Abstract
Breeding crops with the quality of broad-spectrum disease resistance using genetic resources is one of the principal goals of crop improvement. However, the molecular mechanism of broad-spectrum resistance remains largely unknown. Here, we show that GH3-2, encoding an indole-3-acetic acid (IAA)-amido synthetase, mediates a broad-spectrum resistance to bacterial Xanthomonas oryzae pv oryzae and Xanthomonas oryzae pv oryzicola and fungal Magnaporthe grisea in rice (Oryza sativa). IAA, the major form of auxin in rice, results in rice more vulnerable to the invasion of different types of pathogens, which is at least partly due to IAA-induced loosening of the cell wall, the natural protective barrier of plant cells to invaders. X. oryzae pv oryzae, X. oryzae pv oryzicola, and M. grisea secrete IAA, which, in turn, may induce rice to synthesize its own IAA at the infection site. IAA induces the production of expansins, the cell wall-loosening proteins, and makes rice vulnerable to pathogens. GH3-2 is likely contributing to a minor quantitative trait locus for broad-spectrum resistance. Activation of GH3-2 inactivates IAA by catalyzing the formation of an IAA-amino acid conjugate, which results in the suppression of expansin genes. Thus, GH3-2 mediates basal resistance by suppressing pathogen-induced IAA accumulation. It is expected that, regulated by a pathogen-induced strong promoter, GH3-2 alone may be used for breeding rice with a broad-spectrum disease resistance.
Broad-spectrum resistance refers to resistance against two or more types of pathogen species or the majority of races of the same pathogen species (Kou and Wang, 2010). Plants fight against pathogen invasion via two major categories of resistance: qualitative (or vertical or complete) resistance mediated by disease resistance (R) genes, and quantitative (or horizontal or partial) resistance contributed by multiple genes or quantitative trait loci (QTLs). Qualitative resistance is highly efficient, but it is specific for pathogen race; in other words, it has a limited resistance spectrum. In addition, this type of resistance may be easily overcome because of the rapid evolution of pathogens (McDonald and Linde, 2002). In contrast, quantitative resistance is presumably nonspecific for pathogen species or race and is, therefore, durable. Furthermore, it is the only type of resistance for some plants against some pathogens, such as rice (Oryza sativa) against Xanthomonas oryzae pv oryzicola (Xoc), the causal agent of bacterial streak disease (Hu and Wang, 2009). However, it is generally difficult to use resistance QTLs in breeding programs as compared with the use of R genes, because most of these QTLs have small effects, with each explaining less than 10% of phenotypic variation, and because the genes underlying resistance QTLs are largely unknown (Kou and Wang, 2010). Recently, several minor resistance QTLs have been characterized from rice (Hu et al., 2008; Kou et al., 2010) and four major resistance QTLs have been isolated from wheat (Triticum aestivum) and rice (Fu et al., 2009; Fukuoka et al., 2009; Krattinger et al., 2009; Manosalva et al., 2009). These studies are shedding light on the molecular mechanisms of quantitative disease resistance. GH3-8 is one of the genes that contribute to the minor resistance QTLs from rice, and it regulates rice resistance by suppressing auxin signaling (Ding et al., 2008; Hu et al., 2008).
Except for its well-known roles in plant growth and development, auxin has been known for a long time to be a virulence factor in some host-pathogen systems (Sequeira and Kelman, 1962; Glickmann et al., 1998), but it is only recently that the molecular basis of auxin in host-pathogen interactions has been clarified. Flg22 is a bacterium-derived peptide that belongs to an evolutionarily conserved pathogen-associated molecular pattern. The peptide induces a microRNA, which in turn cleaves the RNAs of auxin receptors and results in an immune response in Arabidopsis (Arabidopsis thaliana; Navarro et al., 2006). Arabidopsis GH3-type proteins functioning in auxin signaling positively regulate bacterial resistance, associated with a salicylic acid (SA)-dependent pathway (Jagadeeswaran et al., 2007; Nobuta et al., 2007; Park et al., 2007; Zhang et al., 2007). SA is involved in the suppression of auxin signaling during resistance to bacterial disease in Arabidopsis (Wang et al., 2007a). Indole-3-acetic acid (IAA) is the major form of auxin in most plants, including rice. Rice GH3-8 is an IAA-amido synthetase and inactivates IAA by conjugating it to amino acids. GH3-8 mediates bacterial resistance by suppressing the loosening of the cell wall caused by auxin signaling (Ding et al., 2008). Rice GH3-1 also positively regulates resistance to fungal pathogens in a manner similar to GH3-8 (Domingo et al., 2009). These findings indicate that auxin may have complicated roles in plant-pathogen interactions.
The normal physiologic function of auxin depends on the appropriate concentration in the right place at the right time. Plants have evolved a wide variety of effective ways to regulate IAA homeostasis, one of which is to conjugate IAA to other molecules such as amino acids, sugars, peptides, or even proteins to activate or inactivate it (Seidel et al., 2006; Bari and Jones, 2009; Ludwig-Müller et al., 2009). Some GH3 proteins are IAA-, SA-, or jasmonic acid (JA)-amido synthetases, which modify the action of IAA, SA, or JA by conjugating them to amino acids (Staswick et al., 2005; Ludwig-Müller et al., 2009).
IAA can rapidly and transiently induce the expression of many genes functioning in the IAA-dependent pathway, including some GH3s (Woodward and Bartel, 2005). There are 19 GH3 paralogues in Arabidopsis; they are classified into three groups, and the paralogues in group II are IAA-amido synthetases that remove excess IAA by inactivating it (Staswick et al., 2005). The rice GH3 family consists of 12 active paralogues. Among them, GH3-1 and GH3-8 are activators of rice disease resistance (Ding et al., 2008; Domingo et al., 2009) and are classified into the same group with Arabidopsis GH3 group II proteins (Terol et al., 2006). These results suggest that other orthologs of GH3 group II in the plant kingdom may also have roles in plant-pathogen interactions due to their putative roles in the regulation of IAA homeostasis.
Bacterial blight caused by Xanthomonas oryzae pv oryzae (Xoo), bacterial streak caused by Xoc, and fungal blast caused by Magnaporthe grisea bring disastrous yield losses in rice worldwide every year. Although R genes are major genetic resources for the improvement of rice resistance against Xoo and M. grisea in breeding programs currently, the numbers of R genes against the two types of pathogens are limited (Chu and Wang, 2007; Ballini et al., 2008), along with the shortcomings of R genes mentioned above. In addition, no R gene against Xoc has been reported so far. Thus, disease resistance QTLs are available and valuable resources for breeding rice against these diseases (Wisser et al., 2005; Zhang et al., 2005). A previous study reported that a rice EST, EI5P11, is mapped to two loci, EI5P11a and EI5P11b, which overlap with the regions harboring two resistance QTLs, respectively (Wen et al., 2003). Locus EI5P11b on chromosome 7 is GH3-8. It is confirmed to contribute to a minor resistance QTL against Xoo by the strategy of validation and functional analysis of the QTL (Ding et al., 2008; Hu et al., 2008). Locus EI5P11a on chromosome 1 corresponds to GH3-2, a paralogue of the rice GH3 family, based on the nomenclature of Terol et al. (2006). GH3-2 belongs to group II of this family (Terol et al., 2006). These results suggest that GH3-2 may be involved in disease resistance.
To ascertain whether GH3-2 played a role in rice-pathogen interactions, we performed functional complementary and QTL analyses of GH3-2. These analyses suggest that GH3-2 encodes an IAA-amido synthetase and may contribute to a minor resistance QTL. GH3-2 positively regulates rice disease resistance by suppressing pathogen-induced accumulation of IAA in rice. Activation of GH3-2 confers to rice a broad-spectrum and partial resistance against Xoo, Xoc, and M. grisea.
