Abstract
R7BP (RGS7 family-binding protein) has been proposed to function in neurons as a palmitoylation-regulated protein that shuttles heterodimeric, Gi/oα-specific GTPase-activating protein (GAP) complexes composed of Gβ5 and RGS7 (R7) isoforms between the plasma membrane and nucleus. To test this hypothesis we studied R7BP palmitoylation and localization in neuronal cells. We report that R7BP undergoes dynamic, signal-regulated palmitate turnover; the palmitoyltransferase DHHC2 mediates de novo and turnover palmitoylation of R7BP; DHHC2 silencing redistributes R7BP from the plasma membrane to the nucleus; and Gi/o signaling inhibits R7BP depalmitoylation whereas Gi/o inactivation induces nuclear accumulation of R7BP. In concert with previous evidence, our findings suggest that agonist-induced changes in palmitoylation state facilitate GAP action by (i) promoting Giα depalmitoylation to create optimal GAP substrates, and (ii) inhibiting R7BP depalmitoylation to stabilize membrane association of R7-Gβ5 GAP complexes. Regulated palmitate turnover may also enable R7BP-bound GAPs to shuttle between sites of low and high Gi/o activity or the plasma membrane and nucleus, potentially providing spatio-temporal control of signaling by Gi/o-coupled receptors.
Keywords: G Protein-coupled Receptors (GPCR), G Proteins, Neuron, Neurotransmitters, Nuclear Translocation, Protein Palmitoylation, Protein Targeting, Protein Translocation, RGS Proteins
Introduction
Sensory stimuli, including light and olfactants, and many modulatory neurotransmitters, psychotherapeutic agents and drugs of abuse elicit their biological effects by activating receptors coupled to pertussis toxin (PTX)2-sensitive G proteins of the Gi/o class (1, 2). Signaling by Gi/o-coupled receptors is regulated by the R7 family (RGS6, RGS7, RGS9, and RGS11) of RGS (regulator of G protein signaling) proteins (3, 4). R7 proteins form obligatory heterodimers with Gβ5 (5–7), the most diverged member of the Gβ family, producing complexes that regulate signaling kinetics by functioning as GTPase-activating proteins (GAPs) selective for Gi/oα subunits (6, 8). Indeed, RGS9 regulates phototransduction kinetics in photoreceptor cells (9, 10), and opioid and cocaine action in striatum (11, 12). RGS7 and 11 regulate photoresponse kinetics in retinal ON bipolar cells (13–15). RGS6 regulates muscarinic receptor-evoked IKACh gating kinetics in cardiomyocytes (16).
R7-Gβ5 heterodimers associate with R9AP (RGS9 anchor protein) or R7BP (RGS7 family-binding protein), a pair of related SNARE-like proteins (17). R9AP is a transmembrane protein that binds RGS9 or RGS11 and is expressed in retinal photoreceptors and ON bipolar cells (18, 19). R9AP is required for stable expression of RGS9 in photoreceptor disk membranes (20), and RGS11 in ON bipolar cell dendrites (21). Thus, R9AP is essential for normal photoresponse kinetics in both cell types (10, 20, 21). In contrast, R7BP is expressed widely in the nervous system, binds each R7 isoform, lacks a transmembrane domain, and associates with membranes by covalent attachment of the fatty acid palmitate to two C-terminal cysteine residues (22–24). In striatum, R7BP stabilizes RGS9 expression and function as a regulator of motor coordination and locomotor response to morphine (25). Intriguingly, blockade of R7BP palmitoylation in cultured cells enables the protein to use a conserved C-terminal polybasic sequence resembling a classical nuclear localization sequence to import R7-Gβ5 complexes into the nucleus (23, 24). Because palmitate attachment to proteins occurs via reversible thioester bonds, R7BP potentially functions as a palmitoylation-regulated protein that shuttles R7-Gβ5 heterodimers between the plasma membrane and nucleus.
However, whether endogenous R7BP functions in neuronal cells as a palmitoylation-regulated shuttling protein remains unclear. On the one hand, R7BP is palmitoylated efficiently because at steady state it associates nearly exclusively with membrane fractions of brain extracts (24, 26). On the other, steady state nuclear accumulation of RGS7 and Gβ5 in neurons appears to depend partially on R7BP (27). These potentially discordant lines of evidence derived from steady-state localization measurements underscore the importance of determining whether R7BP palmitoylation is stable or undergoes dynamic and reversible turnover. In principle, either outcome is a possibility because palmitoylation of certain proteins is dynamic and regulatory whereas palmitoylation of others is stable and non-regulatory (28).
Here we have tested the shuttling hypothesis by examining R7BP palmitoylation and localization in neuronal cells. We show that R7BP palmitoylation undergoes extensive turnover, identify palmitoyltransferase activity involved in de novo and turnover palmitoylation of R7BP, and find that G protein signaling regulates R7BP palmitoylation cycling and intracellular localization. Besides supporting the hypothesis that R7BP functions in neuronal cells as a palmitoylation-regulated protein, our findings deepen understanding of mechanisms whereby signal-regulated palmitate turnover controls the action of GAP complexes toward Gi/oα subunits that mediate the action of modulatory neurotransmitters and drugs of abuse.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Neuro2A and HEK293 cells were cultured at 37 °C in DMEM (Dulbecco's modified Eagle's medium) with HEPES (10 mm), penicillin (100 units/ml), streptomycin (100 μg/ml) and fetal bovine serum (10% v/v). Neuro2A cells stably transfected with FLAG-R7BP were maintained in medium supplemented with geneticin (2 mg/ml, Washington University tissue culture support center). Cells were treated as indicated with cycloheximide (CHX, Sigma), WIN 55,212–2 (WIN, Sigma), or puromycin (Sigma) in serum-containing medium at 37 °C unless otherwise noted. Cells were transfected with Fugene HD reagent (Roche Applied Science) according to the supplier's recommendations.
Cortical Neuron Isolation and Culture
Cortical primary neuron isolation was performed according to previous publications with minor modifications (29). All experimental protocols involving vertebrate animals were approved by the Animal Studies Committee at Washington University. Female pregnant rats (Sprague-Dawley) were purchased from Charles River Laboratories. Rat pups were euthanized at day P1. The brain was exposed and cortices were dissected and cut into pieces. Cortical tissue was digested in trypsin/EDTA solution for 15 min at 37 °C, followed by neutralization with FBS. After washing, cortical tissues were dissociated by gentle trituration with a fire-polished Pasteur pipette. Dissociated cells were collected by centrifugation at 500 × g and suspended in Neurobasal/10%FBS medium. Cells were plated into 6-well plates on poly-d-lysine-coated coverslips. After 3 h, the medium was replaced with culture medium (neurobasal medium/B27/glutamine/penicillin-streptomycin). A mixture of antimitotics (5-fluoro-2′-deoxyuridine, uridine, and cytosine β-d-arabinofuranoside) was used to inhibit glial cell propagation at DIV5. For palmitate-labeling experiments, cells were cultured for 14 days and labeled as described below. Otherwise, neuronal cells were cultured for 19 d prior to overnight treatment with 0.2 μg/ml pertussis toxin (30) and paraformaldehyde fixation.
