Abstract
The tumor suppressor protein BRCA1 is a constituent of several different protein complexes and is required for homology-directed repair (HDR) of DNA double strand breaks (DSBs). The most recently discovered BRCA1-RAP80 complex is recruited to ubiquitin structures on chromatin surrounding the break. Deficiency of any member of this complex confers hypersensitivity to DNA-damaging agents by undefined mechanisms. In striking contrast to other BRCA1-containing complexes that are known to promote HDR, we demonstrate that the BRCA1-RAP80 complex restricts end resection in S/G2 phase of the cell cycle, thereby limiting HDR. RAP80 or BRCC36 deficiency resulted in elevated Mre11-CtIP-dependent 5′ end resection with a concomitant increase in HDR mechanisms that rely on 3′ single-stranded overhangs. We propose a model in which the BRCA1-RAP80 complex limits nuclease accessibility to DSBs, thus preventing excessive end resection and potentially deleterious homology-directed DSB repair mechanisms that can impair genome integrity.
Keywords: BRCA BRCB, Deubiquitination, DNA Recombination, DNA Repair, Ubiquitin, BRCA1, BRCC36, RAP80
Introduction
Mutations in the BRCA1 (breast cancer early onset 1) tumor suppressor gene confer strong breast and ovarian cancer susceptibility that tightly correlates with loss of its known DNA repair function, including cell cycle checkpoint execution and homologous recombination (HR)2 (1–4). In order to repair DSBs, cells have evolved different forms of homology-directed and non-homology-directed DSB repair mechanisms, including HR, single strand annealing (SSA), and non-homologous end joining (NHEJ) (5, 6). HR and SSA both require sequence homology and 5′ to 3′ end resection of the break to generate a 3′ single-stranded DNA (ssDNA) overhang. During HR, Rad51 nucleofilaments form on the resected 3′ ssDNA overhang, allowing strand invasion into the sister chromatid duplex and copying of the homologous stretch of DNA. A biochemical association of BRCA1 and BRCA2 is necessary for efficient Rad51 nucleofilament formation (7–9). SSA also uses homology for repair; however, it does not involve copying of the sister chromatid but rather relies on homology-directed pairing within a broken chromatid, deletion of the intervening sequence, and ligation. Although SSA may lead to large genomic deletions, HR is often considered error-free because it occurs in the S and G2 phases of the cell cycle when a sister chromatid is present to be used as a template for repair. However, several studies indicate that HR is also associated with a high incidence of sequence alterations, including loss of heterozygosity, to repaired loci (10, 11). Alternatively, NHEJ is the major pathway used for DSB repair in G1 and does not require the use of end resection or a homologous sequence. NHEJ also can be error-prone and lead to loss of genetic material due to processing of the break or, alternatively, can join discontiguous breaks within the genome to create nonreciprocal translocations. Although NHEJ predominates in G1, it can also be used for repair in G2, giving the cell a choice between homologous and non-homologous DNA repair mechanisms. Although the molecular events determining how a cell chooses between HR and NHEJ are not fully understood, end resection is thought to play a pivotal role in this decision (12).
Recent studies indicate that the process of end resection is highly regulated by cooperation between multiple enzymes that exert their activities on DNA. The MRN complex, composed of the Mre11 nuclease, Rad50, and Nbs1, is one of the initial complexes recruited to DSBs and is considered to be a primary sensor of DNA damage (13, 14). Mre11 itself possesses nuclease activity, and, in addition, MRN is physically associated with the CtIP endonuclease. Together these proteins are thought to initiate 5′ to 3′ resection of the break (15–17). BRCA1 forms a complex with CtIP that is mutually exclusive from BRCA1 complexes containing RAP80 or the BACH1 helicase. The BRCA1-CtIP complex interacts with the MRN complex in a DNA damage-dependent manner (18). This interaction is necessary for end resection and HR in human cells and in a chicken B cell lymphoma cell line (18, 19). DNA is then further resected by the ExoI exonuclease and the BLM helicase to create the larger tracts of ssDNA necessary to initiate homology-directed repair (20). Consequently, deficiency of any of these components impairs ssDNA formation and homology-dependent repair. Interestingly, aberrant end resection impairs NHEJ, leading to gross chromosomal rearrangements and IR sensitivity (16). Thus, maintaining the correct balance between HR and NHEJ is critical to ensure accurate DNA repair and genetic stability. Additional molecular and signaling events controlling the restriction of resection have yet to be described.
In parallel to resection activities at DSB termini, a series of phosphorylation and ubiquitination events on chromatin in cis to the DSB play a central role in recruitment of repair complexes (21, 22). The RNF8 and RNF168 E3 ubiquitin ligases are recruited to chromatin surrounding the break in a γH2AX- and MDC1-dependent manner. RNF8/RNF168 conjugate Lys63-linked ubiquitin polymers on substrates at these sites (23–28). The Lys63 ubiquitin species serve as a signaling platform to recruit repair proteins. Among these is the BRCA1-RAP80 complex, which is specifically targeted to the chromatin surrounding the DSB by recognition of Lys63 ubiquitin by the RAP80 tandem ubiquitin interaction motifs (29–32). The BRCA1-RAP80 complex also contains the deubiquitinating enzyme BRCC36, which requires interactions with other members of the complex for Lys63 ubiquitin-specific deubiquitinating enzyme activity (30, 33–36).
BRCA1 is present in four distinct biochemical complexes (18, 37), three of which have been implicated in promoting HR. Although deficiency of RAP80 or BRCC36 leads to IR sensitivity (29–31), the contribution of the BRCA1-RAP80 complex to DSB repair has remained elusive. In this study, we present a role for the BRCA1-RAP80 complex in preserving the balance of DNA repair pathways by restricting excessive end resection. Loss of the BRCA1-RAP80 complex results in elevated homology-mediated repair processes through an increase in end resection during S and G2 phases of the cell cycle. This excessive end resection is mediated by an increase in Mre11 and CtIP recruitment to sites of DNA damage, suggesting a role for the BRCA1-RAP80 complex in protecting the break from aberrant access to nucleases. These results demonstrate a novel function for the BRCA1-RAP80 complex in limiting HDR and suggest that a balance between the activities of different BRCA1-containing protein complexes influences DSB repair pathway choice.
EXPERIMENTAL PROCEDURES
Cell Culture
All cells were cultured in DMEM (Invitrogen) containing 10% calf serum. Alternatively, HCC1937 cells were cultured in RPMI (Invitrogen) containing 10% calf serum. U2OS FokI reporter cells were maintained in 100 μg/ml hygromycin B. HeLa cells expressing a hairpin to luciferase or RAP80 were maintained in 200 μg/ml hygromycin B. HeLa DR and HeLa SSA cell lines were made using DR-GFP and SA-GFP constructs obtained from Drs. Koji Nakanishi and Maria Jasin. Plasmids were linearized using KpnI and SacI and transfected into HeLa cells. Cells were grown in puromycin, and single clones were selected. NHEJ reporter cells were obtained from Dr. Matthew Porteus.
HR and SSA Assays
HeLa DR or HeLa SSA cells were treated with the indicated siRNAs in 6-well plates (38, 39). The following day, cells were transfected with 1 μg of I-SceI per well. Alternatively, GFP was used as a control for transfection efficiency. 72 h after I-SceI transfection, cells were trypsinized and analyzed by flow cytometry for GFP positivity.
Non-homologous End Joining Assay
NHEJ reporter cells were transfected with the indicated siRNAs as described previously (40). On the following day, they were transfected with I-SceI, and 48 h later, they were assayed for RFP-positive cells by flow cytometry.
Transfections
siRNA transfections were done using Lipofectamine RNAiMax (Invitrogen), and experiments were performed 72 h after transfection. Alternatively, siRNA experiments were done using Oligofectamine (Invitrogen). Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen) or Fugene (Roche Applied Science). All transfections were done according to the manufacturer's protocols.
