Abstract
Eukaryotic cells monitor and maintain protein quality through a set of protein quality control (PQC) systems whose role is to minimize the harmful effects of the accumulation of aberrant proteins. Although these PQC systems have been extensively studied in the cytoplasm, nuclear PQC systems are not well understood. The present work shows the existence of a nuclear PQC system mediated by the ubiquitin-proteasome system in the fission yeast Schizosaccharomyces pombe. Asf1-30, a mutant form of the histone chaperone Asf1, was used as a model substrate for the study of the nuclear PQC. A temperature-sensitive Asf1-30 protein localized to the nucleus was selectively degraded by the ubiquitin-proteasome system. The Asf1-30 mutant protein was highly ubiquitinated at higher temperatures, and it remained stable in an mts2-1 mutant, which lacks proteasome activity. The E2 enzyme Ubc4 was identified among 11 candidate proteins as the ubiquitin-conjugating enzyme in this system, and San1 was selected among 100 candidates as the ubiquitin ligase (E3) targeting Asf1-30 for degradation. San1, but not other nuclear E3s, showed specificity for the mutant nuclear Asf1-30, but did not show activity against wild-type Asf1. These data clearly showed that the aberrant nuclear protein was degraded by a defined set of E1-E2-E3 enzymes through the ubiquitin-proteasome system. The data also show, for the first time, the presence of a nuclear PQC system in fission yeast.
Keywords: Protein Degradation, Ubiquitin, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitin Ligase, Yeast Metabolism
Introduction
In eukaryotic cells, the ubiquitin-proteasome system (UPS)3 has a pivotal role in multiple cellular events, including the cell cycle, signal transduction, and receptor-mediated endocytosis (1–3). This system is essential for the selective degradation of many cellular proteins. The substrate proteins destined for degradation are first recognized by the ubiquitination machinery, triggering the covalent attachment of a polyubiquitin chain to the target protein. Polyubiquitinated proteins are recognized and degraded by the 26 S proteasome. The formation of the polyubiquitin chain is accomplished by a series of enzymatic reactions catalyzed by three enzymes: an E1 (ubiquitin-activating enzyme), an E2 (ubiquitin-conjugating enzyme), and an E3 (ubiquitin ligase). The step catalyzed by the E3 is crucial in determining substrate selectivity and timing of degradation, which implies that the identification and understanding of E3s is important to elucidate the mechanisms of specific substrate selection.
The UPS also contributes to cellular protein quality control (PQC) (4). Aberrant or misfolded proteins are produced in the cell by mutation or environmental stress. The intracellular accumulation of these proteins causes proteotoxic or harmful effects. In humans, for example, the accumulation of aberrant proteins is thought to be associated with diseases such as Alzheimer, Huntington, Parkinson, and Creutzfeldt-Jakob diseases (5). The removal of these harmful proteins and the maintenance of homeostasis are accomplished through the selective degradation of aberrant or misfolded proteins by the UPS. The involvement of the UPS in PQC in the cytoplasm and in the endoplasmic reticulum (ER) have been well studied, and several E3s for PQC have been identified, including Hrd1 and Doa10, which are involved in ER-associated degradation (6), as well as CHIP (7) and Ubr1 (8), both of which are associated with cytoplasmic PQC.
In contrast, the study of nuclear PQC has been very limited to date. The lack of activity of the autophagy-lysosome system in the nucleus (9) suggests that nuclear PQC depends entirely on the UPS. Supporting this hypothesis, studies demonstrated that the San1 E3 ligase, which shows specificity for nuclear aberrant proteins, plays a pivotal role in nuclear PQC in the budding yeast Saccharomyces cerevisiae (10, 11). Furthermore, recent studies in mammalian cells and primary neurons suggest that the UHRF-2 E3 ligase is an essential molecule for nuclear polyglutamine degradation as a component of the nuclear PQC machinery (12). The high cellular toxicity of aberrant nuclear protein aggregates and their likely association with the neurodegenerative pathology of Huntington disease underscores the importance of investigating the role of nuclear PQC (5).
In the present study, the existence of a nuclear PQC system that functions through the UPS is demonstrated in the fission yeast Schizosaccharomyces pombe. A mutant form of Asf1 (Asf1-30) was used as a substrate for the study of the nuclear PQC system. Asf1 (anti-silencing function 1) was originally identified as a protein whose overexpression inhibits the silencing of chromatin in S. cerevisiae (13) and subsequently as a complex with histones H3/H4 that could facilitate the replication-coupled assembly of nucleosomes by CAF1 in vitro (14). Asf1 is now recognized as a histone chaperone in a wide variety of eukaryotes, including humans (15). The results of the present study show that Ubc4 (E2) and San1 (E3) are the enzymes catalyzing the degradation of the Asf1-30 mutant protein. This is the first report identifying the molecules responsible for nuclear PQC in S. pombe.
EXPERIMENTAL PROCEDURES
Strains, Media, and Genetic Manipulation
The S. pombe strains used in this study are listed in Table 1. Standard yeast culture medium preparation and genetic manipulations of S. pombe were performed as described previously (16, 17). The S. pombe strains were grown in complete YES medium or EMM with the addition of the appropriate auxotrophic supplements (75 mg/liter adenine, leucine, uracil, histidine, lysine) when necessary. SPA medium was used to induce sporulation (16). G418 disulfate (Sigma), Hygromycin B (Wako), and ClonNAT (Sigma) were used as selection agents at a concentration of 100, 150, and 100 mg/liter, respectively. Escherichia coli DH5α was grown in LB medium as a host strain for all plasmid manipulations using the standard methods described previously (18).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| L972 | h− | Laboratory stock |
| PR110 | h+, leu1-32,ura4-D18 | Laboratory stock |
| UMP0 | h+, leu1-32,ura4-D18,his2 | This study |
| SKP561-15 | h−, asf1+-13myc-kanMX6,leu1-32,ura4-D18 | This study |
| SKP593-30 | h−,asf1-30-13myc-kanMX6,leu1-32,ura4-D18 | This study |
| SKP593-33 | h−,asf1-33-13myc-kanMX6,leu1–32,ura4-D18 | This study |
| SKP634-1 | h+, mts2-1, leu1-32,his2,ura4-D18 | Lab stock |
| UMP1 | h+, mts2-1, asf1-13myc-kanMX6,leu1–32,his2,ura4-D18 | This study |
| UMP2 | h+, mts2-1, asf1-30-13myc-kanMX6,leu1,his2 | This study |
| UMP3 | h−, asf1+-GFP-hphMX6,leu1,ura4 | This study |
| UMP4 | h−, asf1-30-GFP-hphMX6,leu1,ura4 | This study |
| Δubc2(1331) | h+, Δubc2::ura4+,leu1-32,ura4-D18,ade6-M210,his7-366 | T. Toda |
| ts ubc4 | h−, ubc4-P61S-ura4+,leu1-32,ura4-D18,ade6-M216 | F. Yamao |
| ts ubc11 | h−, ubc11-140-ura4+,leu1-32,ura4-D18,ade6-M210 | F. Yamao |
| KSP1346 | h+, Δubc1::ura4+,leu1-32,ura4-D18,ade6-M210,his7-366 | T. Toda |
| KSP1347 | h+, Δubc6::ura4+,leu1–32,ura4-D18,ade6-M216,his7-366 | T. Toda |
| KSP1348 | h+, Δubc7::ura4+,leu1-32,ura4-D18,ade6-M210,his7-366 | T. Toda |
| KSP1307 | h+, Δubc8::ura4+,leu1-32,ura4-D18,ade6-M210,his7-366 | T. Toda |
| KSP1309 | h+, Δubc13::ura4+,leu1-32,ura4-D18,ade6-M210,his7-366 | T. Toda |
| KSP1310 | h+, Δubc14::ura4+,leu1-32,ura4-D18,ade6-M216,his7-366 | T. Toda |
| KSP1349 | h+, Δubc15::ura4+,leu1-32,ura4-D18,ade6-M216,his7-366 | T. Toda |
| KSP1350 | h+, Δubc16::ura4+,leu1-32,ura4-D18,ade6-M210,his7-366 | T. Toda |
| UMP5 | h−, asf1-30-13myc-kanMX6,ubc4-P61S-ura4+,leu1-32,ura4-D18,ade6-M216 | This study |
| UMP6 | h−, ubc4+-5FLAG-kanMX6 | This study |
| HY95 | h+, cdc10-V50,leu1-32,ura4-D18 | H. Yamano |
| HYY184 | h+, cdc10-V50,cut9-665,leu1-32 | H. Yamano |
| UMP7 | h−, asf1+-13myc-hphMX6,leu1-32,ura4-D18 | This study |
| UMP8 | h−, asf1-30-13myc-hphMX6,leu1-32,ura4-D18 | This study |
| UMP9 | h+, asf1-30-13myc-natMX6,leu1-32,ura4-D18,ade6 | This study |
| UMP10 | h−, asf1-30-13myc-hphMX6,ΔSPAC167.07c::kanMX6,leu1-32,ura4-D18 | This study |
| UMP11 | h−, asf1-30-13myc-hphMX6,ΔSPBP8B7.23::kanMX6,leu1-32,ura4-D18 | This study |
| UMP12 | h−, asf1-30-13myc-hphMX6,ΔSPBC29A.3.03c::kanMX6,leu1-32,ura4-D18 | This study |
| UMP13 | h−, asf1-30-13myc-hphMX6,Δsan1::kanMX6,leu1-32,ura4-D18 | This study |
| UMP14 | h−, asf1-30-13myc-hphMX6,Δhrd1::kanMX6,leu1-32,ura4-D18 | This study |
| UMP15 | h−, asf1–30-13myc-hphMX6,Δdoa10::kanMX6,leu1–32,ura4-D18 | This study |
| UMP16 | h+, asf1-30-13myc-kanMX6,Δubr1::hphMX6,leu1-32,ura4-D18,his2 | This study |
| UMP17 | h+, asf1-30-13myc-kanMX6,Δubr11::hphMX6,leu1-32,ura4-D18,his2 | This study |
| UMP18 | h−, Δsan1::kanMX6 | This study |
| UMP19 | h+, Δsan1::kanMX6, leu1-32,ura4-D18,his2 | This study |
| UMP20 | h−, san1+-13myc-kanMX6 | This study |
| UMP21 | h−, kanMX6-P3nmt1-GFP-san1+ | This study |
| UMP22 | h−, asf1-30-13myc-natMX6, kanMX6-P3nmt1-GFP-san1+ | This study |
| UMP23 | h−, san1+-13myc-kanMX6, cdc25-22 | This study |
| MY2354 | h−,sad1-1, leu1-32 | M. Yanagida |
| Orc5-H37 | h−,orc5-H37-ura4+,leu1-32,ura4-D18 | H. Kato |
| MY566 | h−,mis12-537-GFP-LEU2+,leu1-32 | M. Yanagida |
| MY3517 | h−,pim1-46,leu1-32 | M. Yanagida |
| MY3285 | h−,cnp1-1,leu1-32 | M. Yanagida |
| UMP24 | h−,Δsan1::kanMX6,mis12-537-GFP-LEU2,leu1-32,ura4-D18,his2 | This study |
| UMP25 | h−,Δsan1::kanMX6,pim1-46,leu1-32 | This study |
| UMP26 | h+, Δsan1::kanMX6,cnp1-1,leu1-32,ura4-D18,his2 | This study |
| UMP27 | h−, Δsan1::kanMX6,sad1-1,leu1-32 | This study |
| UMP28 | h−, Δsan1::kanMX6,orcΔH37-ura4+,leu1-32,ura4 | This study |
| MY624 | h−,mis12-537 | M. Yanagida |
| MY3236 | h−,cnp1+-3HA-His6(lys1+),leu1-32,ura4-D18 | M. Yanagida |
| MY3280 | h−,cnp1-1–3HA-His6(lys1+),leu1-32,ura4-D18 | M. Yanagida |
| UMP29 | h−,mis12-537, Δsan1::kanMX6 | This study |
| UMP30 | h−,cnp1-1-3HA-His6(lys1+),Δsan1::kanMX6, leu1-32,ura4-D18 | This study |
DNA Manipulation and Plasmids
Restriction enzymes (BamHI or SalI) were purchased from TOYOBO. The plasmids pREP1, pREP2, pREP41, and pREP42 (19–21) were used as vectors. The plasmids pFA6a-kanMX6, pFA6a-GFP-kanMX6, pFA6a-13myc-kanMX6, pFA6a-3HA-kanMX6, and pFA6a-kanMX6-P3nmt1-GFP (26); pCR2.1-hphMX6 and pCR2.1-natMX6 (22); and pFA6a-5FLAG-kanMX6 (23) were used as PCR templates to amplify DNA fragments for gap repair cloning or strain construction. Plasmids pREP1-san1–5FLAG, pREP41-san1-GFP, pREP41-ubc4-GFP, pREP2-asf1-5FLAG, pREP2-asf1-30–5FLAG, pREP41-asf1-13myc, pREP41-asf1-30-13myc, pREP42-asf1-13myc, pREP42-asf1-30-13myc, and pREP41-asf1-30-3HA were constructed using the primers listed in (supplemental Table S1) by the gap repair cloning method as described previously (24).