Antibodies
The following commercially produced antibodies were used: rabbit polyclonal Goα (Santa Cruz Biotechnology) and GFP (AbCam) antibodies, and mouse monoclonal anti-FLAG (Sigma) antibodies. Our affinity-purified rabbit and chicken polyclonal anti-R7BP antibodies have been described previously (26). Goat anti-mouse IR800 and Goat anti-rabbit IR 680 were obtained from LI-COR. Alexa Fluor 488-labeled goat anti-rabbit IgG and Alexa Fluor 568-labeled goat anti-mouse IgG were obtained from Invitrogen. Pertussis toxin holoenzyme was purchased from Calbiochem.
Plasmids
Plasmids expressing shRNAs were made by inserting hairpin-encoding sequences between the AgeI and BamHI sites of pLKO.1. The following target sequences were used to knock-down expression of mouse DHHC2: GGAGGAATGGAACAACCTTCC (SH1), GGAAGGCACCATTTGCCAATT (SH2), and GAAATCAGAGGGAACACATGC (SH3). The target sequences are located in the non-conserved 3′-UTR of DHHC2 mRNA. A scrambled shRNA sequence (CCTAAGGTTAAGTCGCCCTCG) provided a control. pEGFP-DHHC2, pEGFP-DHHC3, pEGFP-DHHC5, pEGFP-DHHC6, pEGFP-DHHC7, pEGFP-DHHC11, pEGFP-DHHC13, and pEGFP-DHHC17 were constructed by subcloning cDNAs for each enzyme into pEGFP-N1 or -C1. pEGFPmDHHC2 + 3′-UTR was constructed by cloning the DHHC2 coding region and 1169 bp 3′-UTR from mouse brain cDNA between the NcoI and BamHI sites of pEGFP-C3. cDNA was synthesized using a high capacity cDNA reverse transcriptase kit (Applied Biosciences). pTR-UF-tdRFP-R7BP was made by inserting the wild-type R7BP coding region into pTR-UF-tdRFP. Plasmids expressing GFP-N-Ras, GFP-Syntaxin 7, constitutively active GoαQ204L, pEGFP-R7BP, and p3xFLAG-R7BP clones have been described previously (23, 31, 32).
Quantitative RT-PCR
Total RNA was extracted from Neuro2A cells or brain tissues using Nucleospin RNA II kits (Clontech). Quantitative PCR was performed using an Applied Biosystems 7000 system (Applied Biosciences) and the QTaqTM one-step qRT-PCR SYBR kit (Clontech). Expression values were normalized to expression of GAPDH. Primer sequences will be provided upon request.
Metabolic Labeling
To detect new protein synthesis, Neuro2A cells stably expressing FLAG-R7BP (26) (6 × 105 cells/well in 6-well plates) were incubated with methionine-free DMEM for 5 min and then labeled 1 h with medium containing [35S]methionine (50 μCi/ml; Perkin Elmer). Labeled cells were lysed and subjected to anti-FLAG immunoprecipitation and SDS-PAGE analysis as described previously (23). To detect palmitoylation of FLAG-tagged R7BP protein, Neuro2A cells stably transfected with FLAG-R7BP (6 × 105 cells/well in 6-well plates) were incubated 5 min in 2 ml of serum-free DMEM medium containing pyruvate (1 mm) and non-essential amino acids (0.1 mm each). Cells were metabolically labeled 1 h in medium containing [3H]palmitate (0.5 mCi/ml; Perkin Elmer) and dialyzed fetal calf serum (10%). Cells were lysed and subjected to anti-FLAG immunoprecipitation, as follows. Labeled cells were lysed with 500 μl of MCLB buffer (50 mm Tris-HCl pH 7.5, 100 mm NaCl, 5 mm EDTA, 0.75% Igepal, and 1 mm phenylmethylsulfonyl fluoride, 1 mm sodium orthovanadate, complete protease inhibitor mixture (Roche)). Supernatant fractions obtained following centrifugation at 12,000 × g for 10 min were immunoprecipitated with 12.5 μl of FLAG-agarose beads (Sigma). After washing, each immunoprecipitate was eluted and separated on three SDS-polyacrylamide gels: one for anti-FLAG Western blotting, a second for scintillation counting of the FLAG-R7BP band after treating gel slices with Soluvin (Perkin Elmer), and a third for fluorography at −80 °C after soaking with salicylic acid in methanol and drying. To detect palmitoylation of GFP-tagged N-Ras and syntaxin 7 proteins, anti-GFP antibody and protein G-coupled agarose beads were used for immunoprecipitation in the above protocol. For pulse-chase experiments, Neuro2A cells stably transfected with FLAG-R7BP were metabolically labeled 3 h in media containing [3H]palmitate. Cells were washed and incubated in DMEM containing unlabeled palmitate (100 μm), pyruvate (1 mm), non-essential amino acids (0.1 mm each) and dialyzed fetal calf serum (10%). At various time points, labeled cells were harvested, lysed and subjected to anti-FLAG immunoprecipitation. Samples were analyzed on three gels as described above. To detect palmitoylation of endogenous R7BP in rat cortical neurons, we metabolically labeled neuronal cultures (DIV14) 6 h in Neurobasal medium containing [3H]palmitate (0.5 mCi/ml) and pyruvate (1 mm), nonessential amino acids (0.1 mm each) and dialyzed fetal calf serum (10%). Cells were lysed and immunoprecipitated as described above except that affinity-purified rabbit anti-R7BP antibodies were used (26). Immunoprecipitates were resolved by SDS-PAGE, transferred to PVDF membranes, and visualized by immunoblotting with affinity-purified chicken anti-R7BP antibodies (26), and fluorography to detect labeled R7BP.
Fluorescence Confocal Microscopy
Neuro2A cells were transfected as indicated with pEGFP-DHHC2, pEGFP-DHHC11, pEGFP-DHHC17, pTR-UF-tdRFP R7BP, pEGFP-R7BP, and/or pLKO.1-based plasmids for DHHC2 knock down. At 24 h post-transfection, cells were seeded at low density on poly-d-lysine (10 μg/ml)-coated 18-mm glass coverslips (Fisher) and serum starved for an additional 24–48 h. Cells were then fixed with paraformaldehyde (4%) for 10 min at room temperature. Neurons were stained with rabbit anti-R7BP and mouse anti-NeuN, antibodies followed by Alexa Fluor-488-labeled goat anti-rabbit IgG and Alexa Fluor-568-labeled goat anti-mouse IgG. Cells were mounted in VECTASHIELD mounting medium (Vector Laboratories). Fluorescence images were captured at room temperature on an Olympus BX52 microscope equipped with a 1.35NA 100× UPlanApo objective, spinning disc confocal scanner unit (CSU10), Picarro Cyan (488 nm; Sunnyvale, CA) and Cobolt Jive (561 nm; Solna, S.E.) lasers and a Stanford Photonics XR MEGA-10 CCD camera (Palo Alto, CA), by using In Vivo software (Media Cybernetics). Only brightness, contrast, and color balance were adjusted for the whole image using ImageJ software for image analysis and assembly as montages.