Metaphase Spreads
Cells were treated with 0.5 μm nocodazole for 3 h and lysed with 75 mm KCl. Cells were fixed on ice with a 3:1 methanol/acetic acid solution. Metaphases were dropped onto slides at 65 °C, allowed to dry, and stained with Giemsa. The numbers of sister chromatid breaks and chromosome breaks per metaphase were counted. For sister chromatid exchange analysis, cells were pretreated for 48 h with 10 μm BrdU and collected as above. After dropping metaphases onto slides, they were stained with 10 μg/ml Hoechst 33258 in PBS for 20 min. The slides were rinsed in McIlvaine solution for 20 min and treated with 365-nm UV for 30 min. Slides were incubated in 1.5× SSC at 55 °C for 1 h. Slides were then stained with Giemsa and analyzed for the number of sister chromatid exchanges/metaphase.
Clonogenic Survival Assay
Cells were treated with siRNA and the next day were seeded at a density of 500 cells/6-cm dish. The following day, cells were treated with etoposide at the indicated dose for 1 h. The drug was washed out, and cells were allowed to recover for 14 days. Plates were fixed with methanol for 20 min at −20 °C and stained with Giemsa, and colonies were counted. Each siRNA and drug dose was done in triplicate.
Antibodies
Antibodies were used at the following dilutions: RPA2 9H8 (Novus NB600-565), 1:500 (IF, WB); 53BP1 (Novus NB100-904), 1:500 (IF); CENPF (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) 22791), 1:200 (IF); Rad51 (Santa Cruz Biotechnology, Inc. 8349), 1:100 (IF); γH2AX (Upstate JBW301), 1:2000 (IF); CtIP (Santa Cruz Biotechnology, Inc. 5970), 1:100 (IF) and 1:200 (WB); RAP80 (in house) 1:500 (IF, WB); Mre11 (Novus), 1:200 (IF); CtIP (Abcam 70163), 1:500 (WB); pRPA Ser4/Ser8 (Bethyl A300-245A), 1:1000 (WB).
siRNA Sequences
siRNA sequences were as follows: luciferase, 5′-gccauucuauccucuagaggaug; RAP80-1, 5′-ccaguuggagguuuaucaa; RAP80-2, 5′-uuugucuccgagucaauac; BRCC36, 5′-aacaucaacaugugaaggc; 53BP1, 5′-uauuaccgucuccucguuc; CtIP, 5′-acacacucauggugauaaa; BRCA1, 5′-agauaguucuaccaguaaa; Abraxas, 5′-cguuuagagagaggcugcuucacaa.
Laser-generated DNA DSBs
DNA DSBs were generated using a P.A.L.M. MicroBeam laser microdissection system as described previously (13). Cells were seeded onto glass coverslips and incubated with 10 μm BrdU for 24 h before laser-induced DSBs. 80–100 cells/coverslip were subjected to laser-induced DSBs using the ×40 objective at 62% power, at a fixed wavelength of 337 nm. After laser treatment, cells were incubated at 37 °C for the indicated times, pre-extracted with 0.2% Triton solution for 5 min at 4 °C, and then fixed using fixation buffer containing 3% paraformaldehyde, 2% sucrose for 10 min at room temperature. Cells were stained using the immunofluorescence protocol described below.
Immunofluorescence
Cells were washed with PBS and then fixed in solution containing 3% paraformaldehyde, 2% sucrose for 10 min at room temperature. Alternatively, cells were fixed with methanol at −20 °C for 20 min. Cells were subsequently permeabilized with 0.5% Triton solution for 5 min at 4 °C and then incubated with primary antibody for 20 min at 37 °C. Cells were then washed with PBST and next incubated with secondary antibody for 20 min at 37 °C. After four washes with PBST, coverslips were mounted onto glass slides using Vectashield mounting medium containing DAPI (Vector Laboratories) and visualized using a Nikon Eclipse 80i fluorescent microscope.
Image Quantification
Images were obtained using a QImaging Retiga-SRV camera, and all images for each experiment were taken on the same day at the same exposure time. Image quantification was performed using ImageJ software (National Institutes of Health). For laser stripe analyses, the area of damage based upon 53BP1 staining was circled to measure replication protein A (RPA) intensity. Nuclear background was subtracted for each stripe measured. For FokI reporter cells, the area of damage was identified by the mCherry signal, and RPA, Mre11, or CtIP intensity at this location was measured. Background was subtracted for each nucleus. All quantification was performed on unprocessed images.
Nuclear Extract Preparation and Western Blot
Cells were lysed with NETN 100 (0.5% Nonidet P-40, 1 mm EDTA, 20 mm Tris-Cl, pH 8, 100 mm NaCl) for 20 min at 4 °C. The insoluble fraction was then lysed with NETN 420 (0.5% Nonidet P-40, 1 mm EDTA, 20 mm Tris-Cl, pH 8, 420 mm NaCl), resulting in the nuclear fraction. For immunoprecipitation studies, lysates were collected as above and incubated with FLAG beads for 3 h. Beads were washed four times with lysis buffer and eluted using FLAG peptide. Western blot was performed and analyzed using the indicated antibodies.
Cell Cycle Analysis
Cells were trypsinized, washed with PBS, and fixed with 70% ethanol at −20 °C overnight. Fixed cells were washed with PBS and incubated with propidium iodide (50 μg/ml) and RNase A (100 μg/ml) in PBS at 37 °C for 30 min. Flow cytometry for propidium iodide was done to analyze cell cycle distribution.
FokI Assay
Cells were treated with the indicated siRNAs. The following day, cells were trypsinized and transduced with FokI lentivirus. 36 h later, coverslips were fixed and analyzed using immunofluorescence.
Statistical Analysis
Graphs were created, and statistics were calculated using Prism software (GraphPad). Significant differences (p ≤ 0.05) are marked by an asterisk in each figure. Comparisons were performed using Student's t test without assuming equal variances.
RESULTS
The BRCA1-RAP80 Complex Influences the Balance between Homology-directed and Non-homology-directed DSB Repair
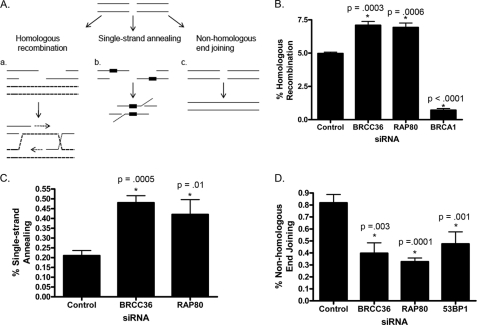
To determine if the BRCA1-RAP80 complex plays a role in a specific DSB repair pathway, we first analyzed how knockdown of RAP80 or BRCC36 affected utilization of HR, SSA, and NHEJ in repair of nuclease-derived DSBs (Fig. 1A). To analyze rates of HR, we used a HeLa cell line containing the DR-GFP repair substrate (supplemental Fig. S1A) (38). This substrate contains two incomplete copies of the GFP allele, where the first copy contains an I-SceI recognition site. Expression of the I-SceI enzyme creates DSBs that, if repaired by strand invasion into a downstream 3′ fragment of a GFP allele, will lead to creation of a functional GFP gene. Thus, GFP-positive cells can be measured as a proxy for rates of HR. As expected, knockdown of BRCA1 resulted in a significant decrease in the percentage of DSBs repaired by HR (Fig. 1B). Surprisingly, knockdown of either RAP80 or BRCC36 caused an increase in HR, indicating that the BRCA1-RAP80 complex plays a unique role in regulating DSB repair. Next, we investigated the effect of RAP80 and BRCC36 loss on the HDR process of SSA. To do so, we utilized a reporter cell line containing a 5′ portion of the GFP allele upstream of a 3′ region of the GFP allele containing an I-SceI recognition site (supplemental Fig. S1B) (39). These two alleles contain 266 base pairs of homology that can be used during repair by SSA to restore a functional GFP gene. Similar to HR, knockdown of RAP80 or BRCC36 resulted in an increase in SSA (Fig. 1C and supplemental Fig. S1D). Interestingly, knockdown of BRCA1 results in a decrease in SSA, as described previously (supplemental Fig. S1E) (39). It has been shown that disruption of the BRCA1-CtIP interaction results in decreased ssDNA formation and homologous recombination (19). SSA requires a significant amount of DSB end resection, and these results suggest that BRCA1 may play a role in SSA repair through ssDNA formation. Together, these data indicate that the BRCA1-RAP80 complex may specifically play a role in homology-directed repair processes that necessitate end resection.