RNA Preparation and RT-PCR
Total RNAs were prepared for RT-PCR as described previously (25). S. pombe cells were grown in YES medium at 36 °C to a density of 1.0 × 107 cells/ml. The cells were precipitated by centrifugation, washed with DEPC-treated H2O, and suspended in 1 ml of the RNA isolation reagent ISOGEN (Nippon Gene), followed by vigorous vortexing for 3 min with glass beads. After centrifugation (10,000 × g for 30 min at 4 °C), the supernatant was precipitated with isopropyl alcohol, and the samples containing total RNA were treated with RQ1 RNase-free DNase (Promega) for 60 min at 37 °C. Samples containing 1.0 μg of total RNA were reverse transcribed by PrimeScript RTase (Takara Bio), followed by semiquantitative PCR using Ex-taq polymerase.
Gene Disruption, Tagging, and Marker Switch
Chromosomal genes were disrupted using PCR generated fragments (26). The 1.6 kb of the kanMX6 module was amplified with flanking homology sequences corresponding to the 5′- and 3′-ends of the target genes. G418-resistant colonies were selected on YES plates containing G418. Correct disruption of the gene of interest was verified by colony PCR. N- and C-terminal tagging of genes with 5FLAG, 13Myc, and GFP was also carried out using PCR-generated fragments, and efficient incorporation of tags was confirmed by colony PCR and immunoblotting with specific antibodies. One-step marker switch from kanMX6 to hphMX6 or natMX6 was performed as described previously (22).
Isolation of asf1 Temperature-sensitive Mutants
To create S. pombe asf1 temperature-sensitive mutants, genomic DNA was prepared from a strain in which the 13myc gene (linked to kanMX6) was inserted into the 3′-end of asf1 driven by its endogenous promoter. The created 3.7-kb asf1+-13myc-kanMX6 fragment was amplified with error-prone PCR (27), followed by transformation of a wild-type strain. G418-resistant colonies were selected at 26 °C, and temperature sensitive transformants were isolated by replica plating at 36 °C. The asf1 ts mutants were transformed with a plasmid containing the asf1+ gene to confirm that the temperature sensitivity was asf1-dependent.
Measurement of Protein Stability
For the analysis of the half-life of proteins, wild-type S. pombe, mutants, and transformants were grown at 26 or 36 °C to a mid-log phase, and then cycloheximide (100 μg/ml) and/or thiamine (2 μm) was added to the cultures. Cells were harvested at the indicated time points, and whole cell extracts were prepared as described previously (28). Protein levels were detected by immunoblotting. ImageJ software (version 1.43, National Institutes of Health) was used to quantify Asf1-30 protein levels. Tubulin was used as a loading control.
Production of Antibodies against the Asf1 Protein
To raise a polyclonal antibody against the Asf1 protein, a purified glutathione S-transferase (GST)-fused Asf1 expressed in E. coli was used according to the following procedure. The 465-bp DNA fragment corresponding to the N terminus of Asf1 (155 amino acids) was amplified by PCR, cloned into pGEX4T-3 (Amersham Biosciences) (a GST fusion protein expression vector), and expressed in E. coli BL21. The soluble GST-Asf1 fusion protein was purified from E. coli extracts by glutathione-Sepharose 4B (Amersham Biosciences) and separated using 12% SDS-PAGE. The purified GST-Asf1 protein eluted from the gel was used as an antigen for the generation of a polyclonal antibody in rabbits. About 1.0 mg of protein was used to raise the antibody. Steps from the injection of antigen into a rabbit to create antiserum were carried out by SCRUM Inc. (Japan). Immunoblotting was performed with the affinity-purified anti-Asf1 antibody. The bacterially expressed GST-Asf1 fusion protein was bound to a PVDF membrane in advance and was used to purify an anti-Asf1 antibody from crude serum by absorbing and releasing as described previously (29).
Indirect Immunofluorescence Staining
S. pombe cells (about 1.0 × 108) were fixed with 3.7% formaldehyde for 1h at room temperature and washed three times with PEM buffer (100 mm PIPES (pH 6.9), 1 mm EGTA, 1 mm MgCl2). Cells were suspended in 1 ml of PEMS (100 mm PIPES (pH 6.9), 1 mm EGTA, 1 mm MgCl2, and 1.2 m sorbitol), and cell walls were digested with 0.3 mg/ml zymolyase100T (Seikagaku) at 37 °C for 30–40 min. Cell pellets were permeabilized by treatment with PEMS containing 1.0% Triton X-100. After washing three times with PEM buffer, cells were incubated with PEMBAL buffer (100 mm PIPES (pH 6.9), 1 mm EGTA, 1 mm MgCl2, 1% BSA, 0.1% NaN3, and 1% lysine-HCl) at 26 °C for 1 h. Then cells were incubated with PEMBAL buffer containing the primary antibodies (c-Myc (9E10) diluted 1:1000 or anti-FLAG(M2) diluted 1:1000) at 26 °C overnight. Cells were washed three times with PEMBAL buffer, followed by incubation with secondary antibodies diluted in PEMBAL buffer (Alexafluor 488 goat anti-mouse IgG (Molecular Probes) diluted 1:1000) at 26 °C overnight. Cells were then washed three times with PEMBAL buffer and mounted on polylysine-coated glass slides with p-phenylenediamine, 90% glycerol containing 0.1% 4′,6′-diamidino-2-phenylindole (DAPI).
Immunoprecipitation of the Asf1 Protein
The S. pombe wild type, mutants, and transformants were grown in YES or EMM medium to the mid-log phase, and cells (3.0 × 108) were harvested by centrifugation and washed once with ice-cold stop buffer (150 mm NaCl, 50 mm NaF, 10 mm EDTA, and 1 mm NaN3, pH 8.0). For immunoprecipitation, HB buffer (25 mm MOPS (pH 7.2), 15 mm MgCl2, 60 mm β-glycerophosphate, 15 mm EGTA, 1 mm DTT, 0.2% Triton X-100, 100 mm NaCl, 1 mm PMSF, and one-fiftieth volume of protease inhibitor mixture (Sigma)) was used to prepare protein extracts. MG132 (Sigma) was also added to the HB buffer to prevent the degradation of polyubiquitinated proteins. The cells were suspended in 0.3 ml of ice-cold HB buffer and then vortexed vigorously with 0.5 ml of glass beads (500 μm; Sigma) using a bead homogenizer at 2500 rpm for 3 min. After centrifugation (10,000 × g for 20 min at 4 °C), the protein concentration in the supernatant was determined by the Bradford assay (Bio-Rad). The resulting lysate was the whole cell extract. A polyclonal antibody against Asf1 and Dynabeads protein A (DYNAL) was used in the immunoprecipitation of the Asf1 protein. A total of 1.0 mg of each whole cell extract was incubated with 20 μl of Dynabeads protein A coated with anti-Asf1 polyclonal antibody for 1 h at 4 °C. The beads were washed three times with 1.0 ml of HB buffer, suspended in SDS loading buffer, and immediately boiled for 3 min. Samples of the immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting.
