Abstract
MicroRNAs play important roles in tumor metastasis. Recently, we reported that the level of miR-520b is inversely related to the metastatic potential of breast cancer cells. In this study, we investigated the role of miR-520b in breast cancer cell migration. We found that miR-520b suppressed the migration of breast cancer cells with high metastatic potential, including MDA-MB-231 and LM-MCF-7 cells, although the inhibition of miR-520b enhanced the migration of low metastatic potential MCF-7 cells. We further discovered that miR-520b directly targets the 3′-untranslated region (3′UTR) of either hepatitis B X-interacting protein (HBXIP) or interleukin-8 (IL-8), which has been reported to contribute to cell migration. Surprisingly, tissue array assays showed that 75% (38:49) and 94% (36:38) of breast cancer tissues and metastatic lymph tissues, respectively, were positive for HBXIP expression. Moreover, overexpression of HBXIP was able to promote the migration of MCF-7 cells. Interestingly, HBXIP was able to regulate IL-8 transcription by NF-κB, suggesting that the two target genes of miR-520b are functionally connected. In addition, we found that miR-520b could indirectly regulate IL-8 transcription by targeting HBXIP. Thus, we conclude that miR-520b is involved in regulating breast cancer cell migration by targeting HBXIP and IL-8 via a network in which HBXIP promotes migration by stimulating NF-κB-mediated IL-8 expression. These studies point to HBXIP as a potential therapeutic target for breast cancer.
Keywords: Breast Cancer, Cell Migration, Cellular Regulation, Gene Transcription, MicroRNA, HBXIP, IL-8, miR-520b
Introduction
MicroRNAs (miRNAs)3 are a class of small noncoding RNAs that act as post-transcriptional regulators of gene expression. It is currently estimated that the human genome may contain at least 700 miRNAs (1, 2). Emerging evidence has demonstrated that miRNAs can function as oncogenes or tumor suppressors involved in neoplasm development, progression, diagnosis, and prognostication (3, 4). It was recently reported that miRNAs are mutated or differentially expressed in breast cancer, with profound positive or negative effects on breast cancer cell migration. For instance, miR-10b is highly expressed in human and mouse metastatic breast cancer cell lines and positively regulates cell migration (5), and MiR-27b stimulates cell migration by targeting ST14 (6). Let-7g targets collagen type I α2 and inhibits cell migration in hepatocellular carcinoma (7). Although a large number of miRNAs have been identified to date, their roles in breast cancer cell migration and the underlying mechanisms of this regulation remain to be determined.
Hepatitis B X-interacting protein (HBXIP) was originally identified by its interaction with the C terminus of the hepatitis B virus X protein (8). It forms a complex with survivin, leading to the suppression of apoptosis initiated through the mitochondrial cytochrome c pathway (9). HBXIP was shown to interact with the hSuv3 protein, which encodes an NTP-dependent DNA/RNA DEXH box helicase predominantly localized in mitochondria (10). Furthermore, HBXIP also plays a critical role in mitosis, serving as a regulator of centrosome duplication in HeLa human carcinoma cells (11). Our previous studies demonstrated that HBXIP could promote cell proliferation by regulating the transcriptional activity of nuclear factor-κB (NF-κB) (12), which plays a role in the regulation of cell migration (13). However, the role of HBXIP in cell migration is still unclear.
Interleukin-8 (IL-8), alternatively known as CXCL8, is a well characterized member of the CXC chemokine family (14). Overexpression of IL-8 has been observed in a subset of tumors, including breast cancer cells (15, 16), and promotes breast cancer cellular migration and invasion (17, 18). IL-8 gene transcription is induced by various factors, such as NF-κB (19–22). However, the role of miRNAs in regulating IL-8 expression remains poorly understood.
We recently showed that miR-520b is down-regulated in breast cancer cells with high metastatic potential relative to the cells with low metastatic potential (23). Furthermore, miR-520b can decrease the sensitivity of breast cancer cells to complement dependent cytotoxicity and circumvent the host immune system by directly targeting the 3′UTR of CD46 (23). However, the function of miR-520b in the regulation of breast cell migration remains unclear.
In this study, we show that down-regulation of miR-520b contributes to the migration of breast cancer cells with high metastasis via a network that involves two target genes, HBXIP and IL-8, in which HBXIP promotes migration by up-regulation of IL-8 mediated by NF-κB.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF-7 cell line and LM-MCF-7 cell line (a metastatic subclone from the MCF-7 breast cancer cell line) (24) were cultured in RPMI 1640 medium (Invitrogen). Breast cancer cell line MDA-MB-231 was cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 100 units/ml penicillin, 100 units/ml streptomycin, and 1% glutamine. Cultures were incubated at 37 °C in a humidified atmosphere with 5% CO2.
Plasmid Construction
The target sites of miR-520b in HBXIP 3′UTR and IL-8 3′UTR were amplified by PCR. We included specific primers for IL-8 3′UTR (forward primer, 5′-CGT TCT AGA AAT GGG TTT GCT AGA ATG TG-3′, and reverse primer, 5′-GGG GGC CGG CCC AAT GAC AAG ACT GGG AGT A-3′) and HBXIP 3′UTR (forward primer, 5′-CGT TCT AGA GAA CAT TAT GAT CCA GAA R-3′, and reverse primer, 5′-GGG GGC CGG CCC CTC CAA ACA GAT TTA TTG-3′). Both of the target fragments were inserted into the XbaI site, downstream of the luciferase gene in the pGL3-control vector (Promega, Madison, WI). The resulting vectors were sequenced and named pGL3-IL-8–3′UTR and pGL3-HBXIP-3′UTR. Site-directed mutants of the miR-520b target sites in pGL3-IL-8–3′UTR and pGL3-HBXIP-3′UTR were named mu-pGL3-IL-8–3′UTR and mu-pGL3-HBXIP-3′UTR, using pGL3-IL-8–3′UTR and pGL3-HBXIP-3′UTR as templates, respectively. Mutagenesis primers used were as follows: 5′-GTG AGG ACA TGT GGA AGA TGC TTA AGT TTT TTC-3′ and 5′-GCA TCT TCC ACA TGT CCT CAC AAC ATC ACT GTG-3′ for mu-pGL3-IL-8–3′UTR; 5′-GCA GCA GGT CCA GGT ACT CTT GTA TAT AGA AT-3′ and 5′-GAG TAC CTG GAC CTG CTG CTT CAA AAC AT-3′ for mu-pGL3-HBXIP-3′UTR.
