Abstract
Selective therapeutics for nuclear receptors would revolutionize treatment for endocrine disease. Specific control of nuclear receptor activity is challenging because the internal cavities that bind hormones can be virtually identical. Only one highly selective hormone analog is known for the thyroid receptor, GC-24, an agonist for human thyroid hormone receptor β. The compound differs from natural hormone in benzyl, substituting for an iodine atom in the 3′ position. The benzyl is too large to fit into the enclosed pocket of the receptor. The crystal structure of human thyroid hormone receptor β at 2.8-Å resolution with GC-24 bound explains its agonist activity and unique isoform specificity. The benzyl of GC-24 is accommodated through shifts of 3–4 Å in two helices. These helices are required for binding hormone and positioning the critical helix 12 at the C terminus. Despite these changes, the complex associates with coactivator as tightly as human thyroid hormone receptor bound to thyroid hormone and is fully active. Our data suggest that increased specificity of ligand recognition derives from creating a new hydrophobic cluster with ligand and protein components.
All metazoan life depends on transcription control by the family of nuclear receptors. Nuclear receptors regulate development and differentiation as well as metabolism and physiology, and their dysfunction contributes to disorders such as diabetes, obesity, cardiovascular disease, and cancer (1). Synthetic hormone analogs have therapeutic potential for altering the function of many nuclear receptors, provided that they are receptor and isoform selective. Agonist ligands of peroxisome proliferator-activated receptor γ are currently used to treat type II diabetes (2–4). Estrogen analogs called selective estrogen receptor modulators that selectively block or activate estrogen receptor isoforms are applied in the therapy of breast cancer and osteoporosis (5, 6).
Although investigations on structure–function relationships show that nuclear receptors possess unique features in regulation, their three-dimensional structures are similar. The ligand-binding domain (LBD) binds hormone and is interdependent on other domains that bind to DNA and coregulators or respond to posttranslational modifications (7). Within the LBD, the critically placed C-terminal helix 12 changes its position and binding surface in an allosteric response to hormone binding (8). The function of this conformational change is to shape the surface for binding of coregulators (9, 10). The coactivator complex attracts further cofactors, which are required for activation of the transcription of target genes (11, 12). The shape and size of the hormone-binding pocket, usually completely buried inside the protein, place severe restrictions on the design of ligands. Any subtle changes in the chemical structure of the hormone might alter the position of helix 12 and so determine the fate of the receptor as repressed or activated.
The synthesis and evaluation of ligands for thyroid hormone receptor (TR), before the structure of the receptor was known, led to the discovery of compounds larger than 3,5,3′-triiodothyronine (T3) that functioned as thyromimetics. In these molecules, the iodine at the 3′ site of T3 was replaced with large rigid groups (13, 14). When the structure of TR bound to thyroid hormone was solved (8), it showed that T3 was completely buried, surrounded by protein and tightly packed without room for chemical groups larger than iodine at the 3′ position. GC-24 is not unlike these T3 analogs that were discovered earlier, possessing a benzyl at the 3′ position of the hormone core moiety. The mystery, in light of the structure of the LBD, is how such compounds bind with normal affinity.
Thyroid hormone influences growth, development, and homeostasis, with important effects on general metabolism, lipid levels, heart rate, and mood (15). Pharmacologic thyroid hormone treatment could be used to combat obesity and lower cholesterol and triglyceride levels but fails in practice because of associated symptoms of hyperthyroidism, in particular, elevated heart rate and arrhythmia (16). Thyroid hormone signals are transduced by two related thyroid receptor subtypes, TRα and TRβ, which are encoded by different genes (17, 18). Studies of TR isoform-specific knockout mice and patients with resistance to thyroid hormone syndrome suggest that TRα mediates the effects of thyroid hormone on heart rate, whereas analogs that exclusively stimulate TRβ might have desirable effects without causing cardiac distress. Indeed, animal studies using thyroid receptor agonists with modest TRβ selectivity have validated this hypothesis (14, 19, 20). Nevertheless, structure-based approaches to develop ligands with further improvements in isoform specificity are limited by the fact that the LBDs of TRα and TRβ are ≈75% identical in amino acid sequence, and that the internal hydrophobic cavities that hold the hormone differ by just one amino acid (Ser-277 in TRα versus Asn-331 in TRβ).
TR ligand design was revolutionized by creating an easily synthesized and potent T3 analog called GC-1. It was reported as a TR agonist with modest β-selectivity (21). The crystal structure of TRβ in complex with GC-1 suggested that this specificity may have been achieved through interactions of the carboxylate tail of GC-1 with the polar part of the hormone-binding pocket (including the TRβ isoform-specific residue Asn-331) (17). It was concluded that a single amino acid difference in the hormone-binding pockets of the TR subtypes could account for the selectivity of GC-1. In this study, we show that a large chemical substitution, appropriately attached to the hormone analog GC-1, improves TRβ selectivity by a factor of 40–60 while retaining normal binding affinity and receptor function. Unlike the case for the modest TRβ-specificity of GC-1 and other TRβ-isoform-specific agonists (22), the marked TRβ specificity of GC-24 is not dependent on the sequence composition of the ligand-binding pocket. Instead, GC-24 moves two framework helices and partners with additional amino acids outside the T3 hormone-binding pocket to sculpt additional surfaces providing further opportunities for specific agonist–receptor interaction.