FIGURE 1.
RAP80 and BRCC36 affect DSB repair pathway utilization. A, double strand breaks can be repaired by homologous recombination (a), single-strand annealing (b), or non-homologous end joining (c). Homologous recombination and single strand annealing involve 5′–3′ resection of the break and use of homologous sequences (heavy dashed lines) for repair, whereas non-homologous end joining does not. B, increased HR utilization in cells deficient for either RAP80 or BRCC36. Cells were treated with the indicated siRNAs and 72 h later were assayed for HR using a DR-GFP reporter cell line. Error bars, S.E. of three independent experiments. p values were calculated in comparison with control siRNA-treated cells. C, increase in SSA utilization in cells deficient for RAP80 or BRCC36. Cells were treated with the indicated siRNAs and assayed for SSA using flow cytometry. Data are representative of three independent experiments. Error bars, S.D. of replicates in one experiment. See supplemental Fig. S1D for an independent replicate experiment. p values were calculated in comparison with control siRNA-treated cells. D, decreased NHEJ utilization in cells deficient for either RAP80 or BRCC36. Cells were treated with the indicated siRNAs and assayed for NHEJ 72 h later. Data are representative of three independent experiments. Error bars, S.D. of replicates in one experiment. See supplemental Fig. S1E for an independent replicate experiment. p values were calculated in comparison with control siRNA-treated cells.
NHEJ does not involve the use of a homologous sequence, and because HR and NHEJ are potentially competing processes of DSB repair, we hypothesized that NHEJ frequency may decrease with loss of the BRCA1-RAP80 complex. To test this, we used a cell line with an integrated NHEJ reporter containing a CMV/CBA promoter driving expression of a GFP gene, flanked by I-SceI sites (supplemental Fig. S1C) (40). Downstream of the GFP allele is an RFP allele that is not expressed. Expression of I-SceI results in two DSBs and loss of the GFP allele in a percentage of cells that have repaired the break by NHEJ-dependent ligation of the I-SceI sites. When this occurs, RFP will be expressed, and the NHEJ frequency can be assessed in cells that are both GFP-negative and RFP-positive. As predicted, 53BP1 knockdown resulted in a decrease in NHEJ. Deficiency of either RAP80 or BRCC36 also caused a similar decrease in NHEJ (Fig. 1D and supplemental Fig. S1F). Considering the increases observed in HDR with RAP80 or BRCC36 deficiency, this indicates that the BRCA1-RAP80 complex may play a role in DNA repair by regulating the choice between homology-driven repair and NHEJ. Given the strong connection between cell cycle phase and DNA repair mechanism choice, we wanted to ascertain if the changes seen in the repair pathway used with RAP80 or BRCC36 knockdown were a result of cell cycle profile alteration. Flow cytometry analysis revealed that RAP80- or BRCC36-deficient cells had no detectable alteration in cell cycle distribution (supplemental Fig. S2A). These results suggest that the BRCA1-RAP80 complex directly affects the choice of DSB repair pathway by restricting the use of homology-directed repair.
Genomic Instability in Cells Deficient in the BRCA1-RAP80 Complex
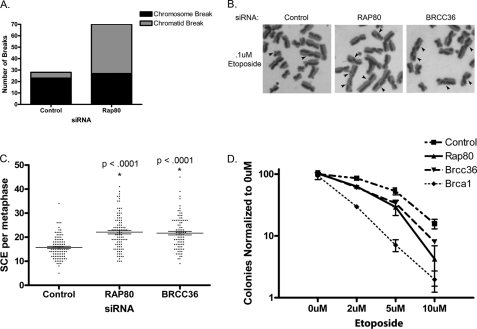
Knockdown of RAP80 has previously been shown to cause mild sensitivity to ionizing radiation (IR) (29–31). We thus hypothesized that loss of the BRCA1-RAP80 complex may lead to an increase in unrepaired DNA breaks during S/G2 when resection processes are active. We analyzed metaphase spreads prepared from cells treated with control or RAP80 siRNA and indeed found that cells with RAP80 knockdown had significantly more spontaneous breaks than cells treated with control siRNA (supplemental Fig. S2B). Interestingly, we observed that there was a specific increase in sister chromatid breaks with RAP80 knockdown when compared with chromosome breaks (Fig. 2A and supplemental Fig. S2C). Chromosome breaks are most likely to occur in G1, forming two broken pieces of DNA following replication in S phase. Alternatively, chromatid breaks are generated in S or G2 cell cycle phases after DNA has been replicated. The BRCA1-RAP80 complex is recruited to chromatin after the advent of a DSB. Therefore, it is likely that the absence of BRCA1-RAP80 does not cause more DSBs but rather disrupts accurate repair of breaks, leading to their accumulation. Because loss of RAP80 leads to an increase in detectable chromatid breaks, this suggests that the BRCA1-RAP80 complex plays a role in specifically maintaining genome stability in S and G2 phases.
FIGURE 2.
Genomic instability and sister chromatid exchange in cells with BRCA1-RAP80 deficiency. A, metaphases were collected from cells treated with RAP80 siRNA and analyzed for chromosome and chromatid breaks. Cells treated with RAP80 siRNA show an increase in chromatid breaks. Data are representative of three independent experiments. See supplemental Fig. S2C for an independent replicate experiment. B, representative images of SCE following knockdown of RAP80 or BRCC36 and subsequent etoposide treatment. C, quantification of images as in B. Cells treated with RAP80 or BRCC36 siRNA show a significant increase in etoposide-induced SCE formation. Error bars, S.E. from three independent experiments. p values were calculated in comparison with control siRNA-treated cells. D, clonogenic survival assay of cells treated with the indicated siRNAs after exposure to different doses of etoposide. Error bars, S.D. See supplemental Fig. S2F for an independent replicate experiment.
A complementary approach to evaluate DNA repair mechanism utilization is sister chromatid exchange (SCE). SCEs form as a result of Holliday junction processing during HR (41). SCEs, therefore, only form as a result of HR repair and not NHEJ or SSA. Because we observed an increase in HR with RAP80 or BRCC36 knockdown using the I-SceI reporter assay, we hypothesized that an increase in SCEs would result from knockdown of RAP80 or BRCC36. As predicted, cells treated with either RAP80 or BRCC36 siRNA exhibited a significantly greater number of spontaneous SCEs (supplemental Fig. S2D). We next examined SCE formation after treatment with etoposide, a topoisomerase II inhibitor that creates DSBs in S and G2 phases of the cell cycle. Consistent with increased HR utilization, cells treated with RAP80 or BRCC36 siRNA showed a significant increase in the number of etoposide-induced SCEs (Fig. 2, B and C). These results support our findings in Fig. 1 and are again consistent with an increase in the frequency of HR after DSB induction with loss of the BRCA1-RAP80 complex. Should this elevated usage of HR lead to errors in DSB repair, a prediction would be that RAP80- or BRCC36-deficient cells would exhibit genomic instability and hypersensitivity to etoposide. To test this hypothesis, we treated cells with etoposide and quantified the number of chromosome breaks per metaphase. RAP80 or BRCC36 siRNA caused a significant increase in the amount chromatid breaks after etoposide treatment (supplemental Fig. S2E). Furthermore, a clonogenic cell survival assay revealed that cells treated with RAP80 or BRCC36 siRNA are hypersensitive to etoposide, albeit less sensitive than BRCA1-deficient cells (Fig. 2D and supplemental Fig. S2F). Together these results stress the importance of maintaining a balance between HR, SSA, and NHEJ to preserve genomic integrity.