Full-length HBXIP plus 3′UTR was amplified from MCF-7 cells using primers (forward primer, 5′-GTC GGA TCC ATG GAG CCA GGT GCA GG-3′, and reverse primer, 5′-GGC CTC GAG AAA CAG ATT TAT TGA TAC AG-3′) and then cloned into pCMV-Tag 2B vector. The resulting vector was sequenced and named pCMV-hbxip-3′UTR.
A 180-bp fragment of the human IL-8 promoter was amplified by PCR using forward primer, 5′-GGG CTC GAG GAA GTG TGA TGA CTC AGG-3′, and reverse primer, 5′-GTG AAG CTT GTG TGC TCT GCT GTC TCT-3′ (21) and cloned into the pGL3-basic vector (Promega), allowing transcription of the firefly luciferase gene under the control of this fragment. The corresponding plasmid was used for site-directed mutagenesis as described by others to mutate the NF-κB site (21). Primers used for mutagenesis was 5′-GGA TGG GCC ATC AGT TGC AAA TCG Tta AcT TTC CTC TGA CAT AATG-3′. All constructs were confirmed by DNA sequencing.
miRNA, Small Interfering RNA, and Plasmid Transfection
miR-520b (5′-AAA GUG CUU CCU UUU AGA GCG-3′), anti-miR-520b (5′-CCC UCU AAA AGG AAG CAC UUU-3′), small interfering RNAs (siRNAs) targeting human IL-8 mRNA (NM_000584.2 68 to 83), human NF-κB-p65 mRNA (NM_001145138, 781 to 801), and control siRNA were designed and synthesized by RiboBio (Guangzhou, China). Transfection with miR-520b, anti-miR-520b, RNAi reagents, and different doses of plasmids were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Establishment of Stable Cell Lines
Stable cell lines were generated by transfecting plasmids (pCMV-Tag 2B, pCMV-hbxip, pSilencer-random, and pSilencer-hbxip vectors) (11) into breast cancer cells with Lipofectamine 2000. Stable cell lines were generated by selection in 800 μg/ml G418 (Invitrogen) and could be maintained in culture with 400 μg/ml G418. The engineered cell lines were named as follows: MCF-7-pCMV (stably transfected pCMV-Tag 2B empty vector), MCF-7-hbxip (stably transfected pCMV-hbxip plasmid), LM-MCF-7 (or MDA-MB-231)-pSilencer-random (stably transfected pSilencer vector containing a random fragment), and LM-MCF-7-pSilencer-hbxip (or MDA-MB-231) (stably transfected pSilencer vector containing the HBXIP RNAi fragment).
Treatment of Breast Cancer Cells
Cells were cultured in serum-free medium for 12 h. In brief, the cells were treated with PDTC (20, 40, or 60 μm) (an inhibitor of NF-κB, Sigma) for 2 h, respectively. The treated cells were subjected to RT-PCR.
Luciferase Reporter Gene Assay
Luciferase activity was determined using a luciferase reporter assay system (Promega) on a luminometer (TD-20/20; Turner Designs, Sunnyvale, CA) according to the manufacturer's instructions. Briefly, cells grown in 24-well plates were co-transfected with 0.2 μg of a plasmid encoding firefly luciferase under the control of wild-type or mutated promoter and 0.1 μg of pRL-TK encoding Renilla luciferase. The pCMV-Tag 2B empty vector, pGL3-basic plasmid, and pGL3-control plasmid were used as controls. Cells were harvested 48 h after transfection and lysed in 1× passive lysis buffer. The luciferase readings of each sample were normalized against the Renilla luciferase levels. All of the data shown in this study were obtained from at least three independent experiments.
Total RNA Isolation, RT-PCR, and Real Time PCR
Total RNA of cells was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized with PrimeScript reverse transcriptase (Takara Bio, China) and oligo(dT). The conditions of PCR were as follows: 95 °C for 5 min, 95 °C for 30s, 52 °C annealing for 30 s, and 72 °C for 30 s followed by 30 cycles. Real time PCR was performed as described previously (23). The conditions of real time PCR were as follows: 95 °C for 10 s, 95 °C for 5 s, and 53 °C for 1 min to anneal and 72 °C for 15 s for 45 cycles. Melt curves did not detect nonspecific amplification. We included specific primers for IL-8 (forward primer, 5′-GGT CTT GAG AGA TGG CTT GC-3′, and reverse primer, 5′-AAT TGG CAA AGC CGT AGT TG-3′). As a control, GAPDH was amplified with specific primers (forward primer, 5′-CAT CAC CAT CTT CCA GGA GCG-3′, and reverse primer, 5′-TGA CCT TGC CCA CAG CCT TG-3′).
Chromatin Immunoprecipitation Assay (ChIP)
The ChIP assay was performed using the EpiQuikTM chromatin immunoprecipitation kit from Epigentek Group Inc. (Brooklyn, NY). Protein-DNA complexes were immunoprecipitated with NF-κB-p65 antibodies, with anti-RNA polymerase II as a positive control antibody, and normal mouse IgG as a negative control antibody. DNA from these samples was subjected to PCR analyses with the following: NF-1, 5′-CAT CAG TTG CAA ATC GTG GA-3′, and NF-2, 5′-GAA GCT TGT GTG CTC TGC TG-3′; GAPDH-1, 5′-GTA TTC CCC CAG GTT TAC AT-3′, and GAPDH-2, 5′-TTC TGT CTT CCA CTC ACT CCT-3′, followed by sequencing. Amplification of soluble chromatin prior to immunoprecipitation was used as an input control.
Western Blot
Methods for Western blot were described previously (23). Briefly, cells were washed with cold PBS three times and lysed in RIPA cell lysis buffer (10 mmol/liter HEPES, pH 7.4, 0.15 mol/liter NaCl, 1 mmol/liter MgCl2, 1 mmol/liter CaCl2, 1 mmol/liter dithiothreitol, 0.1% SDS, 0.1% Nonidet P-40, and 20 μg/ml leupeptin). Protein concentrations were estimated by the Bradford assay (Bio-Rad). Equal amounts of total protein were loaded for immunoblotting. Following SDS-PAGE, resolved proteins were electrotransferred to PVDF membranes (Millipore, Billerica, MA). The membranes were blocked overnight in TBS containing 0.1% Tween 20 (TBST) and 5% skim milk. Membranes were then probed with primary antibody in TBST for 2 h at room temperature or overnight at 4 °C, followed by three 15-min TBST washes at room temperature. Blots were incubated with HRP-conjugated secondary antibody for 1 h and washed three times for 10 min with TBST prior to chemiluminescence detection using ECL substrate (Amersham Biosciences). The primary antibodies used for Western blot were rabbit anti-human HBXIP (1:2000 dilution, Sigma) monoclonal anti-human β-actin (1:1000 dilution, Sigma), and secondary goat anti-rabbit HRP (1:2000 dilution, Bio-Rad). All experiments were repeated three times.