Materials and Methods
Protein Expression and Purification. Human TRβ LBD was produced in BL21 cells (Stratagene) at 22–23°C by using the pET28 (Novagen) TR-E202 plasmid with an N-terminal six-His tag as described (17). To induce protein expression, 0.5 mM isopropyl β-d-thiogalactoside was added when an OD600 of ≈1.2 was reached, and the incubation continued for 6 h. For the purification protocol of TRβ, see ref. 17. Briefly, cells were lysed and sonicated on ice, and the clear lysate (15,000 rpm, 30 min in a Beckman SS 34 rotor) was loaded onto Talon resin (Clontech) and incubated for 1 h at 4°C. The Talon beads with the bound TR were washed to exclude unspecific binding, and TR was eluted with 100 mM imidazole. The protein, which at this point was ≈85% homogenous, was incubated with GC-24 at a molar ratio of 1.5:1 (ligand:protein) and dialyzed against 20 mM Hepes (pH 8)/1 mM DTT/0.2 mM PMSF overnight at 4°C. To isolate the liganded receptor, TSK-phenyl HPLC (Tosoh) was performed. Fractions containing the receptor/ligand complex were pooled, washed with 20 mM Hepes buffer, concentrated to 10.5 mg/ml, and immediately used for crystallization experiments. The routine overall yield was ≈15 mg/liter of protein/ligand complex that was purified to 99% homogeneity.
Crystallization and X-Ray Analysis. Crystals were grown at 4°C in sitting drops by using compact clover crystallization plates (Emerald Biostructures, Bainbridge Island, WA) by mixing 2 μl of TR/GC-24 complex (10.5 mg/liter) and 2 μl of well solution. The reservoir contained 200 μl of 0.1 M sodium acetate/10% polyethylene glycol 3350/0.15 M ammonium sulfate. Five percent cymal-2 (final concentration, detergent screen 3, No. 22, Hampton Research) and 1.5% glucose (final concentration, additive screen 2, No. 6, Hampton Research) were added to the drop. Hexagonal crystals grew within 2 weeks to a size of ≈0.075 mm × 0.075 mm × 0.035 mm. The space group was determined as P6522 with unit cell dimensions a = 56.57 Å and c = 390.05 Å. Each asymmetric unit contained one monomer of TR complexed with GC-24 and 60% solvent. For cryoprotection, ethylene glycol was added directly to the drop in steps up to a final concentration of 25%, before freezing the crystals in liquid nitrogen. Data were measured to 2.8 Å at –170°C at beamline 8.3.1 at the Advanced Light Source, Berkeley, at a wavelength of 1.1 Å, and the observed reflections were reduced, merged, and scaled with denzo and scalepack (23).
Structure Determination and Refinement. The structure of TRβ/GC-24 was solved by molecular replacement (cns package) using the atomic coordinates for the TRβ/GC-1 complex as a model. The phases from the molecular replacement were used to calculate electron density maps based on 2Fo – Fc and Fo – Fc coefficients. For accuracy, the disordered regions K211–T215 and H238–V264 were removed from the model and rebuilt manually. Alternate cycles of model building in quanta and refinement in cns (rigid body refinement, simulated annealing, individual B-factor refinement, gradient minimization refinement, composite annealed omit maps) produced a model with R/Rfree values of 21.6/26.4%. Part of the loop region between H2 and H3, E248–E261, was disordered and could not be included in the structural model. Q241, E324, and H441 were modeled as alanines. The final model consists of human thyroid hormone receptor β LBD residues G209–P247, G262–D461, and 40 water molecules per asymmetric unit.
Ligand-Binding Assays. Competition assays to determine ligand-binding affinities for wild-type and mutant receptor proteins were performed as described (24, 25). Fifteen femtomoles of purified or in vitro translated receptor LBDs was incubated overnight with 0.5 nM l-[125I]T3 (NEN) and various concentrations of competitor ligand in 100 μl of E400 buffer [400 mM NaCl/20 mM potassium phosphate (pH 8.0)/0.5 mM EDTA/1 mM MgCl2/10% glycerol, along with 1 mM monothioglycerol and 50 μg of calf thymus histones added fresh]. The receptor/[125I]T3 complex was isolated through a Sephadex G-25 column and quantified on a γ counter. Ki values were calculated by using the one-site competition model contained in the prism 3.0 program using known Kd values for T3 binding to receptor or receptor mutant LBDs.
Transfection Assays. HeLa cells were cultured in DME/H21 supplemented with 10% fetal bovine serum (HyClone) and penicillin/streptomycin. Transient transfections were performed using electroporation. Approximately 5 million HeLa cells were resuspended in PBS supplemented with 0.1% glucose and 10 μg/ml BioBrene (Applied Biosystems) in a 0.4-cm gap electroporation cuvette, mixed with DNA, including 2 μg of TRE-luciferase and cytomegalovirus promoter-β-galactosidase internal control to estimate transfection efficiency and, where indicated, 1 μg of TRβ expression vector. The cells were then exposed to 960 μF and 0.25 kV delivered by a Bio-Rad electroporation unit, recovered in standard growth medium, and plated on 12-well dishes. The following day, cells were washed in PBS, re-fed with DME/H21 without serum, and treated with the appropriate concentrations of T3 or GC-24 or control vehicle. Luciferase and β-galactosidase activities were measured in cell extracts prepared by standard methods 24 h later by using luciferase (Promega) and Galacto-Light assay systems (Tropix, Bedford, MA). For the purposes of these studies, maximal T3 induction (usually ≈15-fold) was set at 100%.