Next, we wished to assess the contribution of BRCA1 to the function of the entire BRCA1-RAP80 complex. An Abraxas S406A phosphodeficient mutant is not phosphorylated and cannot interact with the BRCT domains of BRCA1, yet it maintains interaction with the rest of the core RAP80 complex (31, 42, 43). Consequently, Abraxas S406A expression results in DSB recruitment of the core RAP80 complex in the absence of BRCA1. We knocked down endogenous Abraxas and evaluated etoposide-induced SCEs in cells expressing siRNA-resistant ectopic Abraxas WT or S406A. We observed a small but consistent increase in SCEs in cells expressing S406A (supplemental Fig. S3A). Additionally, we observed a small increase in DSBs in S406A compared with WT cells after etoposide treatment but saw no detectable sensitivity to etoposide in a clonogenic survival assay (supplemental Fig. S3, B and C). Because cells expressing the S406A mutant are not IR-sensitive, it is likely that there is not a significant impairment of the HR, NHEJ, and SSA repair pathways. To specifically test this, we utilized the Isce-I reporter assays in which we analyzed HR or SSA. Cells expressing S406A showed a slight increase in HR compared with those expressing WT Abraxas (supplemental Fig. S3D). These results are consistent with a small increase in SCE with S406A expression (supplemental Fig. S3A). However, there was no difference in SSA with expression of Abraxas WT or S406A (supplemental Fig. S3E).
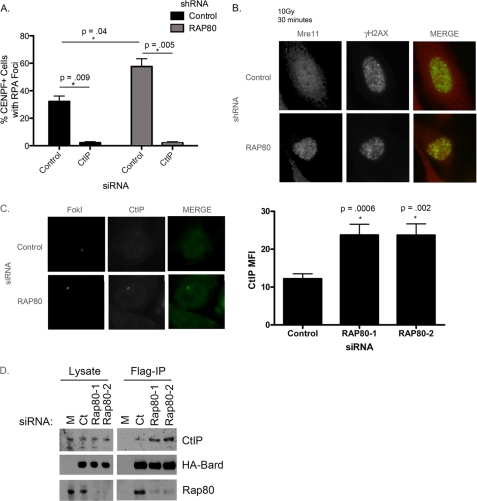
To evaluate NHEJ in a physiologic setting, we analyzed resolution of γH2AX foci in G1, where NHEJ is the only form of DSB repair (supplemental Fig. S3, F and G). G1 cells were identified by lack of CENPF staining. CENPF is a centromeric binding protein that is not expressed in G1 but is readily detectable in S and G2 phase cells (44, 45). Cells were treated with 2 grays of ionizing radiation and G1 (CENPF-negative) cells with γH2AX foci were counted 30 min and 8 h after damage. 100% of cells had γH2AX foci 30 min after irradiation. Conversely, greater than 75% of control cells had resolved the γH2AX foci at 8 h after damage, indicating efficient DSB repair had occurred. Knockdown of either RAP80 or BRCC36 did not delay the kinetics of γH2AX resolution, further suggesting that their role in DNA repair primarily occurs in S/G2 phase of the cell cycle. As a positive control for G1 phase NHEJ repair deficiency, knockdown of 53BP1 impaired γH2AX resolution in G1 cells. Finally, expression of Abraxas WT or S406A did not have a discernable effect on γH2AX resolution, suggesting that disrupting BRCA1-RAP80 interaction does not negate NHEJ repair in G1.
Loss of RAP80 or BRCC36 Causes an Increase in DSB End Resection
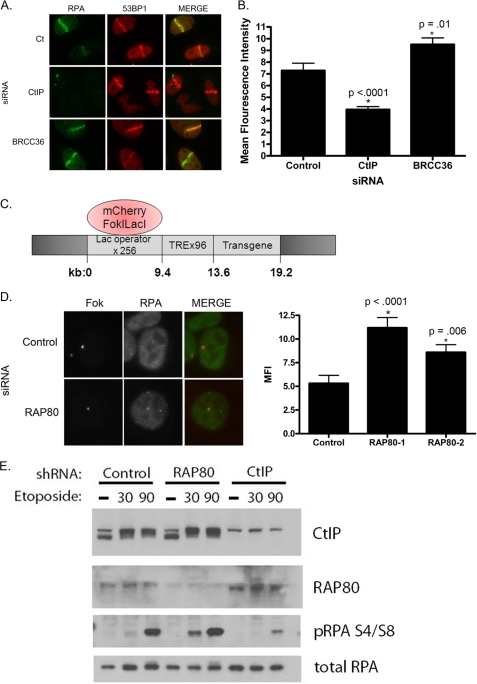
Given the increase in HR and SSA with loss of BRCA1-RAP80 (Fig. 1), we hypothesized that RAP80 or BRCC36 deficiency would cause an increase in DSB end resection. To test this idea, we used RPA as a marker of end resection following RAP80 or BRCC36 knockdown in comparison with control siRNA-treated cells. RPA is an ssDNA-binding protein that coats the DNA overhang formed as a result of resection (13, 15). Thus, RPA is not visible at breaks undergoing NHEJ but can been seen in cells using HR. A 337-nm laser was used to create a linear array of DSBs and dual color immunofluorescence performed with antibodies directed against RPA and 53BP1. Knockdown of CtIP, an endonuclease required for end resection, caused a significant decrease in RPA recruitment to damage as previously shown (15). Alternatively, BRCC36 or RAP80 knockdown resulted in an increase in RPA fluorescence at sites of damage (Fig. 3, A and B). Knockdown of RAP80 provided similar results (supplemental Fig. S4A). These results indicated that BRCC36 and RAP80 negatively regulate the process of end resection and that its absence leads to an aberrant elevation of this process.
FIGURE 3.
Increased end resection in cells with loss of RAP80 or BRCC36. A, cells were treated with the indicated siRNAs, and IF was performed for the indicated proteins 30 min after laser microirradiation. B, quantification of RPA mean fluorescence intensity from images as in A using ImageJ software. Loss of BRCC36 resulted in an increase in RPA fluorescence at breaks. Error bars, S.E. from three independent experiments. p values were calculated in comparison with control siRNA-treated cells. C, schematic of U2OS reporter containing Lac operator repeats and a downstream transgene. Expression of a mCherryFokILacI results in binding of lacI to the operator where the FokI endonuclease creates double strand breaks. D, U2OS reporter cells were treated with control or RAP80 siRNA 24 h prior to transduction with FokI. RPA accumulation was analyzed at the locus by immunofluorescence ∼60 h after siRNA transfection. RPA intensity from these experiments was quantified using ImageJ. Error bars, S.E. from three independent experiments. p values were calculated in comparison with control siRNA-treated cells. E, HeLa cells containing control, RAP80, or CtIP shRNA were untreated (−) or treated with 100 μm etoposide for 30 or 90 min. Nuclear extracts were analyzed for pRPA formation by immunoblot. Cells expressing the RAP80 shRNA show an increase in pRPA in comparison with controls, which is most prominent at the 30 min time point.
To independently assess these findings, we utilized a U2OS cell line that contains a stably integrated reporter containing several hundred repeats of the Lac operator and a downstream transgene (46). Expression of an mCherry-tagged FokI nuclease domain fused to the lac repressor (mCherryFokILacI) results in the binding of lacI to the lac operator where the nonspecific FokI endonuclease domain creates DSBs (Fig. 3C) (47). Reporter cells were treated with control or RAP80 siRNA, transduced with the FokI construct, and subsequently analyzed for RPA accumulation at the site of damage. RAP80 knockdown resulted in an increase in RPA recruitment to sites of DNA damage, again indicating an increase in the process of end resection (Fig. 3D). Additionally, knockdown of BRCC36 also resulted in an increase in RPA recruitment to damage, whereas knockdown of CtIP abrogated this response (supplemental Fig. S4B). Once RPA is loaded onto the ssDNA at a DSB, it is phosphorylated (pRPA), making it a convenient marker of end resection (48). In further support of our immunofluorescence data, loss of RAP80 caused an increase in pRPA after etoposide treatment compared with controls (Fig. 3E). Conversely, CtIP deficiency resulted in impaired pRPA formation after DNA damage as reported previously (20). Interestingly, the increase in pRPA with RAP80 knockdown was more pronounced at the early 30 min time point after damage. These results suggest that loss of RAP80 affects the initial, early processing of the break.