RESULTS
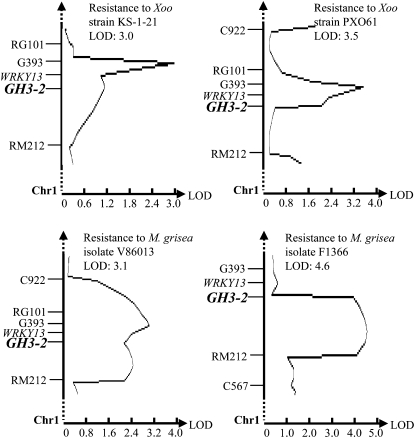
GH3-2 Colocalizes with a Disease Resistance QTL
Rice var Minghui 63 (in subsp. indica) carries R genes Xa3/Xa26 and Xa25(t) against Xoo (Chen et al., 2002; Sun et al., 2004) and rbr2 against M. grisea (Yang et al., 2008). It has resistance to Xoo strain JL691 and M. grisea isolate V86013, moderate resistance to Xoo strain PXO61 and M. grisea isolates F1814 and F1366, and susceptibility to Xoo strain KS-1-21. Rice var Zhenshan 97 (in subsp. indica), carrying no known R gene, is susceptible to these pathogens (Chen et al., 2003; Yang et al., 2003). We mapped GH3-2 in a recombinant inbred line (RIL) segregation population that was developed from a cross between Zhenshan 97 and Minghui 63. This population had been used to screen rice resistance to M. grisea and Xoo (Chen, 2001; Chen et al., 2003). The mapping showed that GH3-2 colocalized with the curves of resistance QTLs against Xoo strains KS-1-21 and PXO61 and M. grisea isolates V86013 and F1366 in chromosome 1, respectively (Fig. 1). GH3-2 also colocalized with two putative QTLs, which had a logarithm of odds of less than 2.5 against M. grisea isolate F1814 and Xoo strain JL691 (Supplemental Fig. S1). These QTLs appear to represent one QTL according to their flanking markers and the specificity of the GH3-2 probe. This QTL explained 5.2%, 6.4%, 3.0%, 5.9%, 9.6%, and 3.9% of the phenotypic variation of resistance to KS-1-21, PXO61, JL691, V86013, F1366, and F1814 in the mapping population, respectively. The defense-responsive gene OsWRKY13, which contributes to a minor resistance QTL (Hu et al., 2008; Qiu et al., 2009), also colocalized with this QTL (Fig. 1; Supplemental Fig. S1). A recent study revealed that several physically clustered defense-responsive genes function in cooperation as a resistance QTL in rice (Manosalva et al., 2009). The present results suggest that GH3-2 and OsWRKY13 may collectively contribute to this resistance QTL.
Figure 1.
Colocalization of GH3-2 and disease resistance QTLs. LOD, Logarithm of odds.
The resistance alleles at the QTLs and the putative QTL against all three Xoo strains were from Minghui 63, and the alleles at the QTLs and the putative QTL against the three M. grisea isolates were from Zhenshan 97 in this population. Comparative analysis of the genomic and cDNA sequences of GH3-2 (GenBank accession no. GU001814) from Minghui 63 showed that the gene was 3,756 bp in length and had a coding region interrupted by two introns (Supplemental Fig. S2A). The GH3-2 alleles from Minghui 63 and Zhenshan 97 encode an identical protein consisting of 614 amino acids. However, a distinct difference was noted in the promoter regions of the two alleles. Except for four single-base changes, a 430-bp insertion and a 51-bp deletion existed in the promoter region of GH3-2 in Minghui 63 in comparison with the GH3-2 promoter region in Zhenshan 97 (Supplemental Fig. S3). These results suggest that the GH3-2 allele putatively contributing to the resistant locus may result from an expressional difference during rice-pathogen interaction, as compared with its susceptible allele.
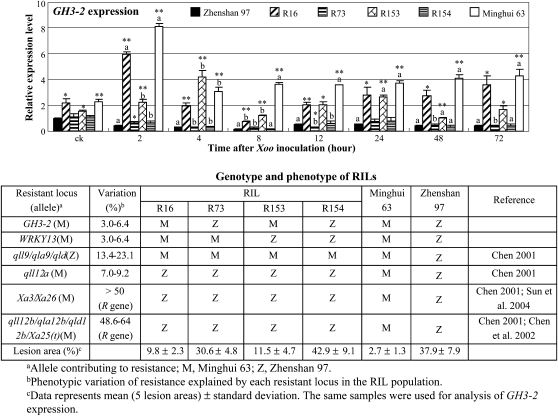
Pathogen Infection Influences GH3-2 Expression
To test whether the promoter difference of the two alleles resulted in their transcriptional discrepancy, we analyzed GH3-2 expression patterns in the resistant (Minghui 63) and susceptible (Zhenshan 97) parents of the RIL population and several RILs that had relatively consistent genetic backgrounds. RILs R16 and R153 showed significantly (P < 0.01) enhanced Xoo resistance compared with RILs R73 and R154, which had the same genotypes at R gene loci and resistance QTLs except for the GH3-2 locus, respectively (Fig. 2). GH3-2 expression was rapidly induced in Minghui 63 and the RILs (R16 and R153) that carried the GH3-2 allele from Minghui 63 but was rapidly suppressed in Zhenshan 97 and the RILs (R73 and R154) that carried the GH3-2 allele from Zhenshan 97 at 2 h after Xoo infection (Fig. 2). In addition, the transcript levels of GH3-2 were significantly higher in Minghui 63, R16, and R153 than in Zhenshan 97, R73, and R154, both without and with Xoo infection (Fig. 2).
Figure 2.
GH3-2 expression was responsive to Xoo infection in RILs and their parents, Zhenshan 97 and Minghui 63. Plants were infected with Xoo strain JL691 at the six-leaf stage. ck, Before infection. Bars represent means (three replicates) ± sd. The “a” or “b” indicates that a significant difference was detected between noninfected and Xoo-infected plants of the same rice line at P < 0.01 or P < 0.05, respectively. Two asterisks or one asterisk indicate that a significant difference was detected between susceptible Zhenshan 97 and other rice lines of the same treatment at P < 0.01 or P < 0.05, respectively.
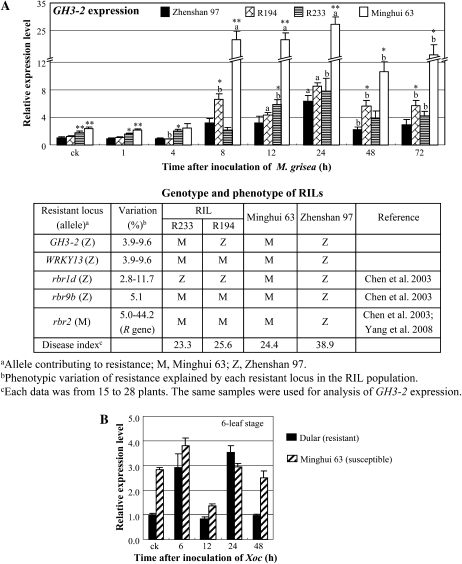
In contrast, M. grisea infection induced GH3-2 expression in both Minghui 63 and Zhenshan 97 and the RILs carrying the GH3-2 alleles from either of the parents (Fig. 3A). However, GH3-2 expression patterns in the two RILs, R233 and R194, that had the same genotypes at the R gene locus and resistance QTLs except for the GH3-2 locus were relatively different compared with the pattern in Zhenshan 97, although the two lines showed the same level of disease phenotypes based on the 0-to-9 scale rating system (International Rice Research Institute, 2002). The expression level of the GH3-2 allele from Minghui 63 in R233 was significantly higher than that in Zhenshan 97 before and at some time points after M. grisea infection, whereas the expression level of the GH3-2 allele from Zhenshan 97 in R194 was significantly higher than that in Zhenshan 97 at other time points after infection (Fig. 3A).
Figure 3.
GH3-2 expression was responsive to pathogen infection. Bars represent means (three replicates) ± sd. A, GH3-2 expression was induced by infection with M. grisea isolate 9-17-2 in RILs and their parents, Zhenshan 97 and Minghui 63, at the four- to five-leaf stage. The “a” or “b” indicates that a significant difference was detected between noninfected and pathogen-infected plants of the same rice line at P < 0.01 or P < 0.05, respectively. Two asterisks or one asterisk indicate that a significant difference was detected between susceptible Zhenshan 97 and other rice lines of the same treatment at P < 0.01 or P < 0.05, respectively. B, GH3-2 expression was influenced by infection with Xoc strain RH3. ck, Before infection.
Rice var Dular (in subsp. indica) is resistant to Xoc strain RH3 (Chen et al., 2006). Compared with Dular (lesion length of 0.22 ± 0.06 cm), Minghui 63 (lesion length of 2.21 ± 0.10 cm) was susceptible to RH3. The expression level of GH3-2 was approximately 3-fold higher in Minghui 63 than in Dular when without pathogen infection (Fig. 3B). However, the GH3-2 expression level increased 2.9- to 3.5-fold in resistant Dular at 6 and 24 h after RH3 infection but was reduced 52% in susceptible Minghui 63 at 12 h after infection (Fig. 3B).