Nuclear Extraction
To examine the nuclear translocation of NF-κB, the nuclear extraction assay was performed by using NE-PER® nuclear and cytoplasmic extraction kit (Pierce). The MCF-7 cells stably transfected with pCMV empty vector or pCMV-hbxip (2 × 106) were harvested, respectively. According to the manufacturer's instructions, the nuclear proteins and cytoplasmic ones were extracted, respectively, followed by Western blot analysis to examine the level of NF-κB in the nuclear extract as shown above under “Western Blot.” Rabbit anti-histone H3 (1:100 dilution, Santa Cruz Biotechnology) was used as loading control.
Tissue Microarray Slides and Evaluation of Immunohistochemical Staining
The tissue array with breast cancer tissues, lymph node tissues with breast cancer metastasis, and normal breast tissues was obtained from the Xi'an Aomei Biotechnology Co., Ltd. (Xi'an, China), which includes duplicate core biopsies (1 mm in diameter) from fixed, paraffin-embedded tumors. Immunohistochemical staining of samples was performed as reported previously (25) using a rabbit anti-human HBXIP monoclonal antibody (Sigma). Light microscopic images were documented using a Nikon Eclipse Ti-U fluorescence microscope (Nikon Corp., Japan) with an attached SPOT RT digital camera (Diagnostic Instruments, Inc., Sterling Heights, MI). The number of cells showing positive nuclear and/or cytoplasmic staining for HBXIP was determined by reviewing the entire slide (25). On the basis of the percentage of cells with positive nuclear and/or cytoplasmic staining, staining patterns were classified on a six-grade scale: 0, absence of any nuclear or cytoplasmic staining; 1+, <10% cells with positive nuclear and/or cytoplasmic staining; 2+, 10–25% positive cells; 3+, 26–50% positive cells; 4+, 51–75% positive cells; 5+, >75% positive cells. For statistical analysis, breast carcinoma patients were subgrouped into negative staining (0, absence of cell nuclear and/or cytoplasmic staining), HBXIP low staining (1–3, nuclear and/or cytoplasmic staining less than 50%), and HBXIP high staining (4 and 5, nuclear and/or cytoplasmic staining >51%) groups. These values were multiplied to provide a single HBXIP scale for each case. The assessment of immunostaining intensity was performed by two or three independent observers.
ELISA for IL-8
IL-8 levels was determined using a commercially available assay (human IL-8 ELISA kit, ADL) according to the manufacturer's instructions. Breast carcinoma cells were seeded in 6-well plates at a density of 2 × 105 cells per well. When the cultures reached 70–80% confluence, fresh medium was applied and collected after 24 h of incubation. IL-8 levels were determined by ELISA reader system (Labsystem, Multiskan Ascent). Triplicate cultures of cells were tested for each experimental condition, and the results are given in pg/ml per 24 h per 106 cells. The protein concentrations in these extracts were determined using a standard protein assay (Bio-Rad).
Immunofluorescence Analysis
Cells were plated on coverslips and fixed using 4% paraformaldehyde for 15 min. Coverslips were rinsed with PBS, and cells were permeabilized with 0.2% Triton X-100 for 30 min. Following three PBS washes, the cells were incubated with anti-NF-κB-p65 antibody (1:100, Santa Cruz Biotechnology) for 1 h. After being washed with PBS, cells were incubated with rhodamine-labeled goat anti-rabbit secondary antibody (1:100, Sigma) for 1 h and 4′,6-diamidino-2-phenylindole (30 nm) for 15 min in the dark. After three washes with PBS, images were captured under a microscope using a digital camera and processed using Spot Advanced 4.5 software.
Wound Healing Assay
Cells were seeded in a 6-well plate and cultured for 24 h to form confluent monolayers. A wound was created by dragging a pipette tip through the monolayer, and plates were washed using pre-warmed PBS to remove cellular debris. Wound images were photographed when the scrape wound was introduced (0 h) and 24 h after wounding. The wound gaps were measured at each time point.
Boyden's Chamber Assay
96-Well Boyden's chambers was used to measure the migratory ability of each cell line. Cells were trypsinized and resuspended in the culture medium 36 h post-transfection. Cells (2 × 104) were plated into the upper chambers on uncoated 8-μm pores in serum-free medium. Medium with 10% FBS was added to the lower chambers as a chemoattractant. After 12 h of incubation, cells remaining on the upper surface of the membrane were removed with a cotton swab, and cells that migrated through the membrane filter were fixed and stained with Giemsa solution for 2 h. The migrated cells were visualized under a microscope, and five random fields were counted.
MTT Assay
Cell growth assays were carried out using MTT reagent (Sigma) as described previously (26). In brief, transfected cells were trypsinized, counted, and plated into duplicate wells of 96-well plates at a density of 103 cells per well. After incubation for different time periods (24, 48, 72, or 96 h), MTT was added directly to each well. Incubation was continued for 4 h, and then the supernatant was removed and 100 μl of dimethyl sulfoxide (DMSO) was added to stop the reaction. Absorbance at 570 nm was measured using an ELISA reader system (Labsystem, Multiskan Ascent). All experiments were performed in triplicate.
Statistical Analysis
Statistical analysis was performed using Sigma Plot 2001 (Systat Software Inc., Richmond, CA). Statistical significance was assessed by comparing mean values (±S.D.) using Student's t test. p < 0.05 was considered significant.
RESULTS
miR-520b Is Responsible for the Migration of Breast Cancer Cells
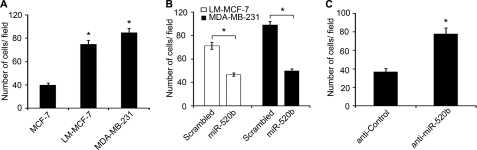
Cell migration is an essential process in cancer metastasis. We previously showed that the expression levels of miR-520b were significantly lower in highly metastatic breast cancer cell lines, including MDA-MB-231 and LM-MCF-7, than in poorly invasive MCF-7 cells (23). Consistent with their metastatic potential, MDA-MB-231 and LM-MCF-7 cells have high migratory potentials compared with MCF-7 cells (Fig. 1A), so we speculated that miR-520b might be involved in breast cancer migration. To test this hypothesis, miR-520b mimics and a commercially synthesized anti-miR inhibitor (anti-miR-520b) were used to alter the levels of miR-520b in breast cancer cells. Overexpression of miR-520b resulted in a significant decrease in cell migration compared with scrambled negative control (Scrambled) in LM-MCF-7 and MDA-MB-231 cells in Boyden's chamber assays (Fig. 1B). Transfection of MCF-7 cells with miR-520b resulted in the decrease of migration capability that was similar to the data in LM-MCF-7 and MDA-MB-231 cells (supplemental Fig. 1A). However, anti-miR-520b significantly promoted the migration of MCF-7 cells relative to anti-control (Fig. 1C). Similar results were observed for LM-MCF-7 and MDA-MB-231 cells (supplemental Fig. 1A). Moreover, cell migration capacity in wound healing assays provided similar data to the Boyden's chamber assay data (supplemental Fig. 1B). We also examined cell viability by MTT assay. Expression of miR-520b inhibited the growth of LM-MCF-7 and MDA-MB-231 cells. Conversely, anti-miR-520b was able to increase the growth of MCF-7 cells (supplemental Fig. 1C), suggesting that miR-520b can inhibit cell growth.