Consistent with excessive end resection driving an increase in HR rather than simply a persistence of RPA on ssDNA, Rad51 levels were also increased following depletion of RAP80 by either of two different siRNAs (supplemental Fig. S4C). Because Rad51 polymers displace RPA on ssDNA in a BRCA1- and BRCA2-dependent manner, these results suggest that RAP80/BRCC36 deficiency increases end resection to allow elevated coating of ssDNA by RPA and Rad51 and an increase in the use of HDR mechanisms.
Loss of BRCA1-RAP80 Leads to an Increase in End Resection in S/G2
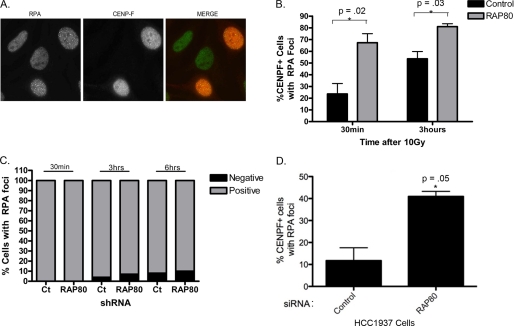
End resection is normally restricted to S and G2 phases; however, it is also possible that breaks could be aberrantly resected in G1. To distinguish between these possibilities, we used the centromeric protein CENPF as a cell cycle marker. CENPF is not expressed in G1, is detectable in S phase, and shows strong pan-nuclear staining in G2 (44, 45). Therefore, the presence of RPA at breaks exclusively in CENPF-positive cells would indicate resection in S/G2. HeLa cells were treated with IR and stained for RPA foci and CENPF expression. As expected, RPA foci only formed in CENPF-positive cells (Fig. 4A). Next, we examined the relationship between RPA focus formation and CENPF expression in HeLa cells that stably express hairpins to a control sequence or RAP80. RAP80 knockdown caused an increase in the number of CENPF-positive cells with RPA foci, indicating that the previously observed increases in end resection are primarily occurring in S/G2 (Fig. 4B). Interestingly, we found that the increase in cells with RPA foci was more pronounced at earlier time points after damage (30 min) as opposed to later time points (3 h) (Fig. 4B). This strongly suggests that a difference in the initial processing of the break accounts for the increased RPA localization in RAP80 knockdown cells as opposed to an accumulation of unrepaired breaks at later time points. Finally, to determine if RAP80 loss led to end resection in G1, we identified cells with RPA foci and analyzed whether they were CENPF-negative (G1) or CENPF-positive (S/G2). RAP80 knockdown had no effect on RPA formation in G1 cells (Fig. 4C). RPA foci were rarely, if ever, found in CENPF-negative cells at multiple time points after damage, indicating that G1 cells are not undergoing aberrant resection by detectable RPA focus formation in the absence of BRCA1-RAP80.
FIGURE 4.
Loss of RAP80 causes an increase in DSB resection in S/G2 phase of the cell cycle. A, cells were irradiated with 10 grays and fixed after 3 h. Cells were stained for RPA and CENPF to demonstrate CENPF cell cycle specificity. B, cells stably expressing hairpins to luciferase or RAP80 were irradiated and stained for RPA and CENPF 30 min and 3 h later. Cells treated with RAP80 shRNA showed an increase in RPA focus formation in CENPF (S/G2)-positive cells. Error bars, S.E. of three independent experiments. p values were calculated in comparison with control siRNA-treated cells. C, cells expressing the indicated shRNAs were irradiated and subsequently fixed at 30 min, 3 h, or 6 h after DNA damage. Cells were stained for RPA and CENPF. Cells containing RPA foci were scored for CENPF positivity. D, HCC1937 cells, expressing a truncated BRCA1 protein that does not interact with RAP80 or accumulate at DSBs, were treated with control or RAP80 siRNA and analyzed for RPA focus formation after ionizing radiation. Loss of RAP80 resulted in an increase in RPA focus formation in BRCA1-mutated cells. Error bars, S.E. from two independent experiments. p values were calculated in comparison with control siRNA-treated cells.
Next, we evaluated the role of the RAP80 complex in limiting end resection independent of its interaction with BRCA1. We examined RPA focus formation following RAP80 knockdown in the breast cancer HCC1937 cell line, which harbors a germ line frameshift at codon 1755 in one BRCA1 allele and has deleted the other wild type BRCA1 gene. The truncated BRCA1 protein expressed in these cells does not interact with RAP80 (30) and does not efficiently accumulate at DSBs (2). Loss of RAP80 caused an increase in RPA foci 30 min after damage in BRCA1-mutated HCC1937 cells (Fig. 4D), indicating that RAP80 limits end resection even in the absence of interaction with BRCA1.
Mre11 and CtIP Are Required for Increased DSB End Resection in the Absence of RAP80
We next asked whether Mre11 and CtIP were required for the increase in resection seen in the absence of RAP80 or BRCC36. Mre11-mutated ataxia telangiectasia-like disorder cells are compromised in RPA focus formation (50, 51). RAP80 knockdown did not restore RPA focus formation in ataxia telangiectasia-like disorder cells, indicating a requirement for Mre11 (data not shown). To investigate the dependence of increased end resection on CtIP, cells expressing control or RAP80 shRNA were transfected with siRNA targeted to CtIP and then evaluated for RPA focus formation 30 min after damage. CtIP deficiency reduced RPA focus formation with similar efficacy in cells expressing either control or RAP80 shRNA (Fig. 5A). Together, these results indicate that the increase in end resection seen with loss of BRCA1-RAP80 is dependent on both Mre11 and CtIP.
FIGURE 5.
Loss of RAP80 results in increased accumulation of Mre11- and CtIP-dependent nuclease activity at breaks. A, cells expressing control or RAP80 shRNA were treated with control or CtIP siRNA, analyzed for RPA focus formation in CENPF-positive cells. CtIP is necessary for the increase in RPA focus formation observed following RAP80 knockdown. Error bars, S.E. of two independent experiments. p values were calculated in comparison with control siRNA-treated cells. B, cells expressing control or RAP80 shRNA were irradiated, fixed after 30 min, and evaluated for Mre11 focus formation following IF. Cells treated with RAP80 knockdown showed an increase in Mre11 focus formation. C, U2OS reporter cells were treated with control or RAP80 siRNA, transduced with FokI, and analyzed for CtIP recruitment to FokI-induced DSBs. Loss of RAP80 led to an increase in CtIP accumulation at damage sites, quantified on the right. Error bars, S.E. of two independent experiments. D, S3 cells expressing FLAG- and HA-tagged BARD1 were transfected with control or RAP80 siRNA. Cell lysates were subjected to FLAG immunoprecipitation (IP) and immunoblotted for CtIP to analyze the BRCA1-CtIP interaction. Loss of RAP80 resulted in increased BRCA1-CtIP complex formation.
These results suggested that loss of BRCA1-RAP80 could result in increased access of Mre11 or CtIP to damage sites. Cells expressing RAP80 shRNA were assessed for Mre11 focus formation kinetics compared with controls. We found that, like RPA, Mre11 recruitment to IRIF was increased in cells with RAP80 knockdown (Fig. 5B). This increase was most striking at early time points (30 min), indicating an effect on the initial recruitment of Mre11 to DSBs after onset of damage. By 3 h, both control cells and cells with RAP80 knockdown showed similar levels of Mre11 foci (supplemental Fig. S4D). Concordant results were obtained using reporter cells and the FokI endonuclease. Cells were treated with RAP80 siRNA and evaluated for Mre11 accumulation at the site of damage. Again, loss of RAP80 resulted in a significant increase in Mre11 recruitment to DSBs (supplemental Fig. S4E). Additionally, we examined CtIP accrual at DSBs with RAP80 knockdown. RAP80 deficiency caused an increase in CtIP recruitment to sites of damage (Fig. 5C), consistent with the previous results involving Mre11 DSB recruitment.
BRCA1 is present in four distinct biochemical complexes, one of which contains CtIP. Due to an increase in CtIP recruitment in the absence of RAP80, we asked if loss of RAP80 led to an increase in BRCA1-CtIP complex formation. We treated cells with control or RAP80 siRNA and performed FLAG immunoprecipitation of HA-FLAG-tagged BARD1, the constitutive binding partner of BRCA1. We then examined the BRCA1-CtIP interaction by immunoblot for CtIP. Interestingly, knockdown of RAP80 with two different siRNAs resulted in increased interaction between BRCA1 and CtIP (Fig. 5D).
