Abstract
Macroautophagy is a catabolic process by which cytosolic components are sequestered by double membrane vesicles called autophagosomes and sorted to the lysosomes/vacuoles to be degraded. Saccharomyces cerevisiae has adapted this mechanism for constitutive transport of the specific vacuolar hydrolases aminopeptidase I (Ape1) and α-mannosidase (Ams1); this process is called the cytoplasm to vacuole targeting (Cvt) pathway. The precursor form of Ape1 self-assembles into an aggregate-like structure in the cytosol that is then recognized by Atg19 in a propeptide-dependent manner. The interaction between Atg19 and autophagosome-forming machineries allows selective packaging of the Ape1-Atg19 complex by the autophagosome-like Cvt vesicle. Ams1 also forms oligomers and utilizes the Ape1 transport system by interacting with Atg19. Although the mechanism of selective transport of the Cvt cargoes has been well studied, it is unclear whether proteins other than Ape1 and Ams1 are transported via the Cvt pathway. We describe here that aspartyl aminopeptidase (Yhr113w/Ape4) is the third Cvt cargo, which is similar in primary structure and subunit organization to Ape1. Ape4 has no propeptide, and it does not self-assemble into aggregates. However, it binds to Atg19 in a site distinct from the Ape1- and Ams1-binding sites, allowing it to “piggyback” on the Ape1 transport system. In growing conditions, a small portion of Ape4 localizes in the vacuole, but its vacuolar transport is accelerated by nutrient starvation, and it stably resides in the vacuole lumen. We propose that the cytosolic Ape4 is redistributed to the vacuole when yeast cells need more active vacuolar degradation.
Keywords: Autophagy, Hydrolases, Membrane Trafficking, Protein Targeting, Yeast
Introduction
Macroautophagy (hereafter autophagy) is a dynamic membrane trafficking process responsible for bulk degradation of cytoplasmic components, including organelles, and is evolutionarily conserved in eukaryotic cells. In this process, a double membrane vesicle, termed an autophagosome, selectively or non-selectively sequesters cytoplasmic materials and fuses to the lysosome/vacuole to deliver the cargo for degradation. Until recently, autophagy has been thought to be a catabolic pathway in response to nutrient deprivation; however, there is growing evidence that it is also involved in various biological events such as development, aging, pathogen resistance, and tumor suppression (1, 2). More recently, it has been recognized that selective types of autophagy contribute to innate immunity and prevent neurodegenerative disorders such as Alzheimer and Parkinson diseases (3–7).
In the yeast Saccharomyces cerevisiae, 34 autophagy-related (ATG)2 genes have been identified as important genes for selective and non-selective autophagy. One of the selective autophagic pathways in yeast is the cytoplasm to vacuole targeting (Cvt) pathway that delivers two hydrolytic enzymes, aminopeptidase I (Ape1) and α-mannosidase (Ams1), to the vacuole. Although the genetic and morphological features of the Cvt pathway are similar to those of autophagy, their biological roles seem to be opposed to each other: the Cvt pathway is a constitutive biosynthetic route for vacuolar enzymes, whereas autophagy is a catabolic process primarily induced in nutrient starvation conditions (8–10). Ape1, the major Cvt cargo, is synthesized on unattached ribosomes in the cytosol as a precursor form (prApe1) with a cleavable propeptide consisting of 45 amino acid residues at the N terminus. The Ape1 propeptide differs in function from those typically found in proteases such as carboxypeptidase Y and proteinase A that play roles in helping protein folding and inactivating proteases until they reach the appropriate site; Ape1 lacking its propeptide properly folds and assembles into a homododecamer as fast as prApe1 (11). Moreover, the prApe1 propeptide is unlikely to regulate its enzymatic activity (12). Instead, the propeptide plays an important role in selective incorporation of prApe1 into Cvt vesicles (13). The prApe1 dodecamer self-assembles into a higher order structure called the Ape1 complex (13–16). In the context of the Ape1 complex, the propeptide links prApe1 to Atg19. The latter serves as a molecular tag that is further recognized by Atg11, allowing the Ape1-Atg19 complex (termed the Cvt complex) to localize at the pre-autophagosomal structure/phagophore assembly site (PAS), a putative site of autophagosome/Cvt vesicle formation (13, 17). Because Atg11 and Atg19 further associate with Atg9 and Atg8, PAS components required for phagophore expansion, the Cvt complex acts as a scaffold for the formation of a double membrane-bound vesicle so that the vesicle excludes cytosolic components (13, 18–22). This mechanism could also ensure efficient packaging with a limited membrane supply in nutrient-rich conditions. Although Atg9 and Atg11 seem to be released from the sequestering vesicles prior to completion, Atg19 travels to the vacuole as a part of the Cvt complex together with Atg8 and is degraded (13, 23). Meanwhile, the prApe1 propeptide is removed by vacuolar proteases, and the Ape1 complex disassembles back into dodecamers. Ams1, the second Cvt cargo protein, is also synthesized in the cytosol as a homo-oligomeric enzyme but does not form a higher order structure by itself in contrast to prApe1 (24). Instead, it is concentrated on the Ape1 complex through its interaction with Atg19, and thus its import to the vacuole is largely dependent on prApe1 in nutrient-rich conditions (13). As Ams1 is dispensable for prApe1 transport to the vacuole, Ams1 just exploits the prApe1 import system to achieve its efficient transport.
Recently, aspartyl aminopeptidase from S. cerevisiae was purified, characterized, and identified as a product of the YHR113W gene; the protein is closely related to mammalian aspartyl aminopeptidases (EC 3.4.11.21) (25, 26). Both Yhr113w and Ape1 belong to the M18 family of metalloproteases and share similar structural features: their amino acid sequences show 32% identity, and both enzymes form 12-subunit homo-oligomeric complexes. Yhr113w, however, lacks the N-terminal domain present in prApe1, thus leading to the proposition that it might function in the cytosol (26). In parallel with the biochemical studies, comprehensive yeast two-hybrid analyses suggest that Yhr113w can associate with Atg19 and prApe1. This raises the possibility that Yhr113w could be transported to the vacuole through the Cvt pathway.
In this study, we found that a portion of Yhr113w is selectively transported to the vacuole via the Cvt pathway in nutrient-rich conditions and that nitrogen starvation accelerates its vacuolar transport. We also address the mechanism of selective transport of Yhr113w and discuss the biological significance of its transport from the cytoplasm to the vacuole.