Statistical Analysis
The results are expressed as means ± S.E. of the mean (S.E.). Statistical comparisons between groups were done using Student's t test.
RESULTS
R7BP Undergoes Palmitate Turnover in Neuronal Cells
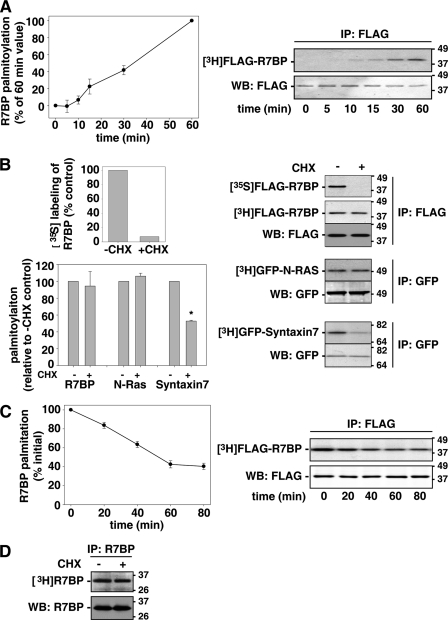
To determine whether R7BP palmitoylation is stable or undergoes turnover in neuronal cells that endogenously express its obligatory R7-Gβ5 binding partners, we analyzed a Neuro2A neuroblastoma cell line stably transfected with FLAG-R7BP at levels that match those of endogenous R7BP in brain (26), and primary cultures of rat cortical neurons. Cells were labeled with [3H]palmitate, harvested over time, lysed in detergent, immunoprecipitated with anti-FLAG or -R7BP antibodies, resolved by SDS-PAGE and analyzed by scintillation counting to detect label incorporated into R7BP and quantitative Western blotting to determine the relative stoichiometry of label incorporation into immunoprecipitated R7BP. Results shown in Fig. 1A indicated that [3H]palmitate labeling of FLAG-R7BP in Neuro2A cells was detected within 10 min and increased over 1 h. To determine the extent that palmitate labeling occurred on newly synthesized versus pre-existing FLAG-R7BP (i.e. whether palmitate turnover occurs), we then added vehicle (DMSO) as a control or CHX 5 min before and during labeling to block new protein synthesis. Blockade of protein synthesis was confirmed by showing that CHX-treated cells failed to incorporate [35S]methionine into FLAG-R7BP (Fig. 1B, top panel). In contrast, CHX treatment did not significantly reduce the relative efficiency of FLAG-R7BP labeling with [3H]palmitate (Fig. 1B), indicating that extensive palmitate cycling on pre-existing FLAG-R7BP occurred during the 1 h labeling period. Palmitate cycling on FLAG-R7BP was not due to CHX treatment, as shown by [3H]palmitate pulse-chase labeling experiments in the absence of CHX. In this experiment, label associated with FLAG-R7BP declined dramatically during the 1 h chase, indicating that extensive palmitate cycling occurred (Fig. 1C).
FIGURE 1.
R7BP undergoes palmitoylation turnover. A, metabolic labeling of Neuro2A cells stably expressing FLAG-R7BP with [3H]palmitate. FLAG-R7BP was immunoprecipitated at the indicated times from [3H]palmitate-labeled cells and resolved by SDS-PAGE. Immunoprecipitates were analyzed by fluorography (right upper panel), scintillation counting to quantify R7BP-incorporated radiolabel and immunoblotting to quantify immunoprecipitated R7BP (right lower panel). Palmitoylation efficiency over time is expressed as the radiolabeled R7BP:immunoprecipitated R7BP ratio (left panel). Results shown are the average of four experiments. B, palmitate turnover assays. Right upper panel: Neuro2A cells stably expressing FLAG-R7BP were metabolically labeled 1h with [35S]methionine or [3H]palmitate in the absence (−) or presence (+) of CHX. FLAG-R7BP was immunoprecipitated, resolved, visualized, and quantified as described in panel A. Lower panels: GFP-N-Ras or GFP-syntaxin 7-transfected Neuro2A cells were metabolically labeled with [3H]palmitate in the absence or presence of CHX. Palmitoylation efficiency of R7BP, N-Ras and syntaxin 7 palmitoylation was quantified as described in panel A. Results shown are the average of three experiments. *, p < 7e-7. C, R7BP depalmitoylation. Neuro2A cells stably expressing FLAG-R7BP were labeled 3h with [3H]palmitate and chased with unlabeled palmitate for the indicated time periods. Palmitoylation of immunoprecipitated R7BP was quantified as described in panel A. Results shown are the average of three experiments. D, R7BP palmitoylation turnover in cortical neurons. Neonatal rat cortical neurons were cultured for 14 days and labeled 6 h with [3H]palmitate in the absence (−) or presence (+) of CHX. R7BP was immunoprecipitated, resolved by SDS-PAGE and detected by fluorography and immunoblotting. Results shown are representative of five experiments. Error bars in panels A–C indicate S.E.
To determine whether palmitate turnover on FLAG-R7BP in Neuro2A cells is specific or occurs on many other proteins, we compared the palmitate labeling efficiency of transfected N-Ras and syntaxin 7 with or without CHX treatment. Like FLAG-R7BP, N-Ras was labeled similarly under both conditions (Fig. 1B), consistent with prior evidence that this protein undergoes extensive and rapid palmitate turnover (33). In contrast, syntaxin 7 was labeled ∼50% less efficiently in the presence versus absence of CHX (Fig. 1B), consistent with previous evidence indicating that palmitate turnover on this protein occurs relatively slowly (32). These differences indicated that rapid palmitoylation turnover on R7BP in Neuro2A cells is an intrinsic property of this protein rather than a general property of this neuronal cell line.
Consistent with this conclusion, we found that palmitate turnover also occurred on R7BP expressed endogenously in primary neurons. We analyzed [3H]palmitate labeling efficiency of R7BP in cortical neurons (14 DIV) isolated from neonatal rats. Results indicated that R7BP was labeled similarly in the absence or presence of CHX (Fig. 1D). Thus, all lines of evidence indicated that R7BP undergoes extensive and rapid palmitate turnover in neuronal cells, and that results from Neuro2A cells and primary cortical neurons are comparable, indicating that Neuro2A cells provide a suitable model to study R7BP palmitoylation.