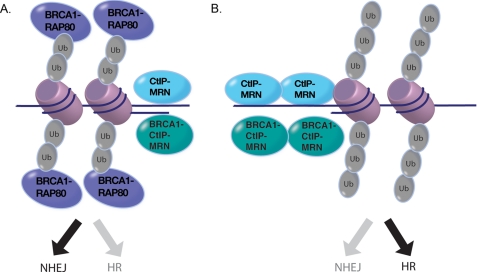
We propose a model in which the BRCA1-RAP80 complex protects DSBs from excessive resection by Mre11 and CtIP, favoring repair by NHEJ (Fig. 6A). Excessive end resection occurs in a largely BRCA1-independent manner, as evidenced by enhanced single-stranded DNA formation at DSBs following RAP80 knockdown in BRCA1-mutated cells (Fig. 4D). A smaller contribution to this phenomenon may be due to BRCA1-RAP80 preventing elevated formation of the BRCA1-CtIP complex, again limiting end resection. Collectively, RAP80 deficiency is predicted to drive a greater percentage of S/G2 breaks into homology-directed repair pathways (Fig. 6B).
FIGURE 6.
A, the BRCA1-RAP80 complex is recruited to Lys63-linked ubiquitin chains on chromatin surrounding DSBs. Occupancy of BRCA1-RAP80 at DSB chromatin prevents excessive recruitment of the MRN and CtIP nucleases and formation of the BRCA1-CtIP-MRN complex, thereby limiting end resection and maintaining an appropriate balance between HR DSB repair and NHEJ in S and G2 cell cycle phases. B, BRCA1-RAP80 complex deficiency enables increased access of MRN and CtIP nucleases to chromatin surrounding the DSB. Additionally, in the absence of RAP80, there is a compensatory increase in BRCA1-CtIP-MRN complex formation that may have a minor contribution to excessive end resection. Increased accumulation of MRN and CtIP leads to excessive end resection, with commensurate increases in single-stranded DNA, to favor utilization of HR at the expense of NHEJ in S and G2 cell cycle phases.
DISCUSSION
The choice between DSB repair pathways is strongly influenced by end processing of the break. Although NHEJ predominates in G1, CDK1-driven end resection in S and G2 permits cells to use either HR or NHEJ. How this choice is governed has yet to be determined, although end resection appears to be a pivotal step in dictating this choice. Here, we demonstrate a role for the BRCA1-RAP80 complex as a negative regulator of DSB end resection in S/G2. Loss of RAP80 leads to an increase in the homology-mediated processes of HR and SSA and a resultant decrease in NHEJ. The BRCA1-RAP80 complex is therefore instrumental in maintaining the balance between homology- and non-homology-directed repair processes by limiting extensive end resection in S/G2. Disruption of this balance leads to genomic instability and sensitivity to DNA-damaging agents.
It is intriguing that an increase in the potentially error-free process of HR leads to an increase in genome instability. However, there are several possible explanations for this occurrence. In response to IR, a biphasic DSB repair response has been documented with NHEJ dominating the fast component of DSB repair (30 min) and HR contributing to the slower phase (up to 7 h) (52). Loss of HR-specific proteins does not substantially affect the percentage of breaks that are normally repaired in the fast phase but rather delays the slow component of repair. Conceivably, disruption of the BRCA1-RAP80 complex would cause persistent breaks by reducing NHEJ efficiency, resulting in the accumulation of more slowly repaired lesions by HDR. Indeed, it has been shown that cells lacking the BRCA1-RAP80 complex component, BRCC36, have slower resolution of γH2AX foci after IR (34). Alternatively, excessive end resection may lead to an increase in genomic deletions from SSA repair or the use of homologous sequences as a template during HR in repetitive regions of the genome. The inability to accurately repair a break due to extensive resection may account for the increase in sister chromatid breaks and etoposide sensitivity seen with the loss of the BRCA1-RAP80 complex.
BRCA-RAP80 deficiency results in an increase in the accumulation of Mre11 and CtIP at DSBs at early time points after damage, suggesting increased accessibility of nucleases acutely after damage in S/G2. Because these factors accumulate at early time points after damage, this indicates that there is a difference in the initial processing of the break. Although it is not entirely clear how the BRCA1-RAP80 complex protects DSB ends, it is intriguing that γH2AX deficiency also results in an increase in the HDR process of SSA (53). Considering that BRCA1-RAP80 complexes depend on γH2AX for DSB recruitment, this raises the possibility that γH2AX recruits BRCA1-RAP80 and RAP80 complexes not bound to BRCA1 to limit access of resecting nucleases. Due to the increase in Mre11 observed with BRCA1-RAP80 complex deficiency, it is tempting to speculate that there is a competition for DSB binding between these two complexes.
The intense interest in understanding how BRCA1 maintains genome integrity to suppress malignancy leads one to wonder about the clinical implications of disrupting the BRCA1-RAP80 complex in vivo. Indeed, rare instances of RAP80 alleles deficient in ubiquitin binding and IR focus formation have been discovered in breast cancer patients (54). Additionally, recent reports reveal that single nucleotide polymorphisms present in the MERIT40 gene, which encodes an essential member of the BRCA1-RAP80 complex (55–57), confer susceptibility to breast and ovarian cancer (58, 59). Moreover, mutations to unique members of the other three BRCA1-containing complexes show that each of these complexes is involved in tumor suppression (60–63). Deficiency in BACH1, CtIP, or PalB2 impairs HR (8, 9, 15, 64, 65). In stark contrast, RAP80 or BRCC36 deficiency promotes HR. These differences may be accounted for by residency of each of these complexes at different locales along a DSB. The BRCA1 complexes containing BACH1, CtIP, and PalB2 are predicted to act directly on DNA near the actual DSB. Conversely, BRCA1-RAP80 instead forms larger DSB foci along the length of ubiquitinated chromatin. It thus appears that characteristic IR-induced BRCA1 foci modulate the repair mechanism by limiting resection and are not essential for all BRCA1-dependent repair function.
Although the RAP80 complex plays a role in delivering BRCA1 to sites of DSBs, we have demonstrated here that it also has BRCA1-independent functions. First, expression of an Abraxas mutant that no longer binds BRCA1 has less substantial effects on DSB repair than RAP80 deficiency, indicating that the RAP80 complex still facilitates DSB repair independent of BRCA1 interaction. Additionally, knockdown of RAP80 in BRCA1-mutated cells continues to cause an increase in RPA focus formation. These results indicate that loss of RAP80 complex proteins in a BRCA1 mutant background may have additional effects on tumorigenesis (58). Finally, it has been recently published that the combined loss of RAP80 and BRCA1 in chicken DT40 cells results in increased sensitivity to etoposide compared with singular loss of either gene (66). These results support a model where RAP80 binding to ubiquitinated chromatin limits excessive end resection. In the absence of BRCA1, the core RAP80 complex can continue to prevent access of nucleases to DNA.
The simultaneous presence of multiple distinct BRCA1 complexes at damage sites also suggests the potential for cooperative DNA repair activities or cross-talk between complexes. We have demonstrated that the BRCA1-RAP80 complex mitigates excessive resection by Mre11 and CtIP, revealing that at some level, these complexes influence the outcome of repair in a coordinated manner. Aberrant resection and loss of NHEJ may have detrimental effects on genomic stability by introducing deletions and mutations into the genome. Alternatively, recent literature has demonstrated potentially clinically relevant benefits to disrupting the balance of repair pathways. Loss of 53BP1 has been shown to rescue the embryonic lethality and cancer susceptibility of BRCA1−/− mice by allowing resection of DSBs and HDR repair (67, 68). Additionally, disruption of NHEJ through deletion of Ku70 rescues cross-linking sensitivity in Fanconi anemia cells by allowing HR to perform accurate repair (69, 70). Therefore, insights into the maintenance of DNA mechanism utilization are critical for understanding both the causes of genome instability and potential means to exploit these processes for clinical purposes.