Pull-Down Assays. GST-fusions to the nuclear receptor interaction domains of glucocorticoid receptor-interacting protein (GRIP)1 (amino acids 563-1121) and nuclear receptor corepressor (NCoR) (amino acids 1944–2453) were expressed in Escherichia coli BL21. Cultures were grown to OD600 1.2–1.5 at ≈22°C and induced with 1 mM isopropyl β-d-thiogalactoside for 4 h. The cultures were centrifuged, and bacterial pellets were resuspended in 20 mM Hepes (pH 7.9)/80 mM KCl/6 mM MgCl2/1 mM DTT/1 mM ATP/0.2 mM phenylmethylsulfonyl fluoride and protease inhibitors and sonicated. Debris was pelleted by centrifugation in a Beckman SS34 rotor for 1 h at 12,000 rpm. The supernatant was incubated with glutathione-Sepharose 4B beads (Amersham Biosciences) and washed as previously described. Protein preparations were stored at –20°C in 20% glycerol until use. Interactions between [35S]methionine-labeled TRβ and GST-fusion proteins were produced by using coupled in vitro transcription–translation (TNT kit, Promega). The binding reactions were carried out on ice in a volume of 150 μl composed of 137.5 μl of protein-binding buffer (PBB) along with 10 μl of GST-bead slurry corresponding to 3 μg of fusion protein, 1 μl of in vitro translated protein, and 1.5 μl of ligand or vehicle. PBB was freshly prepared in 24-ml aliquots composed of 20 ml of A-150 (20 mM Hepes/150 mM KCl/10 mM MgCl2/1% glycerol) and 2 ml each of PBS supplemented with 1% Triton X-100 and 1% Nonidet P-40. PMSF, DTT, BSA, and protease inhibitor mixture (Novagen) were added to 0.1 mM, 1 mM, 2 μg/ml, and 1/1,000 dilution respectively. The mix was incubated at 4°C with gentle agitation; the beads were pelleted, washed four times with PBB containing no BSA, and dried under vacuum for 20 min. The sample was taken up in SDS/PAGE loading buffer and then subjected to SDS/PAGE and autoradiography.
Results
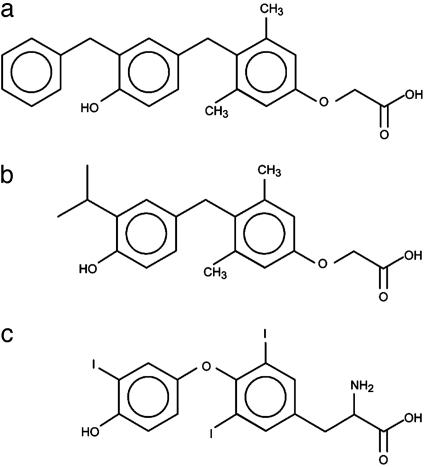
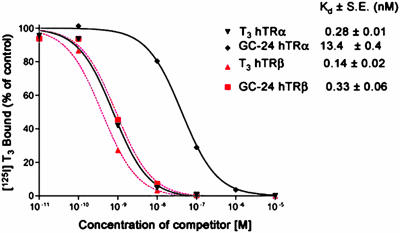
GC-24 Binds with High Affinity and Selectivity to TRβ. GC-24 [3,5-dimethyl-4-(4′-hydroxy-3′-benzyl)benzylphenoxyacetic acid] (Fig. 1a) belongs to a series of analogs that were designed from a thyroid hormone scaffold that was easier to synthesize than the natural hormone. Potentially, compounds in this series could be agonists or antagonists or possibly show selectivity toward the α- or β-subtypes of TR. The first substance that was synthesized in this group and indeed showed β-specificity is GC-1 (21, 26) (Fig. 1b). The major differences between T3 (Fig. 1c) and the GC series are a methylene instead of the ether to bridge the two phenyl rings; the substitution of hydrocarbon residues for the 3, 5, and 3′ iodine atoms; and replacement of the 1-aminopropionic acid with oxyacetic acid. Whereas GC-1, with a 3′-isopropyl substitution at the distal ring, was shown to moderately favor binding to TRβ over TRα (≈3- to 5-fold) (21), GC-24 was designed to test the environs of the LBD for accommodating a larger 3′ substituent. Competition binding studies with radiolabeled T3 revealed that GC-24 has both a high affinity and strong selectivity for the β-subtype (Fig. 2). GC-24 bound TRβ with a Kd that was slightly weaker than that of T3 but showed an average preference for TRβ of ≈40-fold (Fig. 2). Thus, addition of a bulky phenyl extension to the 3′ position of the first aryl ring of GC-1 improves the specificity of binding to TRβ with no reduction in binding affinity.
Fig. 1.
Chemical formulas for TR modulators. (a) GC-24. (b) GC-1. (c) Thyroid hormone (T3).
Fig. 2.
Competition of GC-24 for [125I]T3 binding to TRα and TRβ. Binding affinity was measured by competition of various concentrations of T3 and GC-24 against a fixed concentration of [125I]T3. GC-24 favors binding to TRβ over TRα by a factor of 40, as indicated by Kd values. The affinity of GC-24 for TRβ is comparable with the affinity of T3.
