Abstract
We developed a helper-dependent adenoviral vector for cystic fibrosis lung gene therapy. The vector expresses cystic fibrosis transmembrane conductance regulator (Cftr) using control elements from cytokeratin 18. The vector expressed properly localized CFTR in cultured cells and in the airway epithelia of mice. Cftr RNA and protein were present in whole lung and bronchioles, respectively, for 28 days after a vector dose. Acute inflammation was minimal to moderate. To test the therapeutic potential of the vector, we challenged mice with a clinical strain of Burkholderia cepacia complex (Bcc). Cftr knockout mice (but not Cftr+/+ littermates) challenged with Bcc developed severe lung histopathology and had high lung bacteria counts. Cftr knockout mice receiving gene therapy 7 days before Bcc challenge had less severe histopathology, and the number of lung bacteria was reduced to the level seen in Cftr+/+ littermates. These data suggest that gene therapy could benefit cystic fibrosis patients by reducing susceptibility to opportunistic pathogens.
Keywords: cystic fibrosis, cytokeratin 18, Burkholderia cepacia complex
Cystic fibrosis (CF) is a debilitating and relatively common genetic disorder (≈1 in 2,500 Caucasian births). CF is caused by recessive mutations in the CF transmembrane conductance regulator (Cftr) gene, which encodes a membrane chloride channel present in the epithelium of many organs (1). Morbidity and mortality in CF patients is chiefly due to lung disease and is characterized by inflammation, obstructive mucus, and persistent infection. Staphylococcus aureus or Haemophilus influenzae are early colonizers, whereas Pseudomonas aeruginosa or Burkholderia cepacia complex (Bcc) often occur later, resulting in progressive loss of lung function and premature death. The relationship between the Cftr mutation and CF lung disease, however, is not clear (2).
The CF airway epithelium is defective in mucociliary clearance, antimicrobial activity, and the regulation of inflammatory cytokines (2). Cftr gene replacement therapy has the potential to rescue these defects. The airway epithelium should be accessible by an inhaled vector, and sustained expression of Cftr in a fraction of cells may be sufficient to benefit patients (3, 4). However, lung gene delivery in experimental animals has been hampered by weak or transient expression and/or significant inflammation (5). It is also unknown whether increased Cftr expression or improved electrophysiology in airway epithelium constitutes successful gene therapy. Clinical trials have yielded evidence of limited, transient gene transfer with variable inflammation (6, 7).
We developed vectors for airway expression, using control elements from the human cytokeratin 18 (K18) gene. Expression of K18 is restricted to the epithelium of internal organs (8) in a pattern similar to that of Cftr (9, 10). In the lung, K18, like Cftr, is expressed in the epithelium of large airways and bronchioles and in submucosal glands, but little is expressed in the alveoli (10, 11). Unlike Cftr, K18 is strongly expressed, and the controlling DNA sequences are well characterized (12). K18lacZ cassettes express β-galactosidase in a Cftr-like pattern in transgenic mice (10, 11) and in gene delivery studies (13). Unlike commonly used viral promoters, human promoters may be less susceptible to transcription attenuation by host cytokines (14). In addition, the specificity of K18 elements could reduce immune stimulation resulting from transgene expression in antigen-presenting cells.
Adenoviral vectors (Ads) are used for a variety of gene therapy applications. Unlike earlier generations of Ads, helper-dependent Ads (HD-Ads) lack all viral coding sequences, which reduces inflammation and extends transgene expression (15–18). We recently developed HD-Ads with K18 control elements driving reporter genes. These vectors caused little inflammation in mouse lung and expressed reporter genes for periods of 28 days (bacterial β-galactosidase) to 15 weeks (human α-fetoprotein) (19). We have now developed a K18-based HD-Ad for Cftr gene therapy and tested the vector in a mouse model of CF lung infection.
Methods
HD-Ad Preparation. The expression cassette K18mTECFTag2-i6x7pA contains 2.5 kb of genomic sequence upstream from the human K18 gene, the promoter plus first intron of K18, followed by a translational enhancer, the hCftr cDNA (containing a silent mutation to eliminate cryptic splicing with the K18 intron), and a 3′ fragment of the K18 gene functioning as a 3′ untranslated region and polyadenylation signal (20). The Cftr cDNA was modified to replace the last amino acid residue (L) of CFTR with the peptide RWEGPPGPYTDIEMNRLGK, which incorporates a vesicular stomatitis virus G glycoprotein (VSV-G) epitope tag (20). The epitope tag was added to enhance detection of CFTR produced by the vector and does not effect the stability or activity of CFTR (Y.-H.C., unpublished data).
To prepare HD-Ad expressing Cftr, we cloned K18mTECFTag2-i6x7pA (20) into pC4HSU (21) by removing 4.5 kb of “stuffer” DNA from pC4HSU (AscI to NotI) and replacing it with the 8.8-kb cassette K18mTECFTag2-i6x7pA (MunI to SalI). This plasmid then was cotransfected with H14 helper virus (21) into 293Cre4 cells (22). The resulting HD-Ad, designated K18CFTR-HD-Ad, was expanded through eight culture passages and purified by CsCl gradient centrifugation as described (23). We similarly prepared intact pC4HSU to produce an “empty” control HD-Ad. To measure helper virus contamination, we cut viral DNA with PstI, performed Southern blotting, and probed with adenovirus ITR sequence as described (24).