To summarize these results, GH3-2 appeared to be more rapidly or efficiently induced in resistant reactions than in susceptible reactions. These results suggest that GH3-2 may positively regulate rice resistance against different pathogens.
Activating GH3-2 Enhances Rice Resistance to Xoo, Xoc, and M. grisea
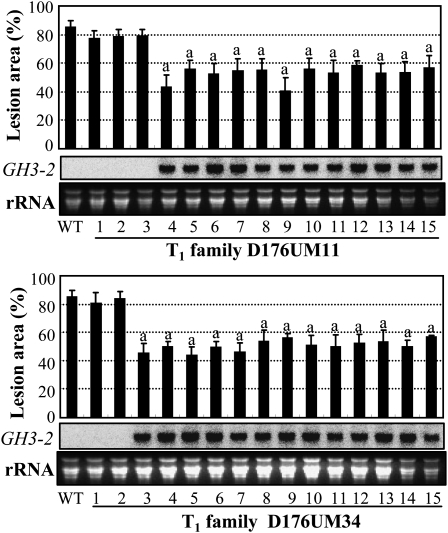
To examine the role of GH3-2 in disease resistance, we manipulated GH3-2 expression. GH3-2 was overexpressed in susceptible var Mudanjiang 8 (in subsp. japonica). Fourteen independent transformants, named D176UM, were developed to the booting stage for further analyses. At this stage, 13 of the 14 T0 plants showed significantly enhanced resistance to Xoo strain PXO61, with the lesion area ranging from 47.8% to 58.9% versus 81.9% for the wild type (Supplemental Fig. S4A). The enhanced resistance of these plants was associated with an increased expression of GH3-2. Plants in three T1 families from resistant T0 plants, D176UM11, D176UM14, and D176UM34, were further analyzed individually at the booting stage for their resistance to PXO61 and the GH3-2 transcript level. All the T1 plants showing significantly enhanced resistance to PXO61 had increased expression of GH3-2, whereas other plants showing no significant difference from the wild type in response to PXO61 inoculation had no detectable increased GH3-2 expression (Fig. 4; Supplemental Fig. S4B). The growth rates of PXO61 on the leaves of resistant transgenic plants were reduced 72% to 77% compared with the susceptible wild-type plants at 14 d after infection (Supplemental Fig. S4C). The cosegregation of enhanced resistance and increased GH3-2 expression suggest that GH3-2 positively regulates rice resistance to Xoo by inhibiting the growth of this bacterium.
Figure 4.
Enhanced resistance of GH3-2-overexpressing T1 plants (D176UM) to Xoo strain PXO61 was associated with increased GH3-2 expression. The “a” indicates a significant difference (P < 0.01) from the wild type (WT). Data represent means (five replicates) ± sd.
GH3-2-suppressing plants were generated using the RNA interference (RNAi) strategy in rice var Zhonghua 11 (in subsp. japonica). The GH3-2 transcript levels in T0-RNAi plants were 23% to 66% of that in wild-type plants (Supplemental Fig. S5). The GH3-2 transcript levels in the positive RNAi plants of two T1 families were 21% to 29% and 31% to 43% of that in the wild type, respectively (Supplemental Fig. S6). However, all the transgenic plants showed a similar level of susceptibility to Xoo as the wild type. This result could be explained by the incomplete suppression of GH3-2 and the functional redundancy among the paralogues. Thus, only GH3-2-overexpressing plants were used for further analyses.
Activation of GH3-2 also increased rice resistance against Xoc and M. grisea. The positive transgenic plants from two T1 families (D176UM11 and D176UM34) showed significantly enhanced resistance to Xoc strain RH3 (Table I); the lesion lengths of the positive transgenic plants were reduced 76% and 80% compared with the negative transgenic plants in the same family, respectively. The homozygote T2 lines of D176UM11 and D176UM34 were also tested for blast resistance by inoculation with M. grisea isolate CHL358. The GH3-2-overexpressing plants showed markedly enhanced resistance (Table II). These results suggested that GH3-2 mediates a broad-spectrum resistance.
Table I. Performance of two GH3-2-overexpressing T1 families after inoculation with Xoc strain RH3 at the booting stage.
| Rice Material | PCRa | Lesion Lengthb | P |
| cm | |||
| D176UM11 | + | 0.31 ± 0.10 | 0.0000 |
| D176UM11 | − | 1.29 ± 0.24 | 0.2505 |
| D176UM34 | + | 0.28 ± 0.09 | 0.0000 |
| D176UM34 | − | 1.40 ± 0.29 | 0.0542 |
| Mudanjiang 8 (wild type) | 1.18 ± 0.21 |
PCR primers used for identification of positive transgenic plants were GUSF and GUSR (Supplemental Table S3).
Data represent means (10–12 lesions) ± sd.
Table II.
Performance of two GH3-2-overexpressing lines after inoculation with M. grisea isolate CHL358 at the four-leaf stage
| Rice Material | Disease Indexa | Resistance/Susceptibility |
| CO39 (susceptible control) | 75.6 | Highly susceptible |
| Mudanjiang 8 (wild type) | 40.0 | Moderately susceptible |
| D176UM11 | 14.4 | Resistant |
| D176UM34 | 13.3 | Resistant |
Disease index was calculated with the individual leaf ratings using the following formula: disease index = [sum of numerical ratings from all leaves/(number of leaves assessed × maximum lesion rating)] × 100. Data were from 38 and 37 plants in lines D176UM11 and D176UM34, respectively.
GH3-2 Encodes an IAA-Amido Synthetase and Modulates Auxin Homeostasis in Rice
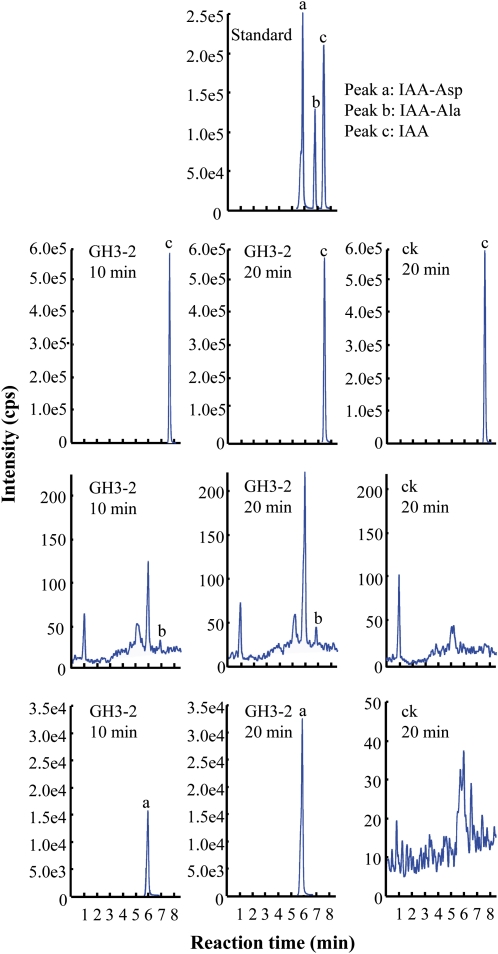
GH3-2 belongs to group II of the GH3 proteins (Terol et al., 2006). This group of GH3 proteins contains IAA-amido synthetases functioning to inactivate auxin by conjugating excess IAA to amino acids in both Arabidopsis and rice (Staswick et al., 2005; Ding et al., 2008). GH3-2 shows 72% sequence identity and 82% sequence similarity to rice GH3-8, which is an IAA-amido synthetase (Supplemental Fig. S7; Ding et al., 2008). To ascertain the biochemical function of GH3-2, we tested the enzyme activity of recombinant GH3-2 in the reaction mixture containing IAA and Asp or Ala. The reactions produced new products that had the same retention times as IAA-Asp and IAA-Ala (Fig. 5). However, more IAA-Asp than IAA-Ala was generated in the reaction mixtures. These results suggest that GH3-2 has amido synthetase activity and is more capable of catalyzing the synthesis of IAA-Asp than IAA-Ala.
Figure 5.
Enzyme activity of GH3-2 in vitro. UFLC-MS/MS analysis of amino acid conjugates of IAA synthesized by recombinant GH3-2 protein in different time courses. ck, Proteins from Escherichia coli transferred with the null vector pET28a. cps, Counts per second. [See online article for color version of this figure.]