Detection of Polyubiquitinated Asf1 Protein
Polyubiquitination analysis was performed as described previously (30). For detection of the polyubiquitinated Asf1 protein, wild-type S. pombe or mutants were transformed with the plasmid pREP1-His6-Ubi (provided by Dr. H. Seino), which contains a gene encoding His6-ubiquitin under the nmt1 promoter. Cells were cultured at 26 °C in the presence or absence of thiamine and shifted to 36 °C for 3 h. Then cells (3.0 × 108) were harvested by centrifugation and washed once with ice-cold stop buffer (150 mm NaCl, 50 mm NaF, 10 mm EDTA, and 1 mm NaN3, pH 8.0). Whole cell extracts were prepared in denaturing Buffer G (6 m guanidine HCl, 0.1 m sodium phosphate, and 50 mm Tris-HCl, pH 8.0) and incubated for 1 h with Ni2+-NTA-agarose beads (Qiagen) at room temperature. The beads were then washed four times with Buffer U (8 m urea, 0.1 m sodium phosphate, and 50 mm Tris-HCl, pH 8.0). Precipitated proteins were immunoblotted with anti-Myc antibody (9E10) to examine whether Asf1–13Myc was polyubiquitinated. Blots were also probed against an anti-ubiquitin antibody as a loading control.
Protein Extraction and Immunoblotting
Rapid protein extraction from S. pombe was performed as described previously (28). A 10-μl sample of each protein extract was analyzed by SDS-PAGE using a 10% polyacrylamide gel and then transferred to Immobilon transfer membranes (Millipore) using a wet transfer system. Mouse monoclonal anti-HA antibody (diluted 1:1000; Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), anti-GFP antibody (diluted 1:1000; Roche Applied Science), anti-Myc antibody (diluted 1:2500; Santa Cruz Biotechnology, Inc.), anti-FLAG antibody (diluted 1:1000; Sigma), anti-tubulin antibody (diluted 1:5000; Sigma), and rabbit polyclonal anti-ubiquitin antibody (diluted 1:100; Sigma) were used diluted in 5% dry milk in PBS-T (137 mm NaCl, 8 mm Na2HPO4·12H2O, 2.7 mm KCl, 1.5 mm KH2PO4, and 0.1% Tween 20). Horseradish peroxidase-conjugated anti-mouse IgG (diluted 1:1000; Cell Signaling) and horseradish peroxidase-conjugated anti-rabbit IgG (diluted 1:1000; Cell Signaling) were used as secondary antibodies in 5% dry milk in PBS-T. The secondary antibodies were detected with the ECL system as described by the manufacturer (Amersham Biosciences).
Fluorescence Microscopy
Fluorescence microscopy was carried out with a Leica TCS-SP5 confocal laser scanning microscope (Leica) using ×630 magnification. GFP and Alexafluor 488 fluorescences were observed by illumination at 485 nm. Cells were counterstained with DAPI to visualize the nuclei.
RESULTS
The Asf1-30 Mutant Protein Was Dramatically Reduced at Restrictive Temperatures
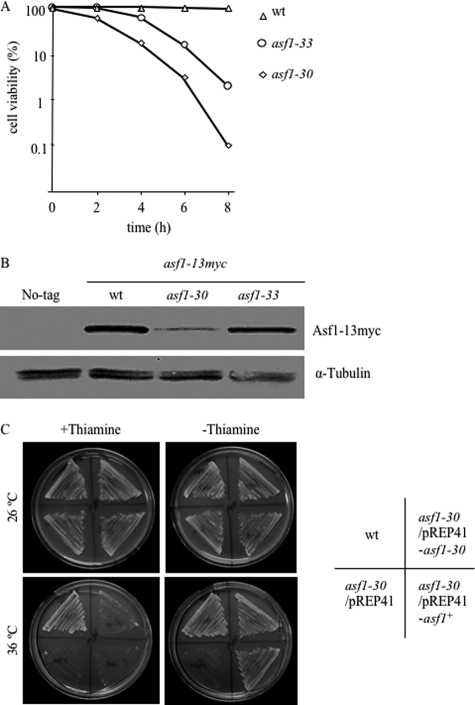
The original objective of this study was to investigate the physiological function of Asf1/Cia1, a histone H3/H4 chaperone of S. pombe. Because the asf1 gene is essential for cell viability (31), temperature-sensitive asf1 mutants were generated using random mutagenic PCR as described under “Experimental Procedures.” This procedure resulted in the isolation of two temperature-sensitive asf1 alleles, which were named asf1-30 and asf1-33. The asf1-30 mutant showed marked temperature sensitivity at 36 °C, whereas the asf1-33 showed moderate temperature sensitivity at 36 °C (Fig. 1A). The asf1-30 mutant contained three missense mutations that resulted in amino acid substitutions at F24S, G120S, and D173G. The asf1-33 allele contained six missense mutations that resulted in the amino acid substitutions A16T, L61P, E119K, L121P, N155S, and E180G. Surprisingly, the Asf1 protein showed a significant decrease in asf1-30 cells at restrictive temperatures, which was not observed in wild-type or asf1-33 cells based on the immunoblotting results (Fig. 1B). These data indicated that the mutations in asf1-30 affected the stability of the Asf1-30 protein, which could be associated with the ts phenotype. To investigate this possibility, the effect of asf1+ or asf1-30 overexpression on the temperature sensitivity of the asf1-30 mutant was examined. The temperature sensitivity was clearly suppressed by overexpression of asf1+ and also by asf1-30 at 36 °C (Fig. 1C). This suppression of the temperature-sensitive phenotype of the asf1-30 mutant by a gene dose effect of asf1-30 further suggested that the instability of the Asf1-30 protein might be a cause of the ts phenotype. Because these results suggested that the phenotype of the asf1-30 mutant could be a result of low Asf1 protein concentrations, the molecular mechanism causing the reduction of the Asf1-30 protein at restrictive temperatures was examined.
FIGURE 1.
Creation of temperature-sensitive asf1 mutants and instability of the Asf1-30 mutant at restrictive temperatures. A, cell viability of wild-type, asf1-30, or asf1-33 ts mutants at the indicated times after a temperature shift to 36 °C from 26 °C. Cell viability was calculated by counting cells (n = 300 cells) for each time point. Each experiment was repeated three times, and the average is shown. B, the protein level of Asf1-30 was reduced at the restrictive temperature. Whole cell extracts were prepared after a 6-h incubation at 36 °C. The Asf1–13Myc protein was detected with an anti-Myc antibody. Tubulin was used as a loading control. C, the asf1-30 mutant showed temperature sensitivity, which was rescued by the plasmid carrying the wild-type asf1+ or asf1-30 genes. Wild-type (wt) and asf1-30 mutants transformed with either a vector control (pREP41) or the same vector carrying asf1+ or asf1-30 were pregrown in EMM medium with 2 μm thiamine. Cells were washed with thiamine-free EMM medium and plated on medium containing 2 μm thiamine or control medium and incubated at 26 or 36 °C for 3 days.
The Mutant Asf1-30 Protein Was Unstable at Restrictive Temperatures
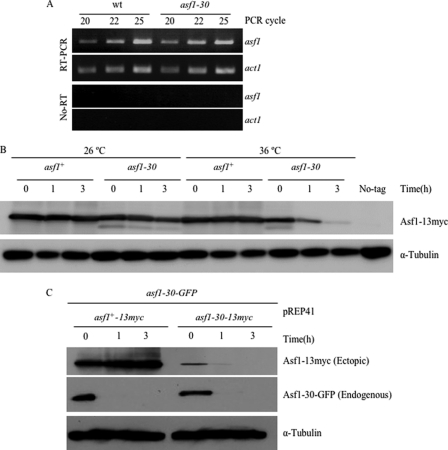
To assess whether the reduction of the Asf1 protein in the asf1-30 mutant was due to a down-regulation of gene expression, the mRNA levels of the asf1 gene were compared between wild-type and asf1-30 mutants at the restrictive temperature by RT-PCR. As shown in Fig. 2A, the mRNA levels of asf1 were not changed in the asf1-30 mutant at 36 °C, indicating that the down-regulation of Asf1-30 in the asf1-30 mutant did not occur at the transcriptional level. To test whether the reduction of the Asf1-30 protein at restrictive temperatures was due to protein instability, the stability of the Asf1 protein was examined at a permissive temperature (26 °C) or a restrictive temperature (36 °C) in the presence of the protein synthesis inhibitor cycloheximide (CHX) by monitoring the amounts of the Asf1 protein tagged with 13myc epitope tags. Whereas the wild-type Asf1 protein was stable at both 26 and 36 °C, the Asf1-30 protein was stable at 26 °C but not at 36 °C (Fig. 2B).
FIGURE 2.
The Asf1-30 mutant protein was unstable at the restrictive temperature. A, the mRNA level of asf1 as monitored by RT-PCR was unchanged between wild type and the asf1-30 mutant. Samples were obtained from wild type and the asf1-30 mutant as indicated. Experiments were done with reverse transcriptase (top two panels) and without reverse transcriptase (bottom two panels). The act1 gene was used as a control. PCR was done through three different cycles (20, 22, and 25). B, instability of the Asf1-30-13Myc protein. The stability of the Asf1 protein was examined at 26 or 36 °C for 0–3 h in the presence of CHX. Wild type and asf1-30 mutant strains carrying the Asf1 protein tagged with the 13Myc epitope on the C terminus were incubated with 100 μg/ml CHX. Asf1 was detected by immunoblotting with an anti-Myc antibody (top). Tubulin was used as a loading control (bottom). C, Asf1-30 was still unstable even in the presence of wild-type Asf1. The asf1+-13myc and asf1-30-13myc genes were placed under the inducible nmt1 promoter in pREP41. These genes were first expressed in the asf1-30-GFP mutant strains at 26 °C in the absence of thiamine. Then these cells were shifted up to 36 °C for 0–3 h in the presence of 2 μm thiamine and 100 μg/ml CHX. Both wild-type Asf1-13Myc and mutant Asf1-30-13Myc proteins were detected by immunoblotting with an anti-Myc antibody (top). The Asf1-GFP protein was detected by immunoblotting with an anti-GFP antibody (middle). Tubulin was used as a loading control (bottom).
Next, the stability of the Asf1-30 protein was examined when wild-type Asf1 was co-expressed in the same cells at 36 °C. For this purpose, the asf1+ gene or the asf1-30 mutant gene was inserted under the thiamine-repressible and middle-strength promoter nmt41 in the 13myc epitope-containing plasmids, and created plasmids (pREP41-asf1+-13myc and pREP41-asf1-30-13myc) were introduced into a strain that expressed asf1-30-gfp. The stability of endogenously expressed Asf1-30-GFP and that of ectopically expressed Asf1-13Myc or Asf1-30-13Myc were examined by immunoblotting against the anti-GFP antibody or the anti-Myc antibody after the addition of thiamine (promoter shutoff) and CHX. As shown in Fig. 2C, the Asf1-30 protein was unstable, whereas its wild-type counterpart was stable at 36 °C. These results strongly suggest that the Asf1-30 protein was selectively degraded in the cells at the restrictive temperature.
