There is a growing impetus in developing novel strategies to address global concerns regarding food security. As crop productivity gains through traditional breeding begin to lag and arable land becomes scarcer, it seems that we are heading for unsustainable global populations. It has been foreshadowed that global food production will need to rise more than 50% before 2050 to meet the ever-increasing demand. Compounding the problem are the uncertainties of climate change and its impact on agriculture. Strategies to improve crop yield potential have begun to examine aspects of supercharging photosynthesis to drive a new “green revolution.” Central to many of these strategies is addressing the limitation of nature’s CO2-fixing enzyme, Rubisco.
The catalytic incorporation of CO2 into ribulose 1,5-bisphosphate (RuBP) by Rubisco is the first step in the production of carbohydrates by plants, which are used to build biomass and produce energy during growth and development. Despite the pivotal role of Rubisco in linking the inorganic (CO2) and the organic (biomass) phases of the global carbon cycle, it is a slow and confused catalyst, limiting productivity and resource (e.g. water and nutrients) use efficiency in many plants (Long et al., 2006). Understandably, Rubisco has been studied intensively and is a prime target for genetic engineering to improve photosynthetic efficiency (Raines, 2006; Parry et al., 2007). Although the challenge of making a “better Rubisco” has exceeded the grasp and career of many scientists, recent advances indicate that it is not insurmountable. Here, we examine conceptual and technological breakthroughs over the last decade that have identified new and unconventional members of the Rubisco family, revealed molecular aspects of Rubisco biogenesis in plastids and cyanobacteria, advanced our understanding of its catalytic chemistry, and widened our appreciation of the challenges we face to improve Rubisco activity and plant productivity.
RUBISCO IN THE GENOMIC ERA
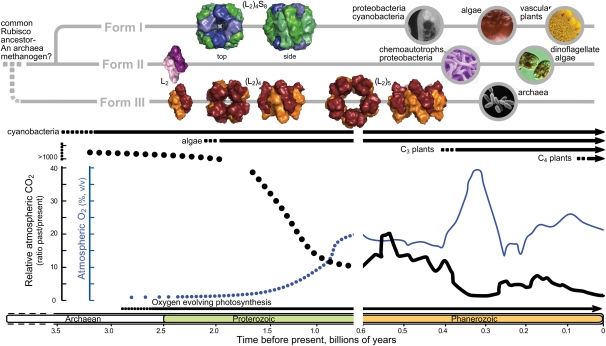
Rubisco is an ancient enzyme, its history beginning more than 3.5 billion years ago, when Earth’s atmosphere was high in CO2 and before the origins of oxygen (O2)-producing photosynthesis (Fig. 1). Modern genomic sequencing projects have identified Rubisco and Rubisco-like proteins (RLPs) in organisms from three kingdoms of life, with some microorganisms possessing multiple forms of these enzymes (Andersson and Backlund, 2008). Phylogenetic analysis of these sequences supports the existence of three different clades of Rubisco (denoted forms I, II, and III), which, together with the more diverse RLPs (or form IV Rubisco), probably share a common ancestor, most likely that of a methanogenic archaea (Fig. 1; Tabita et al., 2008). Structurally, the RLPs lack key conserved active-site residues of Rubiscos and, therefore, do not inherently bind RuBP or catalyze CO2 fixation. Detailed studies of some RLPs indicate that they participate in thiosulfate oxidation, catalyzing the enolization of a RuBP analog (Saito et al., 2009). The increasing variety of sequenced genomes has led to the identification of a growing number of RLPs in multiple bacterial lineages, suggesting that these proteins may have other catalytic functions. Clearly, our perception of RLP biology is still in its infancy.
Figure 1.
Hypothetical profiles of Rubisco phylogeny, the evolutionary timelines of different photosynthetic organisms, and variation in atmospheric CO2 (thicker line) and O2 levels during earth’s history. Hypothetical atmospheric CO2 and O2 levels prior to 0.6 billion years ago are represented by dotted lines. Quaternary structures of each Rubisco were drawn with Pymol using Protein Data Bank coordinates for the spinach (Spinacia oleracea) (L2)4S8 (8RUC), R. rubrum L2 (5RUB), Pyrococcus horikoshii (L2)4 (2CWX), and Thermococcus kodakaraensis (L2)5 (1GEH) enzymes. Structures for larger form II (L2)n Rubiscos are unavailable. Circular images depict types of organisms where the different Rubisco forms are found. Figure details were adapted from Tabita et al. (2008) and Badger et al. (2002).