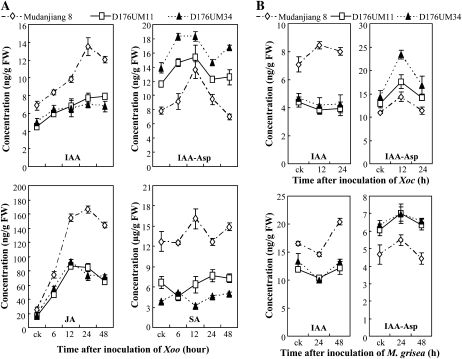
This conclusion is supported by the quantification of IAA and IAA-amino acids in planta. We quantified the concentrations of IAA, IAA-Asp, and IAA-Ala in transgenic plants after infection with Xoo, Xoc, or M. grisea. After Xoo infection, the IAA levels in GH3-2-overexpressing lines were maximally reduced (24 h after infection) 42.9% and 48.6% compared with wild-type plants, whereas the IAA-Asp levels in GH3-2-overexpressing lines were maximally increased (48 h after infection) 1.8- and 2.4-fold compared with wild-type plants (Fig. 6A). No IAA-Ala was detected in transgenic and wild-type plants. The IAA and IAA-Asp levels were also reduced and increased, respectively, in GH3-2-overexpressing lines compared with wild-type plants after infection with Xoc and M. grisea (Fig. 6B). Thus, GH3-2 appears to be involved in the regulation of auxin homeostasis in rice. Activation of GH3-2 may influence the action of auxin.
Figure 6.
Hormone concentrations in wild-type Mudanjiang 8 and GH3-2-overexpressing lines D176UM11 and D176UM34. Bars represent means (three replicates) ± sd. ck, Before infection; FW, fresh weight. A, Concentrations of IAA, IAA-Asp, JA, and SA after inoculation of Xoo strain PXO61 at the booting stage. B, Concentrations of IAA and IAA-Asp after inoculation of Xoc strain RH3 at the booting stage and inoculation of M. grisea isolate CHL358 at the four-leaf stage.
Activation of GH3-2 Suppresses Auxin Signaling
The dwarf and tufted morphology of GH3-2-overexpressing plants also reflected the function of GH3-2 in controlling auxin concentration (Supplemental Fig. S8). This abnormal morphology was also observed in auxin-deficient GH3-8- and GH3-1-overexpressing rice (Ding et al., 2008; Domingo et al., 2009).
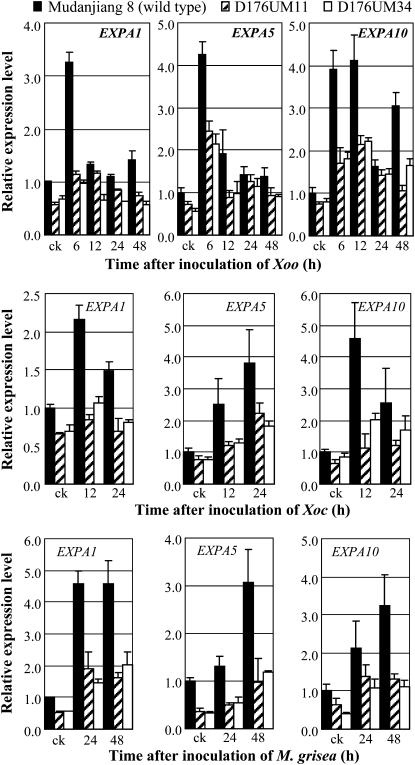
In addition, activation of GH3-2 also suppressed the expression of expansin genes (Fig. 7), which function on the auxin signaling pathway (Ding et al., 2008). The expression of expansin genes EXPA1, EXPA5, and EXPB10 was markedly induced in wild-type plants but only slightly induced in GH3-2-overexpressing lines after Xoo, Xoc, or M. grisea infection (Fig. 7). Furthermore, the expression levels of the three genes were significantly lower (P < 0.05) in GH3-2-overexpressing plants than in wild-type plants in at least one time point examined (Fig. 7).
Figure 7.
Expression of expansin (EXP) genes in GH3-2-overexpressing lines D176UM11 and D176UM34. Plants were inoculated with Xoo strain PXO61 or Xoc strain RH3 at the booting stage or inoculated with M. grisea isolate CHL358 at the four-leaf stage. Bars represent means (three replicates) ± sd. ck, Before infection.
Expansins are cell wall-loosening proteins and function in auxin-regulated growth (McQueen-Mason et al., 1992). Xoo infection and IAA treatment induce the expression of multiple paralogues in this family in rice; overexpression of rice expansin genes EXPA1, EXPA5, and EXPA10 results in increased susceptibility to Xoo (Ding et al., 2008). EXPA1-, EXPA5-, and EXPA10-overexpressing plants also showed significantly increased (P < 0.01) susceptibility to Xoc strain RH3 (Supplemental Table S1). The average lesion lengths caused by RH3 infection were 0.80 ± 0.19, 0.64 ± 0.09, and 0.76 ± 0.17 cm for EXPA1-, EXPA5-, and EXPA10-overexpressing plants, respectively, compared with 0.51 ± 0.07 cm for wild-type Zhonghua 11. EXPA1-, EXPA5-, and EXPA10-overexpressing plants also showed slightly increased susceptibility to M. grisea isolate CHL358 (Supplemental Table S2). The disease index caused by CHL358 infection ranged from 35.6 to 40 for EXPA1-, EXPA5-, and EXPA10-overexpressing plants as compared with 31.1 for wild-type plants. These results suggest that GH3-2 mediates resistance to Xoc and M. grisea, possibly through inhibition of expansin genes by suppressing auxin signaling.
Pathogen-Secreting IAA Is Associated with Host Local Accumulation of IAA
Some plant pathogens secrete auxin as a virulence factor during infection of hosts (Comai and Kosuge, 1982; Surico et al., 1985). To ascertain whether Xoo, Xoc, and M. grisea also possess this feature, we quantified IAA accumulation in the liquid medium used for culturing these pathogens. Three Xoo strains, PXO61, PXO99, and PXO347, were analyzed. After culturing the bacteria in liquid medium until an optical density at 600 nm (OD600) exceeded 1.0, the amounts of IAA in the culture medium were 5.5 ± 0.4, 14.6 ± 1.6, and 9.6 ± 1.5 ng mL−1 OD600−1 for PXO61, PXO99, and PXO347, respectively. The amount of IAA in the medium after culturing Xoc strain RH3 was 5.9 ± 1.1 ng mL−1 OD600−1. The amounts of IAA in the medium after culturing M. grisea isolates CHL358 and RB11 were 15.9 ± 3.1 and 17.8 ± 2.9 ng mL−1 OD600−1, respectively. These results suggest that Xoo, Xoc, and M. grisea can secret IAA and that they may use IAA as a virulence factor to infect rice.
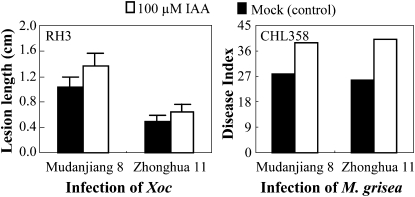
The inference that Xoo uses IAA as a virulence factor is supported by the evidence that pretreating rice with IAA increased rice susceptibility to Xoo infection (Ding et al., 2008). To test whether auxin could also aggravate rice susceptibility to Xoc and M. grisea, we treated rice plants with IAA before infection. Compared with mock-treated control plants, IAA pretreatment significantly (P < 0.01) promoted disease symptoms caused by Xoc strain RH3 in rice varieties Mudanjiang 8 and Zhonghua 11 (Fig. 8). IAA-pretreated plants were also more susceptible to M. grisea isolate CHL358 (Fig. 8). These results suggest that auxin can also help Xoc and M. grisea to infect rice.
Figure 8.
Exogenous application of IAA increased susceptibility to Xoc strain RH3 and M. grisea isolate CHL358 in rice varieties Mudanjiang 8 and Zhonghua 11. Bars represent means (12 replicates for Xoc infection and 20 replicates for M. grisea infection) ± sd. ck, Before infection.