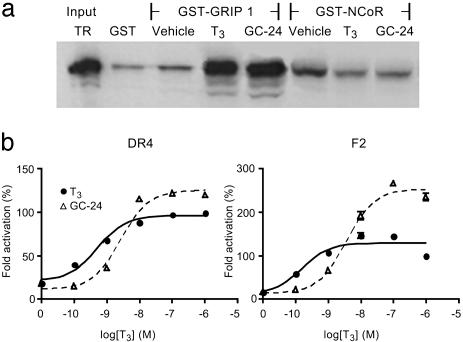
GC-24 Is a Full Agonist for TRβ. The ability of GC-24 to induce the active TR conformation was determined by GST pull-down assays (Fig. 3a). Both T3 and GC-24 showed equivalent ability to promote TRβ interactions with the prototype nuclear receptor coactivator GRIP1. Similar results were obtained with other coactivators, including SRC-1 and TRAP220 (not shown). T3 and GC-24 also showed equivalent ability to promote TRβ dissociation from NCoR. Thus, GC-24 promotes formation of an active receptor conformation required for coactivator binding and corepressor release in vitro. We performed transient transfection assays to test GC-24 for its potential to stimulate transcriptional activation by using reporter genes with two distinct types of consensus thyroid hormone response elements. In the first, TR binding sites (AGGTCA) were arranged as a direct repeat (DR-4); the second used TR binding sites arranged as an inverted palindrome (F2) (Fig. 3b). In both cases, GC-24 showed activity that modestly exceeded that of T3 at saturating doses. Thus, GC-24 behaves as a full agonist in cellular assays.
Fig. 3.
GC-24 is an agonist for TR and activates transcription as well as T3.(a) Autoradiograph of SDS/PAGE gels showing [35S]methionine-labeled in vitro-translated TR bound to bacterially expressed GST fusions to GRIP and NCoR. Pull-down assays show TR input controls (10% of total) and TR bound to GST alone, or TR/buffer, TR/T3, and TR/GC-24 in the presence of GST-GRIP1 or GST-NCoR. TR binds poorly to NCoR when incubated with either T3 or GC-24. (b) Fold activation of transcription of luciferase by TRβ on DR4 and F2 response elements plotted versus concentration of hormone. Luciferase activities were measured in cell extracts, and maximal T3 induction was set at 100%. The superactivation on the F2 element by GC-24 is unexplained.
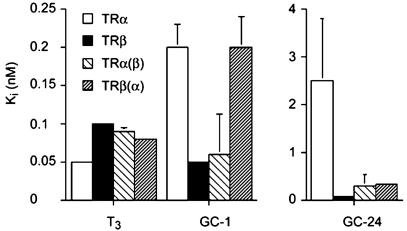
Basis for β-Selectivity of GC-24. We examined the effect of TR hormone-binding pocket mutations on ligand binding to understand the mechanism for the marked TRβ specificity of GC-24 binding by using competition assays (Fig. 4). As expected, T3, the natural hormone, is a high-affinity ligand for both subtypes, GC-1 has a weak TRβ selectivity, and GC-24 strongly favors TRβ. When the hormone-binding pockets of TRα and TRβ are mutated to resemble the other subtype (TRα Ser-277 → Asn to mimic TRβ and TRβ Asn-331 → Ser to represent TRα), there is no effect on T3 binding, but the original preference of GC-1 for TRβ is reversed in favor of TRα. This result confirms our previous assertion that ligand specificity of GC-1 is highly influenced by a single amino acid difference in the hormone-binding pockets of TRα and TRβ and arises from a network of hydrogen bonds that are formed depending on whether Ser or Asn is present (17). A comparable ligand arrangement has been reported for compound 15, another β-selective thyromimetic (22).
Fig. 4.
Binding of T3, GC-1, and GC-24 to TRα and TRβ. Ki values are measured in competition assays to determine ligand-binding affinities for wild-type and mutant receptor proteins. Receptor LBDs were incubated with [125I]T3 and various concentrations of competitor ligand. Ki values relate to relative apparent Kd values. TRα(β) refers to TRα mutated to mimic TRβ, and TRβ(α) refers to TRβ mutated to mimic TRα.
By contrast, the strong preference of GC-24 for TRβ is weakly dependent on the composition of the TR pocket. When we replace Ser-277 in TRα with Asn to resemble the TRβ subtype, binding to TRα is improved, but the specificity is not reversed as for GC-1. The converse is also true for the TRβ Asn-331 → Ser mutation, where binding is weakened but not reduced to the levels observed with TRα. Thus, the disparate amino acid in the hormone pocket alone cannot explain the preferential binding of GC-24 to TRβ. This result indicates that other determinants in TRβ must be responsible for accommodation of GC-24 by TRβ.
Crystallography of GC-24 with Human TRβ. Crystals of the human TRβ LBD complexed to GC-24 were grown and analyzed by x-ray diffraction to provide structural insights (see Materials and Methods). The structure of the TRβ LBD/GC-24 complex was solved by using molecular replacement and refined to an R factor of 21.6%.
We expected few changes relative to T3 or GC-1 but were surprised to find that the crystallization conditions and crystal parameters changed (Table 1). These changes likely relate to a new configuration of the framework helices 3 and 11. The 2Fo – Fc maps contained electron density for a second GC-24 molecule, placed in the potential dimer interface of TR. It is bound in a surface pocket formed by helices 9, 10, and 11 (data not shown).
