyl-CoA, resulting in a decrease in histone H3 Lys-23 propionylation. Ceramides inhibit protein kinase Cβ, which phosphorylates and activates p300/CBP for histone H3 Lys-18 acetylation. In addition, p300 degradation by the ubiquitin-proteasome complex causes a decrease in the expression and activity of p300 and consequent histone H3/H4 hypoacetylation. Meanwhile, essential proteins in the DNA replication fork, chromatin remodeling complex, and RNA polymerase II mediators are down-regulated in differentiated cells, regulating DNA replication, DNA methylation, histone de novo acetylation, and H2B ubiquitination.
Acknowledgments
We thank Hua Xu, Michael Freitas, Guoliang Xu, and Siavash Kurdistani for advice in improving the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK082779 (to Y. W.), R01 CA84487 (to L. C. S.), and 1S10RR027643-01 (to K. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Figs. S1–S6.
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- PMA
- phorbol-12-myristate 13-acetate
- MRM
- multiple reaction monitoring
- HAT
- histone acetyltransferase
- PIP2
- phosphatidylinositol 4,5-bisphosphate.
REFERENCES
- 1. Fischle W., Wang Y., Allis C. D. (2003) Curr. Opin. Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 2. Horwitz G. A., Zhang K., McBrian M. A., Grunstein M., Kurdistani S. K., Berk A. J. (2008) Science 321, 1084–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nordick K., Hoffman M. G., Betz J. L., Jaehning J. A. (2008) Eukaryot. Cell 7, 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ng H. H., Dole S., Struhl K. (2003) J. Biol. Chem. 278, 33625–33628 [DOI] [PubMed] [Google Scholar]
- 5. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. (2003) J. Biol. Chem. 278, 34739–34742 [DOI] [PubMed] [Google Scholar]
- 6. Warner M. H., Roinick K. L., Arndt K. M. (2007) Mol. Cell. Biol. 27, 6103–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. (2005) Mol. Cell 20, 601–611 [DOI] [PubMed] [Google Scholar]
- 8. Pavri R., Zhu B., Li G. H., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Cell 125, 703–717 [DOI] [PubMed] [Google Scholar]
- 9. Selth L. A., Sigurdsson S., Svejstrup J. Q. (2010) Annu. Rev. Biochem. 79, 271–293 [DOI] [PubMed] [Google Scholar]
- 10. Sharma S., Kelly T. K., Jones P. A. (2010) Carcinogenesis 31, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rivenbark A. G., Strahl B. D. (2007) Science 318, 403–404 [DOI] [PubMed] [Google Scholar]
- 12. Singh R. P., Shiue K., Schomberg D., Zhou F. C. (2009) Cell Transplant. 18, 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blüml S., Zupkovitz G., Kirchberger S., Seyerl M., Bochkov V. N., Stuhlmeier K., Majdic O., Zlabinger G. J., Seiser C., Stöckl J. (2009) Blood 114, 5481–5489 [DOI] [PubMed] [Google Scholar]
- 14. Liu B., Lin Y. H., Darwanto A., Song X. H., Xu G. L., Zhang K. L. (2009) J. Biol. Chem. 284, 32288–32295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee J. Y., Kim N. A., Sanford A., Sullivan K. E. (2003) J. Leuk. Biol. 73, 862–871 [DOI] [PubMed] [Google Scholar]
- 16. Adachi S., Rothenberg E. V. (2005) Nucleic Acids Res. 33, 3200–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Follows G. A., Tagoh H., Lefevre P., Hodge D., Morgan G. J., Bonifer C. (2003) EMBO J. 22, 2798–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma Z., Shah R. C., Chang M. J., Benveniste E. N. (2004) Mol. Cell. Biol. 24, 5496–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang N., Xu M. G., Wang P., Li L. (2010) Anal. Chem. 82, 2262–2271 [DOI] [PubMed] [Google Scholar]