Cell Culture. We incubated COS-1 cells with K18CFTR-HD-Ad (2,000 particles per cell) and performed an iodide efflux assay 4 days later. We incubated COS cells for 1 h in isotonic buffer containing 136 mM iodide, washed them extensively with iodide-free buffer, and then added and removed iodide-free buffer at 1-min intervals. After 3 min, we gave the “stimulated” group of cells a mixture consisting of 0.5 mM 3-isobutyl-1-methylxanthine, 0.5 mM 8-(4-chlorophenylthio)adenosine-3′, 5′-cyclic monophosphate, and 20 μM forskolin in iodide-free buffer (11). Iodine concentration in the collected buffer samples was measured with an iodine-specific electrode (Orion, Boston).
We obtained bronchial epithelial cells from CF patients (CftrΔF508/ΔF508) undergoing lung transplant at the University Health Network (Toronto) and cultured these cells for 3 weeks at an air–liquid interface (Transwell plates, Costar-Corning) as described (25). The culture consisted of well differentiated ciliated, nonciliated, and goblet cells in a pseudostratified arrangement with intercellular tight junctions (data not shown). To infect with HD-Ad, we first placed culture medium containing 6 mM EGTA on the apical cell surface for 1 h (26), then replaced it with PBS containing K18CFTR-HD-Ad (5,000 particles per cell).
Ad and Bacteria Delivery to Mice. We anesthetized adult (10- to 12-week-old) CD-1 mice (Charles River Breeding Laboratories) by methoxyflurane inhalation and applied 25 μl of 0.4 M EGTA (pH 7.4) in small drops to the nostrils, where it was aspirated. The animal's mouth was held shut to maximize lung delivery (19, 27). Thirty minutes later, we similarly delivered K18CFTR-HD-Ad in 50 μl of PBS.
Cftr knockout mice (Cftrtm1UNC) (28) were raised on a liquid diet to adulthood (10–12 weeks). We delivered 12.5 μl of EGTA followed 30 min later by K18CFTR-HD-Ad in 25 μl of PBS, as above. Seven days later we challenged mice with aspirated Bcc [5 × 107 colony-forming units (cfu), clinical isolate BC7, belonging to genomovar III ET12 lineage (27, ††)] in 40 μl of PBS. Five days later we excised the lungs under aseptic conditions and fixed them or homogenized and plated them on Bcc-selective agar (27) to determine bacterial persistence. Mice were maintained in microisolators under Level 2 containment, according to protocols approved by the Animal Care Committee of The Hospital for Sick Children.
Immunofluorescence. COS cells were fixed in 3% paraformaldehyde for 10 min, followed by 0.2% Triton X-100 for 4 min, and then blocked with 2% BSA/5% goat serum. Human bronchial cells on Transwell membranes were fixed in 10% formalin, embedded in 2% agar, dehydrated, embedded in paraffin, and cross-sectioned. Deparaffinized slides were boiled (5 min, twice) in 10 mM citrate (pH 6.0), blocked for 1 h with 2% BSA/5% goat serum, and then incubated for 90 min with monoclonal antibody against human CFTR (hCFTR) (MAB1660, R & D Systems) or VSV-G (Boehringer Mannheim, Indianapolis) diluted 1:500 in 1% BSA/3% goat serum. This incubation was followed by one with anti-mouse goat IgG conjugated with fluorescein or rhodamine (diluted 1:300 or 1:100, respectively, in 1% BSA/3% goat serum) (Calbiochem) for 1 h.
To obtain cryosections, we inflated mouse lungs with PBS/optimum cutting temperature (OCT) compound (1:1), removed them, and embedded them in OCT compound (Tissue-Tek, Elkhart, IN). Dry 5-μm cryosections were fixed in methanol for 10 min at –20°C, soaked for 4 min in 0.2% Triton X-100 in PBS, blocked for 1 h, and then incubated for 2 h with antibody against VSV-G or hCFTR (1:1,000 dilution), by using a “mouse-on-mouse” kit according to directions (Vector Laboratories). This procedure was followed by rhodamine-conjugated secondary antibody, as above, and a fluorescent counterstain of sodium 3,3′-dioctadecyl-5,5′-di(4-sulfophenyl)oxacarbocyanine (DiOC, Molecular Probes) (15 μg/ml in PBS for 15 min). Laser scanning confocal microscopy was performed with a Zeiss LSM 510.
Bcc in paraffin sections of mouse lung was detected by using polyclonal antibody R418 (diluted 1:1,000), followed by anti-rabbit IgG conjugated with Cy3 (diluted 1:250) (Jackson Immuno Research), as described previously (27). The counterstain was Mayer's hematoxylin.
RNase Protection Assay. We removed mouse lungs to TRIzol (Invitrogen) and extracted RNA. We made antisense RNA probes complementary to hCftr cDNA (nucleotides 3714–4049) or mouse β-actin sequence, followed by unrelated plasmid sequence, labeled with [α-32P]UTP by in vitro transcription with T3 RNA polymerase (Hoffmann–La Roche). We hybridized gel-purified probes with 20 μg of lung RNA for 18 h at 56°C, then added RNase T1 (Ambion, Austin, TX) and incubated for 45 min at 37°C. We separated probes on a polyacrylamide denaturing gel. Undigested and protected digested hCftr probes were 432 and 336 nt, respectively, whereas undigested and protected digested β-actin probes were 276 and 245 nt, respectively.