Table 1. Crystallographic data and refinement statistics.
| Data collection | |
| Data set | hTRβ/GC-24 |
| Resolution, Å | 2.8 (2.98-2.8) |
| Space group | P6522 |
| Cell dimensions, Å | |
| a | 56.57 |
| b | 56.57 |
| c | 390.05 |
| Observed reflections | 113,019 |
| Unique reflections | 9,368 |
| Completeness, % | 93 (81.5) |
| Rsym, %* | 6.9 (16.1) |
| 〈I/O〉 | 15.8 (5.1) |
| Refinement | |
| Rcryst, %† | 21.6 (28.1) |
| Rfree, %‡ | 26.4 (36.9) |
| rmsd for bond lengths, Å | 0.011 |
| rmsd for angles, Å | 1.6 |
| Average B factor, Å2 | 59.8 |
Values in parentheses refer to the highest-resolution shell. rmsd, rms deviation.
Rsym = ΣhΣi|Ih,i - 〈Ih〉|/ΣIh for the intensity (I) of i observations of reflection h.
Rcryst = Σ|Fobs - Fcalc|/Σ|Fobs|.
Rfree is calculated in the same way as Rcryst, using 5% of the reflections that were set aside for cross-validation and not used in refinement.
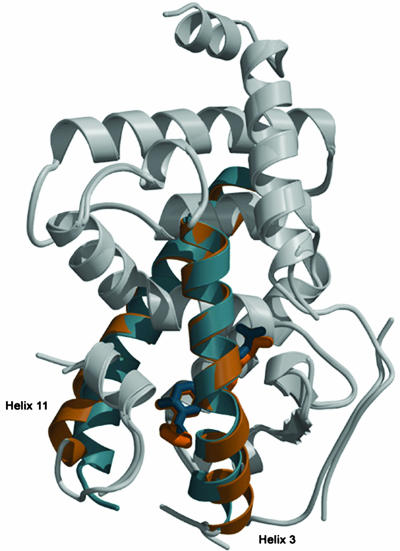
Fig. 5 shows the superimposed structures of the TRβ LBD with GC-24 and GC-1. Of the 12 α-helices and 4 β-strands and their linkages, only helices 3, 11, and a part of the loop between helix 1 and helix 2 show differences. Helix 3 and helix 11 are shifted, and both move outward by 3–4 Å compared with the GC-1 structure. The likely reason for this rearrangement relates to the position of the ligand. The core of GC-24 occupies the hormone-binding pocket formed by helices 3, 5/6, 8, and 11 in approximately the same position as GC-1. However, the aromatic 3′-benzyl extension of GC-24 occupies much more space than the alkyl chain of GC-1 (Fig. 5), and the additional volume required for placing this extension is created by the large shifts in the positioning of helices 3 and 11. Thus, the TR ligand-binding pocket has expanded to accommodate the extra bulk of the ligand.
Fig. 5.
Superimposed structures of TRβ in complex with GC-24 (beige) and GC-1 (blue). The overall structures (gray areas) align very well (rmsd 0.5 Å). The N terminus of helix 3 and the C terminus of helix 11 shift by 3 Å and 4 Å when GC-24 binds. Both helix 3 and helix 11 are straighter in the GC-24 complex. The GC-1 structure has weak density represented as gaps in this figure at positions after helix 1, before helix 3, and after helix 11. The GC-24 structure differs in having weak density only before helix 3.
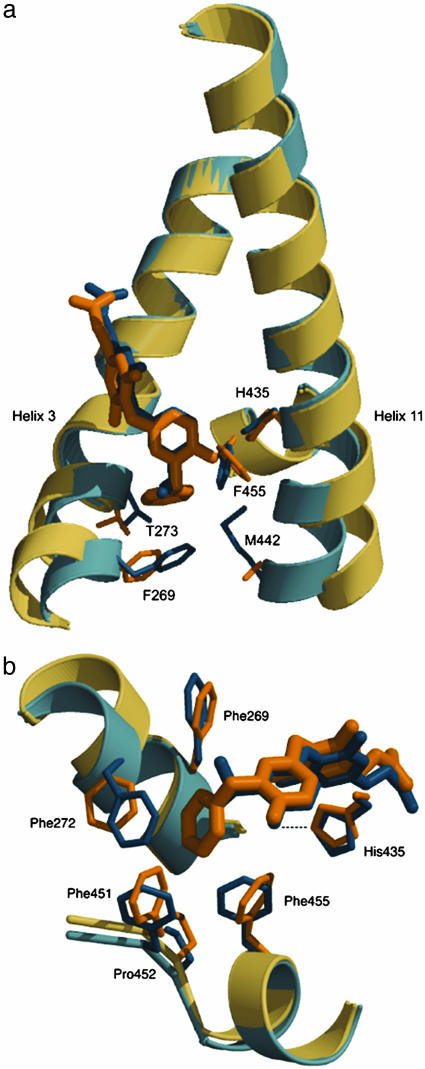
Analysis of the binding of GC-24 reveals changes in the precise network of hydrogen bonds in the polar region of the binding pocket relative to GC-1 and additional contacts to GC-24 in the hydrophobic part of the pocket. GC-24 is shifted by 0.6 Å away from the polar region and toward the hydrophobic region of the pocket relative to GC-1 when the two structures are aligned using the core α-helices (rmsd of 0.5 Å for 120 α-carbons). The carboxylate of the GC-24 molecule, despite the 0.6-Å translation, forms hydrogen bonds with the charged residue Arg-320 and the backbone nitrogen of Asn-331. The polar tail of GC-1 obtains stabilization from additional hydrogen bonds between Arg-282 and the carboxylate. Furthermore, Arg-316 interacts with the carboxylate oxygen of GC-1 through a water-mediated hydrogen bond. Neither contact is available to GC-24. The hydrophobic part of GC-24 is buried in the nonpolar region of the pocket, interacting with Phe-269, Phe-272, and Ile-276 from helix 3; Leu-330 from S3; Leu-341 and Leu-346 from helix 7/8; and Ile-353 from helix 8. The benzyl extension on the ligand makes further contact with Phe-451 (H11) and Phe-455 (H12), two residues that are not involved in binding either T3 or GC-1, which are now used to form part of the hormone-binding pocket. Residues Phe-269, Thr-273, His-435, Met-442, and Phe-455 shift outward to accommodate the GC-24 benzyl moiety (Fig. 6a). To maintain the hydrophobic core for an energetically favorable ligand alignment, Phe-272, Phe-451, and Pro-452 are pulled toward the agonist (Fig. 6b). The benzyl is buried in a hydrophobic pocket formed by four phenylalanines, a proline, and an isoleucine (Fig. 6b). It is likely that the additional hydrophobic contacts in the nonpolar region of the pocket compensate for the poorer fit in the polar part of the cavity.