- 20. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Second T. P., Blethrow J. D., Schwartz J. C., Merrihew G. E., MacCoss M. J., Swaney D. L., Russell J. D., Coon J. J., Zabrouskov V. (2009) Anal. Chem. 81, 7757–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Darwanto A., Curtis M. P., Schrag M., Kirsch W., Liu P., Xu G. L., Neidigh J. W., Zhang K. L. (2010) J. Biol. Chem. 285, 21868–21876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herring J. L., Rogstad D. K., Sowers L. C. (2009) Chem. Res. Toxicol. 22, 1060–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hattori T., Pack M., Bougnoux P., Chang Z. L., Hoffman T. (1983) J. Clin. Investig. 72, 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dreyling M. H., Martinez-Climent J. A., Zheng M., Mao J., Rowley J. D., Bohlander S. K. (1998) Proc. Natl. Acad. Sci. U.S.A. 93, 4804–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tebar F., Bohlander S. K., Sorkin A. (1999) Mol. Biol. Cell 10, 2687–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meyerholz A., Hinrichsen L., Groos S., Esk P. C., Brandes G., Ungewickell E. J. (2005) Traffic 6, 1225–1234 [DOI] [PubMed] [Google Scholar]
- 28. Planque N. (2006) Cell Commun. Signal. 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Donaldson J. G., Jackson C. L. (2000) Curr. Opin. Cell Biol. 12, 475–482 [DOI] [PubMed] [Google Scholar]
- 30. Dasgupta U., Bamba T., Chiantia S., Karim P., Abou Tayoun A. N., Yonamine I., Rawat S. S., Rao R. P., Nagashima K., Fukusaki E., Puri V., Dolph P. J., Schwille P., Acharya J. K., Acharya U. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20063–20068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plutzky J. (2009) Nat. Med. 15, 618–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ecker J., Liebisch G., Englmaier M., Grandl M., Robenek H., Schmitz G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7817–7822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mashek D. G., McKenzie M. A., Van Horn C. G., Coleman R. A. (2006) J. Biol. Chem. 281, 945–950 [DOI] [PubMed] [Google Scholar]
- 34. Olefsky J. M. (2008) Cell 134, 914–916 [DOI] [PubMed] [Google Scholar]
- 35. Cao H. M., Gerhold K., Mayers J. R., Wiest M. M., Watkins S. M., Hotamisligil G. S. (2008) Cell 134, 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kolak M., Westerbacka J., Velagapudi V. R., Wågsäter D., Yetukuri L., Makkonen J., Rissanen A., Häkkinen A. M., Lindell M., Bergholm R., Hamsten A., Eriksson P., Fisher R. M., Oresic M., Yki-Järvinen H. (2007) Diabetes 56, 1960–1968 [DOI] [PubMed] [Google Scholar]
- 37. Sandhoff K., Kolter T., Van Echten-Deckert G. (1998) Ann. N.Y. Acad. Sci. 845, 139–151 [DOI] [PubMed] [Google Scholar]
- 38. Takeda D. Y., Dutta A. (2005) Oncogene 24, 2827–2843 [DOI] [PubMed] [Google Scholar]
- 39. Collins N., Poot R. A., Kukimoto I., Garcia-Jiménez C., Dellaire G., Varga-Weisz P. D. (2002) Nat. Genet. 32, 627–632 [DOI] [PubMed] [Google Scholar]
- 40. Falbo K. B., Shen X. T. (2009) Mol. Cells 28, 149–154 [DOI] [PubMed] [Google Scholar]