Acknowledgments
We thank members of the Greenberg laboratory and Dr. David Livingston for helpful discussions. We thank Drs. Koji Nakanishi and Maria Jasin for the DR-GFP and SA-GFP constructs, Dr. Matthew Porteus for the NHEJ reporter cell line and constructs, and Drs. Susan Janicki and David Spector for the lac operator-containing cell line.
Addendum
Similar findings from Drs. Yiduo Hu and David M. Livingston are currently in press (49). Findings from both studies are in agreement that RAP80 and associated proteins Abraxas and BRCC36 serve to limit excessive Mre11/CtIP-dependent end resection and homology-directed DNA double strand break repair.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grant 1R01CA138835-01 and K08 Awards 1K08CA106597-01 and 3K08CA106597-04S1 (to R. A. G.) and National Institutes of Health Grants T32-GM-007229 and T32-CA-115299 (to K. A. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- HR
- homologous recombination
- DSB
- double strand break
- SSA
- single strand annealing
- NHEJ
- non-homologous end joining
- ssDNA
- single-stranded DNA
- IF
- immunofluorescence
- WB
- Western blot
- SCE
- sister chromatid exchange
- RPA
- replication protein A
- pRPA
- phosphorylated RPA.
REFERENCES
- 1. Rahman N., Stratton M. R. (1998) Annu. Rev. Genet. 32, 95–121 [DOI] [PubMed] [Google Scholar]
- 2. Scully R., Ganesan S., Vlasakova K., Chen J., Socolovsky M., Livingston D. M. (1999) Mol. Cell. 4, 1093–1099 [DOI] [PubMed] [Google Scholar]
- 3. Stratton M. R., Rahman N. (2008) Nat. Genet. 40, 17–22 [DOI] [PubMed] [Google Scholar]
- 4. Venkitaraman A. R. (2002) Cell 108, 171–182 [DOI] [PubMed] [Google Scholar]
- 5. Karran P. (2000) Curr. Opin. Genet. Dev. 10, 144–150 [DOI] [PubMed] [Google Scholar]
- 6. Shrivastav M., De Haro L. P., Nickoloff J. A. (2008) Cell. Res. 18, 134–147 [DOI] [PubMed] [Google Scholar]
- 7. Davies A. A., Masson J. Y., McIlwraith M. J., Stasiak A. Z., Stasiak A., Venkitaraman A. R., West S. C. (2001) Mol. Cell. 7, 273–282 [DOI] [PubMed] [Google Scholar]
- 8. Sy S. M., Huen M. S., Chen J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., Yu X. (2009) Curr. Biol. 19, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hicks W. M., Kim M., Haber J. E. (2010) Science 329, 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sung P., Klein H. (2006) Nat. Rev. Mol. Cell. Biol. 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 12. Huertas P. (2010) Nat. Struct. Mol. Biol. 17, 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M. B., Bartek J., Lukas J. (2006) J. Cell Biol. 173, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mirzoeva O. K., Petrini J. H. (2001) Mol. Cell. Biol. 21, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. (2007) Nature 450, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huertas P., Jackson S. P. (2009) J. Biol. Chem. 284, 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yun M. H., Hiom K. (2009) Nature 459, 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greenberg R. A., Sobhian B., Pathania S., Cantor S. B., Nakatani Y., Livingston D. M. (2006) Genes Dev. 20, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen L., Nievera C. J., Lee A. Y., Wu X. (2008) J. Biol. Chem. 283, 7713–7720 [DOI] [PubMed] [Google Scholar]
- 20. Gravel S., Chapman J. R., Magill C., Jackson S. P. (2008) Genes Dev. 22, 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang T. T., D'Andrea A. D. (2006) Nat. Rev. Mol. Cell. Biol. 7, 323–334 [DOI] [PubMed] [Google Scholar]
- 22. Messick T. E., Greenberg R. A. (2009) J. Cell. Biol. 187, 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 26. Wang B., Elledge S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 28. Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 29. Kim H., Chen J., Yu X. (2007) Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 30. Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., Greenberg R. A. (2007) Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., Elledge S. J. (2007) Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan J., Kim Y. S., Yang X. P., Li L. P., Liao G., Xia F., Jetten A. M. (2007) Cancer Res. 67, 6647–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cooper E. M., Cutcliffe C., Kristiansen T. Z., Pandey A., Pickart C. M., Cohen R. E. (2009) EMBO J. 28, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shao G., Lilli D. R., Patterson-Fortin J., Coleman K. A., Morrissey D. E., Greenberg R. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3166–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng L., Wang J., Chen J. (2010) J. Biol. Chem. 285, 30982–30988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patterson-Fortin J., Shao G., Bretscher H., Messick T. E., Greenberg R. A. (2010) J. Biol. Chem. 285, 30971–30981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu X., Chen J. (2004) Mol. Cell. Biol. 24, 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pierce A. J., Hu P., Han M., Ellis N., Jasin M. (2001) Genes Dev. 15, 3237–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stark J. M., Pierce A. J., Oh J., Pastink A., Jasin M. (2004) Mol. Cell. Biol. 24, 9305–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Potts P. R., Porteus M. H., Yu H. (2006) EMBO. J. 25, 3377–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson D. M., 3rd, Thompson L. H. (2007) Mutat Res. 616, 11–23 [DOI] [PubMed] [Google Scholar]
- 42. Kim H., Huang J., Chen J. (2007) Nat. Struct. Mol. Biol. 14, 710–715 [DOI] [PubMed] [Google Scholar]
- 43. Liu Z., Wu J., Yu X. (2007) Nat. Struct. Mol. Biol. 14, 716–720 [DOI] [PubMed] [Google Scholar]
- 44. Kao G. D., McKenna W. G., Yen T. J. (2001) Oncogene 20, 3486–3496 [DOI] [PubMed] [Google Scholar]
- 45. Beucher A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., Goodarzi A. A., Krempler A., Jeggo P. A., Löbrich M. (2009) EMBO. J. 28, 3413–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Janicki S. M., Tsukamoto T., Salghetti S. E., Tansey W. P., Sachidanandam R., Prasanth K. V., Ried T., Shav-Tal Y., Bertrand E., Singer R. H., Spector D. L. (2004) Cell 116, 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A. (2010) Cell 141, 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anantha R. W., Vassin V. M., Borowiec J. A. (2007) J. Biol. Chem. 282, 35910–35923 [DOI] [PubMed] [Google Scholar]
- 49. Hu Y., Livingston D. M. (2011) Genes Dev., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stewart G. S., Maser R. S., Stankovic T., Bressan D. A., Kaplan M. I., Jaspers N. G., Raams A., Byrd P. J., Petrini J. H., Taylor A. M. (1999) Cell 99, 577–587 [DOI] [PubMed] [Google Scholar]
- 51. Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., Lukas J., Jackson S. P. (2006) Nat. Cell. Biol. 8, 37–45 [DOI] [PubMed] [Google Scholar]