FIGURE 1.
miR-520b affects the migration of breast cancer cells. A, migration potential of MCF-7, LM-MCF-7, and MDA-MB-231 cells was measured by Boyden's chamber assays (*, p < 0.05 versus MCF-7 cells, Student's t test). Graphs indicate the average number of cells per field, 48 h after transfection. B, cell migration potential of LM-MCF-7 or MDA-MB-231 treated with 100 nm miR-520b mimics or scrambled negative control (Scrambled) was measured by Boyden's chamber assay (*, p < 0.05 versus control, Student's t test). C, cell migration potential of MCF-7 cells transfected with 100 nm miR-520b inhibitor (anti-miR-520b) or anti-control was measured by Boyden's chamber assay (*, p < 0.05 versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error.
miR-520b Directly Targets HBXIP
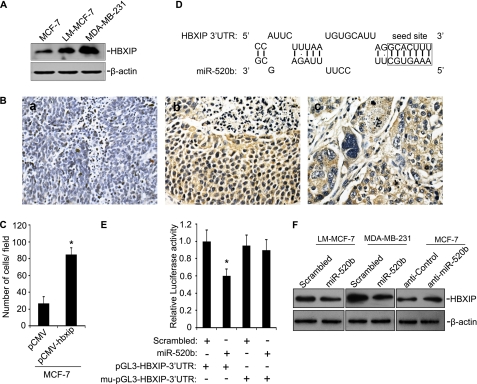
To explore the mechanism underlying the effect of miR-520b on breast cancer cell migration, we searched for targets using an miRNA target prediction program (DIANA-microT version 3.0) and the data base (CapitalBio, Beijing, China). Among the predicted genes, we were particularly interested in HBXIP because it has been the subject of our research in recent years. Previously, we found that HBXIP enhanced cell proliferation via NF-κB (12, 27), which prompted us to speculate that it might also be involved in breast cancer cell migration. We therefore examined the expression levels of HBXIP in different breast cancer cells. LM-MCF-7 and MDA-MB-231 cells exhibited higher levels of HBXIP relative to MCF-7 cells (Fig. 2A). HBXIP expression was assessed by immunohistochemistry on tissue arrays that contained 49 cases of breast cancer, 38 cases of breast carcinoma metastatic to lymph nodes, and 11 samples of normal tissue microarray samples. Clinical characteristics are listed in supplemental Table 1. We found that positive rates of HBXIP in primary tumor tissues and metastatic lymph tissues were 75% (38:49) and 94% (36:38), respectively (Fig. 2B). However, HBXIP expression was not detectable in normal breast tissues (0:11), suggesting that HBXIP may be a novel oncoprotein. We further investigated whether HBXIP influenced migration of breast cancer cells by Boyden's chamber assay. As expected, MCF-7-hbxip cells showed an 2.8-fold increase in migration compared with the control (Fig. 2C). To confirm that HBXIP enhances growth of breast cancer cells, we used MTT assays to examine cell viability. HBXIP was able to enhance cell viability from 48 to 96 h (supplemental Fig. 2).
FIGURE 2.
HBXIP is an miR-520b target gene. A, expression of HBXIP protein in MCF-7, LM-MCF-7, and MDA-MB-231 cells was determined by Western blot. B, immunohistochemical staining shows the expression levels of HBXIP in metastatic lymph node tissues and breast cancer tissues using tissue arrays. Panel a, negative control; panel b, breast cancer tissues with HBXIP-positive staining; panel c, metastatic lymph node tissues. Nuclei were counterstained with hematoxylin. Imaged at ×400 magnification. C, cell migration potential of stable cell lines, including MCF-7-pCMV and MCF-7-hbxip, was measured by Boyden's chamber assay (*, p < 0.05 versus MCF-7-pCMV, Student's t test). D, bioinformatics analysis showed the predicted interaction between miR-520b and its binding site in the 3′UTR of HBXIP. E, luciferase reporter gene assays show the interaction of the 3′UTR of HBXIP and miR-520b by co-transfection with 100 nm miR-520b mimics (or control) and plasmid pGL3-HBXIP-3′UTR (or mu-pGL3-HBXIP-3′UTR) 48 h post-transfection (*, p < 0.05 versus control, Student's t test). F, MDA-MB-231 and LM-MCF-7 cells were transfected with 100 nm miR-520b mimics or control. MCF-7 cells were transfected with 100 nm miR-520b inhibitor (anti-miR-520b) or anti-control. After 48 h, HBXIP protein levels were analyzed by Western blot. The data presented are from three independent experiments; error bars represent standard error.
To test whether miR-520b directly targets the predicted binding site in the HBXIP 3′UTR (Fig. 2D), wild-type or mutant 3′UTRs of HBXIP were cloned into the pGL3-control reporter construct. Co-transfection of pGL3-HBXIP-3′UTR and the miR-520b mimics resulted in a significant decrease of luciferase activity of the reporter in LM-MCF-7 cells, whereas expression of the mu-pGL3-HBXIP-3′UTR did not (Fig. 2E). Next, we examined the effect of miR-520b on the regulation of HBXIP. We found that the miR-520b mimics were able to markedly decrease the expression of endogenous HBXIP in LM-MCF-7 and MDA-MB-231 cells, whereas HBXIP expression was significantly up-regulated in MCF-7 cells with anti-miR-520b (Fig. 2F). Taken together, these data suggest that miR-520b is able to regulate breast cancer cell migration by directly targeting HBXIP.