EXPERIMENTAL PROCEDURES
Strains, Media, and Antibodies/Antisera
The yeast S. cerevisiae strains used in this study are listed in Table 1. Gene disruptions were performed by the method described by Gueldener et al. (27). The entire open reading frame was replaced by the Escherichia coli kanr, Kluyveromyces lactis LEU2, or Schizosaccharomyces pombe his5+ gene using PCR primers containing ∼50 nucleotides identical to the regions flanking the open reading frame. Cells were grown at 30 °C in either YPD (1% yeast extract, 2% peptone, and 2% glucose) or in synthetic complete (SC) medium (0.67% yeast nitrogen base without amino acids, 2% glucose, and 0.2% dropout mixture) as described by Amberg et al. (28). For nitrogen starvation, SD(-N) medium (0.17% yeast nitrogen base without ammonium sulfate and amino acids and 2% glucose) was used. Antiserum to Ape1 has been described (9). Antibody to carboxypeptidase Y (Prc1) was raised against the purified protein (Oriental Yeast, Osaka, Japan) deglycosylated with endoglycosidase Hf (New England Biolabs, Tokyo, Japan). For production of anti-Yhr113w/Ape4 antibody, a recombinant Yhr113w/Ape4 protein was expressed in E. coli BL21 (DE3), purified from an SDS-polyacrylamide gel, and used for immunization of a Japanese White rabbit. Anti-Yhr113w/Ape4 antibody was affinity-purified with antigen immobilized on Sepharose. Anti-YFP (JL-8), anti-phosphoglycerate kinase (Pgk1) (22C5D8), and anti-HA (F-7) were purchased from Takara Bio (Otsu, Japan), Invitrogen, and Santa Cruz Biotechnology (Santa Cruz, CA), respectively.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| SEY6210 | MATa his3-Δ200 leu2–3,112 lys2–801 trp1-Δ901 ura3–52 | |
| suc2-Δ9 GAL | Robinson et al. (47) | |
| WHY1 | SEY6210; atg1Δ::his5+ | Shintani et al. (13) |
| YTS147 | SEY6210; atg11Δ::LEU2 K.l. | This study |
| SSY31 | SEY6210; atg19Δ::his5+ | Scott et al. (43) |
| YMY103 | SEY6210; ape1Δ::LEU2 K.l. | This study |
| YMY105 | SEY6210; pep4Δ::kanr prb1Δ::LEU2 K.l. | This study |
| YMY106 | SEY6210; pep4Δ::kanr prb1Δ::LEU2 K.l. atg19Δ::his5+ | This study |
| YMY109 | SEY6210; atg1Δ::his5+atg19Δ::his5+ | This study |
| YMY111 | SEY6210; ape4Δ::kanr | This study |
| YMY112 | SEY6210; ape4Δ::kanr atg1Δ::his5+ | This study |
| PJ69-4A | MATa trp1-Δ901 leu2–3,112 ura3–52 his3-Δ200 gal4Δ | |
| gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (29) | |
| YTS110 | PJ69-4A; ape1Δ::his5+ | Shintani et al. (13) |
| YTS111 | PJ69-4A; atg19Δ::his5+ | Shintani et al. (13) |
Plasmids Used in This Study
The plasmids and oligonucleotides used in this study are listed in supplemental Tables S1 and S2, respectively. The following procedure was used to make green fluorescent protein (GFP)- and triple hemagglutinin (HA)-tagged constructs for Yhr113w. The entire open reading frame of YHR113W/APE4 with 5′- and 3′-untranslated regions was PCR-amplified using oligonucleotides oTAKA35 and oTAKA36 and then ligated to SacI/SalI-digested pRS416 and pRS424 to generate pTS547 and pTS622, respectively. The BamHI restriction site was introduced just after the first codon of APE4 on pTS547 with a QuikChange site-directed mutagenesis kit (Stratagene) using oligonucleotides oTAKA37 and oTAKA38 to generate pTS548. The DNA fragment encoding GFP or HA cassette with BamHI (for GFP) or BglII (for HA) sites on both sides was then ligated to the newly generated BamHI site to make pGFP-Ape4 (pTS551) and p3HA-Ape4 (pTS549) constructs. The plasmid pTS551 was digested with SacI and SalI to obtain the DNA fragment encoding GFP-Ape4, which was then subcloned into pRS414 to generate pTS572. Similarly, the DNA fragment encoding HA3-Ape4 was subcloned into pRS415 to construct pMY101. pTS548 was digested with BamHI and SalI, and the resulting APE4 fragment was subcloned into pGAD-C1, pGBDU-C1 (29), pCuGFP(416) (30), and pUC18 to generate pGAD-Ape4 (pTS577), pGBDU-Ape4 (pTS578), pTS555, and pTS575, respectively. The DNA fragment encoding AMS1 was cloned into pGBDU-C1 to make pTS617. The SalI fragment of YHR113W/APE4 from pTS575 was subcloned into pET15b to generate pTS576. For the C-terminal deletion of Atg19, the DNA fragments were PCR-amplified with primers oTAKA11 and MY127 for Atg19(1–269) and oTAKA11 and MY128 for Atg19(1–203). The resulting PCR products were digested with EcoRI and SalI followed by ligation to pRS416-CuProtA to generate pTS620 and pTS621, respectively. Alternatively, primers oTAKA1 and oTAKA5 for Atg19(1–269) and oTAKA1 and oTAKA14 for Atg19(1–203) were used to amplify the DNA fragments that were ligated to pGAD-C2 (29) to obtain pTS433 and pTS443. For internal deletion of Atg19, the overlap extension method was used followed by in vivo gap repair to construct pMY102, pYM103, pYM104, pYM105, pYM106, pYM107, pYM108, and pYM109. The combinations of primers MY136 and MY137 (Atg19(Δ204–269)), MY138 and MY139 (Atg19(Δ204–225)), MY140 and MY141 (Atg19(Δ226–247)), and MY142 and MY143 (Atg19(Δ248–269)) were used. Plasmids p3HA-Ams1 (pTS544), pGBDU-Ape1 (pTS425), and pRFP-Ape1 (pTS559) have been described elsewhere (13, 31).
Vacuole Preparation and Protease Protection Assay
Vacuoles were isolated as described previously (24). The protease protection assay was essentially performed as described previously (32). In brief, 100 μl of the vacuole preparations in PS200 (20 mm PIPES-KOH, pH 6.8, and 0.2 m sorbitol) were diluted 2-fold in a solution containing either 100 μg/ml proteinase K, 100 μg/ml proteinase K plus 1% Triton X-100, or 1% Triton X-100 or in distilled water. The samples were incubated for 30 min on ice, and then the reactions were stopped by adding 200 μl of 20% TCA. After centrifugation, the resulting pellets were washed with acetone twice, dissolved in 100 μl of MURB (50 mm sodium phosphate, 25 mm MES-NaOH, 1% SDS, 3 m urea, and 1 mm sodium azide) plus 5% 2-mercaptoethanol, and treated at 55 °C for 15 min. The samples were subjected to immunoblot analysis with anti-Prc1, anti-Ape1, anti-Ape4, anti-Pgk1, and anti-HA antisera.
Protein A Affinity Isolation
Protein A (PA)-Atg19 affinity isolation was carried out essentially as described previously (13). In brief, 50 A600 nm units of yeast cells were lysed with glass beads in 1 ml of lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm EDTA, 0.5% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and Complete EDTA-free protease inhibitor (Roche Applied Science)). After centrifugation at 10,000 × g for 10 min at 4 °C, the resulting supernatant fraction was incubated with IgG-Sepharose 6 Fast Flow (GE Healthcare) for 1 h at 4 °C. After washing with lysis buffer four times, the bound materials were eluted by incubating the beads at 55 °C for 15 min in MURB plus 5% 2-mercaptoethanol. The samples were subjected to immunoblot analysis and probed with anti-Ape1 and anti-HA antisera.
Fluorescence Microscopy
Cells expressing fluorescent protein-fused chimeras were grown in SC medium and, if necessary, further incubated in SD(-N) medium for 3 h. Fluorescence microscopy analysis was performed with an Olympus IX71 fluorescence microscope equipped with ORCA-Flash2.8 (Hamamatsu Photonics K.K., Hamamatsu, Japan).