Histopathological Evaluation. Mouse lungs were formalin-fixed as described previously (27). A veterinary pathologist, blinded to the treatments and genotypes, examined paraffin sections of left and right lung stained with hematoxylin/eosin (H&E), Giemsa, or periodic acid Schiff plus alcian blue. The pathologist scored lungs on a semiquantitative scale from 0 (no inflammatory change) to 5 (severe inflammation with tissue destruction) in the categories of bronchiolitis, respiratory epithelial cell hyperplasia, goblet cell hyperplasia and mucus retention, pneumonia (alveolitis, exudation, and consolidation), and edema (27).
Statistics. Statistical significance (P < 0.05) was determined by one-way ANOVA, followed by post hoc analysis using Duncan's multiple range test, by which significant differences were found among groups (30).
Results
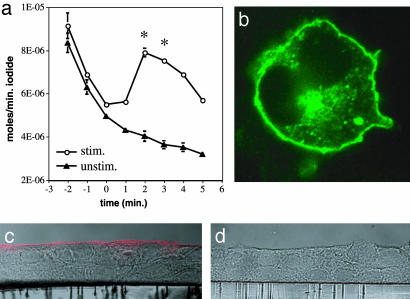
Analysis of K18CFTR-HD-Ad in Cultured Cells. We constructed an HD-Ad for the purpose of delivering hCftr to the lung. This vector, designated K18CFTR-HD-Ad, uses a K18 cassette to express hCftr with a VSV-G epitope tag (20). This vector and an empty control vector (C4HSU-HD-Ad) each contained ≤1% helper virus as determined by Southern blot analysis (data not shown). We used K18CFTR-HD-Ad to infect COS-1 cells, which do not normally express Cftr (11, 20), and tested for halide channel function. An iodide efflux observed in response to a CFTR agonist mixture (Fig. 1a) was consistent with Cftr expression. Antibody against hCFTR (Fig. 1b) or VSV-G (data not shown) yielded a signal at the plasma membrane and in the perinuclear region, as expected for CFTR in nonpolarized cells. K18CFTR-HD-Ad was also able to transduce primary, differentiated human CF bronchial epithelial cells cultured at an air–liquid interface (25). The culture was first exposed to EGTA to disrupt intercellular tight junctions (31), thereby allowing the virus access to its major receptor, present mainly on the basolateral membrane. A signal observed at the apical plasma membrane was consistent with Cftr expression in polarized epithelium (Fig. 1 c and d).
Fig. 1.
Infection of cultured cells with K18CFTR-HD-Ad. (a) Release of iodide from COS cells infected with K18CFTR-HD-Ad in the presence (stim.) or absence (unstim.) of a CFTR agonist mixture delivered at time 0. Bars represent the standard error of the mean (n = 3). Asterisks indicate statistically significant differences between stimulated and unstimulated groups. (b) Immunofluorescence of a COS cell infected with K18CFTR-HD-Ad, by using antibody against hCFTR. (c and d) Infection of primary human CF bronchial epithelial cells with K18CFTR-HD-Ad. Cross sections of cells on culture membranes were probed with antibody against hCFTR, detected with rhodamine-conjugated secondary antibody. Fluorescent and differential interference contrast microscopy (DIC) images are merged. (c) K18CFTR-HD-Ad-infected cells. (d) Mock-infected cells.
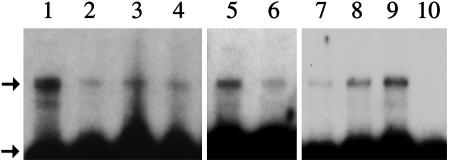
hCftr RNA Expression in Mouse Lung. We delivered EGTA to mice by nasal aspiration, followed 30 min later by 2 × 1011 particles of aspirated K18CFTR-HD-Ad. Total lung RNA then was harvested for an RNase protection assay to detect hCftr RNA. Probe protected by hCftr RNA was seen in assays of mice taken 3, 6, and 28 days after vector delivery (Fig. 2, upper arrow). RNA from a K18Cftr transgenic mouse (Y.-H.C., unpublished data) (lane 1) and a mouse receiving saline (lane 10) served as positive and negative controls, respectively. Mouse β-actin probe was used as an internal control to verify complete digestion of unprotected sequences (Fig. 2, lower arrow).
Fig. 2.
RNase protection assay to detect hCftr RNA in mouse lung. Lane 1, RNA from a transgenic mouse expressing the K18Cftr cassette (positive control). Lanes 2–4, RNA from three mice 3 days after receiving K18CFTR-HD-Ad. Lanes 5 and 6, RNA from two mice 6 days after receiving K18CFTR-HD-Ad. Lanes 7–9, RNA from three mice 28 days after receiving K18CFTR-HD-Ad. Lane 10, RNA from a mouse receiving saline. The upper arrow indicates protected digested hCftr probe; the lower arrow indicates protected digested mouse β-actin probe (internal control).