Fig. 6.
The environment of the hydrophobic benzyl extension of GC-24. GC-24 and surrounding side chains are shown in beige, and GC-1 is shown in blue. (a) Residues most changed by GC-24 binding are found at the start of helix 3 and the C terminus of helix 11. (b) Phe-451, Pro-452, Phe-455, and, to a lesser extent, Ile-276 (residue not shown) enhance the hydrophobic cluster linking helix 11 and helix 12 to the receptor core only in GC-24. The benzyl participates in close packing interactions with six hydrophobic side chains.
Remarkably, the C-terminal H12, which is critical for receptor function, adopts the agonist configuration despite the fact that parts of H3 and H11 are significantly bent. Helix 12 is stabilized by a hydrogen bond between the phenolic hydroxyl of GC-24 and His-435 in H11. His-435 is shifted in the complex of TR/GC-24 to maintain this hydrogen bond, thereby stabilizing the N-terminal portion of H11 and holding H12 in the agonist position.
Discussion
We initiated our study to understand why GC-24 functioned like the natural hormone, T3, rather than inhibiting the action of the receptor. We also wanted to explain why the T3 analog was unexpectedly specific for the β-isoform of the receptor. Placing these observations in a structural context might aid discovery of selective receptor drugs such as α-antagonists or β-agonists, compounds that would have clinical value for the treatment of cardiac disease and hypothyroid-associated ailments such as obesity and hypercholesterolemia.
The design of hormone analogs for nuclear receptors must be guided by the finding that the ligand-binding pocket is inside of the receptor and an integral stabilizing component of its three-dimensional structure. Hormone binding alters the structural elements of the receptor to correctly position helix 12, the register of which is critical to dock the coactivator. Most agonists fit into the hormone-binding pocket like T3 and trigger conformational changes in the LBD correctly for activation (27, 28). Antagonists, on the other hand, must either disrupt the framework structure of the receptor or alter the position of helix 12 that is needed for binding coactivator partners (29, 30). The framework structure for most nuclear receptors is constant. Numerous binding studies of hormone analogs to estrogen receptor, peroxisome proliferator-activated receptor γ, or retinoic acid receptor show that side chains in contact with the ligand may rotate and the position of helix 12 may be altered, but the positions of the other helices of the receptors never change. When the structural framework is changed as is found in some naturally occurring human mutant receptors, activation or stability of the receptor is impaired (31, 32). Point mutations in helix 3 residues that directly contact helix 12, such as T277A in TRβ (33) or V290M in peroxisome proliferator-activated receptor γ (28), weaken the binding of p160-coactivators, because they compromise the interactions of helix 12 with the framework.
Substitutions on hormones can reach toward and alter the position of helix 12, affecting the activity of the receptor. In estrogen receptor, polar extensions to the ligand core displace helix 12 and promote repression (10). Surprisingly, several TR ligands with large substituents, which would be expected to project into helix 3, helix 11, or helix 12 of the receptor, are agonists (34)! The structural basis for the implied correct positioning of helix 12 has been a mystery. Here we have shown that a large group at the 3′ position of the distal ring of the T3 analog GC-24 binds tightly to TRβ and bends helices 3 and 11. The hormone pocket expands by ≈15%, changing its volume from 600 to 700 Å3, and the hormone analog is buried with helix 12 in the correct position to form the coactivator binding surface. Comparing TR/GC-24 with PDB entry 1BSX, which shows TR LBD with the GRIP NR box 2 peptide bound, we find no changes in the positions of the amino acids that bind coactivator within experimental error.