RESULTS
Cvt Pathway Is Responsible for Vacuolar Localization of Ape4
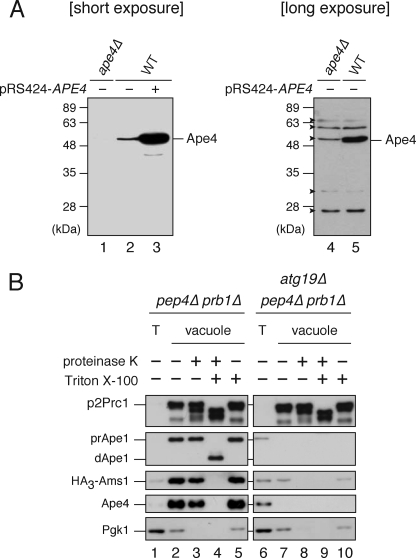
The protein encoded by YHR113W was characterized as a yeast aspartyl aminopeptidase with a substrate specificity and amino acid sequence similar to those of mammalian aspartyl aminopeptidases (EC 3.4.11.21) (25, 26). Therefore, we hereafter refer to it as Ape4. Comprehensive yeast two-hybrid analyses of the yeast interactome network reveal that Ape4 can associate with Atg19 and Ape1 (33, 34). Therefore, we assumed that Ape4 might be transported to the vacuole via the Cvt pathway. For biochemical analyses of Ape4, we generated polyclonal antibody against recombinant full-length Ape4. Immunoblot analysis revealed that the affinity-purified anti-Ape4 antibody specifically recognized a 55-kDa protein, which corresponded well to the predicted molecular mass of Ape4, in crude yeast extracts from wild-type cells (Fig. 1A, lane 2). Introduction of the APE4 gene on a multicopy plasmid resulted in an elevated level of the 55-kDa protein (Fig. 1A, lane 3), confirming that the antibody recognized Ape4 protein. This band was not detected in ape4Δ strain, although faint nonspecific bands, including one close to the 55-kDa band of Ape4, were seen when blots were exposed to x-ray film for a longer time (Fig. 1A, lanes 1, 4, and 5). Using the anti-Ape4 antibody, we examined the vacuolar localization of Ape4 by vacuole isolation followed by a protease protection assay. For this assay, we expressed HA-tagged Ams1, a cargo protein of the Cvt pathway, in a pep4Δ prb1Δ background strain in which two major vacuolar protease genes, PEP4 and PRB1, were deleted to stabilize the triple HA tag in the event that the cargo proteins reached the vacuole. Yeast cells were grown in rich medium, and the vacuoles were isolated by Ficoll flotation. The vacuole preparation from the pep4Δ prb1Δ cells resulted in 39-fold purification of the vacuole fraction as estimated by immunoblot of the precursor 2 (p2) form of the marker protein Prc1 that travels to the vacuole via the vacuolar protein sorting pathway (Fig. 1B, lane 1 versus lane 2), whereas there was relatively little contamination from cytosol (Pgk1, 0.40-fold purification). Similar to p2Prc1, the precursor form of Ape1 was accumulated in the vacuole fraction, indicating that the Cvt pathway occurred normally (Fig. 1B, lane 1 versus lane 2). We observed that both HA3-Ams1 and Ape4 were also concentrated in this fraction (Fig. 1B, lane 2). To verify that Ape4 entered into the vacuolar lumen, we performed a protease protection assay using the isolated vacuoles. The vacuole fraction was treated with proteinase K in the absence or presence of 1% Triton X-100. Pgk1 found in this fraction was sensitive to exogenously added protease even in the absence of detergent, suggesting that a small portion of the cytosol cofractionated with the vacuole rather than that Pgk1 entered into the vacuole lumen (Fig. 1B, lanes 3–5). In contrast, most of the Ape4 was resistant to exogenously added proteases and was fully digested only in the presence of detergent as observed with other vacuolar proteins, suggesting that Ape4 was localized in the vacuolar lumen (Fig. 1B, lanes 3–5).
FIGURE 1.
Aspartyl aminopeptidase Yhr113w/Ape4 is located inside the vacuole. A, wild-type (WT; SEY6210) cells containing either empty vector (pRS424; lanes 2 and 5) or APE4 multicopy plasmid (pRS424-Ape4; lane 3) and the ape4Δ (YMY111) cells containing empty vector (pRS424; lanes 1 and 4) were grown in SC-Trp medium. Protein extracts equivalent to 0.1 A600 nm unit of cells were subjected to SDS-PAGE followed by immunoblot analysis probed with anti-Ape4 antiserum. Arrowheads indicate nonspecific proteins. B, vacuoles isolated from the pep4Δ prb1Δ (lanes 1–5) and atg1Δ pep4Δ prb1Δ (lanes 6–10) cells containing HA3-Ams1 plasmid (pTS544) were treated with or without proteinase K in the absence or presence of 1% Triton X-100. Samples equivalent to 10 μg of proteins were subjected to immunoblot analysis probed with anti-Prc1 (upper panel), anti-Ape1 (upper middle panel), anti-HA (middle panel), anti-Ape4 (lower middle panel), and anti-Pgk1 (lower panel) antisera or antibodies. T (lanes 1 and 6), total cell lysate.
We next isolated vacuoles from atg19Δ pep4Δ prb1Δ cells to examine whether the vacuolar localization of Ape4 was dependent on the Cvt pathway. Atg19 is the receptor only for the Cvt cargo proteins prApe1 and Ams1; thus, Atg19 is required for the Cvt pathway but not for canonical (i.e. starvation-dependent) autophagy or the vacuolar protein sorting pathway. In these cells, p2Prc1, but not prApe1 or HA3-Ams1, was accumulated normally in the vacuole fraction, confirming that the Cvt pathway was specifically blocked (Fig. 1B, lane 6 versus lane 7). Similar to prApe1 and HA3-Ams1, the accumulation of Ape4 in the vacuole fraction was significantly decreased in the atg19Δ pep4Δ prb1Δ cells (Fig. 1B, lane 2 versus lane 7). These results suggested that Ape4 was transported to the vacuole in an Atg19-dependent manner.
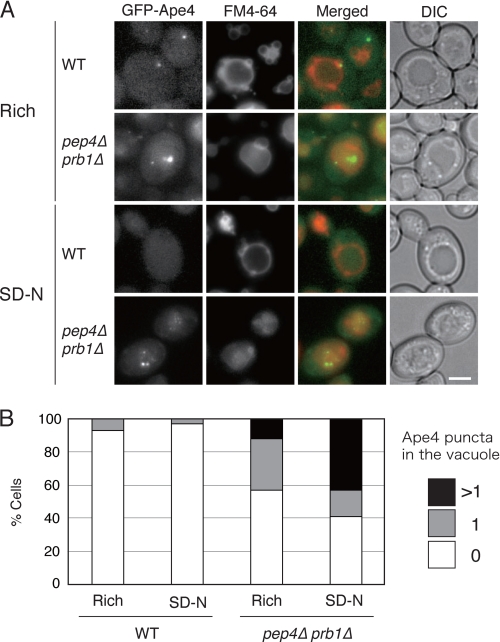
To confirm the vacuolar localization of Ape4, we decided to examine Ape4 localization using fluorescence microscopy. Accordingly, we constructed a chimera with an N-terminal GFP-tagged Ape4 expressed from its endogenous promoter, allowing us to analyze its subcellular localization in physiological conditions. When the wild-type cells were grown in a nutrient-rich medium, the GFP-Ape4 signal was diffuse in the cytosol, but in some cells, it was also observed in the vacuole, which was labeled with FM4-64, a red fluorescent probe for the vacuole membrane (Fig. 2A). There was also a prominent perivacuolar punctum that likely corresponded to the PAS (see below). The vacuolar localization of GFP-Ape4 was not very evident in this condition because the signal was relatively faint and diffuse in the vacuole lumen. Therefore, we examined the pep4Δ prb1Δ strain in which vacuolar protease activities were substantially decreased, thereby accumulating subvacuolar vesicles in the vacuole lumen (this strain is defective in the breakdown of the single membrane autophagic and Cvt bodies that result from the fusion of autophagosomes and Cvt vesicles with the vacuole limiting membrane). As expected, we were able to observe GFP-Ape4 puncta moving within the vacuole lumen, which was delineated by FM4-64 fluorescence, in this strain (Fig. 2A). After induction of autophagy by nitrogen starvation for 3 h, the vacuolar localization of GFP-Ape4 became more evident in the pep4Δ prb1Δ strain; the number of the GFP-Ape4 puncta increased (Fig. 2B).