IL-8 Is a Direct Target of miR-520b
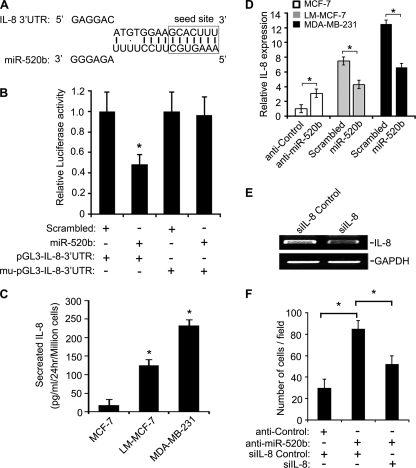
Using on-line tools (DIANA-microT version 3.0, Targetscan 5.1, and miRanda), we also identified IL-8, which is an essential regulator of cell migration (30), as a potential target gene of miR-520b (Fig. 3A). We then confirmed that the 3′UTR of IL-8 is a target of miR-520b. As expected, miR-520b significantly inhibited the luciferase activity of the pGL3-IL-8–3′UTR but not mu-pGL3-IL-8–3′UTR (Fig. 3B). We also examined IL-8 levels in conditioned medium from several breast cancer cells with different metastatic potentials by ELISA. We found a positive association between the level of IL-8 and cell motility (Fig. 3C). Next, we investigated whether miR-520b could regulate endogenous IL-8 protein expression. Ectopic expression of miR-520b (100 nm) resulted in a significant reduction of IL-8 protein levels in conditioned medium from LM-MCF-7 and MDA-MB-231 cells, although knockdown of miR-520b with anti-miR-520b (100 nm) led to an increase of IL-8 protein in conditioned medium from MCF-7 cells (Fig. 3D). To investigate the effect of IL-8 on miR-520b-mediated cell migration, MCF-7 cells were treated with anti-miR-520b and/or IL-8 siRNA, and the levels of IL-8 transcripts were confirmed by RT-PCR (Fig. 3E), and Boyden's chamber assays were used to measure cell migration. Transfection of IL-8 siRNA blocked the migration induced by anti-miR-520b in MCF-7 cells (Fig. 3F). Similar results were observed in wound healing assays (supplemental Fig. 3). Overall, these data strongly suggest that the IL-8 gene is a direct target of miR-520b and is involved in breast cancer cell migration.
FIGURE 3.
miR-520b directly targets IL-8. A, bioinformatics analysis showed the predicted interaction between miR-520b and its binding site in the IL-8 3′UTR. B, luciferase activity assay demonstrated an interaction between the 3′UTR of IL-8 and miR-520b. LM-MCF-7 cells were transfected with 100 nm miR-520b mimics (or control) and plasmid pGL3-IL-8–3′UTR/mu-IL-8 3′UTR. After 48 h, the luciferase activities were determined (*, p < 0.05 versus controls, Student's t test). C, levels of IL-8 in conditioned medium (CM) from MCF-7, LM-MCF-7, and MDA-MB-231 cells were analyzed by ELISA (*, p < 0.05 versus MCF-7, Student's t test). D, MDA-MB-231 and LM-MCF-7 cells were transfected with 100 nm miR-520b mimics or control. MCF-7 cells were transfected with 100 nm miR-520b inhibitor or anti-control. After 48 h, IL-8 protein levels were analyzed by ELISA (*, p < 0.05 versus control, Student's t test). E, effect of IL-8 siRNA on expression of IL-8 mRNA was examined by RT-PCR. F, MCF-7 cells were co-transfected with miR-520b inhibitor (100 nm) and/or siRNA for IL-8 (50 nm), followed by Boyden's chamber assay (*, p < 0.05 versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error.
HBXIP Regulates IL8 by NF-κB
Recent studies have reported associations between gene products that are co-regulated by the same miRNA (28, 29). We therefore investigated the relationship between HBXIP and IL-8. We previously reported that HBXIP was able to activate NF-κB in hepatoma cells (12). IL-8 is a downstream effector of NF-κB that is involved in breast cancer cell migration (17, 30). We hypothesized that HBXIP could up-regulate IL-8 by NF-κB.
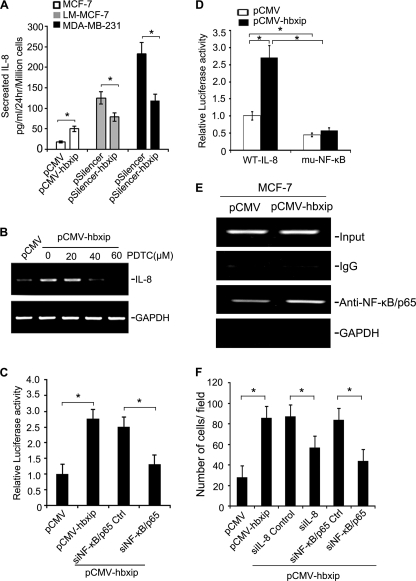
We first investigated the effect of HBXIP on IL-8 protein levels. After 24 h of incubation, conditioned medium from the stable cell lines was collected and examined by ELISA. Interestingly, up-regulation of IL-8 was observed in stable MCF-7-hbxip cells, although down-regulation of HBXIP led to markedly decreased levels of IL-8 in LM-MCF-7 (or MDA-MB-231)-pSilencer-hbxip cell lines (Fig. 4A). The protein levels of HBXIP in each cell line were confirmed in parallel by Western blot analysis (supplemental Fig. 4A). We also examined cell viability in these cells using MTT assays. Cell growth from 48 to 96 h was also affected in cell lines in which IL-8 was up- or down-regulated by HBXIP (supplemental Figs. 2 and 4B), suggesting that during the regulation of IL-8 mediated by HBXIP cell viability was affected by HBXIP as well.
FIGURE 4.
HBXIP stimulates IL-8 by NF-κB. A, ELISAs show the levels of IL-8 in conditioned medium from the engineered breast cancer cells (*, p < 0.05 versus control, Student's t test). B, MCF-7-hbxip cells were treated with increasing concentrations of PDTC, an inhibitor of NF-κB (20, 40, 60 μm) for 2 h. The expression of IL-8 mRNA was analyzed by RT-PCR. C, MCF-7-hbxip cells were transfected with 50 nm siRNAs for NF-κB-p65 (or control siRNA) along with the reporter construct WT-IL-8. After 48 h, cells were lysed for the luciferase assay reporter gene assay (*, p < 0.05 versus control, Student's t test). D, MCF-7-pCMV and MCF-7-hbxip cells were transiently transfected with WT-IL-8 or muNF-κB. After 48 h, cells were lysed for the luciferase reporter gene assay (*, p < 0.05 versus control, Student's t test). E, ChIP assay were performed on cell lysates from MCF-7-pCMV and MCF-7-hbxip cells. The lysates were cross-linked with 1% formaldehyde, and chromatin was immunoprecipitated with anti-NF-κB p65 or normal IgG, which served as the negative control. The genomic fragments associated with immunoprecipitated DNA were isolated and amplified by PCR using specific primers flanking the NF-κB-binding site within the IL-8 promoter. The input was used as a positive control, and equal amounts of the cell lysates were loaded; GAPDH was used as the negative control. F, MCF-7-hbxip cells were transfected with 50 nm siRNAs for IL-8 (or NF-κB-p65). Then Boyden's chamber assays were performed to evaluate the effect of siRNAs on the function of HBXIP (*, p < 0.05 versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error.