When GC-24 binds, we observe that helix 11 bends near His-435, the link to the hydroxyl of the distal ring of the analog. His-435 is the hinge point in helix 11. However, the C terminus of helix 11 moves by 4.0 Å, displacing two of its three side chains contacting helix 12; Phe-394 moves by 1.1 Å, and Met-442 moves by 3.8 Å. Similarly, the N terminus of helix 3 bends near the hinge residue Ile-275 and shifts by nearly 3.0 Å. This bend moves the side chains of two additional residues in contact with helix 12, Phe-269 and Thr-273, by ≈1.6 Å. We conclude that 4 of the 14 side chains responsible for proper registration of helix 12 move by 1.6–4.0 Å without affecting its position in the protein. The unchanged position of helix 12 likely derives from a stable cluster of hydrophobic side chains for which the details of structure are less critical than the nature of the interactions. The 14 side chains form a compact hydrophobic cluster, and the cluster tolerates small changes in the three-dimensional positions of the components with little loss in free energy, as the relative positions of the hydrophobic atoms are not critical as long as solvent is excluded.
The reasons for the β-selectivity of GC-24 are likely due to the same hydrophobic cluster. The hormone analog is translated by ≈0.6 Å in the pocket, with its 3′ benzyl moving closer into the hydrophobic cluster. GC-24 is accepted because of the flexibility of helix 11 in TRβ. About 20% fewer van der Waals contacts in the range of 3.8–4.2 Å between the last three turns of helix 11 and the body of the receptor are cataloged in TRβ compared with TRα. We speculate that helix 11 is better packed in TRα, and that changes in the spatial volume near the ligand substitution are probably less tolerated in the α subtype.
β-Selectivity may alternatively derive from the stability of the TR–retinoid X receptor (RXR) heterodimer in transcription. The changed position of helix 11 might alter TR–RXR dimer stability. Using peroxisome proliferator-activated receptor–RXR as a model, we built a TR–RXR dimer and found that the bend at His-435 might affect contacts with RXR helix 11. The assays for transactivation (Fig. 3b) show normal to enhanced activation, implying that the functional TR–RXR heterodimer is at least as stable with GC-24 as with T3 bound to TR.
Realizing that a hydrophobic cluster may vary in detail yet provide for a stable and well positioned helix 12, we gain freedom for designing new compounds that may have improved isoform or even receptor specificity. TR is not unique in providing this opportunistic venue for engineering ligands. A similar hydrophobic cluster is found in retinoic acid receptor, and modified clusters are present in both estrogen and androgen receptors. For these two, strategic buried polar side chains make contacts with the steroid D-ring OH group, so the hydrophobic character of the ligand must be carefully designed.
Acknowledgments
We thank J. W. Apriletti for performing binding assays for TR. This work was supported by National Institutes of Health Grants DK-53417 (to R.J.F.), DK-58390 (to R.J.F.), and DK-41842 (to J.D.B.).
Abbreviations: GRIP, glucocorticoid receptor-interacting protein; NCoR, nuclear receptor corepressor; LBD, ligand-binding domain; rmsd, rms deviation; RXR, retinoid X receptor; TR, thyroid hormone receptor; T3, 3,5,3′-triiodothyronine.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1Q4X).
References
- 1.Mangelsdorf, D. J., Thummel, C., Beato, M., Herrlich, P., Schutz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P., et al. (1995) Cell 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu, H. E., Lambert, M. H., Montana, V. G., Plunket, K. D., Moore, L. B., Collins, J. L., Oplinger, J. A., Kliewer, S. A., Gampe, R. T., Jr., McKee, D. D., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 13919–13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore, K. J., Rosen, E. D., Fitzgerald, M. L., Randow, F., Andersson, L. P., Altshuler, D., Milstone, D. S., Mortensen, R. M., Spiegelman, B. M. & Freeman, M. W. (2001) Nat. Med. 7, 41–47. [DOI] [PubMed] [Google Scholar]
- 4.Spiegelman, B. M. (1998) Diabetes 47, 507–514. [DOI] [PubMed] [Google Scholar]
- 5.Brzozowski, A. M., Pike, A. C., Dauter, Z., Hubbard, R. E., Bonn, T., Engstrom, O., Ohman, L., Greene, G. L., Gustafsson, J. A. & Carlquist, M. (1997) Nature 389, 753–758. [DOI] [PubMed] [Google Scholar]
- 6.Gustafsson, J. A. (1998) Curr. Opin. Chem. Biol. 2, 508–511. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro, R. C., Kushner, P. J. & Baxter, J. D. (1995) Annu. Rev. Med. 46, 443–453. [DOI] [PubMed] [Google Scholar]
- 8.Wagner, R. L., Apriletti, J. W., McGrath, M. E., West, B. L., Baxter, J. D. & Fletterick, R. J. (1995) Nature 378, 690–697. [DOI] [PubMed] [Google Scholar]
- 9.Darimont, B. D., Wagner, R. L., Apriletti, J. W., Stallcup, M. R., Kushner, P. J., Baxter, J. D., Fletterick, R. J. & Yamamoto, K. R. (1998) Genes Dev. 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiau, A. K., Barstad, D., Loria, P. M., Cheng, L., Kushner, P. J., Agard, D. A. & Greene, G. L. (1998) Cell 95, 927–937. [DOI] [PubMed] [Google Scholar]
- 11.Fondell, J. D., Guermah, M., Malik, S. & Roeder, R. G. (1999) Proc. Natl. Acad. Sci. USA 96, 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, M. (2001) Trends Endocrinol. Metab. 12, 127–134. [DOI] [PubMed] [Google Scholar]
- 13.Stephan, Z. F., Yurachek, E. C., Sharif, R., Wasvary, J. M., Leonards, K. S., Hu, C. W., Hintze, T. H. & Steele, R. E. (1996) Atherosclerosis 126, 53–63. [DOI] [PubMed] [Google Scholar]
- 14.Taylor, A. H., Stephan, Z. F., Steele, R. E. & Wong, N. C. (1997) Mol. Pharmacol. 52, 542–547. [DOI] [PubMed] [Google Scholar]
- 15.Utiger, R. D. (1995) in The Thyroid: Physiology, Thyrotoxicosis, Hypothyroidism, and the Painful Thyroid, eds. Felig, F. P., Baxter, J. D. & Frohman, C. A. (McGraw–Hill, New York), pp. 435–519.