- 52. Mao Z., Bozzella M., Seluanov A., Gorbunova V. (2008) DNA Repair 7, 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xie A., Puget N., Shim I., Odate S., Jarzyna I., Bassing C. H., Alt F. W., Scully R. (2004) Mol. Cell. 16, 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nikkilä J., Coleman K. A., Morrissey D., Pylkäs K., Erkko H., Messick T. E., Karppinen S. M., Amelina A., Winqvist R., Greenberg R. A. (2009) Oncogene 28, 1843–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Feng L., Huang J., Chen J. (2009) Genes Dev. 23, 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shao G., Patterson-Fortin J., Messick T. E., Feng D., Shanbhag N., Wang Y., Greenberg R. A. (2009) Genes Dev. 23, 740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang B., Hurov K., Hofmann K., Elledge S. J. (2009) Genes Dev. 23, 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Antoniou A. C., Wang X., Fredericksen Z. S., McGuffog L., Tarrell R., Sinilnikova O. M., Healey S., Morrison J., Kartsonaki C., Lesnick T., Ghoussaini M., Barrowdale D., Peock S., Cook M., Oliver C., Frost D., Eccles D., Evans D. G., Eeles R., Izatt L., Chu C., Douglas F., Paterson J., Stoppa-Lyonnet D., Houdayer C., Mazoyer S., Giraud S., Lasset C., Remenieras A., Caron O., Hardouin A., Berthet P., Hogervorst F. B., Rookus M. A., Jager A., van den Ouweland A., Hoogerbrugge N., van der Luijt R. B., Meijers-Heijboer H., Gómez García E. B., Devilee P., Vreeswijk M. P., Lubinski J., Jakubowska A., Gronwald J., Huzarski T., Byrski T., Górski B., Cybulski C., Spurdle A. B., Holland H., Goldgar D. E., John E. M., Hopper J. L., Southey M., Buys S. S., Daly M. B., Terry M. B., Schmutzler R. K., Wappenschmidt B., Engel C., Meindl A., Preisler-Adams S., Arnold N., Niederacher D., Sutter C., Domchek S. M., Nathanson K. L., Rebbeck T., Blum J. L., Piedmonte M., Rodriguez G. C., Wakeley K., Boggess J. F., Basil J., Blank S. V., Friedman E., Kaufman B., Laitman Y., Milgrom R., Andrulis I. L., Glendon G., Ozcelik H., Kirchhoff T., Vijai J., Gaudet M. M., Altshuler D., Guiducci C., Loman N., Harbst K., Rantala J., Ehrencrona H., Gerdes A. M., Thomassen M., Sunde L., Peterlongo P., Manoukian S., Bonanni B., Viel A., Radice P., Caldes T., de la Hoya M., Singer C. F., Fink-Retter A., Greene M. H., Mai P. L., Loud J. T., Guidugli L., Lindor N. M., Hansen T. V., Nielsen F. C., Blanco I., Lazaro C., Garber J., Ramus S. J., Gayther S. A., Phelan C., Narod S., Szabo C. I., Benitez J., Osorio A., Nevanlinna H., Heikkinen T., Caligo M. A., Beattie M. S., Hamann U., Godwin A. K., Montagna M., Casella C., Neuhausen S. L., Karlan B. Y., Tung N., Toland A. E., Weitzel J., Olopade O., Simard J., Soucy P., Rubinstein W. S., Arason A., Rennert G., Martin N. G., Montgomery G. W., Chang-Claude J., Flesch-Janys D., Brauch H., Severi G., Baglietto L., Cox A., Cross S. S., Miron P., Gerty S. M., Tapper W., Yannoukakos D., Fountzilas G., Fasching P. A., Beckmann M. W., Dos Santos Silva I., Peto J., Lambrechts D., Paridaens R., Rüdiger T., Försti A., Winqvist R., Pylkäs K., Diasio R. B., Lee A. M., Eckel-Passow J., Vachon C., Blows F., Driver K., Dunning A., Pharoah P. P., Offit K., Pankratz V. S., Hakonarson H., Chenevix-Trench G., Easton D. F., Couch F. J. (2010) Nat. Genet. 42, 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bolton K. L., Tyrer J., Song H., Ramus S. J., Notaridou M., Jones C., Sher T., Gentry-Maharaj A., Wozniak E., Tsai Y. Y., Weidhaas J., Paik D., Van Den Berg D. J., Stram D. O., Pearce C. L., Wu A. H., Brewster W., Anton-Culver H., Ziogas A., Narod S. A., Levine D. A., Kaye S. B., Brown R., Paul J., Flanagan J., Sieh W., McGuire V., Whittemore A. S., Campbell I., Gore M. E., Lissowska J., Yang H. P., Medrek K., Gronwald J., Lubinski J., Jakubowska A., Le N. D., Cook L. S., Kelemen L. E., Brook-Wilson A., Massuger L. F., Kiemeney L. A., Aben K. K., van Altena A. M., Houlston R., Tomlinson I., Palmieri R. T., Moorman P. G., Schildkraut J., Iversen E. S., Phelan C., Vierkant R. A., Cunningham J. M., Goode E. L., Fridley B. L., Kruger-Kjaer S., Blaeker J., Hogdall E., Hogdall C., Gross J., Karlan B. Y., Ness R. B., Edwards R. P., Odunsi K., Moyisch K. B., Baker J. A., Modugno F., Heikkinenen T., Butzow R., Nevanlinna H., Leminen A., Bogdanova N., Antonenkova N., Doerk T., Hillemanns P., Dürst M., Runnebaum I., Thompson P. J., Carney M. E., Goodman M. T., Lurie G., Wang-Gohrke S., Hein R., Chang-Claude J., Rossing M. A., Cushing-Haugen K. L., Doherty J., Chen C., Rafnar T., Besenbacher S., Sulem P., Stefansson K., Birrer M. J., Terry K. L., Hernandez D., Cramer D. W., Vergote I., Amant F., Lambrechts D., Despierre E., Fasching P. A., Beckmann M. W., Thiel F. C., Ekici A. B., Chen X., Johnatty S. E., Webb P. M., Beesley J., Chanock S., Garcia-Closas M., Sellers T., Easton D. F., Berchuck A., Chenevix-Trench G., Pharoah P. D., Gayther S. A. (2010) Nat. Genet. 42, 880–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heikkinen K., Rapakko K., Karppinen S. M., Erkko H., Knuutila S., Lundán T., Mannermaa A., Børresen-Dale A. L., Borg A., Barkardottir R. B., Petrini J., Winqvist R. (2006) Carcinogenesis 27, 1593–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rahman N., Seal S., Thompson D., Kelly P., Renwick A., Elliott A., Reid S., Spanova K., Barfoot R., Chagtai T., Jayatilake H., McGuffog L., Hanks S., Evans D. G., Eccles D., Easton D. F., Stratton M. R. (2007) Nat. Genet. 39, 165–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Seal S., Thompson D., Renwick A., Elliott A., Kelly P., Barfoot R., Chagtai T., Jayatilake H., Ahmed M., Spanova K., North B., McGuffog L., Evans D. G., Eccles D., Easton D. F., Stratton M. R., Rahman N. (2006) Nat. Genet. 38, 1239–1241 [DOI] [PubMed] [Google Scholar]
- 63. Erkko H., Xia B., Nikkilä J., Schleutker J., Syrjäkoski K., Mannermaa A., Kallioniemi A., Pylkäs K., Karppinen S. M., Rapakko K., Miron A., Sheng Q., Li G., Mattila H., Bell D. W., Haber D. A., Grip M., Reiman M., Jukkola-Vuorinen A., Mustonen A., Kere J., Aaltonen L. A., Kosma V. M., Kataja V., Soini Y., Drapkin R. I., Livingston D. M., Winqvist R. (2007) Nature 446, 316–319 [DOI] [PubMed] [Google Scholar]
- 64. Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F. J., Livingston D. M. (2006) Mol. Cell. 22, 719–729 [DOI] [PubMed] [Google Scholar]
- 65. Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P. R., Cantor S. B. (2005) Cancer Cell. 8, 255–265 [DOI] [PubMed] [Google Scholar]
- 66. Iijima J., Zeng Z., Takeda S., Taniguchi Y. (2010) Cancer Res. 70, 8467–8474 [DOI] [PubMed] [Google Scholar]
- 67. Bouwman P., Aly A., Escandell J. M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., Haffty B. G., Tommiska J., Blomqvist C., Drapkin R., Adams D. J., Nevanlinna H., Bartek J., Tarsounas M., Ganesan S., Jonkers J. (2010) Nat. Struct. Mol. Biol. 17, 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bunting S. F., Callén E., Wong N., Chen H. T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C. X., Finkel T., Nussenzweig M., Stark J. M., Nussenzweig A. (2010) Cell 141, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Adamo A., Collis S. J., Adelman C. A., Silva N., Horejsi Z., Ward J. D., Martinez-Perez E., Boulton S. J., La Volpe A. (2010) Mol. Cell. 39, 25–35 [DOI] [PubMed] [Google Scholar]
- 70. Pace P., Mosedale G., Hodskinson M., Rosado I. V., Sivasubramaniam M., Patel K. J. (2010) Science 329, 219–223 [DOI] [PubMed] [Google Scholar]