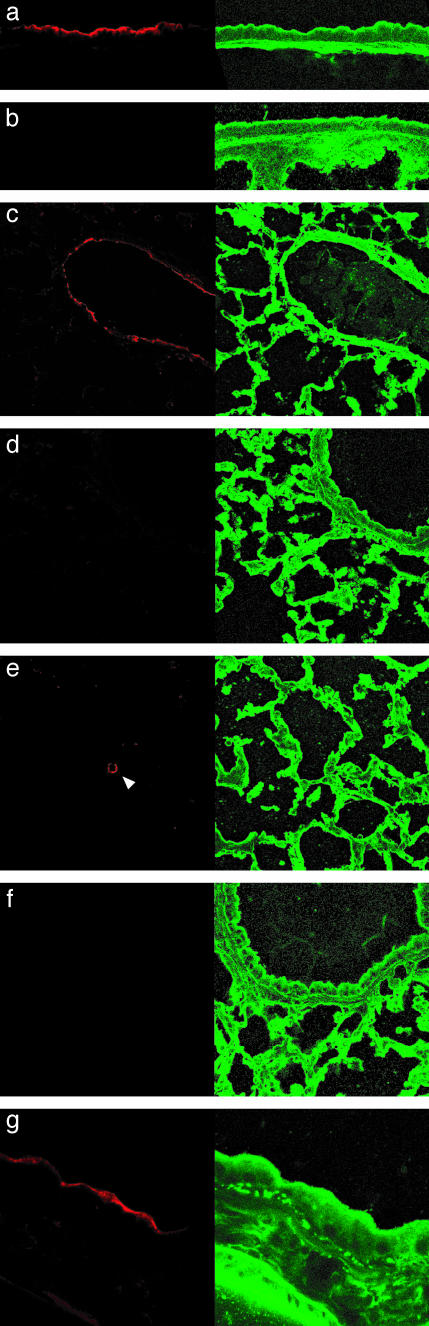
hCFTR Protein Expression in Mouse Lung. The lungs of mice receiving EGTA followed by 2 × 1011 particles of K18CFTR-HD-Ad were sectioned and probed with antibody against VSV-G-tagged hCFTR. Three days after vector delivery, we detected signal in trachea and bronchioles by using antibody against VSV-G (Fig. 3 a and c). Signal was observed in the region of tracheal submucosal glands, but staining to identify positive cells was inconclusive (data not shown). Positive alveolar cells were rare (Fig. 3e). Virtually identical results, not shown, were obtained by using a monoclonal antibody against hCFTR that recognizes an epitope unique to the human protein. No signal was seen (when using either primary antibody) on sections from mice receiving only saline (Fig. 3b) or the control vector C4HSU-HD-Ad (Fig. 3d). Omitting the primary antibody yielded no signal (Fig. 3f). Bronchiolar fluorescence was localized to the apical epithelium (Fig. 3g), as expected for CFTR (9). Twenty-eight days after vector delivery, we still could detect signal in bronchioles (Fig. 4a) at the apical epithelium (Fig. 4b).
Fig. 3.
Expression of hCFTR in mice 3 days after vector delivery. Lung sections from mice 3 days after receiving K18CFTR-HD-Ad (a, c, e, f, and g), saline (b), or C4HSU-HD-Ad (d). (Left) Sections of trachea (a and b), bronchiole (c and d), alveoli (e), and bronchiole (g) were probed with antibody against VSV-G, detected with rhodamine-conjugated secondary antibody. (Right) Sodium 3,3′-dioctadecyl-5,5′-di(4-sulfophenyl)oxacarbocyanine (DiOC) counterstain. (e) The arrowhead indicates a positive alveolar cell. (f) Primary antibody was omitted from bronchiole section. (a–f, ×400; g, ×1,000.)
Fig. 4.
Expression of hCFTR in mice 28 days after receiving K18CFTR-HD-Ad. (Left) Lung sections were probed with antibody against VSV-G, detected with rhodamine-conjugated secondary antibody. (Right) Sodium 3,3′-dioctadecyl-5,5′-di(4-sulfophenyl)oxacarbocyanine (DiOC) counterstain. (a, ×400; b, ×1,000.)
Lung Inflammation. To assess lung inflammation, we first delivered EGTA followed by 4 × 1010 particles of aspirated K18CFTR-HD-Ad, the dose used in our previous study (19). A pathologist examined slides of lung tissue harvested 3 days after delivery of the vector or saline (see Methods for scoring system). Control mice receiving saline had negligible lung inflammation (scores 0–1, n = 2). Inflammation in mice receiving K18CFTR-HD-Ad was minimal to moderate (scores 0–3, n = 4). We then repeated the study with 2 × 1011 particles of K18CFTR-HD-Ad, the higher dose used to detect signal in our RNA and immunofluorescence studies. Inflammation was minimal to moderate (scores 0–3, n = 4) with patchy distribution throughout the lung.
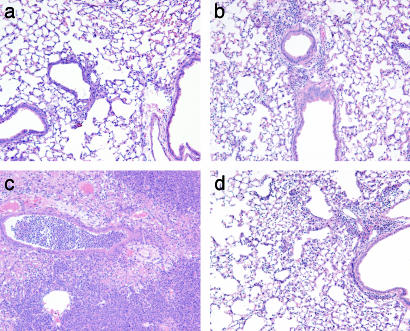
Bacterial Challenge of Cftr Knockout Mice. To determine whether our vector improved the resistance of Cftr knockout mice to acute lung infection, we delivered EGTA followed by a moderate dose of K18CFTR-HD-Ad (2 × 1010 particles) or saline to knockout mice (Cftrtm1UNC) or Cftr+/+ littermates. Mice were challenged 7 days later with an aspirated dose of a clinical isolate of Bcc (27, ††). After 5 days the lungs were fixed for examination by a pathologist (see Methods). Cftr+/+ mice receiving saline before bacterial challenge had minimal to moderate inflammation (scores 1–2, n = 5; Fig. 5a). Cftr+/+ mice receiving K18CFTR-HD-Ad before Bcc also had minimal to moderate inflammation (scores 1–3, n = 5; Fig. 5b). Inflammation was patchy, with bronchiolitis and scattered foci of mild to moderate pneumonia. Cftr–/– mice receiving saline before Bcc had marked inflammation (scores 3–5, n = 4; Fig. 5c),†† seen histopathologically as severe pneumonia and bronchiolitis; inflammatory cell infiltration, exudation, and consolidation; respiratory cell hyperplasia; and necrosis. Cftr–/– mice receiving K18CFTR-HD-Ad before Bcc had minimal to moderate inflammation (scores 1–3, n = 7; Fig. 5d). Inflammation in the lower-scoring mice was similar to that in Cftr+/+ mice receiving K18CFTR-HD-Ad, whereas the higher-scoring mice had scattered foci of moderate pneumonia, bronchiolitis, and respiratory cell hyperplasia.