FIGURE 2.
Fluorescence microscopy reveals perivacuolar and vacuolar localization of GFP-Ape4. A, wild-type (WT; SEY6210) and pep4Δ prb1Δ (YMY105) cells expressing GFP-Ape4 were labeled with vacuolar membrane marker FM4-64 for 20 min in SC-Trp medium and chased for 2 h in the same medium. The labeled cells were further starved in SD(-N) for 3 h. Bar, 2 μm. B, in each condition, 100 cells were counted for the number of subvacuolar vesicles labeled with GFP-Ape4 under fluorescence microscopy. The values show percentages of the population with a single (1) or multiple (>1) GFP-labeled subvacuolar vesicles and without GFP-labeled subvacuolar vesicle (0). DIC, differential interference contrast.
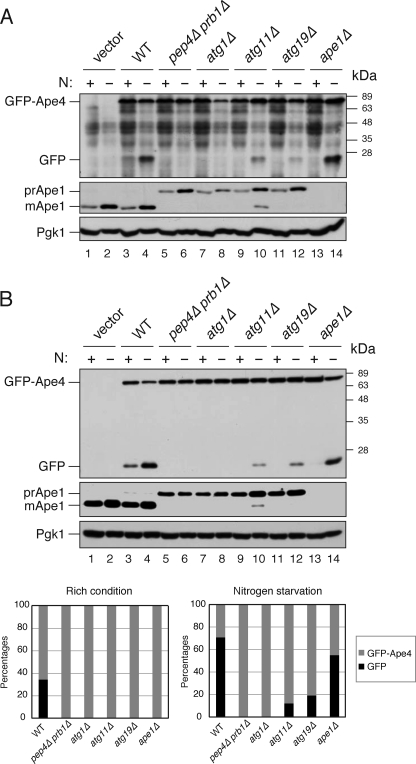
Next, the vacuolar transport of GFP-Ape4 was confirmed by a biochemical GFP processing assay, which relied on release of the GFP moiety from GFP-fused proteins in a vacuolar proteolysis-dependent manner; the generation of free GFP, which is relatively resistant to vacuolar proteases, can be used to monitor vacuolar delivery of the chimera (35). We expressed GFP-Ape4 from a centromeric plasmid under the control of the endogenous promoter in wild-type and pep4Δ prb1Δ cells. In nutrient-rich conditions, a protein band corresponding to the 27-kDa free GFP was detected in the wild-type cells along with that of full-length GFP-Ape4 (Fig. 3A, lane 3). In contrast, no free GFP band was observed in the pep4Δ prb1Δ cells (Fig. 3A, lane 5). These results indicated that a portion of GFP-Ape4 was transported to the vacuole and processed by vacuolar proteases. Furthermore, GFP-Ape4 processing was increased in the wild-type cells but not in the pep4Δ prb1Δ cells in nitrogen starvation conditions, suggesting that its vacuolar transport was enhanced by nitrogen starvation (Fig. 3A, lanes 4 and 6). Because it was difficult to reduce nonspecific background bands due to the low abundance of GFP-Ape4 (Fig. 3A, lanes 1 and 2), we also expressed GFP-Ape4 from the CUP1 promoter to corroborate its processing. We found that basically the same results were obtained: free GFP appeared in a vacuolar proteolysis-dependent manner, and nitrogen starvation activated the processing (Fig. 3B, lanes 3–6).
FIGURE 3.
Biochemical analysis of GFP-Ape4 delivery to vacuole. A, an empty vector (pRS414) was used for transformation of wild-type strain (SEY6210; vector; lanes 1 and 2). Wild-type (WT; SEY6210, lanes 3 and 4), pep4Δ prb1Δ (YMY105; lanes 5 and 6), atg1Δ (WHY1; lanes 7 and 8), atg11Δ (YTS147; lanes 9 and 10), atg19Δ (SSY31; lanes 11 and 12), and ape1Δ (YMY103; lanes 13 and 14) cells were transformed with a plasmid encoding GFP-Ape4 (pTS572). The cells were grown in SC-Trp medium (N+; lanes 1, 3, 5, 7, 9, 11, and 13) and starved in SD(-N) for 3 h (N−, lanes 2, 4, 6, 8, 10, 12, and 14). Protein extracts equivalent to 0.1 A600 nm unit of cells were subjected to SDS-PAGE followed by immunoblot analysis with anti-YFP (upper panel) and anti-Pgk1 (lower panel) antibodies and anti-Ape1 (middle panel) antiserum. B, an empty vector (pRS416) was used for transformation of wild-type strain (SEY6210; vector; lanes 1 and 2). Wild-type (WT; SEY6210; lanes 3 and 4), pep4Δ prb1Δ (YMY105; lanes 5 and 6), atg1Δ (WHY1; lanes 7 and 8), atg11Δ (YTS147; lanes 9 and 10), atg19Δ (SSY31; lanes 11 and 12), and ape1Δ (YMY103; lanes 13 and 14) cells were transformed with a plasmid expressing GFP-Ape4 from the CUP1 promoter (pTS555). The cells were grown and analyzed as described in A. Bands were quantified using ImageJ software, and the percentages of GFP-Ape4 (gray) and GFP (black) were plotted.
We extended this analysis by examining whether the transport of GFP-Ape4 relied on the Cvt pathway. Atg1 is a serine/threonine protein kinase required for both the Cvt pathway and starvation-induced autophagy. As expected, no GFP was released from GFP-Ape4 in atg1Δ cells in either nutrient-rich or starvation conditions (Fig. 3, A and B, lanes 7 and 8). In contrast to Atg1, Atg11 is required for selective types of autophagy in yeast, including the Cvt pathway, pexophagy, and mitophagy, but is not essential for starvation-induced autophagy (36, 37). In the Cvt pathway, Atg11 interacts with Atg19 to localize the Ape1-Atg19-Ams1 complex at the PAS (13). Deletion of ATG11 or ATG19 inhibited the processing of GFP-Ape4 in rich conditions, and this block was partially rescued by nitrogen starvation (Fig. 3, A and B, lanes 9–12). Similarly, the deletion of APE1 fully affected the transport of GFP-Ape4 in nutrient-rich conditions (Fig. 3, A and B, lane 13), although there was only a slight decrease in starvation conditions (Fig. 3, A and B, lane 14). These results indicated that Ape4 was selectively transported to the vacuole via the Cvt pathway. The minor protein band of 60 kDa also appeared in all strains expressing GFP-Ape4 under the control of the endogenous promoter (Fig. 3A, lanes 3–14). This suggested that a minor portion of GFP-Ape4 was cleaved in the cytoplasm, although the mechanism was unknown.