DHHC2 Augments Biosynthetic and Turnover Palmitoylation of R7BP
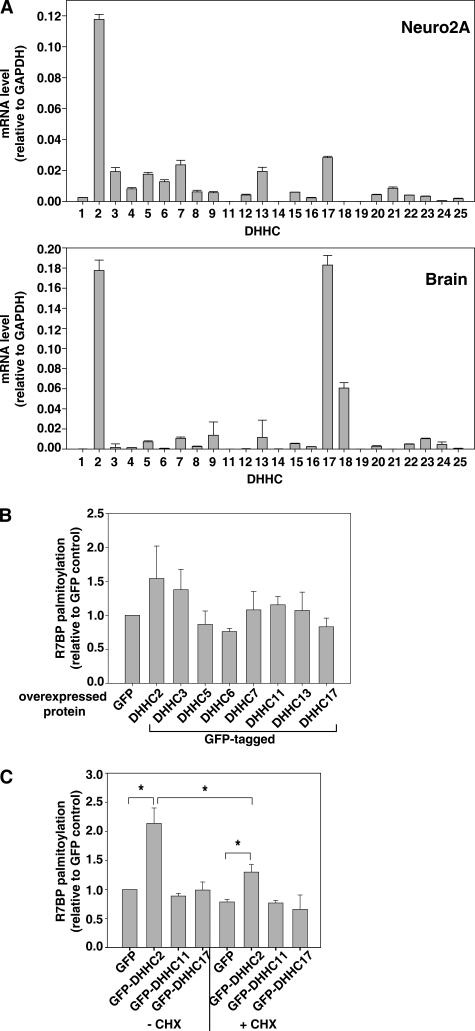
To identify mechanisms responsible for R7BP palmitoylation, we studied members of the 23 mouse or human genes encoding the DHHC family of palmitoyltransferases (34). Analysis of these enzymes would provide tools needed to identify mechanisms responsible for trafficking of R7BP. Results of quantitative RT-PCR experiments (Fig. 2A) indicated that Neuro2A cells stably transfected with FLAG-R7BP significantly express DHHC2, 3, 5, 6, 7, 13, and 17 but not other DHHC isoforms. Furthermore, we found that DHHC2, 17, and 18 mRNAs are highly expressed in mouse brain (Fig. 2A, lower panel). We therefore determined which of several DHHC proteins, when overexpressed in HEK293 cells, augments palmitoylation of co-transfected FLAG-R7BP. This approach has been used recently by several investigators to screen DHHC isoforms that augment palmitoylation of a given substrate (reviewed in Ref. 28). We found that DHHC2 overexpression increased palmitate labeling of FLAG-R7BP (Fig. 2B), whereas overexpression of other DHHC proteins had less effect. The results of expression and palmitoylation experiments therefore suggested that DHHC2 potentially mediates R7BP palmitoylation.
FIGURE 2.
Overexpression of DHHC2 augments de novo and turnover palmitoylation of R7BP. A, DHHC mRNA expression profiles in Neuro2A cells and brain. DHHC gene expression in Neuro2A cells stably expressing FLAG-R7BP (upper panel) and total mouse brain (lower panel) was quantified by RT-qPCR. Results shown are the average of four experiments. B, effects of DHHC isoform overexpression on R7BP palmitoylation in HEK293 cells. HEK293 cells co-transfected with FLAG-R7BP and the indicated GFP-tagged DHHC proteins were metabolically labeled with [3H]palmitate and analyzed as described in Fig. 1. Results shown are the average of 3–8 experiments. C, effects of DHHC2 or DHHC11 overexpression on de novo and turnover palmitoylation of R7BP in Neuro2A cells. GFP, GFP-DHHC2 or GFP-DHHC11 were transfected into Neuro2A cells stably expressing FLAG-R7BP, labeled with [3H]palmitate in the absence (−) or presence (+) of CHX, and analyzed for R7BP palmitoylation efficiency as described in Fig. 1. Results shown are the average of three experiments. *, p < 0.004. Error bars indicate S.E.
To explore the role of DHHC2 in neuronal cells where R7BP and its R7-Gβ5 binding partners are co-expressed, we studied the effects of overexpressing this enzyme or DHHC11 and DHHC17 as a controls in Neuro2A cells stably expressing FLAG-R7BP. Here, DHHC2 overexpression augmented the extent of FLAG-R7BP palmitoylation nearly 2-fold whereas overexpression of DHHC11 or DHHC17 had no significant effect (Fig. 2C and supplemental Fig. S1). Even in the presence of CHX, DHHC2 overexpression increased the palmitoylation of pre-existing FLAG-R7BP by ∼50% whereas DHHC11 or DHHC17 overexpression had no effect under these conditions (Fig. 2C). Therefore, overexpressed DHHC2 but not DHHC11 or DHHC17 augmented palmitoylation of pre-existing FLAG-R7BP. Because DHHC2 overexpression increased palmitoylation of FLAG-R7BP to a greater extent in control versus CHX-treated cells (Fig. 2C), this difference indicated that DHHC2 also augments palmitoylation of newly synthesized FLAG-R7BP. Thus, overexpressed DHHC2 can mediate both de novo and turnover palmitoylation of FLAG-R7BP in Neuro2A cells.
DHHC2 and R7BP Are Co-localized and -expressed
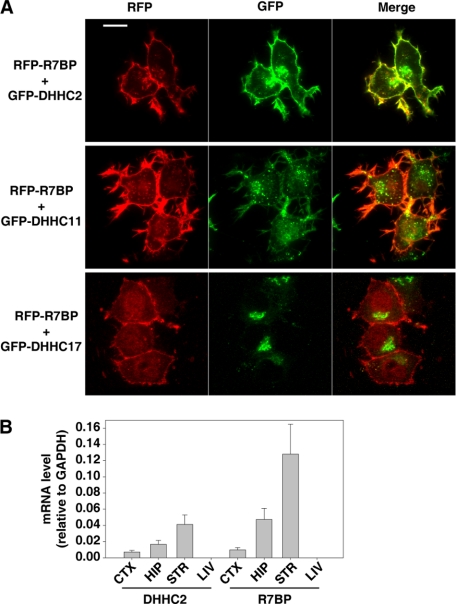
Because DHHC2 augments R7BP palmitoylation, the two proteins may be co-localized and -expressed. To test this possibility we co-transfected Neuro2A cells with GFP-tagged DHHC2 (GFP-DHHC2) and RFP-tagged R7BP (RFP-R7BP). For controls we co-transfected RFP-R7BP with GFP-DHHC11 or GFP-DHHC17, which did not augment R7BP palmitoylation. Results of confocal fluorescence microscopy experiments (Fig. 3A) indicated that GFP-DHHC2 co-localized extensively with RFP-R7BP on the plasma membrane whereas GFP-DHHC11 or -DHHC17 did not. Relatively little co-localization of RFP-R7BP with GFP-DHHC2, -DHHC11, or -DHHC17 was evident in intracellular compartments. Because R7BP is a neuron-specific protein that is highly expressed in several brain regions (23), we determined whether DHHC2 is also widely expressed in brain. Results of quantitative RT-PCR experiments indicated that DHHC2 and R7BP mRNA are expressed in cortex, hippocampus, and striatum (Fig. 3B). Therefore, DHHC2 and R7BP exhibit similar localization and expression patterns, as expected for a bona fide enzyme-substrate pair.