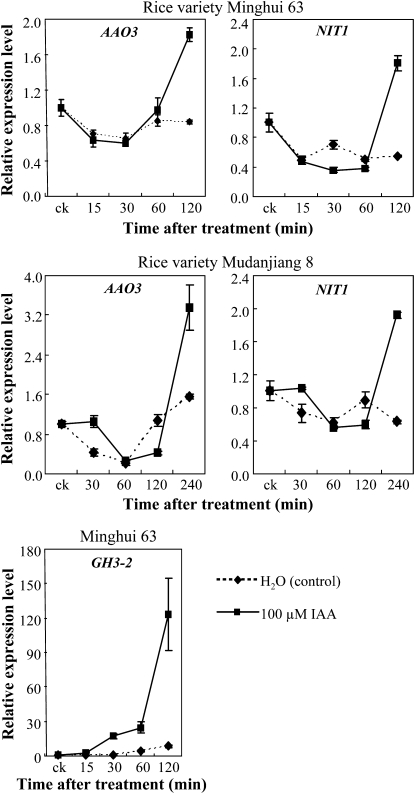
Indole-3-acetaldehyde oxidase (AAO) and nitrilase (NIT) are two protein families involved in the two Trp-dependent pathways for IAA biosynthesis in plants (Woodward and Bartel, 2005). Expression of AAO3 (AK065990) and NIT1 (AK104033) genes, which were putatively involved in IAA biosynthesis, was markedly induced in a susceptible rice line after Xoo infection (Ding et al., 2008). To ascertain whether resistant and susceptible plants had different expression patterns of IAA synthesis-related genes on pathogen infection, we examined the expression of AAO3 and NIT1 in different rice-pathogen interactions (Fig. 9). Xoo, Xoc, and M. grisea infection all induced AAO3 and NIT1 in both resistant and susceptible rice lines. However, the transcript levels of AAO3 and NIT1 were significantly higher in susceptible lines than in resistant lines at more time points examined after Xoo, Xoc, or M. grisea infection. These results suggest that the susceptible reaction may be associated with rapidly increased endogenous biosynthesis of IAA.
Figure 9.
Expression of putative IAA synthesis-related genes AAO3 and NIT1 in different rice varieties after infection with Xoo strain PXO61, Xoc strain RH3, or M. grisea isolate CHL358. Bars represent means (three replicates) ± sd. Two asterisks or one asterisk indicate that a significant difference was detected between susceptible and resistant rice lines of the same treatment at P < 0.01 or P < 0.05, respectively. ck, Before infection.
The expression of AAO3 and NIT1 was also influenced by IAA treatment. IAA treatment first suppressed and then induced AAO3 and NIT1 as compared with water-treated control plants (Fig. 10). Treatment of rice with IAA also rapidly induced GH3-2 (Fig. 10). These results suggest that pathogens secreting IAA may induce rice to synthesize its own IAA, which further helps the infection by pathogens in susceptible rice.
Figure 10.
IAA influenced the expression of AAO3, NIT1, and GH3-2. Bars represent means (three replicates) ± sd. ck, Before IAA treatment.
GH3-2-Mediated Resistance Does Not Require the Activation of JA and SA Signaling
Previous studies reported that enhanced Xoo resistance is associated with increased accumulation of SA, SA synthesis-related genes, and SA-responsive genes or with increased accumulation of JA and JA synthesis-related genes (Qiu et al., 2007; Yuan et al., 2007; Tao et al., 2009; Xiao et al., 2009). The expression patterns of seven defense-responsive genes, pathogenesis-related (PR) genes PR1a (for acidic PR protein 1; AJ278436), PR1b (for basic PR protein 1; U89895), and PR10 (for ribonuclease; D38170) that are responsive to SA and JA signaling, SA synthesis-related genes PAL1 (for Phe ammonia-lyase 1; X16099) and PAD4 (for phytoalexin deficient 4; CX118864), and JA biosynthesis-related genes LOX (for lipoxygenase; D14000) and AOS2 (for allene oxide synthase 2; AY062258), were examined after infection with Xoo strain PXO61 in transgenic plants. The expression of the seven genes was significantly suppressed (P < 0.05) in GH3-2-overexpressing plants before or at some time points after pathogen infection (Supplemental Fig. S9).
Accompanying the suppression of these defense-responsive genes, the SA and JA levels in GH3-2-overexpressing plants were also comparatively lower than those in wild-type plants (Fig. 6A). PXO61 infection dramatically induced JA accumulation and slightly induced SA accumulation in both wild-type and GH3-2-overexpressing plants. However, the JA and SA concentrations in wild-type Mudanjiang 8 were 1.4- to 1.5-fold and 1.9- to 3.3-fold higher than those in GH3-2-overexpressing lines before pathogen infection, respectively, and maximally 2.2- to 2.3-fold and 2.8- to 5.0-fold higher than those in GH3-2-overexpressing lines after pathogen infection, respectively. These results suggest that GH3-2-mediated Xoo resistance does not require the activation of SA or JA signaling.
DISCUSSION
The results presented here suggest that GH3-2, an IAA-amido synthetase, is an activator in rice resistance against different pathogens. Because of the functional redundancy of the GH3 family in rice, this conclusion is mostly based on the analyses of GH3-2-overexpressing plants. However, the following evidence suggests that GH3-2 should play a role in rice-pathogen interaction in physiologic conditions. First, different types of pathogens more rapidly induced GH3-2 expression in resistant rice lines than in susceptible lines, implying its involvement in defense responses. Second, GH3-2 belongs to the group II proteins of the GH3 family; at least three Arabidopsis proteins (GH3.5, GH3.11, and GH3.12) and two rice proteins (GH3-1 and GH3-8) of this group have been reported to positively regulate pathogen-induced defense responses (Staswick et al., 1998, 2005; Jagadeeswaran et al., 2007; Nobuta et al., 2007; Zhang et al., 2007; Ding et al., 2008; Domingo et al., 2009). In addition, this study has revealed a potential candidate for the improvement of rice broad-spectrum resistance for breeding programs.
IAA Secreted by Xoo, Xoc, and M. grisea May Play a Role in Increasing Disease Symptoms
Auxin increases the susceptibility of Arabidopsis, tobacco (Nicotiana tabacum), sweet orange (Citrus sinensis), and rice to biotrophic, hemibiotrophic, or necrotrophic bacterial or fungal infection (Navarro et al., 2006; Chen et al., 2007; Ding et al., 2008; Ferrari et al., 2008; Cernadas and Benedetti, 2009). Xoo and Xoc are biotrophic pathogens, and M. grisea is a hemibiotrophic pathogen. IAA makes rice more vulnerable to Xoo infection (Ding et al., 2008). The results presented here suggest that this hormone also makes rice more vulnerable to Xoc and M. grisea infection.
Many biotrophic, hemibiotrophic, and necrotrophic pathogenic bacteria and fungi, including some species in the Xanthomonas genus, have the ability to produce the plant hormone IAA (Sheldrake, 1973; Fett et al., 1987; Hasan, 2002; Maor et al., 2004; Yang et al., 2007; Reineke et al., 2008). The IAA produced by some pathogens is pivotal for the pathogenicity process (Comai and Kosuge, 1982; Surico et al., 1985). The results presented here add another three pathogens, Xoo, Xoc, and M. grisea, to the list of IAA-producing microorganisms. Although our results cannot answer whether Xoo, Xoc, and M. grisea use IAA as a virulence factor to invade rice, the IAA secreted by these pathogens appears to play an important role in rice accumulating endogenous IAA. Accumulation of IAA is associated with an increase in disease symptoms. This inference is supported by the following evidence. IAA could induce the expression of IAA synthesis-related genes (Fig. 10). Xoo, Xoc, and M. grisea infection all induced these genes (Fig. 9) and resulted in rice, especially susceptible rice, accumulating IAA (Fig. 6; Ding et al., 2008). These results suggest that pathogen infection-induced accumulation of IAA at the infection site may be at least partly due to the activation of rice IAA biosynthesis through the initial IAA produced by these pathogens.