Ape4 Undergoes Limited Proteolysis in Cvt Pathway-dependent Manner
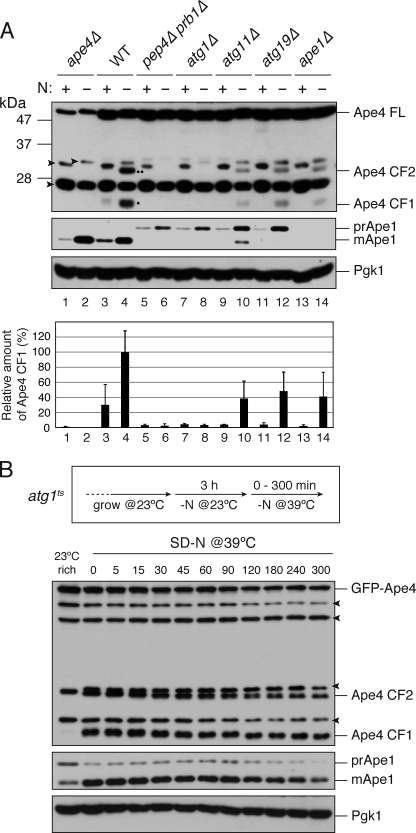
Yokoyama et al. (26) reported that Yhr113w/Ape4 is cleaved by proteases without loss of enzyme activity; albeit no mechanistic insight was provided. Therefore, we examined whether the cleavage of Ape4 was dependent on vacuolar proteases. Immunoblot analysis with anti-Ape4 antibody revealed that 24-kDa (cleaved form 1 (CF1)) and 31-kDa (CF2) protein bands appeared in response to nitrogen starvation in the wild-type cells (Fig. 4A, lane 4). The Ape4 CF1 was also detected during vegetative growth in YPD, although it was very faint (the CF2 was not even detected) (Fig. 4A, lane 3). The cleaved forms of Ape4 did not appear in the pep4Δ prb1Δ and atg1Δ cells and significantly decreased in the atg11Δ and atg19Δ cells (Fig. 4A, lanes 5–12). The appearance of these cleaved forms was more evident when GFP-Ape4 was overexpressed from the CUP1 promoter at a concentration of copper normally contained in SC or SD(-N) medium: both cleaved forms as well as free GFP were generated in an autophagy-dependent manner (supplemental Fig. S1). These results indicated that Ape4 travels to the vacuole primarily via a selective autophagy pathway. A nonselective pathway may contribute to the vacuolar localization of Ape4 in starvation conditions when the selective pathway is impaired (e.g. in atg11Δ or atg19Δ strains).
FIGURE 4.
Ape4 undergoes limited cleavage in vacuolar proteolysis- and autophagy-dependent manner. A, the ape4Δ (YMY111; lanes 1 and 2), wild-type (WT; SEY6210; lanes 3 and 4), pep4Δ prb1Δ (YMY105; lanes 5 and 6), atg1Δ (WHY1; lanes 7 and 8), atg11Δ (YTS147; lanes 9 and 10), atg19Δ (SSY31; lanes 11 and 12), and ape1Δ (YMY103; lanes 13 and 14) cells were grown in YPD medium (lanes 1, 3, 5, 7, 9, 11, and 13) and starved in SD(-N) medium for 3 h (lanes 2, 4, 6, 8, 10, 12, and 14). The cell extracts (0.1 A600 nm cell equivalent) were analyzed by immunoblot probed with anti-Ape4 (upper panel) and anti-Ape1 (middle panel) antisera and anti-Pgk1 (lower panel) antibody. A single dot and double dot indicate CF1 and CF2, respectively. Quantification of CF1 and Pgk1 were done using ImageJ software. The band intensity of CF1 was normalized by that of Pgk1 and plotted relative to WT treated with SD(-N). Error bars indicate the S.D. of three independent experiments. B, the ape4Δ atg1Δ (YMY112) cells were transformed with an atg1ts plasmid (pAtg1ts) and a plasmid expressing GFP-Ape4 from the CUP1 promoter (pTS555). The strain was grown in SC-Ura-Trp medium and starved in SD(-N) at a permissive temperature of 23 °C for 3 h. After shifting to the nonpermissive temperature of 39 °C, samples were collected at the indicated time points and analyzed by immunoblot using anti-Ape1 (upper panel) and anti-Ape4 (middle panel) antisera and anti-Pgk1 (lower panel) antibody. FL, full length. Arrowheads indicate nonspecific proteins.
Although the cleaved form of Ape4 maintains aspartyl aminopeptidase activity (26), it is possible that this form could be destabilized in the vacuolar lumen. To test this hypothesis, we monitored changes in the amounts of the cleaved form of Ape4 after blocking autophagy in an atg1ts mutant strain overexpressing GFP-Ape4. After the atg1ts cells were starved in SD(-N) medium for 5 h at permissive temperature, they were shifted to a nonpermissive temperature of 39 °C to prevent the nascent GFP-Ape4 proteins from entering into the vacuole via autophagy. At several time points, the amounts of Ape4 species and Ape1 proteins were determined by immunoblotting. The protein levels of the cleaved forms of Ape4 were slightly decreased over time, but the decrease was similar to that of Ape1, a bona fide vacuolar protein (Fig. 4B). The cleaved forms generated from endogenous Ape4 basically exhibited the same stability (supplemental Fig. S2). These results suggested that, similar to Ape1, Ape4 stably resides in the vacuole.
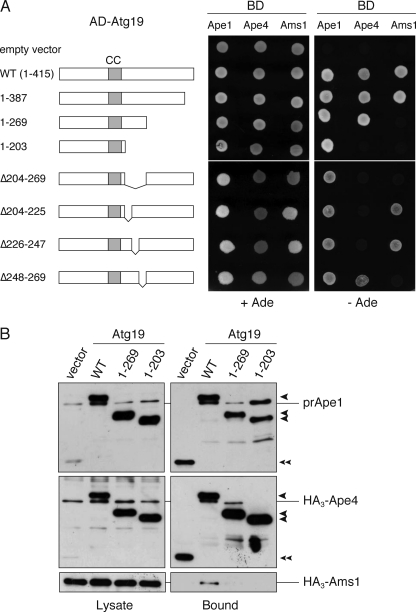
Ape4 Interacts with Atg19 at a Site Distinct from Ape1- and Ams1-binding Sites
Because Atg19 contributes to Ape4 transport to the vacuole, we verified whether Ape4 interacted with Atg19. To this end, we first utilized a yeast two-hybrid system. The atg19Δ test strain was transformed with two-hybrid plasmids encoding Ape4 and full-length or mutant Atg19. The transformed strains were then tested for the GAL4-dependent transcriptional activation of the ADE2 and lacZ genes. Plasmids encoding Ape1 and Ams1 were used for controls. Ape1 and Ams1 interact with the coiled coil and the C-terminal domains of Atg19, respectively (13). As expected, Ape1 exhibited strong interactions with the wild-type and mutant forms of Atg19 used in this study, all of which contained the coiled coil domain (Fig. 5A). Ams1 interacted with the full-length Atg19 and Atg19(1–387) but not with Atg19(1–269) or Atg19(1–203), confirming that the C-terminal region was responsible for Ams1 binding (Fig. 5A). Notably, Ape4 bound to the full-length Atg19, Atg19(1–387), or Atg19(1–269) but not to Atg19(1–203), indicating that the binding sites for Ape4, prApe1, and Ams1 are separable in Atg19. It should be noted that the binding activity of Ape4 with Atg19 was similar to that of Ams1 but was about 4-fold less than that of prApe1 as estimated by β-galactosidase activity (supplemental Fig. S3). The interaction between Ape4 and Atg19 was preserved in the ape1Δ test strain, suggesting that prApe1 is not required for the formation of the Ape4-Atg19 complex (supplemental Fig. S4).