FIGURE 3.
Subcellular localization and expression of DHHC2 and R7BP. A, DHHC2 and R7BP localization in transfected Neuro2A cells. RFP-R7BP was co-transfected with GFP-DHHC2 (upper panel), GFP-DHHC11 (middle panel), or GFP-DHHC17 (lower panel). Cells were imaged by fluorescence confocal microscopy. Scale bar, 5 μm. B, expression of DHHC2 and R7BP mRNA in mouse tissues detected by RT-qPCR. CTX, cortex; HIP, hippocampus; STR, striatum; LIV, liver. Results shown are the average of three experiments. Error bars indicate S.E.
DHHC2 Silencing Blunts Palmitoylation and Induces Nuclear Localization of R7BP
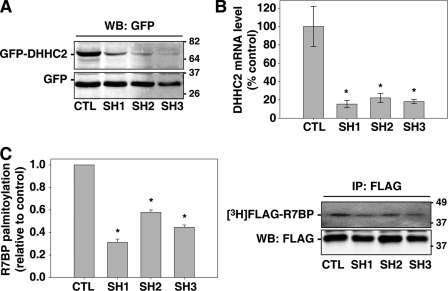
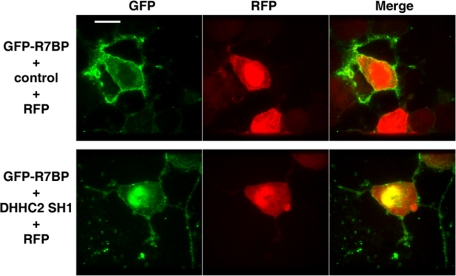
The preceding results suggested that DHHC2 could be an enzyme important for R7BP palmitoylation. We tested this hypothesis by determining the effects of silencing DHHC2 expression in Neuro2A cells. Initially we identified three shRNAs targeted to unique sequences in the 3′-UTR of the mouse DHHC2 mRNA that, relative to a scrambled shRNA control, reduced expression of transiently transfected GFP-tagged DHHC2 protein in HEK293 cells (Fig. 4A). Subsequently we stably expressed these DHHC2-specific shRNAs in Neuro2A cells stably expressing FLAG-R7BP. Each shRNA reduced endogenous DHHC2 mRNA expression by ∼80% (Fig. 4B) relative to the scrambled control shRNA, and decreased palmitate labeling of FLAG-R7BP by 50–70% (Fig. 4C). DHHC2 silencing also caused GFP-tagged R7BP to accumulate within the nucleus of Neuro2A cells (60% of cells; n = 46), whereas few cells (4%) expressing the control shRNA had detectable levels of GFP-R7BP in the nucleus (Fig. 5). Therefore, DHHC2 is critically important in Neuro2A cells for palmitoylation and plasma membrane localization of R7BP.
FIGURE 4.
DHHC2 silencing impairs R7BP palmitoylation. A, identification of shRNAs that silence DHHC2 expression. HEK293 cells were co-transfected with plasmids expressing GFP, GFP-DHHC2, and shRNAs (SH1, SH2, SH3) targeted to three sites within the DHHC2 3′-UTR or a control shRNA. Silencing of GFP-DHHC2 expression was analyzed by immunoblotting with GFP antibodies. B, silencing DHHC2 expression in Neuro2A cells. Neuro2A cells stably expressing FLAG-R7BP were transfected with control or the indicated DHHC2-specific shRNA-expressing plasmids. DHHC2 silencing was quantified by RT-qPCR relative to control shRNA-transfected cells. C, DHHC2 silencing impairs FLAG-R7BP palmitoylation. Neuro2A cells stably expressing FLAG-R7BP and the indicated control or DHHC2-specific shRNAs were analyzed by [3H]palmitate labeling as described in Fig. 1. Results shown are the average of three experiments. Error bars indicate S.E. *, p < e-10.
FIGURE 5.
DHHC2 silencing induces nuclear accumulation of R7BP. Neuro2A cells were transfected with plasmids expressing GFP-R7BP, RFP, and a control or DHHC2-specific shRNA (SH1), and analyzed by confocal fluorescence microscopy. Results shown are representative of three experiments. Scale bar, 5 μm.
Gi/o Signaling Regulates Palmitate Turnover and Trafficking of R7BP
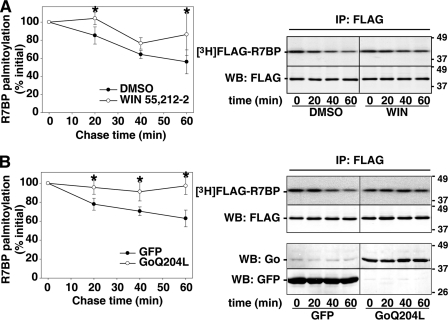
Having demonstrated that R7BP undergoes rapid and extensive palmitate turnover in neuronal cells via DHHC2, our final objective was to determine whether palmitate turnover and trafficking of R7BP occurs in a constitutive or regulated manner. Addressing this question would directly test the hypothesis that R7BP functions as a palmitoylation-regulated plasma membrane-nuclear shuttling protein for Gi/o GAP complexes composed of R7-Gβ5 heterodimers, potentially resolving conflicting evidence from previous studies that simply examined steady-state palmitoylation and localization patterns. We determined whether R7BP palmitoylation and localization are altered when Gi/o signaling is activated or inactivated. Initially we analyzed palmitate turnover on FLAG-R7BP stably expressed in Neuro2A cells that were stimulated by an agonist (WIN 55,212–2) for Gi/o-coupled CB1 cannabinoid receptors, which are expressed endogenously by these cells (35). Results of palmitate pulse-chase labeling experiments indicated that CB1 receptor activation stabilized FLAG-R7BP palmitoylation by inhibiting the rate of depalmitoylation (Fig. 6A). A similar inhibitory effect on FLAG-R7BP depalmitoylation was observed in pulse-chase experiments using Neuro2A cells transiently transfected with constitutively active Goα-Q204L (Fig. 6B).
FIGURE 6.