Although the Asf1-30 mutant protein was found to be unstable at 36 °C, these results cannot be extended to the endogenous Asf1-30 protein because the antibody used to monitor protein amounts recognizes the tagged epitope. To examine the stability of endogenous Asf1-30, a polyclonal antibody against the purified Asf1 protein was generated and used to detect Asf1 levels by immunoblotting. Unfortunately, the antibody raised did not retain sufficient activity to detect endogenous Asf1 by immunoblotting (data not shown). However, this polyclonal antibody effectively immunoprecipitated the Asf1 protein (supplemental Fig. S1A); the half-life of the Asf1-30-13Myc protein was then examined in the presence of CHX by immunoblotting with the anti-Myc antibody. The Asf1-30 protein had a similar half-life in whole cell extracts and in the immunoprecipitated sample (supplemental Fig. S1B). These results indicated that the mutant Asf1-30 itself was really unstable at 36 °C.
The Asf1-30 Mutant Protein but Not the Asf1 Protein Was Selectively Degraded via the Ubiquitin-Proteasome System
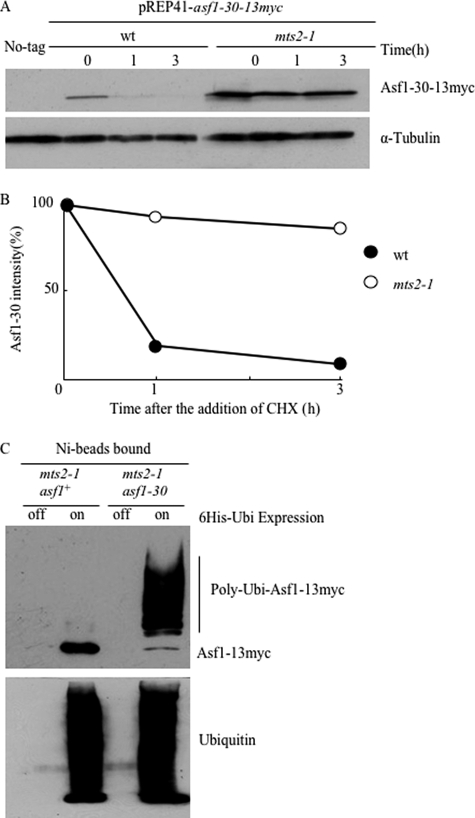
The instability and selective degradation of the Asf1-30 mutant protein at the restrictive temperature suggested the involvement of the ubiquitin-proteasome system. To test this possibility, the temperature-sensitive mts2-1 mutant, which is defective in proteasome activity at restrictive temperatures (32) was used. Ectopically expressed Asf1-30 was greatly stabilized in the mts2-1 mutant in the presence of CHX at the restrictive temperature (Fig. 3A). The half-life of Asf1-30 was estimated to be about 40 min in wild type, whereas it was about 7 h in the mts2-1 mutant (Fig. 3B). To examine whether Asf1-30 is polyubiquitinated in vivo, an asf1-30 mts2-1 double mutant was generated to enable the detection of polyubiquitinated intermediates that are normally undetectable due to rapid proteasome degradation. Unexpectedly, the asf1-30 mts2-1 double mutant showed the synthetic temperature-sensitive phenotype (supplemental Fig. S2A). Despite this temperature-sensitive phenotype, the strain could be used for further testing. Total proteins of the asf1-30 mts2-1 double mutant were prepared at 26 °C or 36 °C and subjected to immunoblot analysis with the anti-Myc antibody. Higher molecular weight forms of Asf1-30-13Myc, which most likely represented the polyubiquitinated protein, were evident at the restrictive temperature in the mts2-1 mutant, although higher molecular weight forms of Asf1-13Myc were not detected under the same conditions (data not shown). To confirm the polyubiquitination of Asf1-30, a His6-tagged ubiquitin (His6-Ubi) was expressed in an mts2-1 strain that also expressed Asf1–13Myc or Asf1-30-13Myc, and polyubiquitinated proteins with His6-Ubi were purified using Ni2+-NTA beads. Immunoblotting with an anti-Myc antibody detected higher molecular weight bands representing polyubiquitinated Asf1-30-13Myc, whereas Asf1–13Myc was hardly polyubiquitinated (Fig. 3C). Control cells that did not express His6-Ubi did not show any bands. These results implied that Asf1-30 but not wild-type Asf1 is selectively polyubiquitinated and degraded via the proteasome in vivo.
FIGURE 3.
The Asf1-30 mutant protein was degraded via the ubiquitin-proteasome system. A, proteasome-dependent degradation of mutant Asf1-30. Wild-type and mts2-1 ts mutants harboring pREP41-asf1-30-13myc were pregrown in EMM medium at 26 °C, followed by a temperature shift to 36 °C. After incubation for 1 h at 36 °C, 2 μm thiamine and 100 μg/ml CHX were added to the medium, and whole cell lysates were prepared at the indicated time points at 36 °C. Asf1-30-13myc was detected by immunoblotting with an anti-Myc antibody (top). Tubulin was used as a loading control (bottom). B, quantification of Asf1-30 protein stability. Band intensities of Asf1-30-13Myc in A were quantified by normalizing relative intensity to tubulin signals. Relative intensity at time 0 was set up as 100% in each case. C, Asf1-30 was polyubiquitinated in vivo, but wild-type Asf1 was not. His6-ubiquitin (pREP1-His6-Ubi) was expressed in asf1+-13myc and asf1-30-13myc strains, and polyubiquitinated proteins were purified with Ni2+-NTA beads in the presence of 6 m guanidine HCl. Immunoblotting was performed with an anti-Myc antibody to examine the polyubiquitination of Asf1 (top). Ubiquitin was also detected with an anti-ubiquitin antibody as a loading control (bottom).
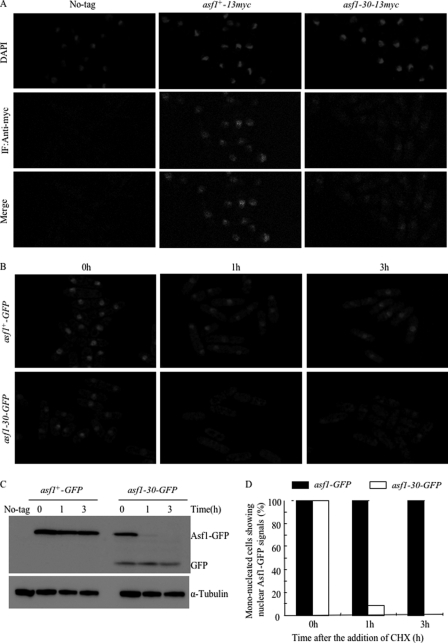
Both Asf1-30 and Asf1 Proteins Are Localized to the Nucleus
Protein localization experiments showed that Asf1 is located in the nucleus (33). The nuclear localization of the Asf1 protein was verified, and the effect of missense mutations in the asf1-30 allele on the nuclear localization of Asf1 at the permissive temperature was examined. To this end, indirect immunofluorescence microscopy was performed on cells fixed with formaldehyde (which preserves nuclear structure) and stained by DAPI and anti-Myc antibodies, using the wild-type, asf1+-13myc, or asf1-30-13myc strains. No nuclear staining was seen when the wild-type strain was used. Although the intensity of the nuclear fluorescent signal in the asf1-30-13myc strain was lower than that in the asf1+-13myc strain, the Asf1-30-13Myc protein was clearly found to localize to the nucleus at the permissive temperature (26 °C) (Fig. 4A). To further observe the localization of Asf1 in living cells, the 13Myc epitope tag at the C terminus of Asf1 was replaced with GFP. Strains that expressed Asf1-GFP grew at a similar rate as wild-type cells and showed a normal appearance when examined microscopically at the permissive temperature. In contrast, the asf1-30-GFP strain exhibited the temperature-sensitive phenotype at the restrictive temperature (36 °C), just like the original asf1-30-13myc strain (data not shown). Cells expressing Asf1-30-GFP were cultured at the permissive temperature (26 °C) and further incubated at the restrictive temperature (36 °C). Cells were observed by microscopy without fixation after adding CHX (100 μg/ml) for 0, 1, and 3 h at the restrictive temperature (36 °C) (Fig. 4B). The Asf1-30-GFP protein at the restrictive temperature was very unstable, whereas Asf1-GFP was stable under this condition (Fig. 4C). The half-life of the Asf1-30-GFP protein at the restrictive temperature was similar to that of the Asf1-30-13Myc protein in the presence of CHX (Fig. 4C). In an asf1+-GFP strain, the nuclear localization of Asf1-GFP was clearly seen before and after the temperature was increased. In contrast, although the nuclear localization of Asf1-30-GFP was clearly seen in all cells (∼100%) before the temperature shift, it disappeared after 3 h of incubation at 36 °C (∼5%) (Fig. 4, B and D). This result indicated that the nuclear localized Asf1-30 protein was unstable at the restrictive temperature. To determine the cellular compartment where polyubiquitinated Asf1-30 is degraded in S. pombe, indirect immunofluorescence microscopy was performed on the cells fixed with formaldehyde. An anti-Myc antibody was used to stain the asf1-30-13myc and asf1-30-13myc mts2-1 strains (supplemental Fig. S3). Although the nuclear fluorescent signal was observed in both asf1-30-13myc and asf1-30-13myc mts2-1 strains at permissive temperature, no nuclear staining was seen in both asf1-30-13myc and asf1-30-13myc mts2-1 strains at restrictive temperature. Fluorescent signal was rather observed in the cytoplasm in asf1-30-13myc mts2-1 strain at restrictive temperature, a condition in which polyubiquitinated Asf1-30 is not degraded. Note that all microscopic observation was carried out at a single focal plane, so that the signals of GFP or indirect immunofluorescence staining could not be seen in a fraction of cells in which the nuclei were out of focus. These data suggested that polyubiquitinated Asf1-30 might not be degraded in the nucleus but instead might be shunted out of the nucleus for degradation in the cytoplasm.