Next, we examined the effect of HBXIP on NF-κB activity. We found that HBXIP increased the promoter activity of NF-κB in a dose-dependent manner in MCF-7 cells by luciferase reporter gene assay (supplemental Fig. 4C). Translocation of NF-κB to the nucleus is preceded by the phosphorylation, ubiquitination, and proteolytic degradation of IκB (31). To determine whether the HBXIP-mediated NF-κB activation was caused by IκB phosphorylation, we examined the level of cytoplasmic IκB protein phosphorylation by Western blot. Interestingly, we observed that overexpression of HBXIP corresponded with an increase in the level of phosphorylated IκB, which corresponded to the level of p65 in the nucleus in a concentration-dependent manner (supplemental Fig. 4D). In addition, HBXIP-induced translocation of p65 from cytoplasm to nucleus was also observed by immunofluorescence staining (supplemental Fig. 4E). These data suggest that HBXIP is able to activate NF-κB in breast cancer cells.
To determine the involvement of NF-κB in the regulation of IL-8 transcription by HBXIP, the effect of NF-κB inhibition on the mRNA levels of IL-8 in the stable MCF-7-hbxip cells was examined using different concentrations of PDTC. We found that PDTC significantly lowered HBXIP-induced IL-8 release in MCF-7-hbxip cells in a dose-dependent manner (Fig. 4B). In addition, we constructed a luciferase reporter containing the wild-type IL-8 proximal promoter (WT-IL-8). As shown in Fig. 4C, co-transfection of pCMV-hbxip and WT-IL-8 resulted in a 2.6-fold increase of luciferase activity in MCF-7 cells relative to control cells, whereas silencing of NF-κB-p65 by siRNA abolished the effect. Western blot analysis showed efficient silencing by siRNA targeting NF-κB-p65 in MCF-7 cells (supplemental Fig. 4F). We next constructed an IL-8 proximal promoter mutated at the NF-κB-binding site (muNF-κB), which was transiently transfected into MCF-7-pCMV cells or pCMV-hbxip cells. Luciferase activity in control cells was significantly reduced (about 53%) for the NF-κB mutated construct compared with the wild-type promoter. Luciferase activity in HBXIP-overexpressing cells transfected with the NF-κB mutated construct was reduced close to 80% compared with cells expressing the wild-type promoter (Fig. 4D).
We further investigated the role of HBXIP on IL-8 regulation by chromatin immunoprecipitation (ChIP). ChIP showed that overexpression of HBXIP resulted in an increase in the occupancy of NF-κB on its binding site in IL-8 promoter (Fig. 4E), confirming that HBXIP up-regulates IL-8 by NF-κB.
To test whether HBXIP regulated breast cancer cell migration involves NF-κB and IL-8, we transiently transfected into MCF-7-hbxip cells siRNAs targeting NF-κB-p65 or IL-8 mRNA. Boyden's chamber assays showed that knockdown of IL-8 or NF-κB-p65 significantly abrogated the effect of HBXIP on cell migration (Fig. 4F). Taken together, these results suggest that up-regulation of IL-8 transcription by HBXIP via NF-κB can enhance the migration of breast cancer cells.
miR-520b Indirectly Regulates IL-8 by HBXIP
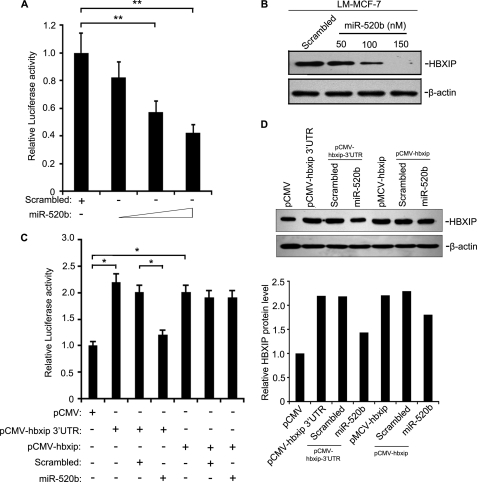
Given the effect of HBXIP on IL-8 transcription, we wondered whether this regulation was influenced by miR-520b. We found that miR-520b mimics decreased the promoter activity of IL-8, as well as the protein levels of HBXIP, in a dose-dependent manner in LM-MCF-7 cells after 48 h of transfection (Fig. 5, A and B). We also examined cell viability by MTT 48 h after transfection with miR-520b. miR-520b inhibited the proliferation of LM-MCF-7 cells in a dose- and time-dependent manner (supplemental Fig. 5). To further investigate the effect of miR-520b on IL-8 transcription by HBXIP, we cloned the HBXIP coding sequence and HBXIP coding sequence with the HBXIP 3′UTR into pCMV-Tag 2B vectors, termed pCMV-hbxip and pCMV-hbxip-3′UTR, respectively. We found that transient transfection of pCMV-hbxip or pCMV-hbxip-3′UTR into MCF-7 cells significantly enhanced the promoter activity of IL-8. In contrast, miR-520b (100 nm) mimics significantly reduced the IL-8 transcriptional activity in pCMV-hbxip-3′UTR-transfected MCF-7 cells relative to controls (Fig. 5C). Western blot analysis verified the expression of HBXIP in the engineered cell lines (Fig. 5D). These data suggest that miR-520b regulates IL-8 indirectly.
FIGURE 5.
miR-520b represses the transcription of IL-8 by HBXIP. A, LM-MCF-7 cells were co-transfected with increasing concentrations of miR-520b mimics (50, 100, and 150 nm) and WT-IL-8. After 48 h, cells were lysed for the luciferase reporter gene assay (**, p < 0.01 versus control, Student's t test). B, in parallel, HBXIP expression was analyzed by Western blot analysis. C, MCF-7 cells were co-transfected with miR-520b (or control) along with plasmid pCMV-hbxip-3′UTR (or pCMV-hbxip) and WT-IL-8. After 48 h, cells were lysed for luciferase reporter gene assays (*, p < 0.05 versus control, Student's t test). D, expression of HBXIP was examined by Western blot assay in above system. The data were analyzed by applying Glyco Band-Scan software. The data presented are from three independent experiments; error bars represent standard error.