GH3-2 Mediates Basal Resistance
The plant cell wall is an important component of basal resistance. The expansin genes encoding cell wall-loosening proteins are induced by IAA treatment and Xoo infection, suggesting that Xoo-induced IAA-stimulated local production of expansins may be one of the mechanisms used by Xoo to infect rice (Ding et al., 2008). The results presented here further support this inference. Xoc and M. grisea infection also induced expansins, and activation of expansins resulted in rice more susceptible to these pathogens (Fig. 7; Supplemental Tables S1 and S2). GH3-2 positively regulates disease resistance by suppressing the action of IAA, which in turn prevents the production of expansins (Fig. 7). Thus, GH3-2 mediates basal resistance. The disease resistance regulated by Arabidopsis GH3-type proteins is SA dependent (Jagadeeswaran et al., 2007; Nobuta et al., 2007; Park et al., 2007; Zhang et al., 2007). Rice GH3-8-mediated Xoo resistance did not require the activation of either the SA- or JA-dependent pathway (Ding et al., 2008). Rice GH3-2 mediates resistance in the same way as GH3-8. This may be the consequence of rice and Arabidopsis adopting different mechanisms to suppress auxin signaling during disease resistance.
Although pathogen-secreted IAA may have a pathogenic role by inducing rice synthesis of IAA, IAA can also induce GH3-2 expression (Fig. 10). GH3-2 in turn suppresses IAA activity in a feedback loop. However, induction of GH3-2 on pathogen infection is more rapid or efficient in R gene-containing rice lines or resistant lines not carrying an R gene than in susceptible rice lines (Figs. 2 and 3), suggesting that other factors in addition to IAA may also influence GH3-2 expression in the rice-pathogen interaction.
GH3-2 May Contribute to a Resistance QTL
Map-based cloning is not the best choice to isolate minor resistance QTLs because of their small effect on disease resistance. The strategy of validation and functional analysis of the QTL has proved to be an applicable approach to isolate minor resistance QTLs (Hu et al., 2008; Kou and Wang, 2010). Through a functional complementary test, QTL mapping, and pathogen-influenced expression analysis, we infer that GH3-2, in cooperation with OsWRKY13, may contribute to a minor resistance QTL. This suggestion is also supported by phenotypic comparison of different RILs with Xoo infection. When R gene loci and other resistance QTLs were fixed, the RILs that carried the putative resistance GH3-2 allele showed significantly enhanced resistance compared with RILs that carried the putative susceptible GH3-2 allele (Fig. 2).
It is interesting that at the GH3-2 locus, the resistant allele for Xoo resistance was from resistant line Minghui 63 and the resistant allele for M. grisea resistance was from susceptible line Zhenshan 97 in the same segregation population. However, the two alleles encode an identical protein but have different expression patterns in rice-pathogen interactions (Figs. 2 and 3). In quantitative resistance, the alleles contributing to resistant loci can be divided into three groups by comparison with their corresponding susceptible alleles (Kou and Wang, 2010). GH3-2 appears to belong to the group whose resistant alleles result from expressional or posttranslational differences during host-pathogen interactions. This inference is supported by the evidence that the GH3-2 alleles have different promoters; the GH3-2 promoter in Minghui 63 carries extra cis-acting elements that are putatively involved in auxin response and disease defense (Supplemental Fig. S3). The induced expression of GH3-2 alleles from Minghui 63 was associated with enhanced Xoo resistance (Fig. 2). However, the expression levels of GH3-2 alleles were differentially induced in RILs that had relatively consistent genetic backgrounds in comparison with that in Zhenshan 97 in the rice-M. grisea interaction (Fig. 3A). This difference may be partially explained by the fact that Minghui 63 and Zhenshan 97 had different background levels of IAA, although differential expression of GH3-2 was associated with different levels of IAA-Asp conjugate in the two rice varieties (Supplemental Fig. S10). Further study is needed to examine whether this expressional difference of GH3-2 alleles in Minghui 63 and Zhenshan 97 resulted in their putatively different contributions in quantitative resistance.
Perspective of GH3-2 in Breeding Programs
The features of broad-spectrum resistance conferred by GH3-2 suggest that this gene is a potential candidate for rice breeding programs. This gene is extremely valuable for improving rice resistance against Xoc because the host genes involved in the rice-Xoc interaction are poorly understood and no R gene for Xoc resistance has been identified. It is expected that GH3-2 may also mediate resistance to other rice pathogens that also secrete IAA as a virulence factor. Marker-assisted selection has been applied in breeding programs for targeted transferring and pyramiding of major resistance QTLs in different crops. However, marker-assisted selection of a single minor QTL is not effective for improving disease resistance because of its small effect on phenotype. The results presented here suggest that, by manipulating its expression, a single minor resistant QTL may be used for improving broad-spectrum resistance in rice. However, GH3-2 is also a regulator for rice growth and development. Thus, the efficient way of using GH3-2 in rice improvement of disease resistance may use a nonspecific pathogen species-induced strong promoter to enhance its expression when infected.
CONCLUSION
The pathogens that cause rice bacterial blight, bacterial streak, and blast diseases secrete IAA. IAA increases rice disease symptoms. GH3-2, encoding an IAA-amido synthetase, confers a broad-spectrum quantitative resistance against bacterial blight, bacterial streak, and blast via suppressing auxin signaling. It is expected that, controlled under a pathogen-induced strong promoter, GH3-2 could be a candidate for rice breeding programs.
MATERIALS AND METHODS
QTL Analysis
A rice (Oryza sativa) recombinant inbred line population was used for comapping of GH3-2 gene and resistance QTLs. This population consisted of 241 lines developed from a cross between Zhenshan 97 and Minghui 63 by single-seed descent. A molecular linkage map containing 221 markers and covering the whole rice genome was developed with this population (Xing et al., 2002). This population had been used to identify resistance QTLs against four Xanthomonas oryzae pv oryzae strains (Chinese strains JL691 and KS-1-21 and Philippine strains PXO61 and PXO339; Chen, 2001) and three Magnaporthe grisea isolates (Chinese isolates F1366 and F1814 and Philippine isolate V86013; Chen et al., 2003). GH3-2 was mapped using a PCR-based cleave amplification polymorphism sequence marker (Supplemental Table S3) designed according to a single nucleotide polymorphism localized at the 3′-untranslated region of GH3-2 (Supplemental Fig. S2A). Mapmaker/Exp 3.0 (Lincoln et al., 1992) was used for linkage analysis. QTL analysis was conducted using the computer program Windows QTL Cartographer Version 2.5 for composite interval mapping at a threshold of logarithm of odds 2.5 (Wang et al., 2007b).
Gene Isolation and Structure Analysis
The sequence (GenBank accession no. CX100320; Zhang et al., 2005) of cDNA clone BI101D22 of the GH3-2 gene from rice var Minghui 63 was used to screen the rice whole genome sequence database (Rice Genome Annotation Project; http://rice.plantbiology.msu.edu) using the BLAST program (Altschul et al., 1997). The cDNA sequence CX100320 was highly homologous to gene locus LOC_Os01g55940. Based on the structure of LOC_Os01g55940, CX100320 harbored the entire coding sequence of GH3-2. A series of PCR primers (Supplemental Table S3) were designed to sequence the genomic sequence of GH3-2 from rice Minghui 63. The genomic and cDNA sequences of GH3-2 from Minghui 63 were compared to determine the structure of this gene.
Transformation
The overexpression construct of GH3-2 was constructed by cutting GH3-2 from cDNA clone BI101D22 using restriction enzymes KpnI and BamHI and ligating it into the transformation vector pU1301 (Supplemental Fig. S2A), which contained a maize ubiquitin gene promoter in the multicloning site (Cao et al., 2007). To construct an RNAi vector of GH3-2, a 442-bp cDNA fragment of GH3-2 was amplified from cDNA clone BI101D22 using primers pEI5P11a(3) and pEI5P11a(4) (Supplemental Table S3) and inserted into the pDS1301 vector (Supplemental Fig. S2B; Yuan et al., 2007). The overexpression and RNAi constructs were transferred into Agrobacterium tumefaciens strain EHA105 by electroporation. Agrobacterium-mediated transformation was performed using calli derived from mature embryos of rice varieties Mudanjiang 8 and Zhonghua 11 (Lin and Zhang, 2005).
Pathogen Infection
For the evaluation of bacterial blight disease, rice plants were inoculated with Chinese Xoo strain JL691 or Philippine Xoo strains PXO61 or PXO347 by the leaf-clipping method (Chen et al., 2002). Disease was scored by calculating the percentage lesion area (lesion length/leaf length) at 2 weeks after inoculation. The growth rate of Xoo in rice leaves was analyzed by counting colony-forming units (Sun et al., 2004).
For evaluating bacterial streak disease, plants were inoculated with Xanthomonas oryzae pv oryzicola strain RH3 at a concentration of 9 × 108 colony-forming units mL−1 using the needle-stab method (Tao et al., 2009). Lesion lengths were measured 3 weeks after inoculation.