FIGURE 4.
Both Asf1 and mutant Asf1-30 proteins localized to the nucleus at the permissive temperature. A, subcellular localization of endogenous wild-type Asf1 and the mutant Asf1-30. The same cells described in the legend to Fig. 2A were cultured at 26 °C, fixed with formaldehyde, and processed for indirect immunofluorescence (IF) staining with an anti-Myc antibody. Wild-type cells (No-tag) were used as specificity controls. Nuclei were stained with DAPI. The same exposure time was applied in each image. B, the nuclear GFP fluorescence of Asf1 was examined at 36 °C for 0–3 h in the presence of CHX. Wild-type and asf1-30 mutant strains carrying the Asf1 protein fused with GFP epitope tags on the C terminus were incubated with 100 μg/ml CHX for the indicated times. GFP fluorescence images were observed at each time point with a fluorescence microscope. The same exposure time was applied in each picture. C, instability of the Asf1-30-GFP protein. The degradation of the Asf1-GFP protein was examined at 36 °C for 0–3 h in the presence of CHX. Wild-type and asf1-30 mutant strains carrying the Asf1 protein fused with GFP epitope tags on the C terminus were incubated with 100 μg/ml CHX. Asf1 was detected by immunoblotting with an anti-GFP antibody (top). Tubulin was used as a loading control (bottom). D, the percentages of mononucleated cells showing nuclear Asf1-GFP signals as in B were calculated by counting cells (n = 300 cells). Each experiment was repeated three times, and the average numbers are shown.
The Instability of Asf1-30 Is Dependent on ubc4, a Ubiquitin-conjugating Enzyme
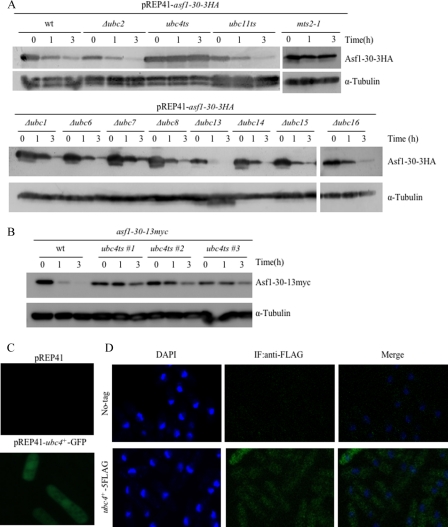
The results described suggest that the Asf1-30 protein was selectively degraded via the ubiquitin-proteasome system. The identification of the E2 ubiquitin-conjugating enzyme (Ubc) and the E3 ubiquitin ligase involved in the degradation of the nuclear located Asf1-30 protein was therefore attempted. To identify the corresponding E2, the stability of Asf1-30 was assessed in mutants defective in various E2 ubiquitin-conjugating enzymes. Among the 11 E2 genes that function in the UPS in S. pombe, those encoding Ubc4 (34) and Ubc11 (35) are essential for cell viability, whereas the genes encoding Ubc1, Ubc2/Rhp6, Ubc6, Ubc7, Ubc8, Ubc13, Ubc14, Ubc15, and Ubc16 are not essential for cell growth (36). Plasmid pREP41-asf1-30-3HA was introduced into these 11 E2-defective mutant cells, and the half-life of ectopically expressed Asf1-30-3HA was examined by immunoblotting with the anti-HA antibody after the addition of thiamine (promoter shutoff) and CHX. An mts2-1 mutant was used as a control. As shown in Fig. 5A, a significant stabilization of nuclear Asf1-30 at the restrictive temperature was only observed in the ubc4 temperature-sensitive mutant. To further confirm that Ubc4 regulates the stability of Asf1-30, an asf1-30 ubc4 double mutant that also endogenously expressed the Asf1-30-13Myc protein was generated. This double ts mutant showed the synthetic temperature-sensitive phenotype for unknown reasons (supplemental Fig. S2B). Using this strain, the stability of endogenously expressed Asf1-30 was examined at the restrictive temperature in the presence of CHX. Consistent with Fig. 5A, the instability of the Asf1-30 protein was suppressed at the restrictive temperature in the ubc4 ts mutant (Fig. 5B). The subcellular localization of Ubc4 was assessed using GFP-tagged Ubc4. A C-terminal GFP-tagged Ubc4 ectopically expressed using a mid-strength nmt1 promoter (nmt41) resulted in GFP fluorescent signals detected in both the cytoplasm and the nucleus (Fig. 5C). To rule out the possibility that overexpression of C-terminal fusion to GFP affects localization, Ubc4 was modified with C-terminal 5FLAG epitope tags at the endogenous genomic locus. Expression of the correct size proteins was confirmed by immunoblotting, and the strain containing Ubc4–5FLAG grew at a rate similar to that of wild-type cells (data not shown). Indirect immunofluorescence staining with FLAG antibodies confirmed that Ubc4 localized to both the cytoplasm and the nucleus (Fig. 5D). This is consistent with the previous finding of Ubc4 localization (33). No staining was seen when wild type (no tag) was used. These data suggested that the instability of Asf1-30 depended on Ubc4 at the restrictive temperature.
FIGURE 5.
The instability of the Asf1-30 mutant protein depends on ubc4, a ubiquitin-conjugating enzyme. A, stabilization and accumulation of Asf1-30 in the ubc4-P61S ts mutant. The Asf1-30-3HA protein was ectopically expressed from pREP41-asf1-30-3HA in the indicated E2 mutants defective in ubiquitin conjugation. Cells were grown in EMM medium at 26 °C and further incubated at 36 °C for 1 h, followed by the addition of 2 μm thiamine and 100 μg/ml CHX and preparation of whole cell lysates at each time point (0, 1, and 3 h). Asf1-30–3HA was detected by immunoblotting with an anti-HA antibody (top). Tubulin was used as a loading control (bottom). B, the instability of Asf1-30 was suppressed in ubc4-P61S mutants. Protein extracts were prepared at each time point (0, 1, and 3 h) from exponentially growing wild type (wt) or ubc4-P61S ts mutants that expressed chromosomally derived Asf1-30-13Myc. Cells were grown at 36 °C in the presence of 100 μg/ml CHX. The Asf1-30-13Myc protein was detected by immunoblotting with an anti-Myc antibody (top). Tubulin was used as a loading control (bottom). C, subcellular localization of Ubc4-GFP by observation of live cells. GFP fusion proteins were expressed under the control of the nmt1 promoter in the vector pREP41-ubc4+-GFP. Wild-type cells carrying either pREP41 (empty) or pREP41-ubc4+-GFP were grown in EMM medium and incubated at 26 °C. Exposure time was the same for both strains. D, subcellular localization of the Ubc4–5FLAG protein by observation of fixed cells. The cells that expressed chromosomally derived Ubc4–5FLAG were cultured and fixed with formaldehyde before processing for indirect immunofluorescence (IF) staining with an anti-FLAG antibody. Wild-type cells (No-tag) were used as specificity controls. Nuclei were stained with DAPI.
San1 Is the E3 Responsible for the Polyubiquitination of the Asf1-30 Mutant Protein
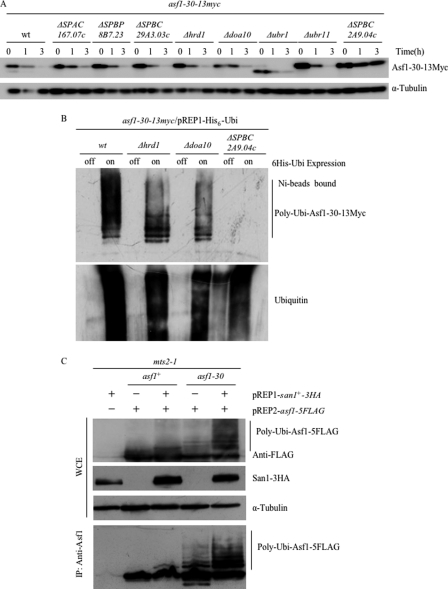
The next experiments were carried out to identify the E3 ubiquitin ligase targeting Asf1-30 for degradation because E3 ligases determine the specificity of ubiquitination. The APC/cyclosome E3 ubiquitin ligase has been shown to cooperate with the ubiquitin-conjugating enzyme Ubc4 (34, 37); therefore, its possible association with Asf1-30 was first examined. However, the instability of Asf1-30 was not suppressed in the cut9-665 ts mutant, which contains a temperature-sensitive mutation in one of the APC/cyclosome components (data not shown). It was then hypothesized that an E3 contributing to nuclear protein quality control would localize to the nucleus, and the gene for E3 would be up-regulated under stress conditions. Based on this hypothesis, the S. pombe genome was searched for genes/proteins meeting the following three criteria: nuclear localization of proteins (33), the presence of the RING or HECT domains that define E3s, and the heat shock-mediated up-regulation of their genes (38). Six E3 candidates were selected in silico, namely ubr1, ubr11, SPAC167.07c, SPBC29A3.03c, SPBP8B7.23, and SPBC2A9.04c. Deletion mutants of those six genes were generated, and the half-life of nuclear Asf1-30 was assessed. Deletion mutants of the hrd1 and doa10 genes (39), which function in ER-associated degradation, were also generated. The instability of nuclear Asf1-30 was significantly suppressed in the SPBC2A9.04c mutant at the restrictive temperature (Fig. 6A). To examine the polyubiquitination of Asf1-30 at the restrictive temperature, His6-Ubi was expressed in these E3 mutants in which Asf1-30-13Myc was endogenously expressed, and proteins polyubiquitinated with His6-Ubi were purified by Ni2+-NTA beads. Immunoblotting with an anti-Myc antibody showed that higher molecular weight bands of Asf1-30-13Myc were absent only in the SPBC2A9.04c mutant (Fig. 6B) (data not shown). These data indicated that the SPBC2A9.04c gene product is responsible for targeting the Asf1-30 nuclear protein for degradation. Interestingly, the SPBC2A9.04c gene is the predicted S. pombe ortholog of the S. cerevisiae SAN1, which encodes a RING finger type E3 ubiquitin ligase and is involved in nuclear protein quality control (11). The SPBC2A9.04c gene was therefore named san1.
FIGURE 6.