DISCUSSION
Accumulating evidence indicates that aberrant expression of miRNAs, such as miR-10b (5), miR-21(32), miR-146 (33), and miR-29a (34), is associated with tumor cell migration. Recently, Li et al. (35) found lower levels of miR-520b in hepatocellular carcinoma HepG2 cells compared with hepatocyte L-O2 by miRNA microarray, which is similar to our previous reports. Cell motility is an essential feature of the cancer cell metastatic process. We previously found that expression levels of miR-520b were significantly lower in highly metastatic breast cancer cell lines, including MDA-MB-231 and LM-MCF-7, than in MCF-7 cells (23). In this study, we focused on the role of miR-520b in controlling the migration of breast cancer cells.
We found that the levels of miR-520b were inversely associated with the migratory behavior of breast cancer cells (Fig. 1 and supplemental Fig. 1). We next set out to identify the targets of miR-520b. We predicted that HBXIP was a direct target of miR-520b by bioinformatics analysis (Fig. 2D) and confirmed this hypothesis by biochemical analysis (Fig. 2E). HBXIP has been the primary research focus in our laboratory in recent years. We previously suggested that HBXIP was an evolutionary conserved protein that could promote cancer cell proliferation via activating NF-κB (12). The association of NF-κB with cancer cell growth and motility has been well documented (36–38). However, the role of HBXIP in cell migration remains unclear. We hypothesized that HBXIP might also be involved in breast cancer migration. The results shown here demonstrate that HBXIP is positively correlated with metastasis in several breast cancer cell lines (Fig. 2A). In particular, HBXIP was highly expressed in clinical breast cancer tissues and metastatic lymph node tissues (Fig. 2B), suggesting that HBXIP may be a novel oncoprotein. We also provide functional evidence of the possible role of HBXIP in cell migration by showing that overexpression of HBXIP in MCF-7 cells was able to promote cell motility (Fig. 2C).
Thus far, two miR-520b targets have been described as follows: NKG2D ligand MHC class I-related chain A (29) and CD46 (23), suggesting a role of miR-520b in cancer immunoediting. Here, we selected IL-8 for further analysis among the predicted targets of miR-520b because it can modulate cancer cell migration and metastasis through several signaling pathways, such as phosphatidylinositol 3-kinase and Rho-GTPases (17, 20, 39). We confirmed the correlation between IL-8 expression and migration of breast cancer cells (Fig. 3B). Moreover, our data showed that miR-520b could suppress the migration of breast cancer cells by targeting IL-8 (Fig. 3). Recently, it has been reported that miRNAs such as miR-21 (40) and miR-27b (28) may target multiple components of certain pathways. O'Day and Lal (41) reported that among the 34 genes known to be altered by 11 miRNAs in breast cancer, 19 formed a well connected gene interaction network. This pattern allows miRNAs to function more effectively on cellular pathways. The following question remains. What is the link between two miRNA targets? It has been shown that the NF-κB site in the IL-8 promoter is the predominant regulator of IL-8 gene transcription by numerous signals (42–44). Here, we showed that HBXIP was able to up-regulate IL-8 and that it required an intact NF-κB-binding site in the promoter region of IL-8 (Fig. 4 and supplemental Fig. 4), suggesting that the two target genes of miR-520b are functionally connected.
To rule out the possibility that cell growth is involved in the changes in migration caused by miR-520b or HBXIP, we used MTT to examine cell growth and viability. We found that either miR-520b or HBXIP was able to influence cell viability at 48 h post-transfection (supplemental Figs 1, 2, and 4B). Thus, both miR-520b and HBXIP are likely to be involved in the regulation of cell growth. However, Boyden's chamber assays showed that both miR-520b and HBXIP were involved in the regulation of cell migration of breast cancer cells from 36 to 48 h. They are unlikely to influence proliferation over these 12 h. Thus, we conclude that miR-520b and/or HBXIP is involved in the regulation of cell migration, which is not simply due to changes in proliferation.
It has been suggested that miRNAs indirectly regulate IL-8 transcription by target-induced NF-κB activation (45, 46). We proposed that miR-520b-regulated IL-8 transcription involves HBXIP. Our data showed that miR-520b was able to regulate IL-8 transcription by targeting HBXIP (Fig. 5). Therefore, we conclude that miR-520b is involved in the promotion of breast cancer cell migration by a regulatory network involving HBXIP and IL-8. Potentially, these target genes of miR-520b, especially HBXIP, may serve as novel therapeutic targets.
In summary, our data demonstrate that miR-520b is involved in the regulation of breast cancer cell migration by targeting the HBXIP and IL-8. Moreover, HBXIP is able to regulate IL-8 production by NF-κB activation. These findings provide an increased understanding of the mechanism of miRNAs in the regulation of breast cancer progression.
This work was supported in part by National Basic Research Program of China 973 Program Grants 2007CB914804, 2007CB914802, and 2009CB521702, Natural Scientific Foundation of China Grants 30770826 and 81071623, and Tianjin Natural Scientific Foundation Grant 08JCZDJC20700.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Table 1.
- miRNA
- microRNA
- HBXIP
- hepatitis B virus X-interacting protein
- HBX
- hepatitis B virus X protein
- NF-κB
- nucleus factor-κB
- MTT
- methyl thiazolyl tetrazolium
- PDTC
- ammonium pyrrolidinedithiocarbamate.