RUBISCO STRUCTURE
From a structural point of view, all Rubisco enzymes comprise at least two large (L-) subunits of approximately 50 kD. Despite there being as little as 30% amino acid identity between the different Rubisco forms, they all show a conserved L-subunit structure comprising an N-terminal domain (approximately 150 amino acids) and a larger C-terminal domain (approximately 320 amino acids) that forms an α/β-barrel (Fig. 2). Paired L-subunits arrange head to tail to form a dimer (L2), with two active sites located at the L-L interface. The highly conserved catalytic residues predominantly reside within the α/β-barrel domain, with a few residues supplied by the N-terminal domain of the adjacent L-subunit.
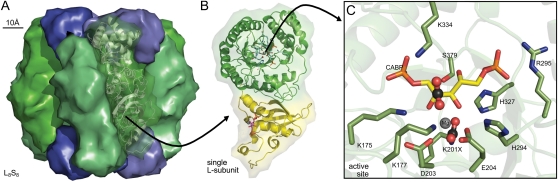
Figure 2.
Conserved structural features of Rubisco. A, Spinach L8S8 Rubisco (Protein Data Bank 8RUC) drawn using Pymol to highlight arrangement of the S-subunits (blue) capping the catalytic core of four L2 subunits (green). B, Structural details for one L-subunit of an L2 pair highlighting one active site within the α/β-barrel of the C-terminal domain (green ribbons) and residues in the N-terminal domain (yellow ribbons) that contribute to the second active site in each L2. C, Arrangement of the conserved Rubisco active-site residues within a L-subunit C-terminal domain relative to carbamylated Lys-201 (K201X), bound Mg2+, and the six-carbon reaction intermediate mimic, 2-carboxyarabinitol 1,5-bisphosphate (CABP). The activating CO2 in K201X and the approximate positioning of substrate CO2 that binds to C-2 of the RuBP enediol are highlighted in ball-and-stick representations. Residues are numbered relative to spinach Rubisco. The conserved active site residues Glu-60 and Asn-123 from the N-terminal domain of the paired L-subunit are not shown.
During evolution, different organisms have developed diverse arrangements of the Rubisco L2 building blocks. Form I Rubiscos are the most abundant form found in plants, algae, and many photosynthetic bacteria (Fig. 1). The L2 subunits in form I Rubiscos are arranged in an (L2)4 core, with two groups of four small (S-) subunits (approximately 13–17 kD) capping the L8 core to form an L8S8 molecule (Fig. 2A). Although not strictly required for CO2 fixation, the S-subunits are essential for maximal activity and provide structural stability (Andersson and Backlund, 2008). Recent evidence suggests that S-subunits in the unicellular green alga Chlamydomonas reinhardtii may also play a structural role in pyrenoid development (Genkov et al., 2010). Rubiscos classed as forms II and III lack S-subunits, containing only L-subunits arranged into L2 to (L2)5 complexes (Fig. 1). These Rubiscos are primarily found in phototrophic proteobacteria, chemoautotrophs, dinoflagellates, and achaea.
Although the classification of Rubisco enzymes into forms I, II, and III is generally supported by sequence phylogenies, quaternary structures, and functional properties, there are exceptions. For example, the structural and biochemical properties of the Rubisco enzyme from the archaea Methanococcoides burtonii correlate to form III, despite closer sequence identity to form II Rubiscos (Alonso et al., 2009). Like the RLPs, further discovery and characterization of divergent Rubisco forms are of great importance if we are to understand the diversity of the Rubisco family, refine its nomenclature, and comprehend its origin and evolution.