Fig. 5.
Lung histopathology after aspirated Bcc challenge. Representative sections (H&E stain) from Cftr+/+ (a and b) and Cftr–/– (c and d) mice receiving saline (a and c) or K18CFTR-HD-Ad (b and d) before bacterial challenge. (×100.)
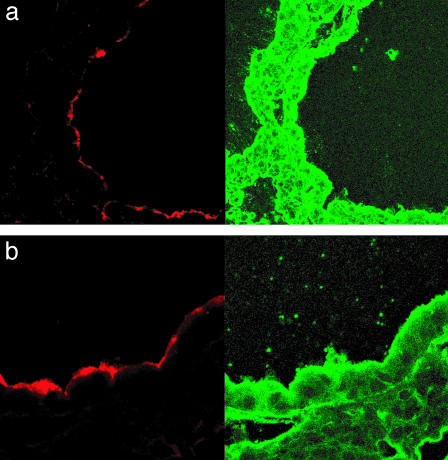
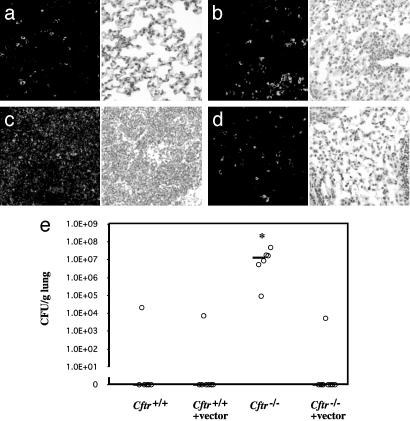
To detect Bcc in the lungs, we probed sections from the above experiment with a polyclonal antibody against Bcc (27). Cftr+/+ mouse lungs contained little fluorescent signal corresponding to Bcc 5 days after bacterial challenge (Fig. 6a). Cftr+/+ mice receiving K18CFTR-HD-Ad before the challenge also had little fluorescent signal in their lungs (Fig. 6b). The lungs of Cftr–/– mice contained abundant fluorescent signal (Fig. 6c), but Cftr–/– mice receiving K18CFTR-HD-Ad before bacterial challenge had little signal (Fig. 6d).
Fig. 6.
Detection of bacteria in mouse lungs after aspirated Bcc challenge. (a–d) (Left) Representative sections probed with antibody against Bcc (fluorescent field). (Right) Corresponding bright field (hematoxylin stain). (a) Cftr+/+ mouse. (b) Cftr+/+ mouse pretreated with K18CFTR-HD-Ad. (c) Cftr–/– mouse. (d) Cftr–/– mouse pretreated with K18CFTR-HD-Ad. (e) Viable Bcc (cfu) recovered from lungs of Cftr+/+ and Cftr–/– mice, with or without K18CFTR-HD-Ad (vector) pretreatment. Each point represents the cfu per g lung of an individual mouse, and the horizontal bar represents the median in each group. The asterisk indicates a statistically significant difference.
Because the antibody used against Bcc does not distinguish between live and dead bacteria, we repeated the experiment but this time homogenized the lungs and plated them on Bcc-selective agar (Fig. 6e). Five of six Cftr+/+ mice had no viable Bcc in their lungs. Six of seven Cftr+/+ mice receiving K18CFTR-HD-Ad also had no viable Bcc in their lungs. In contrast, the lungs of all six Cftr–/– mice contained Bcc, with five of six mice harboring large numbers of the bacteria (median = 1.3 × 107 cfu per g). The lungs of seven of eight Cftr–/– mice receiving K18CFTR-HD-Ad, however, contained no viable Bcc (Fig. 6e). When we delivered the control vector C4HSU-HD-Ad before Bcc challenge, all four mice tested harbored large numbers of the bacteria in their lungs. Three of these mice did not survive to the end of the challenge, succumbing to infection after 3 days (data not shown, median = 4.2 × 107 cfu per g).
Discussion
If airway epithelial cells can be transduced by a vector expressing Cftr, correction of a fraction of defective cells may suffice for effective gene therapy. However, the presumed benefits of Cftr gene therapy for CF patients have not been demonstrated. Various approaches have been tried in animals and humans, but transient, weak expression and/or significant inflammation had hampered efforts. To address some of these difficulties, we developed an HD-Ad with control elements from the K18 gene driving Cftr expression.
CF lung disease begins as inflammation and infection of the bronchioles (2). However, Cftr is expressed in both upper and lower airway epithelia and in submucosal glands (9), suggesting that large and small airways may be important targets for gene therapy. K18 is strongly expressed in a pattern similar to that of Cftr (9–11), so K18 control elements are potentially useful to drive Cftr expression for gene therapy. K18CFTR-HD-Ad expressed hCftr in both large and small airways (submucosal glands could not be confirmed). Neither Cftr nor K18 is strongly expressed in alveoli (9, 11), and this region is not a primary target of CF gene therapy. We found few positive alveolar cells after delivery of K18CFTR-HD-Ad. Our K18-based HD-Ad expressing β-galactosidase similarly transduced tracheal and bronchiolar epithelia (including ciliated, basal, and Clara cells), few alveolar cells (identified as epithelial type II), and also submucosal glands (19). Mouse studies are indispensable, because it is the only animal for which a CF model is available, but significant differences exist between mouse and human airways. These include a lack of submucosal glands in mouse airways, except in the proximal trachea, and a predominance of Clara cells in the bronchioles (up to 90% of total epithelial cells).