Gi/o signaling inhibits R7BP depalmitoylation. A, activation of Gi/o-coupled CB1 cannabinoid receptors inhibits R7BPdepalmitoylation. Neuro2A cells stably expressing FLAG-R7BP were pulse-labeled with [3H]palmitate and chased for the indicated time with unlabeled palmitate in the absence (DMSO) or presence of a cannabinoid receptor agoinst (10 μm WIN 55,212–2) (WIN). R7BP palmitoylation was quantified as described in Fig. 1C. B, activated Goα inhibits R7BP depalmitoylation. Neuro2A cells stably expressing FLAG-R7BP were transfected with a plasmid expressing constitutively active Goα (Q204L), and pulse-chase labeled with [3H]palmitate as described in Fig. 1C. Results shown are the average of three experiments. Error bars indicate S.E. *, p < 0.04.
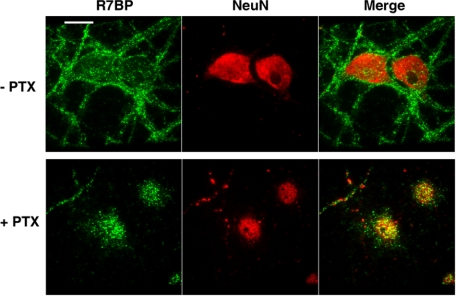
The preceding results indicated that CB1 receptor activation or activated Goα modestly decrease the rate of R7BP depalmitoylation relative to control cells. Potentially, such modest effects could occur because agonists for Gi/o-coupled receptors are present in serum-containing media used for palmitate labeling, thus providing substantial Gi/o signaling that slows the rate of R7BP depalmitoylation. Thus, as an alternative we determined whether blocking Gi/o signaling would have a larger and opposite effect by promoting R7BP depalmitoylation and allowing R7BP to shuttle into the nucleus. To test this idea and extend the analysis to primary neurons, we studied the localization of endogenous R7BP in rat primary cortical neurons that were treated or not with PTX, which blocks receptor-mediated activation of Gi/o-class G proteins. In control neurons stained with affinity-purified R7BP antibodies, endogenously expressed R7BP localized to the plasma membrane of neuronal soma and dendrites (Fig. 7). As expected from previous studies (26), nuclear-localized R7BP was undetectable under these control conditions (n = 114 neurons). In striking contrast, we found in PTX-treated neurons (n = 89) that R7BP is readily detected within nuclei (60% of neurons; Fig. 7). R7BP antibody staining was specific because low, background staining was observed in glia, which do not express R7BP (26), and in neurons stained with antigen-blocked R7BP antibodies (supplemental Fig. S2). Why PTX did not cause R7BP accumulation in the nuclei of all neurons is unknown. Perhaps R7BP localization is regulated differentially in various neuron classes present in primary cultures, or dynamic processes such as R7BP nuclear import/export still occur in PTX-treated neurons and affect the probability of detecting R7BP within the nucleus. Nevertheless, the effects of activating versus blocking Gi/o signaling indicated that palmitoylation of R7BP is a dynamic and regulated process that controls the ability of this protein to shuttle from the plasma membrane to the nucleus in neuronal cells.
FIGURE 7.
Blocking Gi/o signaling in primary neurons induces nuclear accumulation of R7BP. Neonatal rat cortical neurons (DIV19) were treated with or without PTX to block Gi/o activation and stained with affinity-purified antibodies specific for R7BP and a neuron-specific nuclear protein (NeuN). Results shown are representative of four experiments. Scale bar, 5 μm.
DISCUSSION
Here we have shown that R7BP palmitoylation and subcellular localization in neuronal cells are dynamic processes that are controlled in part by the palmitoyltransferase DHHC2 and Gi/o signaling. As discussed below, these findings support the hypothesis that R7BP functions in neuronal cells as a palmitoylation-regulated shuttling protein for Gi/o-specific R7-Gβ5 GAP complexes, and provide new understanding of protein palmitoylation mechanisms and their roles in controlling G protein signaling networks.
R7BP Shuttling of R7-Gβ5 GAP Complexes in Neuronal Cells
Initial evidence suggesting that R7BP functions as a palmitoylation-regulated protein that shuttles GAP complexes (R7-Gβ5 heterodimers) between the plasma membrane and nucleus was based on overexpression studies and steady state measurements of palmitoylation and localization (23, 36). Although subsequent studies of brain extracts or slices failed to detect a significant pool of nuclear R7BP at steady state (19, 26, 27), these investigations did not test the shuttling hypothesis because they did not examine whether R7BP palmitoylation or localization is static or dynamic. Similar factors limit interpretation of recent studies of neurons from R7BP knock-out mice showing that steady state nuclear accumulation of RGS7 and Gβ5 depends in part on R7BP (27). In contrast, here we have provided several lines of evidence directly supporting the hypothesis that R7BP is a palmitoylation-regulated shuttling protein in neuronal cells: R7BP undergoes extensive palmitate turnover; the palmitoyltransferase DHHC2 mediates de novo and turnover palmitoylation of R7BP; DHHC2 silencing impairs R7BP palmitoylation and causes nuclear accumulation of R7BP; and Gi/o signaling inhibits R7BP depalmitoylation whereas blockade of Gi/o signaling enables R7BP to accumulate in the nucleus. These findings suggest that Gi/o signaling in vivo may stabilize R7BP palmitoylation and thus limit steady state accumulation of R7BP within the nucleus. They also suggest that changes in Gi/o signaling activity may alter the dynamics of R7BP palmitoylation and thus affect trafficking.
Because R7-Gβ5 heterodimers are obligate binding partners of R7BP (26), the palmitoylation and trafficking mechanisms reported herein are likely to impact subcellular targeting, shuttling, and function of the R7BP-bound pool of GAP complexes in neuronal cells. Complexes containing RGS9-2 are likely to be regulated particularly effectively by these mechanisms because this striatum-enriched R7 splice variant requires R7BP for proteolytic stability and subcellular targeting (37). However, because other R7-Gβ5 dimers (e.g. RGS7-Gβ5) exist in R7BP-bound and -free pools (26), trafficking and function of only the R7BP-bound pool would be affected by the regulatory mechanisms we have identified here. Indeed, the absence of R7BP only partially affects association of RGS7-Gβ5 dimers with membrane or nuclear fractions of brain extracts (27). Thus, R7BP-independent localization mechanisms of certain R7-Gβ5 complexes in neuronal cells remain to be elucidated.