The E3 ligase San1 is responsible for polyubiquitination of the Asf1-30 mutant protein. A, the instability of Asf1-30 was suppressed in the san1 mutant. Protein extracts were prepared at each time point (0, 1, and 3 h) from exponentially growing wild type (wt) or the indicated E3 mutants expressing the chromosomally derived Asf1-30-13Myc protein. Cells were grown at 36 °C in the presence of 100 μg/ml CHX. Asf1-30-13Myc was detected by immunoblotting with an anti-Myc antibody (top). Tubulin was used as a loading control (bottom). B, no bands of polyubiquitinated mutant Asf1-30 were observed in the san1 mutant. His6-ubiquitin (pREP1-His6-Ubi) was expressed in wild type or the indicated E3 mutant strains expressing chromosomally derived Asf1-30-13myc. Polyubiquitinated proteins were purified with Ni2+-NTA beads in the presence of 6 m guanidine HCl. Immunoblotting was performed using an anti-Myc antibody to examine the polyubiquitination of Asf1-30 (top). Ubiquitin was also detected with an anti-ubiquitin antibody as a loading control (bottom). C, overexpression of san1 accelerated the accumulation of polyubiquitinated mutant Asf1-30 proteins but not that of the wild-type counterparts. The san1+-3HA (or empty) and asf1+-5FLAG (or asf1-30-5FLAG) genes were placed under the inducible nmt1 promoter in pREP1 and pREP2, respectively. These genes were co-expressed in a mts2-1 mutant at 36 °C in the absence of thiamine. Whole cell extracts (WCE) were prepared and immunoprecipitated (IP) with an anti-Asf1 antibody. Immunoblotting was performed to detect Asf1–5FLAG and San1–3HA. Tubulin was used as loading control.
To further examine whether the higher molecular weight forms of Asf1-30-13Myc were increased when the san1 gene was overexpressed, the San1 dependent polyubiquitination of Asf1-30 was examined using the mts2-1 background and ectopically overexpressed San1-3HA and Asf1-5FLAG or Asf1-30-5FLAG. Consistent with the above data, when both Asf1-30 and San1 were overexpressed, polyubiquitinated Asf1-30 was increased (Fig. 6C). Furthermore, we found that San1-mediated polyubiquitination was specific to the mutant Asf1-30 because the stable wild-type Asf1 showed no polyubiquitination. These results indicate that San1 is the E3 ubiquitin ligase that selectively recognizes Asf1-30 and catalyzes its polyubiquitination but shows no activity against wild-type Asf1.
Effects of san1 Deletion or san1 Overexpression on Temperature Sensitivity in the asf1-30 Allele
Next, the effect of the san1 deletion on the temperature sensitivity of the asf1-30 allele was investigated. As shown in Fig. 7A, there was a slight but significant suppression of temperature sensitivity in the san1 mutant at 32 °C but not at 36 °C. This result is consistent with the finding that the instability of Asf1-30 was not fully suppressed in the san1 mutant at 36 °C (Fig. 6A). In addition, the overexpression of the san1 gene increased the temperature sensitivity of the asf1-30 allele (Fig. 7B). Although the asf1-30 allele showed temperature sensitivity at temperatures higher than 32 °C, the asf1-30 nmt1-GFP-san1 mutant showed significant temperature sensitivity even at 30 °C only in the absence of thiamine, a condition in which the san1 gene was overexpressed. This result is consistent with the finding that overexpression of the san1 gene increases the instability of Asf1-30 at 36 °C (data not shown).
FIGURE 7.
Effects of san1 deletion or san1 overexpression on temperature sensitivity of asf1-30 ts mutant. A, deletion of the san1 gene slightly suppressed the temperature sensitivity of the asf1-30 mutant at semirestrictive temperature. Wild type (wt), the asf1-30 mutant, or the indicated asf1-30 mutants that concomitantly lost the indicated E3 genes were pregrown in YES at 26 °C. Cells were serially diluted and spotted on YES at the indicated temperatures for 4 days. B, overexpression of the san1 gene significantly increased the temperature sensitivity of the asf1-30 mutant at the permissive temperature. Wild type, the asf1-30 mutant, wild type expressing nmt1-GFP-san1, or the asf1-30 mutant expressing nmt1-GFP-san1 was pregrown in EMM at 26 °C. Cells were serially diluted and spotted on EMM plates in the presence or absence of thiamine and grown at the indicated temperature for 6 days.
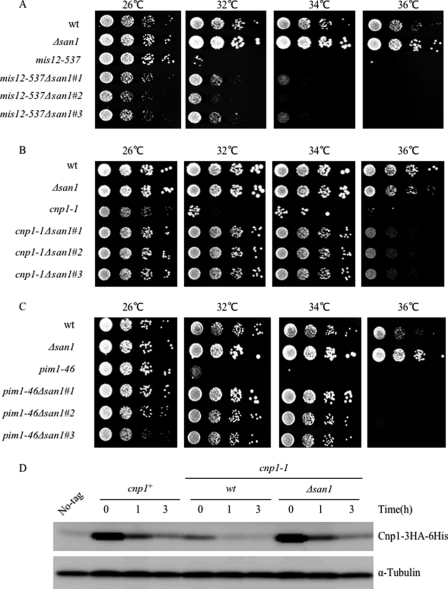
Although the present work only addresses the instability of Asf1-30, other nuclear located mutant proteins, such as Mis12-537, Orc-H37, Cnp1-1, Pim1-46, and Sad1-1, have been reported to be unstable at high temperatures (40–44), suggesting that these proteins could also be processed by the UPS. Therefore, the effect of the san1 deletion on the temperature sensitivity of those temperature-sensitive mutants was investigated. Among those mutants, there was a significant suppression of temperature sensitivity of mis12-537, cnp1-1, or pim1-46 by loss of san1 at 32 or 34 °C (Fig. 8, A–C). In addition, the instability of Cnp1-1 proteins was suppressed at the restrictive temperature (36 °C) in the san1 disruptant (Fig. 8D). Due to technical reasons, we were not sure that the stability of Mis12-537 and Pim1-46 proteins was dependent on San1. Those genetic and biochemical data indicated that San1 is responsible for degradation of the other nuclear mutant proteins (at least Cnp1-1) in addition to Asf1-30.
FIGURE 8.
Effects of san1 deletion on temperature sensitivity of mis12-537, cnp1-1, and pim1-46 ts mutants. A, loss of san1 suppressed the temperature sensitivity of the mis12-537 mutant at semirestrictive temperature. Wild type (wt), the mis12-537 mutant, the san1 disruptant, and the mis12-537 mutant that concomitantly lost san1 were pregrown in YES at 26 °C. Cells were serially diluted and spotted on YES at the indicated temperatures for 3 days. B, loss of san1 suppressed the temperature sensitivity of the cnp1-1 mutant. Wild type, the cnp1-1 mutant, the san1 disruptant, and the cnp1-1 mutants that concomitantly lost san1 were pregrown in YES at 26 °C. Cells were serially diluted and spotted on YES at the indicated temperatures for 3 days. C, loss of san1 suppressed the temperature sensitivity of the pim1-46 mutant. Wild type, the pim1-46 mutant, the san1 disruptant, and the pim1-46 mutant that concomitantly lost san1 were pregrown in YES at 26 °C. Cells were serially diluted and spotted on YES at the indicated temperatures for 3 days. D, the instability of Cnp1-1 was suppressed in the san1 mutant. Protein extracts were prepared at each time point (0, 1, and 3 h) from exponentially growing wild type or cnp1-1 temperature-sensitive mutant, which also expressed the chromosomally derived Cnp1-3HA-His6 protein. Cells were grown at 36 °C in the presence of 100 μg/ml CHX. Cnp1-3HA-His6 was detected by immunoblotting with an anti-HA antibody (top). Tubulin was used as a loading control (bottom).
To further investigate the physiological function of san1, the sensitivity of the san1 mutant toward various stresses, such as 10 mg/liter thiabendazole (TBZ), 0.01% SDS, 10 mm hydroxyurea, 0.005% methyl methanesulfonate (MMS), 2 mm H2O2, 10 mm DTT, 15 mm mercaptoethanol, 10% ethanol, and 2 m sorbitol, was examined. No significant growth differences between wild type and the san1 mutant were detected under those conditions (data not shown). Furthermore, the san1 mutant grew normally at lower (26 °C) and higher (36 °C) temperatures. These results suggest that San1 might not be the only E3 involved in PQC. In contrast to the san1 mutant, the mts2-1 mutant, which contains a mutation in 19 S proteasome regulatory subunit Rpt2 (32), showed sensitivity to several stresses, such as 30 mg/liter l-canavanine, 2 mm H2O2, 0.01% SDS, and 10% ethanol even at the permissive temperature (26 °C) (data not shown). These results indicated that the activity of the proteasome is important for cell growth under stress conditions.
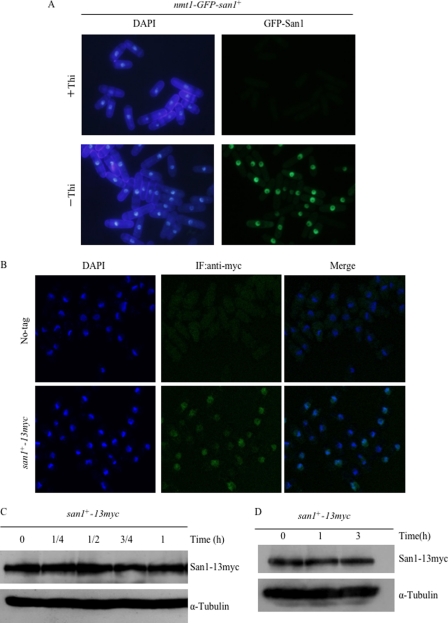
San1 Is a Stable Nuclear Protein
To examine the cellular localization of the San1 protein, GFP-San1 was expressed under the control of the nmt1 promoter, which was integrated in frame in front of the initiation codon of the genomic san1 gene. The resulting strain (nmt1-GFP-san1) grew normally in both the presence and absence of thiamine (Fig. 7B). GFP-San1 localized to the nucleus in all cell cycle stages (Fig. 9A). Furthermore, GFP-San1 localized not only to chromatin regions but also to non-chromatin regions. To rule out the possibility that the overexpression of an N-terminal GFP-tagged San1 could affect its localization, San1 was modified with C-terminal 13Myc epitope tags at the endogenous genomic locus. Immunoblotting showed the expression of correct size proteins, and the strain containing San1–13Myc grew at a rate similar to that of wild-type cells (data not shown). Indirect immunofluorescence staining with anti-Myc antibodies confirmed that San1 was located not only in the chromatin region but also in the non-chromatin region at 26 °C (Fig. 9B). Note that wild type (no tag) was not stained under the same conditions, and microscopic observation was carried out at a single focal plane by a confocal laser-scanning microscope. The subcellular localization of San1 was examined at 36 °C for 0, 3, and 6 h. The results showed that San1 localized to the nucleus before and after the temperature shift to 36 °C for 3 or 6 h (data not shown). Based on prior findings in S. pombe showing that transcription of the san1 gene is up-regulated under heat shock stress (38), the protein levels of San1 were examined under heat shock conditions. As shown in Fig. 9C, there were no differences in San1 protein levels before and after a temperature shift to 36 °C. Furthermore, the stability of San1 was assessed under heat shock conditions, and San1 was found to be a stable protein at 36 °C (Fig. 9D). Assessment of the concentration of the San1 protein during the cell cycle revealed that the levels of San1 did not change significantly (supplemental Fig. S4). These results suggest that San1 is a stable nuclear protein, showing no significant changes in level under the conditions used.