FIGURE 5.
Mapping of Ape4-binding domain within Atg19. A, two-hybrid analysis of the physical interaction between Atg19 mutants and cargo proteins. The atg19Δ test strain (YTS111) was transformed with the activation domain (AD) plasmids containing ATG19 and the DNA-binding domain (BD) plasmid containing APE1, APE4, and AMS1. After transformants grown in SC-Leu-Ura liquid medium were washed with distilled water, 0.01 A600 nm unit of the cells was spotted on SC-Leu-Ura and SC-Ade-Leu-Ura plates to assess the interaction-dependent activation of the ADE2 gene. The cells shown here were grown at 30 °C for 3 days. CC, coiled coil domain. B, total lysates from the atg1Δ atg19Δ cells (YMY109) expressing either wild-type or mutant Atg19 fused with protein A (PA-Atg19) were used to precipitate PA-Atg19 proteins with IgG-Sepharose. The left and right panels show the protein blots of total cell extracts (Lysate) and the IgG precipitates (Bound), respectively, which were probed with anti-Ape1 antiserum (upper panel) and anti-HA antibody (middle and lower panels). Single arrowheads show PA-Atg19 derivatives, and a double arrowhead indicates protein A without a fusion protein.
To verify these results, we performed an affinity isolation experiment in the atg1Δ background strain. In the atg1Δ mutant, Atg19 is blocked at the PAS, will not travel to the vacuole, and hence does not get degraded, thereby maintaining similar levels of wild-type and mutant Atg19 proteins in the cytosol. Therefore, we transformed the atg1Δ atg19Δ cells with protein A-fused Atg19 constructs together with plasmids encoding HA3-Ape4 and HA3-Ams1. PA-Atg19 proteins were isolated from the total cell lysates with IgG-Sepharose, and the resulting samples were analyzed by immunoblotting with anti-HA antibody and anti-Ape1 antiserum (note that PA-Atg19 reacts with both because of the PA moiety). Precursor Ape1 was co-isolated with all of the PA-Atg19 proteins, whereas Ape4 lost the interaction with Atg19(1–203), and only full-length Atg19 could bind with Ams1 (Fig. 5B). Together with the results from the two-hybrid analysis, these results indicated that three cargo proteins associated with Atg19 by binding to distinct sites.
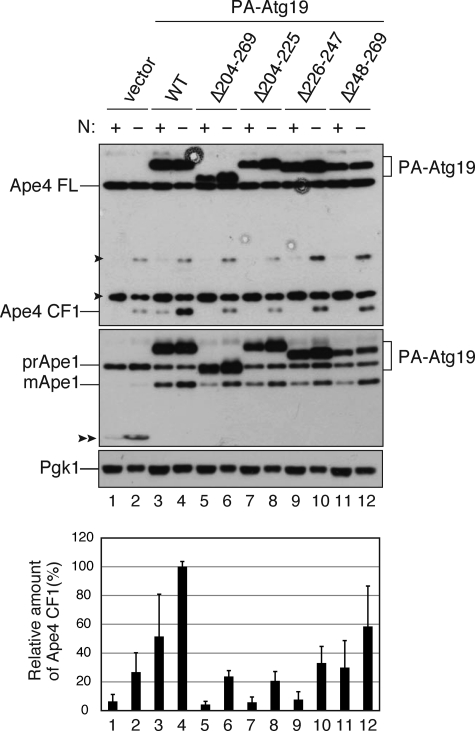
We next attempted to make an Atg19 mutant that was unable to bind Ape4 but could transport prApe1 to examine whether the interaction between Atg19 and Ape4 was important for the transport of the latter. As the C-terminal 28 amino acids of Atg19 are required for the interaction of Atg19 with Atg8 and Atg11 (13) and amino acid residues 204–269 were important for interaction with Ape4, we constructed an Atg19 mutant lacking amino acid residues 204–269 (Atg19(Δ204–269)), which was fused with the Gal4 activation domain or protein A. To narrow down the Ape4-binding domain, Atg19(Δ204–225), Atg19(Δ225–247), and Atg19(Δ248–269) were also constructed (Fig. 5A). Based on the yeast two-hybrid assay, all of these constructs preserved prApe1 binding ability. In contrast, Atg19(Δ204–269) lost the interaction with both Ape4 and Ams1 (Fig. 5A). Furthermore, Atg19(Δ204–225) and Atg19(Δ225–247) lost interaction with Ape4, whereas Atg19(Δ248–269) lost the ability to bind Ams1 (Fig. 5A). These results suggested that the region responsible for Ape4 binding included that encompassed by amino acids 204–247, whereas Ams1 required more distal amino acids, including at least some of those within the 248–269 range. We next expressed these Atg19 mutants as protein A fusion proteins from centromeric plasmids under the control of the CUP1 promoter in atg19Δ cells and determined whether they could complement the defect in prApe1 and Ape4 transport resulting from the absence of endogenous Atg19. Although the expression levels of PA-Atg19(Δ204–269), PA-Atg19(Δ204–225), and PA-Atg19(Δ225–247) were almost identical to that of full-length PA-Atg19 (Fig. 6, lanes 3–10), PA-Atg19(Δ248–269) was somewhat reduced presumably due to degradation caused by a loss of proper folding (Fig. 6, lanes 11 and 12). Nonetheless, similar to full-length Atg19, all mutants were able to complement the defect in prApe1 maturation, suggesting that these mutants were able to function as prApe1 receptors in the Cvt pathway (Fig. 6, lower panel, lanes 3–12). In contrast, the cleaved form of Ape4 was significantly decreased in these mutants in both growing and starvation conditions (Fig. 6, upper panel, lanes 3–12). Although the two-hybrid analysis suggested that Atg19(Δ248–269) interacted with Ape4, PA-Atg19(Δ248–269) could not fully complement Ape4 transport. This discrepancy might be due to relatively weak interaction of Atg19(Δ248–269) with Ape4 (supplemental Fig. S3) and/or the lower expression level of PA-Atg19(Δ248–269) (Fig. 6, lanes 11 and 12). These results suggested that the association of Atg19 and Ape4 is important for the selective transport of Ape4 to the vacuole.
FIGURE 6.
Atg19 mutants lacking Ape4-binding domain specifically affect Ape4 transport to vacuole. The atg19Δ cells (SSY31) expressing either empty vector (pRS416-CuPA; lanes 1 and 2), wild-type (WT; pTS477; lanes 3 and 4), or mutant Atg19 (lanes 5–12) fused with protein A (PA-Atg19) were grown in SC medium and starved in SD(-N) for 3 h. Protein extracts equivalent to 0.2 A600 nm unit of cells were subjected to SDS-PAGE followed by immunoblot analysis and probed with anti-Ape4 (upper panel) and anti-Ape1 (middle panel) antisera and anti-Pgk1 (lower panel) antibody. Quantification of CF1 and Pgk1 was done as described in Fig. 4A. Error bars indicate the S.D. of three independent experiments. Single arrowheads, nonspecific protein bands. Double arrowhead, protein A without fusion. FL, full length.