Functions of DHHC2
To our knowledge, our studies of DHHC2 and R7BP are the first to indicate that a single DHHC isoform can mediate both de novo and turnover palmitoylation of a specific substrate. DHHC2 is likely to have diverse functions because it can palmitoylate several substrates, including PSD-95 (38, 39), endothelial nitric oxide synthase (40), cytoskeleton-associated protein 4 (41), tetraspanins (42), GAP43 (38), and Giα2 (43). It is intriguing that DHHC2 participates in palmitoylation turnover on R7BP and PSD-95, which both localize postsynaptically. DHHC2 localization to Golgi elements in dendrites and activity-sensitive recruitment to postsynaptic sites (39) makes this enzyme well suited to participate in signal-regulated palmitoylation. Furthermore, the ability of DHHC2 to palmitoylate both Giα2 and R7BP suggests that this enzyme has critical roles in regulating the action of Gi/o-coupled modulatory neurotransmitter receptors by mechanisms discussed below. However, because substrate proteins often can be palmitoylated by various DHHC isoforms (28, 34, 44), we anticipate that R7BP could be palmitoylated by enzymes in addition to DHHC2.
Control of GAP-regulated Gi/o Signaling by Palmitate Turnover
Taken together with previous evidence, our findings broaden understanding of mechanisms whereby agonist regulation of palmitate turnover facilitates GAP regulation of Gi/o signaling kinetics. As shown previously, cell stimulation by a Gi/o-coupled receptor agonist augments depalmitoylation of Giα (45), increasing the pool of depalmitoylated Giα subunits that are preferred substrates for several RGS protein GAPs (46). We have found that Gi/o signaling inhibits R7BP depalmitoylation, which is expected to stabilize or recruit R7BP-bound GAP complexes at the plasma membrane. Thus, the concerted yet opposite effects of agonist stimulation on Giα and R7BP depalmitoylation potentially act in concert to facilitate GAP complex regulation of Gi signaling kinetics. In a variation of this theme, constitutive palmitate turnover on Goα (47) would provide the pool of optimal GAP substrate, and agonist stimulation would inhibit R7BP depalmitoylation to recruit GAP complexes to the plasma membrane.
Mechanisms enabling receptor agonist stimulation to inversely regulate Giα and R7BP depalmitoylation are unknown. However, a simple hypothesis is that the accessibility of protein-palmitoylthioesterase activity toward Giα and R7BP is augmented and inhibited, respectively, upon agonist stimulation. APT1, the only thioesterase known thus far to depalmitoylate protein substrates (48), is presumably involved in this process. Interestingly, APT1 can exhibit large differences in activity toward distinct substrates (49, 50). Thus, studies of APT1 action toward Giα and R7BP might reveal how a single stimulus can have opposing effects on depalmitoylation rates of distinct proteins.
A further function of palmitate turnover on R7BP may be to shuttle GAP complexes between sites of low and high Gi/o activity on the plasma membrane, potentially providing spatial control of Gi/o signaling kinetics. R7BP-bound GAP complexes could be recruited to sites of high Gi/o activity because R7BP depalmitoylation would be inhibited there. As Gi/o activity at these sites wanes, R7BP depalmitoylation is expected to increase, releasing R7BP for repalmitoylation and targeting to other sites where relatively high Gi/o activity occurs.
Perhaps most provocatively, as neuronal Gi/o signaling desensitizes, depalmitoylated R7BP may accumulate at levels sufficient to shuttle GAP complexes into the nucleus. Such a process could provide a mechanism of transducing desensitization signals between the plasma membrane and nucleus, or promoting resensitization by sequestering GAP complexes within the nucleus. Indeed, as cells become resensitized these mechanisms could be reversed easily because unpalmitoylated R7BP shuttles in and out of the nucleus (36), enabling it to be repalmitoylated and targeted to the plasma membrane as Gi/o signaling resumes. In summary, our discovery that R7BP palmitoylation is dynamic and regulated reveals a novel and versatile mechanism capable of providing spatio-temporal control of Gi/o-coupled receptor signaling networks mediating the action of scores of modulatory neurotransmitters and drugs of abuse. It will be intriguing to dissect the roles of palmitate cycling once neuron-based assays of R7BP function are established.
Acknowledgments
We thank members of the Blumer and Linder Laboratories, especially Ben Jennings and Kevin Kaltenbronn, for support and discussion. We also thank Drs. Sheila Stewart for providing the pLKO.1 plasmid and John Cooper for access to confocal microscopy facilities.
This work was supported, in whole or in part, by Grants GM044592 and HL075632 (to K. J. B.), and GM051466 (to M. E. L.) from the National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PTX
- pertussis toxin
- CHX
- cycloheximide
- DHHC
- palmitoyltransferase bearing an aspartic acid, histidine, histidine, cysteine motif
- GAP
- GTPase-activating protein
- RGS
- regulator of G protein signaling
- R7BP
- RGS7 family-binding protein
- RT-qPCR
- reverse transcriptase-coupled quantitative polymerase chain reaction.
REFERENCES
- 1. Gainetdinov R. R., Premont R. T., Bohn L. M., Lefkowitz R. J., Caron M. G. (2004) Annu. Rev. Neurosci. 27, 107–144 [DOI] [PubMed] [Google Scholar]
- 2. Wettschureck N., Offermanns S. (2005) Physiol. Rev. 85, 1159–1204 [DOI] [PubMed] [Google Scholar]
- 3. Anderson G. R., Posokhova E., Martemyanov K. A. (2009) Cell. Biochem. Biophys 54, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hooks S. B., Martemyanov K., Zachariou V. (2008) Biochem. Pharmacol. 75, 76–84 [DOI] [PubMed] [Google Scholar]
- 5. Cabrera J. L., de Freitas F., Satpaev D. K., Slepak V. Z. (1998) Biochem. Biophys. Res. Commun. 249, 898–902 [DOI] [PubMed] [Google Scholar]
- 6. Posner B. A., Gilman A. G., Harris B. A. (1999) J. Biol. Chem. 274, 31087–31093 [DOI] [PubMed] [Google Scholar]
- 7. Snow B. E., Krumins A. M., Brothers G. M., Lee S. F., Wall M. A., Chung S., Mangion J., Arya S., Gilman A. G., Siderovski D. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hooks S. B., Waldo G. L., Corbitt J., Bodor E. T., Krumins A. M., Harden T. K. (2003) J. Biol. Chem. 278, 10087–10093 [DOI] [PubMed] [Google Scholar]