A variety of methods has been explored to enhance delivery of Ads to airway epithelium. We exposed the epithelium to EGTA (31), which disrupts tight junctions, allowing the virus access to its major receptor, found predominantly on the basolateral membrane. It might be possible to use EGTA treatment in humans (31) or, alternatively, to modify the tropism of the virus (32). Unlike the normal and Cftr knockout mice used in this study, CF patients have a thickened airway mucus that constitutes an additional vector barrier. Thus, a mucolytic agent may be helpful as adjunct therapy. Adenovirus-neutralizing antibodies in the airway surface liquid also could be problematic, possibly requiring host immunomodulation (33) or shielding the adenovirus coat from antibody recognition through chemical modification (34) or lipid encapsulation (35). Switching serotypes from human adenovirus 5 to 2 has proven useful in allowing a second delivery to mice (15) and baboons (16).
HD-Ads do not express adenovirus genes and therefore cause less inflammation than earlier Ads delivered intravenously (17, 18) or by nasal aspiration (19). Lung inflammation due to aspirated K18CFTR-HD-Ad was minimal to moderate and variable. Inflammation and variability could be reduced further in clinical trials by delivery of the vector in an aerosol rather than a liquid (36).
The use of HD-Ads typically results in transgene expression for longer periods of time than did the use of earlier Ad vectors. Expression from HD-Ads given intravenously has been observed in mouse liver for 1–2 years (15, 17, 18) and >1 year in baboons (16). Expressing hCftr from vectors delivered to rodent lung has proven challenging historically. Previous studies using earlier adenoviral vectors (ΔE1 or ΔE1ΔE3) with viral promoters driving hCftr have shown a limited duration of expression. In these studies, hCftr RNA was detected in rodent lung up to 6 weeks after vector delivery (37–39). We detected hCftr RNA 4 weeks after delivery of K18CFTR-HD-Ad (longer times were not examined). This type of analysis, however, does not allow the relative contributions of different lung epithelia (i.e., conducting versus respiratory) to be determined. Previous studies have used immunofluorescence to localize hCFTR in airway epithelium, for periods up to 7 days after vector delivery (37, 39). We anticipated improved performance from our vector, because our previous analysis of K18-based HD-Ads bearing reporter genes demonstrated long-term expression in mouse airway epithelium and transduction of basal epithelial cells (19), which may have progenitor properties. Indeed, K18CFTR-HD-Ad yielded immunofluorescence evidence of hCFTR in airway epithelium for 28 days. An additional concern is the action of cytotoxic lymphocytes, which can eliminate cells expressing nonself genes (40). hCFTR is not strongly immunogenic in mice (40), but our added VSV-G tag could be problematic, eventually leading to the elimination of cells expressing tagged CFTR. The VSV-G tag was added to assist with anticipated difficulties in immunofluorescence detection in mice and for future studies with human tissue. We think the development of a vector without the tag will rectify this potential problem.
Preclinical evaluation of gene therapy requires a functional test to assess therapeutic potential. We therefore used a pathogen challenge to examine whether our vector could protect mice from lung infection. Chronic infection with mucoid forms of P. aeruginosa is relatively common in CF patients. However, until very recently (41), Cftr knockout mice could not be reliably colonized with P. aeruginosa unless the bacteria were first immobilized in agarose beads, to prevent mucociliary clearance from the lungs. We chose to challenge mice with Bcc, an opportunistic pathogen found in CF patients, which can be used to establish chronic or acute infection in the lungs of Cftr knockout mice without artificial immobilization (27, 42, ††). We used Bcc ET12, the most frequently cultured strain from CF patients in Canada and Great Britain and the strain most often associated with fatal necrotizing pneumonia and septicemia (the cepacia syndrome) (29). Aspirated Bcc ET12 causes severe lung infection in Cftr knockout mice (Cftrtm1UNC or Cftrm1HGU), whereas Cftr+/+ mice can resist infection (27, 42, ††). We chose to use the Cftrtm1UNC mouse (28), which is hardier than inbred strains. Like other outbred strains, the Cftrtm1UNC mouse does not develop lung disease spontaneously. Cftr–/– mice receiving K18CFTR-HD-Ad 7 days before Bcc challenge had less severe lung histopathology than Cftr–/– mice receiving only saline. Furthermore, pretreatment of Cftr–/– mice with K18CFTR-HD-Ad dramatically reduced the number of live Bcc recovered from lungs. These data were statistically significant, but the relatively low numbers of mice in each group dictate that the results should be considered preliminary until further studies can be performed. We did not challenge mice for periods of >7 days after gene therapy, so the duration of acquired protection is unknown. Repeated delivery of adenoviral vectors to extend transgene expression is problematic because of neutralizing antibodies produced by the host.
These data suggest that Cftr gene therapy might benefit CF patients in the future by reducing their susceptibility to pathogens such as Bcc. Because CF is a chronic disease, studies should determine whether K18CFTR-HD-Ad can protect Cftr knockout mice from colonization in a chronic infection model of Bcc (27) or P. aeruginosa (without artificial bacterial immobilization) (41) and the duration of the acquired protection.