- 16.Osman, F., Gammage, M. D. & Franklyn, J. A. (2002) Thyroid 12, 483–487. [DOI] [PubMed] [Google Scholar]
- 17.Wagner, R. L., Huber, B. R., Shiau, A. K., Kelly, A., Cunha-Lima, S. T., Scanlan, T. S., Apriletti, J. W., Baxter, J. D., West, B. L. & Fletterick, R. J. (2001) Mol. Endocrinol. 15, 398–410. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, C. C., Weinberger, C., Lebo, R. & Evans, R. M. (1987) Science 237, 1610–1614. [DOI] [PubMed] [Google Scholar]
- 19.Baxter, J. D., Dillmann, W. H., West, B. L., Huber, R., Furlow, J. D., Fletterick, R. J., Webb, P., Apriletti, J. W. & Scanlan, T. S. (2001) J. Steroid Biochem. Mol. Biol. 76, 31–42. [DOI] [PubMed] [Google Scholar]
- 20.Grover, G. J., Mellström, K., Ye, L., Malm, J., Li, Y. L., Bladh, L. G., Sleph, P. G., Smith, M. A., George, R., Vennström, B., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 10067–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiellini, G., Apriletti, J. W., Yoshihara, H. A., Baxter, J. D., Ribeiro, R. C. & Scanlan, T. S. (1998) Chem. Biol. 5, 299–306. [DOI] [PubMed] [Google Scholar]
- 22.Ye, L., Li, Y. L., Mellström, K., Mellin, C., Bladh, L. G., Koehler, K., Garg, N., Garcia Collazo, A. M., Litten, C., Husman, B., et al. (2003) J. Med. Chem. 46, 1580–1588. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 24.Apriletti, J. W., Baxter, J. D., Lau, K. H. & West, B. L. (1995) Protein Expression Purif. 6, 363–370. [DOI] [PubMed] [Google Scholar]
- 25.Swillens, S. (1995) Mol. Pharmacol. 47, 1197–1203. [PubMed] [Google Scholar]
- 26.Trost, S. U., Swanson, E., Gloss, B., Wang-Iverson, D. B., Zhang, H., Volodarsky, T., Grover, G. J., Baxter, J. D., Chiellini, G., Scanlan, T. S. & Dillmann, W. H. (2000) Endocrinology 141, 3057–3064. [DOI] [PubMed] [Google Scholar]
- 27.Ribeiro, R. C., Apriletti, J. W., Wagner, R. L., Feng, W., Kushner, P. J., Nilsson, S., Scanlan, T. S., West, B. L., Fletterick, R. J. & Baxter, J. D. (1998) J. Steroid Biochem. Mol. Biol. 65, 133–141. [DOI] [PubMed] [Google Scholar]
- 28.Kallenberger, B. C., Love, J. D., Chatterjee, V. K. & Schwabe, J. W. (2003) Nat. Struct. Biol. 10, 136–140. [DOI] [PubMed] [Google Scholar]
- 29.Pissios, P., Tzameli, I., Kushner, P. & Moore, D. D. (2000) Mol. Cell 6, 245–253. [DOI] [PubMed] [Google Scholar]
- 30.Marimuthu, A., Feng, W., Tagami, T., Nguyen, H., Jameson, J. L., Fletterick, R. J., Baxter, J. D. & West, B. L. (2002) Mol. Endocrinol. 16, 271–286. [DOI] [PubMed] [Google Scholar]
- 31.Huber, B. R., Sandler, B., West, B. L., Cunha-Lima, S. T., Nguyen, H. T., Apriletti, J. W., Baxter, J. D. & Fletterick, R. J. (2003) Mol. Endocrinol. 17, 643–652. [DOI] [PubMed] [Google Scholar]
- 32.Huber, B. R., Desclozeaux, M., West, B. L., Cunha-Lima, S. T., Nguyen, H. T., Baxter, J. D., Ingraham, H. A. & Fletterick, R. J. (2003) Mol. Endocrinol. 17, 107–116. [DOI] [PubMed] [Google Scholar]
- 33.Collingwood, T. N., Wagner, R., Matthews, C. H., Clifton-Bligh, R. J., Gurnell, M., Rajanayagam, O., Agostini, M., Fletterick, R. J., Beck-Peccoz, P., Reinhardt, W., et al. (1998) EMBO J. 17, 4760–4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiellini, G., Nguyen, N. H., Apriletti, J. W., Baxter, J. D. & Scanlan, T. S. (2002) Bioorg. Med. Chem. 10, 333–346. [DOI] [PubMed] [Google Scholar]