- 41. You Z. Y., Masai H. (2008) J. Biol. Chem. 283, 24469–24477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chong J. P., Hayashi M. K., Simon M. N., Xu R. M., Stillman B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chong J. P., Mahbubani H. M., Khoo C. Y., Blow J. J. (1995) Nature 375, 418–421 [DOI] [PubMed] [Google Scholar]
- 44. Lee J. K., Hurwitz J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agarwal M. K., Amin A., Agarwal M. L. (2007) Cancer Res. 67, 116–121 [DOI] [PubMed] [Google Scholar]
- 46. Stopka T., Zakova D., Fuchs O., Kubrova O., Blafkova J., Jelinek J., Necas E., Zivny J. (2000) Leukemia 14, 1247–1252 [DOI] [PubMed] [Google Scholar]
- 47. Makowski A. M., Dutnall R. N., Annunziato A. T. (2001) J. Biol. Chem. 276, 43499–43502 [DOI] [PubMed] [Google Scholar]
- 48. Bourbon N. A., Yun J., Berkey D., Wang Y. Z., Kester M. (2001) Am. J. Physiol. 280, C1403–C1411 [DOI] [PubMed] [Google Scholar]
- 49. Lee J. Y., Hannun Y. A., Obeid L. M. (1996) J. Biol. Chem. 271, 13169–13174 [DOI] [PubMed] [Google Scholar]
- 50. Chmura S. J., Nodzenski E., Weichselbaum R. R., Quintans J. (1996) Cancer Res. 56, 2711–2714 [PubMed] [Google Scholar]
- 51. Zhang W., Liu H. T. (2002) Cell Res. 12, 9–18 [DOI] [PubMed] [Google Scholar]
- 52. Housey G. M., Johnson M. D., Hsiao W. L., Obrian C. A., Murphy J. P., Kirschmeier P., Weinstein I. B. (1988) Cell 52, 343–354 [DOI] [PubMed] [Google Scholar]
- 53. Ziboh V. A., Miller C. C., Cho Y. H. (2000) Am. J. Clin. Nutr. 71, 361S–366S [DOI] [PubMed] [Google Scholar]
- 54. Brouillard F., Cremisi C. E. (2003) J. Biol. Chem. 278, 39509–39516 [DOI] [PubMed] [Google Scholar]
- 55. Clarke D. L., Sutcliffe A., Deacon K., Bradbury D., Corbett L., Knox A. J. (2008) J. Immunol. 181, 3503–3514 [DOI] [PubMed] [Google Scholar]
- 56. Poizat C., Sartorelli V., Chung G., Kloner R. A., Kedes L. (2000) Mol. Cell. Biol. 20, 8643–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jin Y. T., Zeng S. X., Lee H., Lu H. (2004) J. Biol. Chem. 279, 20035–20043 [DOI] [PubMed] [Google Scholar]
- 58. Ait-Si-Ali S., Polesskaya A., Filleur S., Ferreira R., Duquet A., Robin P., Vervish A., Trouche D., Cabon F., Harel-Bellan A. (2000) Oncogene 19, 2430–2437 [DOI] [PubMed] [Google Scholar]
- 59. Tasaki T., Mulder L. C. F., Iwamatsu A., Lee M. J., Davydov I. V., Varshavsky A., Muesing M., Kwon Y. T. (2005) Mol. Cell. Biol. 25, 7120–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bostick M., Kim J. K., Esteve P. O., Clark A., Pradhan S., Jacobsen S. E. (2007) Science 317, 1760–1764 [DOI] [PubMed] [Google Scholar]
- 61. Jones P. L., Veenstra G. J. C., Wade P. A., Vermaak D., Kass S. U., Landsberger N., Strouboulis J., Wolffe A. P. (1998) Nat. Genet. 19, 187–191 [DOI] [PubMed] [Google Scholar]
- 62. Kristjuhan A., Walker J., Suka N., Grunstein M., Roberts D., Cairns B. R., Svejstrup J. Q. (2002) Mol. Cell 10, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barman H. K., Takami Y., Nishijima H., Shibahara K., Sanematsu F., Nakayama T. (2008) Biochem. Biophys. Res. Commun. 373, 624–630 [DOI] [PubMed] [Google Scholar]
- 64. Rhee I., Bachman K. E., Park B. H., Jair K. W., Yen R. W., Schuebel K. E., Cui H., Feinberg A. P., Lengauer C., Kinzler K. W., Baylin S. B., Vogelstein B. (2002) Nature 416, 552–556 [DOI] [PubMed] [Google Scholar]