Acknowledgments
We thank Lin Ye, Catherine Luk, Lily Morikawa, Huimin Wang, Cathy Gibson, Ross Ridsdale, Martin Post, Raha Mohammad-Panah, Christine Bear (Hospital for Sick Children), and Gregory Downey (University Health Network, Toronto) for comments and technical assistance; and Arthur Beaudet (Baylor College of Medicine, Houston) and Hugh O'Brodovich (Hospital for Sick Children) for reading the manuscript. We thank Merck & Co. for providing pC4HSU, H14 helper virus, and 293Cre4 cells. This work was funded by operating grants from the Canadian Cystic Fibrosis Foundation (to J.F.F. and J.H.) and the Canadian Institutes of Health Research (to J.H. and A.K.T). D.R.K. held a Canadian Cystic Fibrosis Foundation Postdoctoral Fellowship, and J.H. is a Canadian Cystic Fibrosis Foundation Scholar.
Abbreviations: CF, cystic fibrosis; Cftr, CF transmembrane conductance regulator; Bcc, Burkholderia cepacia complex; Ad, adenoviral vector; HD-Ad, helper-dependent Ad; VSV-G, vesicular stomatitis virus G glycoprotein; cfu, colony-forming units; hCftr, human Cftr.
Footnotes
Sajjan, U., Kent, G. & Forstner, G. (2002) Pediatr. Pulmonol. 34, 232 (abstr.).
References
- 1.Quinton, P. M. (1999) Physiol. Rev. 79, S3–S22. [DOI] [PubMed] [Google Scholar]
- 2.Pilewski, J. M. & Frizzell, R. A. (1999) Physiol. Rev. 79, S215–S255. [DOI] [PubMed] [Google Scholar]
- 3.Johnson, L. G., Olsen, J. C., Sarkadi, B., Moore, K. L., Swanstrom, R. & Boucher, R. C. (1992) Nat. Genet. 2, 21–25. [DOI] [PubMed] [Google Scholar]
- 4.Dorin, J. R., Farley, R., Webb, S., Smith, S. N., Farini, E., Delaney, S. J., Wainwright, B. J., Alton, E. W. & Porteous, D. J. (1996) Gene Ther. 3, 797–801. [PubMed] [Google Scholar]
- 5.Koehler, D. R., Hitt, M. M. & Hu, J. (2001) Mol. Ther. 4, 84–91. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, R. C. (1999) J. Clin. Invest. 103, 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albelda, S. M., Wiewrodt, R. & Zuckerman, J. B. (2000) Ann. Intern. Med. 132, 649–660. [DOI] [PubMed] [Google Scholar]
- 8.Moll, R., Franke, W. W., Schiller, D. L., Geiger, B. & Krepler, R. (1982) Cell 31, 11–24. [DOI] [PubMed] [Google Scholar]
- 9.Engelhardt, J. F., Zepeda, M., Cohn, J. A., Yankaskas, J. R. & Wilson, J. M. (1994) J. Clin. Invest. 93, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, Y. H., Plumb, J., Wen, Y., Steer, B. M., Lu, Z., Buchwald, M. & Hu, J. (2000) Mol. Ther. 2, 359–367. [DOI] [PubMed] [Google Scholar]
- 11.Chow, Y. H., O'Brodovich, H., Plumb, J., Wen, Y., Sohn, K. J., Lu, Z., Zhang, F., Lukacs, G. L., Tanswell, A. K., Hui, C. C., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 14695–14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pankov, R., Neznanov, N., Umezawa, A. & Oshima, R. G. (1994) Mol. Cell. Biol. 14, 7744–7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koehler, D. R., Hannam, V., Belcastro, R., Steer, B., Wen, Y., Post, M., Downey, G., Tanswell, A. K. & Hu, J. (2001) Mol. Ther. 4, 58–65. [DOI] [PubMed] [Google Scholar]
- 14.Qin, L., Ding, Y., Pahud, D. R., Chang, E., Imperiale, M. J. & Bromberg, J. S. (1997) Hum. Gene Ther. 8, 2019–2029. [DOI] [PubMed] [Google Scholar]
- 15.Kim, I. H., Jozkowicz, A., Piedra, P. A., Oka, K. & Chan, L. (2001) Proc. Natl. Acad. Sci. USA 98, 13282–13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morral, N., O'Neal, W., Rice, K., Leland, M., Kaplan, J., Piedra, P. A., Zhou, H., Parks, R., Velji, R., Aguilar-Cordova, E., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 12816–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morral, N., Parks, R. J., Zhou, H., Langston, C., Schiedner, G., Quinones, J., Graham, F. L., Kochanek, S. & Beaudet, A. L. (1998) Hum. Gene Ther. 9, 2709–2716. [DOI] [PubMed] [Google Scholar]
- 18.Schiedner, G., Morral, N., Parks, R. J., Wu, Y., Koopmans, S. C., Langston, C., Graham, F. L., Beaudet, A. L. & Kochanek, S. (1998) Nat. Genet. 18, 180–183. [DOI] [PubMed] [Google Scholar]
- 19.Toietta, G., Koehler, D. R., Finegold, M., Lee, B., Hu, J. & Beaudet, A. L. (2003) Mol. Ther. 7, 649–658. [DOI] [PubMed] [Google Scholar]