For fungal blast disease evaluation, seedlings at the four- to five-leaf stage were inoculated with M. grisea isolate 9-17-2 or CHL358 by the spraying method (Chen et al., 2003). Disease was scored using the 0-to-9 scale rating system at 7 d after inoculation (Tao et al., 2009). In this rating system, disease index was calculated with the individual leaf ratings using the following formula: disease index = [sum of numerical ratings from all leaves/(number of leaves assessed × maximum lesion rating)] × 100. A disease index of ≥0 and ≤5 indicates high resistance, >5 and ≤15 indicates resistance, >15 and ≤30 indicates moderate resistance, >30 and ≤45 indicates moderate susceptibility, >45 and ≤60 indicates susceptibility, and >60 indicates high susceptibility.
The effect of IAA on disease development was applied as described previously (Ding et al., 2008). Rice plants at the tillering stage (for Xoc infection) or at four- to five-leaf stage (for M. grisea infection) were sprayed with a solution containing 100 μm IAA diluted in 0.02% Tween 20. The control plants were mock sprayed with a solution containing 0.02% Tween 20. The plants were inoculated with Xoc strain RH3 or M. grisea isolate CHL358 at 4 h after IAA treatment. The bacterial and fungal inocula were prepared as in the procedures cited above, except that the inocula contained 100 μm IAA.
Quantification of Hormones
Leaf fragments about 5 cm long right next to the inoculation sites were used for analysis. Three replicates of each leaf sample (0.1 g) were used for phytohormone quantification. The samples were prepared as described previously (Ding et al., 2008). SA, JA, IAA, IAA-Asp, and IAA-Ala were quantified using the ultrafast liquid chromatography (UFLC)-electrospray ionization-tandem mass spectrometry (MS/MS) system. The purified sample was diluted using methanol in a ratio of 1:200 (v/v) for SA quantification. The quantitative data of SA and naphthalene acetic acid (as internal standard for SA; Sigma-Aldrich) were obtained using the peaks of the precursor ions 136.9 and 183.0 and the product ions 92.9 and 141.0, respectively. The quantitative data of JA and 10-dihydro-JA (as internal standard for JA; Olchemim) were obtained using the peaks of the precursor ions 209.1 and 211.2 and the product ions 59 and 59, respectively (Tao et al., 2009). The quantitative data of IAA, IAA-Asp, IAA-Ala, and D2-IAA (as internal standard for IAA and IAA-amino acid; Sigma-Aldrich) were obtained using the peaks of the precursor ions 173.9, 289, 244.9, and 175.9 and the product ions 129.9, 131.9, 156, and 131.9, respectively.
To prepare samples for the quantification of secreted IAA by bacterial pathogens, Xoo and Xoc were first grown on potato-agar medium (Chen et al., 2002) for 2 d at 28°C; M. grisea organisms were grown on tomato-oat-agar medium, which was prepared using 20 g of oats, 150 mL of tomato juice, and 14 g of agar and adding deionized water to 1,000 mL for 5 d at 28°C. A loopful of bacterial pathogen cells was inoculated into potato liquid medium and grown at 28°C with shaking (200 rpm) overnight in the dark. Then, 1 mL of the cell suspension was inoculated to 100 mL of the same fresh medium and was grown at 28°C with shaking (200 rpm) in the dark. Finally, a 1.5-mL sample was harvested 14 h later. A 100-μL sample was tested for OD600. The remaining sample was centrifuged at 12,000g for 10 min. The supernatant (600 μL) was mixed with IAA internal standard D2-IAA (Sigma-Aldrich). The sample was purified by filtration (pore size of 0.22 μm). The filtrate was dried under a stream of nitrogen. The dried sample was resuspended into 100 μL of methanol for quantification of IAA. For quantification of secreted IAA by fungal pathogens, a loopful of M. grisea mycelia was inoculated into tomato liquid medium and grown at 28°C with shaking (200 rpm) in the dark for 3 d. Then, a 2-mL sample was harvested and centrifuged at 12,000g for 10 min. The supernatant (600 μL) was treated following the same procedures described above. For control, potato or tomato liquid medium without inoculations of pathogen was shaken and harvested at the same time. The ultimate IAA concentration secreted by pathogens was determined by subtracting the amount of IAA in noninoculated medium from that in pathogen-grown medium.
Enzyme Assay
The entire coding region of GH3-2 was amplified from cDNA clone BI101D22 using primers YHBD3.2-FNdeI and YHBD3.2-R (Supplemental Table S3), and the PCR product was inserted into a pET28a expression vector (EMD Biosciences) to generate His-GH3-2. The fusion protein was expressed and purified, and the reaction for IAA-amino acid conjugate formation was performed as described previously (Staswick et al., 2005; Ding et al., 2008). The sample (20 μL) of reaction product was diluted with 80 μL of methanol. An aliquot (1 μL) of the diluted sample was further diluted with 199 μL of methanol. An aliquot (10 μL) of the finally diluted sample was injected into the UFLC-MS/MS apparatus equipped with a Waters C18 Atlantis column (2.1 × 150 mm). IAA, IAA-Ala, and IAA-Asp were separated using UFLC at a flow rate of 0.25 mL min−1 with linear gradients of solvent A (0.04% acetic acid) and solvent B (0.04% acetic acid in acetonitrile) set according to the following profile: 0 min, 90% A + 10% B; 2.0 min, 10% A + 90% B; and then with isocratic conditions: 13 min, 10% A + 90% B; 16.1 min, 90% A + 10% B; and holding for 3.9 min. IAA and IAA-amino acid conjugates in the reaction mixture were quantified using UFLC-electrospray ionization-MS/MS as described above. The retention times of IAA-Asp, IAA-Ala, and IAA were 5.9, 6.8, and 7.5 min, respectively. Standard IAA-Asp, IAA-Ala, and IAA were purchased from Sigma-Aldrich.
RNA Gel-Blot and Quantitative Reverse Transcription-PCR Analyses
Leaf fragments about 5 cm long right next to the inoculation site were used for analysis. Aliquots (15 μg) of total RNA were used for RNA gel-blot analysis (Zhou et al., 2002). A 442-bp cDNA fragment of GH3-2, which was amplified using primers pEI5P11a(3) and pEI5P11a(4) (Supplemental Table S3), was used as a hybridization probe. Quantitative reverse transcription-PCR was conducted as described previously (Qiu et al., 2007). The PCR primers for gene expression analysis are listed in Supplemental Table S3. The expression level of the rice actin gene was used to standardize the RNA sample for each quantitative reverse transcription-PCR. The assays were repeated at least twice biologically, with each repetition having three replicates; similar results were obtained in repeated experiments. sd was calculated for each data point.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GU001814 (Minghui 63) for GH3-2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Colocalization of GH3-2 and putative disease resistance QTLs.
Supplemental Figure S2. Schematic diagrams of the GH3-2 gene and the transformation constructs of GH3-2.
Supplemental Figure S3. Sequence comparison of promoter regions of GH3-2 alleles in Minghui 63 and Zhenshan 97.
Supplemental Figure S4. Enhanced resistance of GH3-2-overexpressing plants (D176UM) to Xoo strain PXO61 was associated with increased GH3-2 expression.
Supplemental Figure S5. The expression levels of GH3-2 in GH3-2-RNAi T0 plants (D175R).
Supplemental Figure S6. The GH3-2 expression level and responses to Xoo strain PXO347 of two GH3-2-RNAi T1 families.
Supplemental Figure S7. Sequence comparison of rice GH3-2 and GH3-8.
Supplemental Figure S8. GH3-2-overexpressing plants (T2) D176UM showed dwarf morphology.
Supplemental Figure S9. Expression of defense-responsive genes related to SA and JA signaling in a GH3-2-overexpressing line.
Supplemental Figure S10. IAA and IAA-Asp concentrations and GH3-2 expression in Minghui 63 and Zhenshan 97 at the four-leaf stage.
Supplemental Table S1. Performance of EXPA1-, EXPA5-, and EXPA10-overexpressing T1 families after inoculation with Xoc strain RH3 at the booting stage.
Supplemental Table S2. Performance of EXPA1-, EXPA5-, and EXPA10-overexpressing T2 families after inoculation with M. grisea isolate CHL358 at the four-leaf stage.
Supplemental Table S3. Primers used for PCR amplification.