Ape4 Utilizes Ape1 Transport System
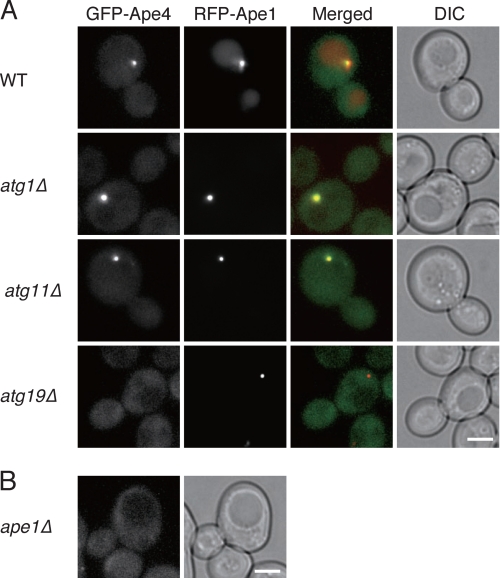
Assembly of the Ape1 complex in the cytosol and its targeting to the PAS activate the Cvt pathway in rich conditions (18). Moreover, the binding of the Ams1 oligomer to the complex through Atg19 allows efficient import of Ams1 to the vacuole via the Cvt pathway (13). Accordingly, we decided to examine whether prApe1 was needed for the efficient vacuolar import of Ape4. In ape1Δ cells, Ape4 transport to the vacuole was significantly decreased as estimated by GFP-Ape4 processing and Ape4 cleavage in rich conditions (Fig. 3, A and B, upper panel, lane 13, and Fig. 4A, upper panel, lane 13). We next used fluorescence microscopy to observe GFP-Ape4 in several atg mutants co-expressing RFP-Ape1. In wild-type cells, the prominent GFP-Ape4 punctum (Fig. 2) was colocalized with RFP-Ape1 in growing conditions (Fig. 7). Because prApe1 localizes at the PAS and Ape1 localizes in the vacuole in the wild-type cells (13), the GFP-Ape4 punctum corresponded to the PAS. In starvation conditions, the punctate localization of GFP-Ape4 disappeared in most of the cells probably because any GFP-Ape4 that transiently localized to the PAS was efficiently transported to the vacuole (supplemental Fig. S5). In atg1Δ cells, the punctate signal became prominent in both growing and starvation conditions, suggesting that Ape4 proteins were blocked at the PAS (Fig. 7 and supplemental Fig. S5). Deletion of ATG11 also led to a strong punctate localization of GFP-Ape4 that colocalized with RFP-Ape1 (Fig. 7 and supplemental Fig. S5). Atg11 is not needed for the interaction of Atg19 with prApe1, and apparently this is also the case with Ape4. However, Atg11 is needed for localization of the prApe1-Atg19 complex to the PAS (13), suggesting that Ape4 could associate with the prApe1-Atg19 complex before arriving at the PAS. Moreover, when ATG19 was deleted, GFP-Ape4 could no longer associate with the Ape1 complex, thereby resulting in the dispersal of GFP-Ape4 in the cytosol even though RFP-Ape1 still formed a complex (Fig. 7 and supplemental Fig. S5). Similarly, the lack of Ape1 resulted in a dispersed localization of GFP-Ape4, which was presumably at least partly in a complex with Atg19 (Fig. 7 and supplemental Fig. S5).
FIGURE 7.
Localization of GFP-Ape4 in atg mutants and ape1Δ mutant in nutrient-rich condition. A, wild-type (WT; SEY6210), atg1Δ (WHY1), atg11Δ (YTS147), and atg19Δ (SSY31) cells expressing GFP-Ape4 and RFP-Ape1 were grown in SC-Ura-Trp medium. B, the ape1Δ cells (YMT103) expressing GFP-Ape4 were grown in SC medium. Bar, 2 μm. DIC, differential interference contrast.
DISCUSSION
Saccharomyces species adapted autophagy as a mechanism to achieve a selective and constitutive transport of the oligomeric vacuolar hydrolases (Ape1 and Ams1 in S. cerevisiae) via the Cvt pathway. The cargo proteins maintain their folding and oligomeric states during membrane translocation with this transport system because they do not need to pass through a transport channel that is used for conventional translocation mechanisms. In this study, we found that aspartyl aminopeptidase (Yhr113w/Ape4) is a novel Cvt cargo protein. Although this enzyme is similar to Ape1 in amino acid sequence and subunit organization, it lacks the N-terminal domain that is used by prApe1 for binding to Atg19, the receptor for the Cvt pathway. Therefore, Ape4 associates with Atg19 through a relatively weak interaction. As the Ape4-binding site in Atg19 is different from those for prApe1 and Ams1, these enzymes are unlikely to compete for binding to Atg19, enabling Ape4 and Ams1 to be simultaneously localized on the Ape1 complex and incorporated into the Cvt vesicle (Figs. 5–7). Recently, a three-dimensional structure of the Ams1-binding domain (amino acid residues 254–367) in Atg19 was determined by NMR spectroscopy, which revealed that the Ams1-binding domain has a typical immunoglobulin fold with eight β-strands (38). Atg19(Δ204–225) and Atg19(Δ226–247) exhibited interaction with Ams1, which is consistent with the fact that both mutants contain an intact Ams1-binding domain. Interestingly, an Atg19 homolog, Yol083w/Atg34, also contains the Ams1-binding domain and is responsible for Ape1-independent vacuolar transport of Ams1 under starvation conditions (38, 39) as predicted from in silico studies (Atg19-B; Ref. 40). As Atg34 did not interact with Ape4, it might not be involved in Ape4 transport (data not shown).
Recently, it has been reported that leucine aminopeptidase III (Lap3) is selectively delivered to the vacuole for degradation (41). This transport is observed only when Lap3 is overproduced in nitrogen starvation conditions and is partially mediated by Atg19 but not prApe1. Lap3 forms a homohexameric complex of ∼220 kDa, which further forms an aggregate independently of prApe1 (41, 42). Thus, it will be interesting to examine whether Lap3 can interact with Atg19 and whether the binding site is different from those for prApe1, Ape4, and Ams1 to understand the relationship between biosynthetic and degradative pathways that utilize autophagy.