- 20.Koehler, D. R., Chow, Y. H., Plumb, J., Wen, Y., Rafii, B., Belcastro, R., Haardt, M., Lukacs, G. L., Post, M., Tanswell, A. K. & Hu, J. (2000) Pediatr. Res. 48, 184–190. [DOI] [PubMed] [Google Scholar]
- 21.Sandig, V., Youil, R., Bett, A. J., Franlin, L. L., Oshima, M., Maione, D., Wang, F., Metzker, M. L., Savino, R. & Caskey, C. T. (2000) Proc. Natl. Acad. Sci. USA 97, 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, L., Anton, M. & Graham, F. L. (1996) Somat. Cell Mol. Genet. 22, 477–488. [DOI] [PubMed] [Google Scholar]
- 23.Ng, P., Parks, R. J. & Graham, F. L. (2002) Methods Mol. Med. 69, 371–388. [DOI] [PubMed] [Google Scholar]
- 24.Zhou, H., Pastore, L. & Beaudet, A. L. (2002) Methods Enzymol. 346, 177–198. [DOI] [PubMed] [Google Scholar]
- 25.Smith, J. J., Travis, S. M., Greenberg, E. P. & Welsh, M. J. (1996) Cell 85, 229–236. [DOI] [PubMed] [Google Scholar]
- 26.Wang, G., Zabner, J., Deering, C., Launspach, J., Shao, J., Bodner, M., Jolly, D. J., Davidson, B. L. & McCray, P. B., Jr. (2000) Am. J. Respir. Cell Mol. Biol. 22, 129–138. [DOI] [PubMed] [Google Scholar]
- 27.Sajjan, U., Thanassoulis, G., Cherapanov, V., Lu, A., Sjolin, C., Steer, B., Wu, Y. J., Rotstein, O. D., Kent, G., McKerlie, C., et al. (2001) Infect. Immun. 69, 5138–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snouwaert, J. N., Brigman, K. K., Latour, A. M., Malouf, N. N., Boucher, R. C., Smithies, O. & Koller, B. H. (1992) Science 257, 1083–1088. [DOI] [PubMed] [Google Scholar]
- 29.Govan, J. R. & Deretic, V. (1996) Microbiol. Rev. 60, 539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snedecor, G. W. & Cochran, W. G. (1980) Statistical Methods (Iowa State Univ. Press, Ames).
- 31.Chu, Q., St. George, J. A., Lukason, M., Cheng, S. H., Scheule, R. K. & Eastman, S. J. (2001) Hum. Gene Ther. 12, 455–467. [DOI] [PubMed] [Google Scholar]
- 32.Wickham, T. J. (2000) Gene Ther. 7, 110–114. [DOI] [PubMed] [Google Scholar]
- 33.Chirmule, N., Raper, S. E., Burkly, L., Thomas, D., Tazelaar, J., Hughes, J. V. & Wilson, J. M. (2000) J. Virol. 74, 3345–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Riordan, C. R., Lachapelle, A., Delgado, C., Parkes, V., Wadsworth, S. C., Smith, A. E. & Francis, G. E. (1999) Hum. Gene Ther. 10, 1349–1358. [DOI] [PubMed] [Google Scholar]
- 35.Yotnda, P., Chen, D. H., Chiu, W., Piedra, P. A., Davis, A., Templeton, N. S. & Brenner, M. K. (2002) Mol. Ther. 5, 233–241. [DOI] [PubMed] [Google Scholar]
- 36.Joseph, P. M., O'Sullivan, B. P., Lapey, A., Dorkin, H., Oren, J., Balfour, R., Perricone, M. A., Rosenberg, M., Wadsworth, S. C., Smith, A. E., et al. (2001) Hum. Gene Ther. 12, 1369–1382. [DOI] [PubMed] [Google Scholar]
- 37.Zabner, J., Petersen, D. M., Puga, A. P., Graham, S. M., Couture, L. A., Keyes, L. D., Lukason, M. J., St. George, J., Gregory, R. J., Smith, A. E., et al. (1994) Nat. Genet. 6, 75–83. [DOI] [PubMed] [Google Scholar]
- 38.Rosenfeld, M. A., Yoshimura, K., Trapnell, B. C., Yoneyama, K., Rosenthal, E. R., Dalemans, W., Fukayama, M., Bargon, J., Stier, L. E., Stratford, P. L., et al. (1992) Cell 68, 143–155. [DOI] [PubMed] [Google Scholar]
- 39.Grubb, B. R., Pickles, R. J., Ye, H., Yankaskas, J. R., Vick, R. N., Engelhardt, J. F., Wilson, J. M., Johnson, L. G. & Boucher, R. C. (1994) Nature 371, 802–806. [DOI] [PubMed] [Google Scholar]
- 40.Kaplan, J. M., Armentano, D., Sparer, T. E., Wynn, S. G., Peterson, P. A., Wadsworth, S. C., Couture, K. K., Pennington, S. E., St. George, J. A., Gooding, L. R. & Smith, A. E. (1997) Hum. Gene Ther. 8, 45–56. [DOI] [PubMed] [Google Scholar]
- 41.Coleman, F. T., Mueschenborn, S., Meluleni, G., Ray, C., Carey, V. J., Vargas, S. O., Cannon, C. L., Ausubel, F. M. & Pier, G. B. (2003) Proc. Natl. Acad. Sci. USA 100, 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davidson, D. J., Dorin, J. R., McLachlan, G., Ranaldi, V., Lamb, D., Doherty, C., Govan, J. & Porteous, D. J. (1995) Nat. Genet. 9, 351–357. [DOI] [PubMed] [Google Scholar]