- 9. Chen C. K., Burns M. E., He W., Wensel T. G., Baylor D. A., Simon M. I. (2000) Nature 403, 557–560 [DOI] [PubMed] [Google Scholar]
- 10. Nishiguchi K. M., Sandberg M. A., Kooijman A. C., Martemyanov K. A., Pott J. W., Hagstrom S. A., Arshavsky V. Y., Berson E. L., Dryja T. P. (2004) Nature 427, 75–78 [DOI] [PubMed] [Google Scholar]
- 11. Rahman Z., Schwarz J., Gold S. J., Zachariou V., Wein M. N., Choi K. H., Kovoor A., Chen C. K., DiLeone R. J., Schwarz S. C., Selley D. E., Sim-Selley L. J., Barrot M., Luedtke R. R., Self D., Neve R. L., Lester H. A., Simon M. I., Nestler E. J. (2003) Neuron 38, 941–952 [DOI] [PubMed] [Google Scholar]
- 12. Zachariou V., Georgescu D., Sanchez N., Rahman Z., DiLeone R., Berton O., Neve R. L., Sim-Selley L. J., Selley D. E., Gold S. J., Nestler E. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13656–13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen F. S., Shim H., Morhardt D., Dallman R., Krahn E., McWhinney L., Rao A., Gold S. J., Chen C. K. (2010) Invest. Ophthalmol. Vis. Sci. 51, 686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mojumder D. K., Qian Y., Wensel T. G. (2009) J. Neurosci. 29, 7753–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J., Jeffrey B. G., Morgans C. W., Burke N. S., Haley T. L., Duvoisin R. M., Brown R. L. (2010) Invest Ophthalmol Vis. Sci. 51, 1121–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Posokhova E., Wydeven N., Allen K. L., Wickman K., Martemyanov K. A. (2010) Circ. Res. 107, 1350–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jayaraman M., Zhou H., Jia L., Cain M. D., Blumer K. J. (2009) Trends Pharmacol. Sci. 30, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu G., Wensel T. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9755–9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song J. H., Song H., Wensel T. G., Sokolov M., Martemyanov K. A. (2007) Mol. Cell. Neurosci. 35, 311–319 [DOI] [PubMed] [Google Scholar]
- 20. Keresztes G., Martemyanov K. A., Krispel C. M., Mutai H., Yoo P. J., Maison S. F., Burns M. E., Arshavsky V. Y., Heller S. (2004) J. Biol. Chem. 279, 1581–1584 [DOI] [PubMed] [Google Scholar]
- 21. Jeffrey B. G., Morgans C. W., Puthussery T., Wensel T. G., Burke N. S., Brown R. L., Duvoisin R. M. (2010) Vis. Neurosci. 27, 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martemyanov K. A., Yoo P. J., Skiba N. P., Arshavsky V. Y. (2005) J. Biol. Chem. 280, 5133–5136 [DOI] [PubMed] [Google Scholar]
- 23. Drenan R. M., Doupnik C. A., Boyle M. P., Muglia L. J., Huettner J. E., Linder M. E., Blumer K. J. (2005) J. Cell. Biol. 169, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song J. H., Waataja J. J., Martemyanov K. A. (2006) J. Biol. Chem. 281, 15361–15369 [DOI] [PubMed] [Google Scholar]
- 25. Anderson G. R., Cao Y., Davidson S., Truong H. V., Pravetoni M., Thomas M. J., Wickman K., Giesler G. J., Jr., Martemyanov K. A. (2010) Neuropsychopharmacology 35, 1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grabowska D., Jayaraman M., Kaltenbronn K. M., Sandiford S. L., Wang Q., Jenkins S., Slepak V. Z., Smith Y., Blumer K. J. (2008) Neuroscience 151, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panicker L. M., Zhang J. H., Posokhova E., Gastinger M. J., Martemyanov K. A., Simonds W. F. (2010) J. Neurochem. 113, 1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iwanaga T., Tsutsumi R., Noritake J., Fukata Y., Fukata M. (2009) Prog. Lipid Res. 48, 117–127 [DOI] [PubMed] [Google Scholar]
- 29. Kim J., Castellano J. M., Jiang H., Basak J. M., Parsadanian M., Pham V., Mason S. M., Paul S. M., Holtzman D. M. (2009) Neuron 64, 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaslow H. R., Burns D. L. (1992) FASEB. J. 6, 2684–2690 [DOI] [PubMed] [Google Scholar]
- 31. Choy E., Chiu V. K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I. E., Philips M. R. (1999) Cell 98, 69–80 [DOI] [PubMed] [Google Scholar]
- 32. He Y., Linder M. E. (2009) J. Lipid Res. 50, 398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P. I. H. (2005) Science 307, 1746–1752 [DOI] [PubMed] [Google Scholar]
- 34. Smotrys J. E., Linder M. E. (2004) Annu. Rev. Biochem. 73, 559–587 [DOI] [PubMed] [Google Scholar]
- 35. He J. C., Neves S. R., Jordan J. D., Iyengar R. (2006) Can. J. Physiol. Pharmacol. 84, 687–694 [DOI] [PubMed] [Google Scholar]
- 36. Drenan R. M., Doupnik C. A., Jayaraman M., Buchwalter A. L., Kaltenbronn K. M., Huettner J. E., Linder M. E., Blumer K. J. (2006) J. Biol. Chem. 281, 28222–28231 [DOI] [PubMed] [Google Scholar]
- 37. Anderson G. R., Semenov A., Song J. H., Martemyanov K. A. (2007) J. Biol. Chem. 282, 4772–4781 [DOI] [PubMed] [Google Scholar]
- 38. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 39. Noritake J., Fukata Y., Iwanaga T., Hosomi N., Tsutsumi R., Matsuda N., Tani H., Iwanari H., Mochizuki Y., Kodama T., Matsuura Y., Bredt D. S., Hamakubo T., Fukata M. (2009) J. Cell. Biol. 186, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fernández-Hernando C., Fukata M., Bernatchez P. N., Fukata Y., Lin M. I., Bredt D. S., Sessa W. C. (2006) J. Cell. Biol. 174, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Planey S. L., Keay S. K., Zhang C. O., Zacharias D. A. (2009) Mol. Biol. Cell. 20, 1454–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sharma C., Yang X. H., Hemler M. E. (2008) Mol. Biol. Cell. 19, 3415–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsutsumi R., Fukata Y., Noritake J., Iwanaga T., Perez F., Fukata M. (2009) Mol. Cell. Biol. 29, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fukata Y., Fukata M. (2010) Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 45. Chen C. A., Manning D. R. (2000) J. Biol. Chem. 275, 23516–23522 [DOI] [PubMed] [Google Scholar]
- 46. Tu Y., Wang J., Ross E. M. (1997) Science 278, 1132–1135 [DOI] [PubMed] [Google Scholar]
- 47. Chisari M., Saini D. K., Kalyanaraman V., Gautam N. (2007) J. Biol. Chem. 282, 24092–24098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeidman R., Jackson C. S., Magee A. I. (2009) Mol. Membr Biol. 26, 32–41 [DOI] [PubMed] [Google Scholar]
- 49. Duncan J. A., Gilman A. G. (2002) J. Biol. Chem. 277, 31740–31752 [DOI] [PubMed] [Google Scholar]
- 50. Yeh D. C., Duncan J. A., Yamashita S., Michel T. (1999) J. Biol. Chem. 274, 33148–33154 [DOI] [PubMed] [Google Scholar]