FIGURE 9.
San1 is a stable nuclear protein at high temperatures. A, subcellular localization of the GFP-San1 protein. A strain containing the integrated nmt1-GFP-san1+ was grown in EMM medium in the presence or absence of thiamine. DAPI was used for nuclear staining. Exposure time was the same in each image. B, subcellular localization of the endogenous San1 protein. The cells expressing chromosomally derived San1-13Myc were fixed with formaldehyde and processed for indirect immunofluorescence (IF) staining with anti-Myc antibody. Wild-type cells that did not contain any Myc-tagged alleles (No-tag) were used as specificity controls. Cell nuclei were stained with DAPI. C, San1 protein levels at high temperatures. Exponentially growing cells containing a tagged San1-13Myc were first precultured at 26 °C and shifted up to 36 °C. Protein extracts were prepared at the indicated time points (h), and San1–13Myc was detected by immunoblotting with an anti-Myc antibody (top). Tubulin was used as a loading control (bottom). D, San1 is a stable protein at high temperatures. The half-life of San1 was examined at 36 °C for 0–3 h in the presence of CHX. Exponentially growing cells containing a tagged San1–13Myc were first precultured at 26 °C and grown at 36 °C in the presence of 100 μg/ml CHX. San1-13Myc was detected by immunoblotting with an anti-Myc antibody.
DISCUSSION
The results of the present study show that the selective degradation of nuclear aberrant proteins mediates nuclear PQC in the fission yeast S. pombe. Although nuclear PQC has been reported in budding yeast and mammals (11, 12, 45), the molecular mechanisms of nuclear PQC in other organisms, including S. pombe, have not been elucidated. The present work demonstrates the instability of the mutant nuclear protein Asf1-30 at higher temperatures and its selective degradation by the ubiquitin-proteasome system. Although the present work only addresses the instability of Asf1-30, other nuclear mutant proteins (Mis12-537, Orc5-H37, Cnp1-1, Pim1-46, and Sad1-1) have also been found to be unstable at high temperatures (40–44), suggesting that these mutant proteins could also be processed by the UPS. In fact, deletion of the san1 gene suppressed the temperature sensitivity of the mis12-537, cnp1-1, and pim1-46 mutants at semirestrictive temperature (Fig. 8, A–C). Furthermore, the instability of Cnp1-1 proteins was suppressed at the restrictive temperature in the san1 disruptant (Fig. 8D). These data suggested that San1 is involved in polyubiquitination and degradation of a variety of nuclear mutant proteins in S. pombe.
The Ubc4 ubiquitin-conjugating enzyme (E2) was required for Asf1-30 degradation, and San1 was identified as the E3 ligase catalyzing the polyubiquitination of the Asf1-30 mutant protein. S. pombe Ubc4 is essential for growth and is responsible for the polyubiquitination of mitotic cyclin Cdc13 (34). Asf1-30 is therefore the second target of the E2 enzyme Ubc4 found to date. One unexpected finding was that different E2s mediate nuclear PQC in S. pombe and S. cerevisiae. In S. cerevisiae, the E2s Cdc34 and Ubc1 are associated with four distinct mutant nuclear proteins in the nuclear PQC (11). In the present work, the instability of Asf1-30 was not suppressed in S. pombe ubc15 (CDC34 orthologue) and ubc1 (UBC1 orthologue) mutants (Fig. 5A). Conversely, Ubc4 and Ubc5 (orthologues of Ubc4) are not required for degradation of mutant nuclear proteins in S. cerevisiae (11). Ubc4 and Ubc5 mediate the selective degradation of short lived and abnormal cytosolic proteins (46). These results suggest that different E2 ubiquitin-conjugating enzymes are involved in the nuclear PQCs of S. cerevisiae and S. pombe.
In contrast to the difference in nuclear PQC E2 enzymes between S. cerevisiae and S. pombe, the E3 enzyme San1 is active in both species. The san1 (sir antagonist) gene was originally identified as an extragenic suppressor of the sir4-9 mutant in S. cerevisiae (47), and it was later shown to possess ubiquitin ligase activity in vitro, and this activity was required for the in vivo function of San1 (10). Recently, it was shown that San1 mediates the UPS involvement in the nuclear PQC of S. cerevisiae (11). In addition, ectopically expressed S. cerevisiae San1 enhanced the degradation of nuclear polyglutamine aggregates in cultured mammalian cells and thereby rescued polyglutamine-induced cytotoxicity (12). Furthermore, recent reports show that the association between San1 and the E3 ligase Ubr1 is important for cytoplasmic PQC (8). The present work used a different strategy to identify San1 as a ligase involved in the nuclear PQC of S. pombe by selecting six gene/proteins among 100 E3 candidates based on three criteria: nuclear localization, proteins containing an E3 domain, and the up-regulation of genes by heat shock. The S. pombe San1 E3 ligase was identified as the enzyme mediating the polyubiquitination of the Asf1-30 protein (Fig. 6, B and C). Although the role of San1 in targeting Asf1-30 was clear, the finding that the instability of Asf1-30 was not completely suppressed in a san1 mutant (Fig. 6A) compared with the ubc4ts mutant (Fig. 5A) suggests that other proteins might be involved in PQC. This would also explain the moderate suppression of temperature sensitivity of the asf1-30 allele by the san1 deletion (Fig. 7A). Intriguingly, the temperature sensitivity of the ubc4ts or mts2-1 mutants was enhanced by a concomitant asf1-30 mutation (Fig. S2). The accumulation of an Asf1-30 protein might have a deleterious effect in those ubc4ts and mts2-1 mutants characterized by an impaired degradation of aberrant and harmful proteins, emphasizing the importance of the nuclear PQC for the removal of damaging proteins and the maintenance of cellular homeostasis. Alternatively, stable Asf1-30 mutant protein in those ubc4ts and mts2-1 mutants might affect the process of other target proteins of UPS.
Although the transcription of the san1 gene was up-regulated under heat shock stress (38), the amount of San1 protein was not dramatically changed under these conditions (Fig. 9C). Because the San1 protein is stable in response to heat shock (Fig. 9D), a supraliminal amount of San1 might be sufficient to function in nuclear PQC. The finding that the san1 mutant did not show any obvious phenotypes under various different stress conditions tested (data not shown) suggests that other E3 ligases might function in nuclear PQC in S. pombe. These results are consistent with the finding that there is no obvious growth difference between wild type and the san1 mutant under various conditions in S. cerevisiae (11). Because San1 is the only E3 reported to function in nuclear PQC in two yeast types, it would be of great interest to find and characterize the other E3 ubiquitin ligases involved in nuclear PQC.
The cellular compartment where polyubiquitinated Asf1-30 is degraded in S. pombe remains unknown. In S. pombe, the 26 S proteasome is enriched in the nucleus and nuclear periphery, both during interphase and the mitotic phase (48–50). The nuclear localization of Asf1-30 and San1 E3 ligase (Figs. 4 and 9) suggests that polyubiquitinated Asf1-30 is degraded by the nuclear proteasome. However, the immunofluorescent signal of polyubiquitinated Asf1-30 was observed in the cytoplasm but not in the nucleus in the mts2-1 mutant at restrictive temperature (supplemental Fig. S3). These data suggested that polyubiquitinated Asf1-30 is not degraded in the nucleus but instead shunted out of the nucleus for degradation in the cytoplasm.
As in S. pombe, the proteasome is enriched inside the nucleus throughout the cell cycle in S. cerevisiae (51). In contrast to S. cerevisiae and S. pombe, the 26 S proteasome has been found in both the cytoplasm and nucleus of higher eukaryotic cells (52). Considering that the nucleus of yeasts is not broken down during cytokinesis, the nuclear localization of the 26 S proteasome is beneficial for the degradation of aberrant proteins generated in the cytoplasm and the nucleus in yeasts.
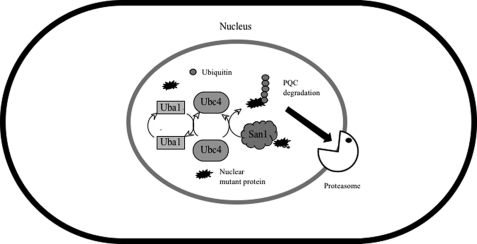
The results presented in this study can be interpreted using the model shown in Fig. 10. At high temperature, the nuclear mutant protein Asf1-30 was selectively polyubiquitinated by the Uba1 E1 (ubiquitin-activating enzyme), Ubc4 E2 (a ubiquitin-conjugating enzyme), and San1 E3 (ubiquitin ligase) and degraded by the proteasome as a result of protein quality control in S. pombe. The present results suggest that the UPS functions in nuclear protein quality control in S. pombe and that nuclear PQC mediated by San1 and the UPS was evolutionarily conserved from S. cerevisiae to S. pombe. Although there are no apparent san1 orthologs in mammalian cells, analogous systems must exist in higher eukaryotes due to the importance of nuclear PQC in protecting non-dividing cells, such as neural and muscle cells, against the deleterious accumulation of nuclear protein aggregates. In fact, recent studies identified UHRF-2 as an essential E3 ubiquitin ligase involved in nuclear polyglutamine degradation as a component of the nuclear PQC in cultured cells and primary neurons (12). This result indicates that nuclear PQC plays a key role in neuroprotection against the deleterious accumulation of nuclear protein aggregates in quiescent (G0) mammalian cells.