REFERENCES
- 1. Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Sharon E., Spector Y., Bentwich Z. (2005) Nat. Genet. 37, 766–770 [DOI] [PubMed] [Google Scholar]
- 2. Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 3. Zhang B., Pan X., Cobb G. P., Anderson T. A. (2007) Dev. Biol. 302, 1–12 [DOI] [PubMed] [Google Scholar]
- 4. Bushati N., Cohen S. M. (2007) Annu. Rev. Cell Dev. Biol. 23, 175–205 [DOI] [PubMed] [Google Scholar]
- 5. Ma L., Teruya-Feldstein J., Weinberg R. A. (2007) Nature 449, 682–688 [DOI] [PubMed] [Google Scholar]
- 6. Wang Y., Rathinam R., Walch A., Alahari S. K. (2009) J. Biol. Chem. 284, 23094–23106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang G., Mao W., Zheng S. (2008) FEBS Lett. 582, 3663–3668 [DOI] [PubMed] [Google Scholar]
- 8. Melegari M., Scaglioni P. P., Wands J. R. (1998) J. Virol. 72, 1737–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marusawa H., Matsuzawa S., Welsh K., Zou H., Armstrong R., Tamm I., Reed J. C. (2003) EMBO J. 22, 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Minczuk M., Mroczek S., Pawlak S. D., Stepien P. P. (2005) FEBS J. 272, 5008–5019 [DOI] [PubMed] [Google Scholar]
- 11. Wen Y., Golubkov V. S., Strongin A. Y., Jiang W., Reed J. C. (2008) J. Biol. Chem. 283, 2793–2803 [DOI] [PubMed] [Google Scholar]
- 12. Wang F. Z., Sha L., Ye L. H., Zhang X. D. (2008) Acta Pharmacol. Sin. 29, 83–89 [DOI] [PubMed] [Google Scholar]
- 13. Prasad S., Ravindran J., Aggarwal B. B. (2010) Mol. Cell. Biochem. 336, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ben-Baruch A. (2006) Cancer Metastasis Rev. 25, 357–371 [DOI] [PubMed] [Google Scholar]
- 15. Bendrik C., Dabrosin C. (2009) J. Immunol. 182, 371–378 [DOI] [PubMed] [Google Scholar]
- 16. Benoy I. H., Salgado R., Van Dam P., Geboers K., Van Marck E., Scharpé S., Vermeulen P. B., Dirix L. Y. (2004) Clin. Cancer Res. 10, 7157–7162 [DOI] [PubMed] [Google Scholar]
- 17. Wu K., Katiyar S., Li A., Liu M., Ju X., Popov V. M., Jiao X., Lisanti M. P., Casola A., Pestell R. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6924–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yao C., Lin Y., Chua M. S., Ye C. S., Bi J., Li W., Zhu Y. F., Wang S. M. (2007) Int. J. Cancer 121, 1949–1957 [DOI] [PubMed] [Google Scholar]
- 19. Chavey C., Mühlbauer M., Bossard C., Freund A., Durand S., Jorgensen C., Jobin C., Lazennec G. (2008) Mol. Pharmacol. 74, 1359–1366 [DOI] [PubMed] [Google Scholar]
- 20. Kim S. W., Hayashi M., Lo J. F., Fearns C., Xiang R., Lazennec G., Yang Y., Lee J. D. (2005) Cancer Res. 65, 8784–8791 [DOI] [PubMed] [Google Scholar]
- 21. Freund A., Jolivel V., Durand S., Kersual N., Chalbos D., Chavey C., Vignon F., Lazennec G. (2004) Oncogene 23, 6105–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin Y., Huang R., Chen L., Li S., Shi Q., Jordan C., Huang R. P. (2004) Int. J. Cancer 109, 507–515 [DOI] [PubMed] [Google Scholar]
- 23. Cui W., Zhang Y., Hu N., Shan C., Zhang S., Zhang W., Zhang X., Ye L. (2010) Cancer Biol. Ther. 10, 232–241 [DOI] [PubMed] [Google Scholar]
- 24. Ye L. H., Wu L. Y., Guo W., Ma H. T., Zhang X. D. (2006) Zhonghua Yi Xue Za Zhi 86, 61–65 [PubMed] [Google Scholar]
- 25. Zhang X., Dong N., Yin L., Cai N., Ma H., You J., Zhang H., Wang H., He R., Ye L. (2005) J. Med. Virol. 77, 374–381 [DOI] [PubMed] [Google Scholar]
- 26. Shan C., Xu F., Zhang S., You J., You X., Qiu L., Zheng J., Ye L., Zhang X. (2010) Cell Res. 20, 563–575 [DOI] [PubMed] [Google Scholar]
- 27. Wang F. Z., Sha L., Zhang W. Y., Wu L. Y., Qiao L., Li N., Zhang X. D., Ye L. H. (2007) Acta Pharmacol. Sin. 28, 431–438 [DOI] [PubMed] [Google Scholar]
- 28. Pan Y. Z., Gao W., Yu A. M. (2009) Drug Metab. Dispos. 37, 2112–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yadav D., Ngolab J., Lim R. S., Krishnamurthy S., Bui J. D. (2009) J. Immunol. 182, 39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin D., Galisteo R., Gutkind J. S. (2009) J. Biol. Chem. 284, 6038–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 32. Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y. Y. (2008) Cell Res. 18, 350–359 [DOI] [PubMed] [Google Scholar]
- 33. Hurst D. R., Edmonds M. D., Scott G. K., Benz C. C., Vaidya K. S., Welch D. R. (2009) Cancer Res. 69, 1279–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gebeshuber C. A., Zatloukal K., Martinez J. (2009) EMBO Rep. 10, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Q., Wang G., Shan J. L., Yang Z. X., Wang H. Z., Feng J., Zhen J. J., Chen C., Zhang Z. M., Xu W., Luo X. Z., Wang D. (2010) J. Gastroenterol. Hepatol. 25, 164–171 [DOI] [PubMed] [Google Scholar]
- 36. Shen H. M., Tergaonkar V. (2009) Apoptosis 14, 348–363 [DOI] [PubMed] [Google Scholar]
- 37. Lerebours F., Vacher S., Andrieu C., Espie M., Marty M., Lidereau R., Bieche I. (2008) BMC Cancer 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Braunstein S., Formenti S. C., Schneider R. J. (2008) Mol. Cancer Res. 6, 78–88 [DOI] [PubMed] [Google Scholar]
- 39. Matsuo Y., Ochi N., Sawai H., Yasuda A., Takahashi H., Funahashi H., Takeyama H., Tong Z., Guha S. (2009) Int. J. Cancer 124, 853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Papagiannakopoulos T., Shapiro A., Kosik K. S. (2008) Cancer Res. 68, 8164–8172 [DOI] [PubMed] [Google Scholar]
- 41. O'Day E., Lal A. (2010) Breast Cancer Res. 12, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. (2002) J. Leukocyte Biol. 72, 847–855 [PubMed] [Google Scholar]
- 43. Cortés Sempere M., Rodríguez Fanjul V., Sánchez Pérez I., Perona R. (2008) Clin. Transl. Oncol. 10, 143–147 [DOI] [PubMed] [Google Scholar]
- 44. Vandercappellen J., Van Damme J., Struyf S. (2008) Cancer Lett. 267, 226–244 [DOI] [PubMed] [Google Scholar]
- 45. Bhaumik D., Scott G. K., Schokrpur S., Patil C. K., Campisi J., Benz C. C. (2008) Oncogene 27, 5643–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strum J. C., Johnson J. H., Ward J., Xie H., Feild J., Hester A., Alford A., Waters K. M. (2009) Mol. Endocrinol. 23, 1876–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]