Precursor Ape1 is tightly packed into a high density complex, which is sequestered by a double membrane-bound vesicle, thereby achieving efficient transport of prApe1 to the vacuole even in growing conditions in which canonical autophagy is not induced (13, 14). In contrast, Ape4 transport is minimal in this condition probably because of its relatively weak interaction with Atg19. Nitrogen starvation accelerates Ape4 transport to the vacuole presumably because of the increased expression of prApe1 and Atg19 that occurs in starvation conditions as well as the induction of general autophagy activity (43). Because both the cytosolic (full-length) and vacuolar (cleaved) forms of Ape4 seem to possess enzymatic activity (26), this protein may contribute to peptide metabolism in both locations. Although the optimal pH of Ape4 activity occurs at pH 7.5–7.9, Ape4 exhibits 60–70% of its enzymatic activity at vacuolar pH (pH 6.2–6.5) compared with that at cytosolic pH (pH 7.2).3 This suggests that Ape4 is still functional in the vacuolar lumen. Uncontrolled proteolysis is harmful for cell function, and cells sequester many proteolytic enzymes in a lytic compartment, the lysosome/vacuole, or highly control their activity. Aspartyl aminopeptidase has very high substrate specificity: it favors short peptides with acidic amino acid residues at the N termini. This suggests that Ape4 does not cause random degradation of native proteins even if it exhibits enzyme activity in the cytosol. Rather, Ape4 might contribute to the degradation of short peptides generated by ordered proteolysis such as occurs through the ubiquitin-proteasome system to clear debris in the cytosol. The vacuolar aminopeptidases (Ape1 and aminopeptidase Y) seem to have difficulty in removing acidic amino acid residues at the N termini of peptides (44, 45). Thus, it might be possible that Ape4 relocates from the cytosol to the vacuole in starvation conditions in which more active vacuolar proteolysis is needed to achieve efficient protein breakdown to generate free amino acids that can serve as nutrient blocks. Recently, it has been reported that Ams1 plays an important role in degrading free oligosaccharides released from misfolded glycoproteins retrotranslocated from the endoplasmic reticulum lumen to the cytoplasm by the endoplasmic reticulum-associated degradation pathway (46). When the Cvt pathway is impaired by deleting ATG19, an increased processing of free oligosaccharides is observed, meaning that Ams1 functions in the cytoplasm before it is targeted to the vacuole. Together with our results, this might indicate that the Cvt pathway is the mechanism by which active cytosolic hydrolases are redistributed to the vacuole.
In this study, we identified aspartyl aminopeptidase Ape4 as the third cargo protein for the Cvt pathway. Although in vivo substrates of Ape4 are not known, it may contribute to peptide degradation both in the cytosol and the vacuole. Yeast cells may respond to environmental conditions by changing the distributions of some cytosolic enzymes to the vacuole. Identification of new cargo enzymes and their substrates will be helpful to understand the role of selective autophagy in cellular responses to environmental changes.
Acknowledgment
We thank Dr. Yoshinori Ohsumi for providing the atg1ts plasmid.
This work was supported by the Sapporo Bioscience Foundation (to T. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
H. Kawasaki, personal communication.
- ATG
- autophagy-related
- Cvt
- cytoplasm to vacuole targeting
- PAS
- pre-autophagosomal structure/phagophore assembly site
- SC
- synthetic complete
- Prc1
- carboxypeptidase Y
- RFP
- red fluorescent protein
- Ape1
- aminopeptidase I
- Ams1
- α-mannosidase
- Pgk1
- phosphoglycerate kinase
- PA
- protein A
- CF
- cleaved form
- Lap3
- leucine aminopeptidase III.
REFERENCES
- 1. Shintani T., Klionsky D. J. (2004) Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 5. Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 6. Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 7. Thurston T. L., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. (2009) Nat. Immunol. 10, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 8. Yoshihisa T., Anraku Y. (1990) J. Biol. Chem. 265, 22418–22425 [PubMed] [Google Scholar]
- 9. Klionsky D. J., Cueva R., Yaver D. S. (1992) J. Cell Biol. 119, 287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. (1992) J. Cell Biol. 119, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim J., Scott S. V., Oda M. N., Klionsky D. J. (1997) J. Cell Biol. 137, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andrei-Selmer C., Knuppel A., Satyanarayana C., Heese C., Schu P. V. (2001) J. Biol. Chem. 276, 11606–11614 [DOI] [PubMed] [Google Scholar]
- 13. Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) Dev. Cell 3, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baba M., Osumi M., Scott S. V., Klionsky D. J., Ohsumi Y. (1997) J. Cell Biol. 139, 1687–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scott S. V., Baba M., Ohsumi Y., Klionsky D. J. (1997) J. Cell Biol. 138, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suzuki K., Kamada Y., Ohsumi Y. (2002) Dev. Cell 3, 815–824 [DOI] [PubMed] [Google Scholar]
- 17. Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shintani T., Klionsky D. J. (2004) J. Biol. Chem. 279, 29889–29894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He C., Song H., Yorimitsu T., Monastyrska I., Yen W. L., Legakis J. E., Klionsky D. J. (2006) J. Cell Biol. 175, 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He C., Baba M., Cao Y., Klionsky D. J. (2008) Mol. Biol. Cell 19, 5506–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakatogawa H., Ichimura Y., Ohsumi Y. (2007) Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- 22. Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008) Genes Cells 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 23. Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. (2004) Dev. Cell 6, 79–90 [DOI] [PubMed] [Google Scholar]
- 24. Hutchins M. U., Klionsky D. J. (2001) J. Biol. Chem. 276, 20491–20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilk S., Wilk E., Magnusson R. P. (1998) J. Biol. Chem. 273, 15961–15970 [DOI] [PubMed] [Google Scholar]
- 26. Yokoyama R., Kawasaki H., Hirano H. (2006) FEBS J. 273, 192–198 [DOI] [PubMed] [Google Scholar]
- 27. Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. (2002) Nucleic Acids Res. 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amberg D. C., Burke D., Strathern J. N. (2005) Methods in Yeast Genetics 2005: a Cold Spring Harbor Laboratory Course Manual, pp. 199–210, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim J., Huang W. P., Klionsky D. J. (2001) J. Cell Biol. 152, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shintani T., Reggiori F. (2008) Methods Enzymol. 451, 43–56 [DOI] [PubMed] [Google Scholar]
- 32. Shintani T., Suzuki K., Kamada Y., Noda T., Ohsumi Y. (2001) J. Biol. Chem. 276, 30452–30460 [DOI] [PubMed] [Google Scholar]
- 33. Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A. L., Tavernier J., Hill D. E., Vidal M. (2008) Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheong H., Klionsky D. J. (2008) Methods Enzymol. 451, 1–26 [DOI] [PubMed] [Google Scholar]
- 36. Kim J., Kamada Y., Stromhaug P. E., Guan J., Hefner-Gravink A., Baba M., Scott S. V., Ohsumi Y., Dunn W. A., Jr., Klionsky D. J. (2001) J. Cell Biol. 153, 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kanki T., Klionsky D. J. (2008) J. Biol. Chem. 283, 32386–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe Y., Noda N. N., Kumeta H., Suzuki K., Ohsumi Y., Inagaki F. (2010) J. Biol. Chem. 285, 30026–30033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzuki K., Kondo C., Morimoto M., Ohsumi Y. (2010) J. Biol. Chem. 285, 30019–30025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meijer W. H., van der Klei I. J., Veenhuis M., Kiel J. A. (2007) Autophagy 3, 106–116 [DOI] [PubMed] [Google Scholar]
- 41. Kageyama T., Suzuki K., Ohsumi Y. (2009) Biochem. Biophys. Res. Commun. 378, 551–557 [DOI] [PubMed] [Google Scholar]
- 42. Enenkel C., Wolf D. H. (1993) J. Biol. Chem. 268, 7036–7043 [PubMed] [Google Scholar]
- 43. Scott S. V., Guan J., Hutchins M. U., Kim J., Klionsky D. J. (2001) Mol. Cell 7, 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Metz G., Röhm K. H. (1976) Biochim. Biophys. Acta 429, 933–949 [DOI] [PubMed] [Google Scholar]
- 45. Yasuhara T., Nakai T., Ohashi A. (1994) J. Biol. Chem. 269, 13644–13650 [PubMed] [Google Scholar]
- 46. Hirayama H., Seino J., Kitajima T., Jigami Y., Suzuki T. (2010) J. Biol. Chem. 285, 12390–12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]