Supplementary Material
Acknowledgments
We thank Prof. Youliang Peng of China Agricultural University and Prof. Qinghua Pan of South China Agricultural University for providing M. grisea isolates.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini E, Morel JB, Droc G, Price A, Courtois B, Notteghem JL, Tharreau D. (2008) A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant Microbe Interact 21: 859–868 [DOI] [PubMed] [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S. (2007) The expression pattern of a rice disease resistance gene xa3/xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics 177: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas RA, Benedetti CE. (2009) Role of auxin and gibberellin in citrus canker development and in the transcriptional control of cell-wall remodeling genes modulated by Xanthomonas axonopodis pv. citri. Plant Sci 177: 190–195 [Google Scholar]
- Chen C, Zheng W, Huang X, Zhang D, Lin X. (2006) Major QTL conferring resistance to rice bacterial leaf streak. Agric Sci China 5: 216–220 [Google Scholar]
- Chen H. (2001) Population structure of Pyricularia grisea from central and southern China and comparative mapping of QTL for blast- and bacterial blight-resistance in rice and barley [in Chinese]. PhD thesis Huazhong Agriculture University, Wuhan, China [Google Scholar]
- Chen H, Wang S, Xing Y, Xu C, Hayes PM, Zhang Q. (2003) Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc Natl Acad Sci USA 100: 2544–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang S, Zhang Q. (2002) New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology 92: 750–754 [DOI] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104: 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z, Wang S. (2007) Isolation, structure, function relationship, and molecular evolution of disease resistance genes. Zhang Q, , Genetics and Improvement of Resistance to Bacterial Blight in Rice. Science Press, Beijing, pp 349–377 [Google Scholar]
- Comai L, Kosuge T. (1982) Cloning characterization of iaaM, a virulence determinant of Pseudomonas savastanoi. J Bacteriol 149: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Andrés F, Tharreau D, Iglesias DJ, Talón M. (2009) Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant Microbe Interact 22: 201–210 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Pontiggia D, Manfredini C, Lionetti V, Bellincampi D, Cervone F, De Lorenzo G. (2008) Transgenic expression of a fungal endo-polygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol 146: 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett WF, Osman SF, Dunn MF. (1987) Auxin production by plant-pathogenic pseudomonads and xanthomonads. Appl Environ Microbiol 53: 1839–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J. (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323: 1357–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, et al. (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325: 998–1001 [DOI] [PubMed] [Google Scholar]
- Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y. (1998) Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 11: 156–162 [DOI] [PubMed] [Google Scholar]
- Hasan HA. (2002) Gibberellin and auxin-indole production by plant root-fungi and their biosynthesis under salinity-calcium interaction. Acta Microbiol Immunol Hung 49: 105–118 [DOI] [PubMed] [Google Scholar]
- Hu K, Wang S. (2009) Rice disease resistance resources and genetic improvement. Zhang Q, , Strategies and Practice for Developing Green Super Rice. Science Press, Beijing, pp 35–57 [Google Scholar]
- Hu KM, Qiu DY, Shen XL, Li XH, Wang SP. (2008) Isolation and manipulation of quantitative trait loci for disease resistance in rice using a candidate gene approach. Mol Plant 1: 786–793 [DOI] [PubMed] [Google Scholar]
- International Rice Research Institute (2002) Reference guide. Standard Evaluation System for Rice (SES). International Rice Research Institute, Los Banos, Philippines, p 56 [Google Scholar]
- Jagadeeswaran G, Raina S, Acharya BR, Maqbool SB, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R. (2007) Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J 51: 234–246 [DOI] [PubMed] [Google Scholar]
- Kou Y, Li X, Xiao J, Wang S. (2010) Identification of genes contributing to quantitative disease resistance in rice. Sci China Life Sci 53: 1263–1273 [DOI] [PubMed] [Google Scholar]
- Kou Y, Wang S. (2010) Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol 13: 181–185 [DOI] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. (2005) Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep 23: 540–547 [DOI] [PubMed] [Google Scholar]
- Lincoln S, Daly M, Lander E. (1992) Constructing Genetics Maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report. Whitehead Institute, Cambridge, MA [Google Scholar]
- Ludwig-Müller J, Decker EL, Reski R. (2009) Dead end for auxin conjugates in Physcomitrella? Plant Signal Behav 4: 116–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosalva PM, Davidson RM, Liu B, Zhu X, Hulbert SH, Leung H, Leach JE. (2009) A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol 149: 286–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor R, Haskin S, Levi-Kedmi H, Sharon A. (2004) In planta production of indole-3-acetic acid by Colletotrichum gloeosporioides f. sp. aeschynomene. Appl Environ Microbiol 70: 1852–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald BA, Linde C. (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40: 349–379 [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436–439 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Okrent RA, Stoutemyer M, Rodibaugh N, Kempema L, Wildermuth MC, Innes RW. (2007) The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol 144: 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM. (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282: 10036–10046 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Cheng H, Li X, Wang S. (2009) Exploring transcriptional signalling mediated by OsWRKY13, a potential regulator of multiple physiological processes in rice. BMC Plant Biol 9: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke G, Heinze B, Schirawski J, Buettner H, Kahmann R, Basse CW. (2008) Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol Plant Pathol 9: 339–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel C, Walz A, Park S, Cohen JD, Ludwig-Müller J. (2006) Indole-3-acetic acid protein conjugates: novel players in auxin homeostasis. Plant Biol (Stuttg) 8: 340–345 [DOI] [PubMed] [Google Scholar]
- Sequeira L, Kelman A. (1962) The accumulation of growth substances in plants infected by Pseudomonas solanacearum. Phytopathology 52: 439–448 [Google Scholar]
- Sheldrake AR. (1973) The production of hormones in higher plants. Biol Rev Camb Philos Soc 48: 509–559 [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC. (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15: 747–754 [DOI] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q. (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37: 517–527 [DOI] [PubMed] [Google Scholar]
- Surico G, Iacobellis NS, Sisto A. (1985) Studies on the role of indole-3-acetic acid and cytokinins in the formation of knots on olive and oleander plants by Pseudomonas syringae pv. savastanoi. Physiol Plant Pathol 26: 309–320 [Google Scholar]
- Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S. (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J, Domingo C, Talón M. (2006) The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371: 279–290 [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. (2007a) Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17: 1784–1790 [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. (2007b) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (April 13, 2007) [Google Scholar]
- Wen N, Chu Z, Wang S. (2003) Three types of defense-responsive genes are involved in resistance to bacterial blight and fungal blast diseases in rice. Mol Genet Genomics 269: 331–339 [DOI] [PubMed] [Google Scholar]
- Wisser RJ, Sun Q, Hulbert SH, Kresovich S, Nelson RJ. (2005) Identification and characterization of regions of the rice genome associated with broad-spectrum, quantitative disease resistance. Genetics 169: 2277–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Liu H, Li Y, Li X, Xu C, Long M, Wang S. (2009) A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS ONE 4: e4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z, Tan F, Hua P, Sun L, Xu G, Zhang Q. (2002) Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet 105: 248–257 [DOI] [PubMed] [Google Scholar]
- Yang H, Chu Z, Fu J, Wang S. (2008) The major blast resistance QTL rbr2 is an allele of Pib gene. Mol Plant Breed 6: 213–219 [Google Scholar]
- Yang S, Zhang Q, Guo J, Charkowski AO, Glick BR, Ibekwe AM, Cooksey DA, Yang CH. (2007) Global effect of indole-3-acetic acid biosynthesis on multiple virulence factors of Erwinia chrysanthemi 3937. Appl Environ Microbiol 73: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Sun X, Wang S, Zhang Q. (2003) Genetic and physical mapping of a new gene for bacterial blight resistance in rice. Theor Appl Genet 106: 1467–1472 [DOI] [PubMed] [Google Scholar]
- Yuan B, Shen X, Li X, Xu C, Wang S. (2007) Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta 226: 953–960 [DOI] [PubMed] [Google Scholar]
- Zhang J, Feng Q, Jin C, Qiu D, Zhang L, Xie K, Yuan D, Han B, Zhang Q, Wang S. (2005) Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J 42: 772–780 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Q, Li Z, Staswick PE, Wang M, Zhu Y, He Z. (2007) Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol 145: 450–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Peng K, Zhaohui C, Wang S, Zhang Q. (2002) The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci China C Life Sci 45: 449–467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.