FIGURE 10.
E1-E2-E3 molecules that recognize a nuclear aberrant protein in S. pombe. At high temperatures, the nuclear localized mutant Asf1-30 protein was selectively polyubiquitinated by Uba1 (E1), Ubc4 (E2), and San1(E3) and degraded by the proteasome as a protein quality control system.
Recent studies have suggested that S. pombe is an excellent model for the study of cellular quiescence, which can be achieved experimentally through nutritional limitation. Studies on the regulatory mechanism of G0 phase in S. pombe showed that the function of the proteasome is required for the maintenance of G0 quiescence (53). The proteasome signal persisted in the nuclear periphery in G0 phase in S. pombe, but it diminished with the increase in cytoplasmic localization (53, 54). Furthermore, dysfunction of the proteasome in G0 phase caused the appearance of aberrant nuclear structures (55). These features of S. pombe could help elucidate the physiological significance of the nuclear PQC in G0 phase, which would lead to a better understanding of the nuclear PQC in G0 phase in higher eukaryotes. In conclusion, nuclear PQC systems are active in budding yeast, fission yeast, and mammals, and they involve different E2s and E3s in each species.
Acknowledgments
We thank Drs. J. Bahler, T. Toda, H. Seino, F. Yamao, H. Yamano, and M. Yanagida and National Bio Resource Project (NBRP)/Yeast Genetic Resource Center (YGRC) for providing the materials and strains used in this study. We also thank Drs. H. Katoh, K. Nishimura, T. Kainou, T. Nakagawa, Y. Nagano, T. Anai, K. Watanabe, and our laboratory members for helpful discussions and experimental support.
This work was supported by Japan Society for the Promotion of Science research fellowships for young scientists.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- UPS
- ubiquitin-proteasome system
- PQC
- protein quality control
- ER
- endoplasmic reticulum
- ts
- temperature-sensitive
- Ubi
- ubiquitin
- CHX
- cycloheximide.
REFERENCES
- 1. Hershko A. (1997) Curr. Opin. Cell Biol. 9, 788–799 [DOI] [PubMed] [Google Scholar]
- 2. Hochstrasser M. (1996) Annu. Rev. Genet. 30, 405–439 [DOI] [PubMed] [Google Scholar]
- 3. Ciechanover A. (1998) EMBO J. 17, 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldberg A. L. (2003) Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 5. Shastry B. S. (2003) Neurochem. Int. 43, 1–7 [DOI] [PubMed] [Google Scholar]
- 6. Hampton R. Y. (2002) Curr. Opin. Cell Biol. 14, 476–482 [DOI] [PubMed] [Google Scholar]
- 7. Murata S., Chiba T., Tanaka K. (2003) Int. J. Biochem. Cell Biol. 35, 572–578 [DOI] [PubMed] [Google Scholar]
- 8. Heck J. W., Cheung S. K., Hampton R. Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1106–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwata A., Christianson J. C., Bucci M., Ellerby L. M., Nukina N., Forno L. S., Kopito R. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13135–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dasgupta A., Ramsey K. L., Smith J. S., Auble D. T. (2004) J. Biol. Chem. 279, 26830–26838 [DOI] [PubMed] [Google Scholar]
- 11. Gardner R. G., Nelson Z. W., Gottschling D. E. (2005) Cell 120, 803–815 [DOI] [PubMed] [Google Scholar]
- 12. Iwata A., Nagashima Y., Matsumoto L., Suzuki T., Yamanaka T., Date H., Deoka K., Nukina N., Tsuji S. (2009) J. Biol. Chem. 284, 9796–9803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le S., Davis C., Konopka J. B., Sternglanz R. (1997) Yeast 13, 1029–1042 [DOI] [PubMed] [Google Scholar]
- 14. Tyler J. K., Adams C. R., Chen S. R., Kobayashi R., Kamakaka R. T., Kadonaga J. T. (1999) Nature 402, 555–560 [DOI] [PubMed] [Google Scholar]
- 15. Munakata T., Adachi N., Yokoyama N., Kuzuhara T., Horikoshi M. (2000) Genes Cells 5, 221–233 [DOI] [PubMed] [Google Scholar]
- 16. Moreno S., Klar A., Nurse P. (1991) Methods Enzymol. 194, 795–823 [DOI] [PubMed] [Google Scholar]
- 17. Kawamukai M., Gerst J., Field J., Riggs M., Rodgers L., Wigler M., Young D. (1992) Mol. Biol. Cell 3, 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sambrook J., Fritsch E. F., Maniatis T. (1989) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19. Maundrell K. (1990) J. Biol. Chem. 265, 10857–10864 [PubMed] [Google Scholar]
- 20. Basi G., Schmid E., Maundrell K. (1993) Gene 123, 131–136 [DOI] [PubMed] [Google Scholar]
- 21. Ozoe F., Kurokawa R., Kobayashi Y., Jeong H. T., Tanaka K., Sen K., Nakagawa T., Matsuda H., Kawamukai M. (2002) Mol. Cell. Biol. 22, 7105–7119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sato M., Dhut S., Toda T. (2005) Yeast 22, 583–591 [DOI] [PubMed] [Google Scholar]
- 23. Noguchi E., Noguchi C., McDonald W. H., Yates J. R., 3rd, Russell P. (2004) Mol. Cell. Biol. 24, 8342–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsuo Y., Kishimoto H., Horiuchi T., Tanae K., Kawamukai M. (2010) Biosci. Biotechnol. Biochem. 74, 685–689 [DOI] [PubMed] [Google Scholar]
- 25. Oowatari Y., Toma K., Ozoe F., Kawamukai M. (2009) Biosci. Biotechnol. Biochem. 73, 1591–1598 [DOI] [PubMed] [Google Scholar]
- 26. Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 27. Lehmann A., Katayama S., Harrison C., Dhut S., Kitamura K., McDonald N., Toda T. (2004) Genes Cells 9, 367–382 [DOI] [PubMed] [Google Scholar]
- 28. Matsuo Y., Asakawa K., Toda T., Katayama S. (2006) Biosci. Biotechnol. Biochem. 70, 1992–1994 [DOI] [PubMed] [Google Scholar]
- 29. Tanabe K., Ito N., Wakuri T., Ozoe F., Umeda M., Katayama S., Tanaka K., Matsuda H., Kawamukai M. (2003) Eukaryot. Cell 2, 1274–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shiozaki K., Russell P. (1997) Methods Enzymol. 283, 506–520 [DOI] [PubMed] [Google Scholar]
- 31. Umehara T., Chimura T., Ichikawa N., Horikoshi M. (2002) Genes Cells 7, 59–73 [DOI] [PubMed] [Google Scholar]
- 32. Gordon C., McGurk G., Dillon P., Rosen C., Hastie N. D. (1993) Nature 366, 355–357 [DOI] [PubMed] [Google Scholar]
- 33. Matsuyama A., Arai R., Yashiroda Y., Shirai A., Kamata A., Sekido S., Kobayashi Y., Hashimoto A., Hamamoto M., Hiraoka Y., Horinouchi S., Yoshida M. (2006) Nat. Biotechnol. 24, 841–847 [DOI] [PubMed] [Google Scholar]
- 34. Seino H., Kishi T., Nishitani H., Yamao F. (2003) Mol. Cell. Biol. 23, 3497–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osaka F., Seino H., Seno T., Yamao F. (1997) Mol. Cell. Biol. 17, 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kitamura K., Katayama S., Dhut S., Sato M., Watanabe Y., Yamamoto M., Toda T. (2001) Dev. Cell 1, 389–399 [DOI] [PubMed] [Google Scholar]
- 37. Javerzat J. P., Cranston G., Allshire R. C. (1996) Nucleic Acids Res. 24, 4676–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bähler J. (2003) Mol. Biol. Cell 14, 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hughes B. T., Nwosu C. C., Espenshade P. J. (2009) J. Biol. Chem. 284, 20512–20521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goshima G., Iwasaki O., Obuse C., Yanagida M. (2003) EMBO J. 22, 2752–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kato H., Matsunaga F., Miyazaki S., Yin L., D'Urso G., Tanaka K., Murakami Y. (2008) Cell Cycle 7, 1085–1096 [PubMed] [Google Scholar]
- 42. Chen E. S., Saitoh S., Yanagida M., Takahashi K. (2003) Mol. Cell 11, 175–187 [DOI] [PubMed] [Google Scholar]
- 43. Matsumoto T., Beach D. (1993) Mol. Biol. Cell 4, 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hagan I., Yanagida M. (1995) J. Cell Biol. 129, 1033–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. von Mikecz A. (2006) J. Cell Sci. 119, 1977–1984 [DOI] [PubMed] [Google Scholar]
- 46. Seufert W., Jentsch S. (1990) EMBO J. 9, 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schnell R., D'Ari L., Foss M., Goodman D., Rine J. (1989) Genetics 122, 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilkinson C. R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J. P., McIntosh J. R., Gordon C. (1998) EMBO J. 17, 6465–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takeda K., Yanagida M. (2005) Cell 122, 393–405 [DOI] [PubMed] [Google Scholar]
- 50. Tatebe H., Yanagida M. (2000) Curr. Biol. 10, 1329–1338 [DOI] [PubMed] [Google Scholar]
- 51. Russell S. J., Reed S. H., Huang W., Friedberg E. C., Johnston S. A. (1999) Mol. Cell 3, 687–695 [DOI] [PubMed] [Google Scholar]
- 52. Wójcik C., DeMartino G. N. (2003) Int. J. Biochem. Cell Biol. 35, 579–589 [DOI] [PubMed] [Google Scholar]
- 53. Takeda K., Yanagida M. (2010) Autophagy 6, 564–565 [DOI] [PubMed] [Google Scholar]
- 54. Laporte D., Salin B., Daignan-Fornier B., Sagot I. (2008) J. Cell Biol. 181, 737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takeda K., Yoshida T., Kikuchi S., Nagao K., Kokubu A., Pluskal T., Villar-Briones A., Nakamura T., Yanagida M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3540–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]