RUBISCO CATALYSIS
A Bifunctional Enzyme That Catalyzes Multistep Chemistry
Despite amino acid sequence variability within the Rubisco family, key active-site residues are absolutely conserved among forms I, II, and III Rubiscos (Andersson and Backlund, 2008). As a result, the activation process and complex catalytic chemistry are also preserved, despite the different biological roles of forms I and II Rubiscos, which initiate primary carbon assimilation, and the catabolic role of archaeal form III enzymes, which remove RuBP produced during purine/pyrimidine metabolism (Sato et al., 2007). Most structure-function studies have focused on Rubiscos involved in carbon assimilation and provide much of our mechanistic understanding of its catalysis (for review, see Parry et al., 2003).
Prior to catalysis, Rubisco needs to be preactivated via the reaction of a CO2 molecule with a conserved active-site Lys (residue 201 in most plant Rubisco L-subunits) to form a carbamate, which is then stabilized by Mg2+ binding (Fig. 2C; Andersson and Backlund, 2008). Following activation, Rubisco can productively bind RuBP and catalyze a complex five-step reaction that adds a CO2 and a water molecule to RuBP, followed by its cleavage and release of two 3-phosphoglycerate (3PGA) molecules (Fig. 3). The complexity of the multistep process can lead to unwanted side reactions that result in the formation of inhibitors such as xyulose-1,5-bisphosphate (Pearce, 2006; Parry et al., 2007). Furthermore, the electrostatic similarity between O2 and CO2 and their disproportionate atmospheric abundance (21% O2, 0.04% CO2) make it hard for Rubisco to totally discriminate between them, resulting in the unwanted oxygenation of RuBP and the production of one molecule of 3PGA and one of 2-phosphoglycolate. In plants, 2-phosphoglycolate is recycled back to 3PGA via photorespiration, an energy-consuming process (e.g. ATP) that liberates fixed carbon as CO2 (Peterhansel et al., 2008; Fig. 3).
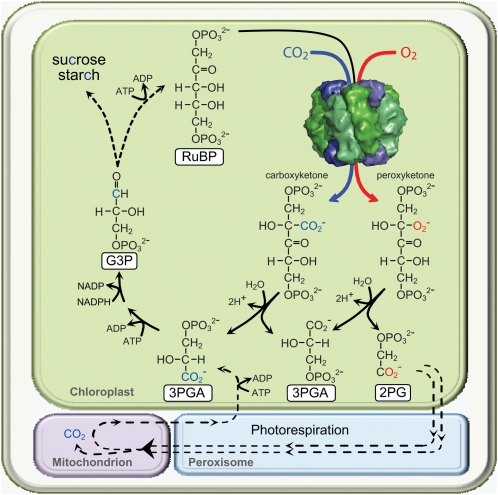
Figure 3.
Simplified scheme illustrating how CO2 fixed to RuBP by Rubisco is distributed among the resulting two molecules of 3PGA that feed into the photosynthetic Calvin cycle to produce triose phosphates (glyceraldehyde 3-phosphate [G3P]) for carbohydrate synthesis or RuBP regeneration. The contrasting oxygenation reaction of Rubiscos produces 2-phosphoglycolate (2PG), which requires the photorespiratory pathway to recycle it back to 3PGA. Photorespiration is a complex pathway that involves four subcellular compartments and multiple enzymatic steps (represented by dashed lines), requires additional energy (ATP), and results in a loss of fixed CO2 in the mitochondria (Maurino and Peterhansel, 2010).
Detailed understanding of Rubisco catalysis has taken advantage of the exponential rise in computer power and the increasing accuracy of theoretical protein models. Several groups have applied up-to-date computational tools to examine the energetics and atomistic details of Rubisco’s carboxylation and oxygenation reactions (Kannappan and Gready, 2008). While these calculations reveal molecular details of the contribution of active-site residues to Rubisco catalysis, the large size of Rubisco prevents the inclusion of all atoms in the calculation. As a result, the design of “better” Rubiscos using in silico modeling tools remains to be demonstrated.
The Natural Catalytic Diversity of Rubisco
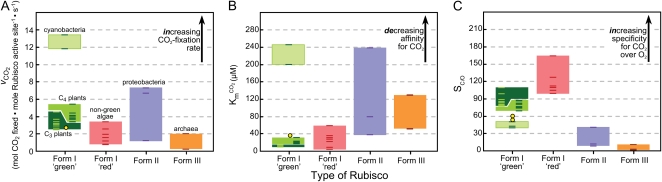
The Rubisco family shows significant catalytic variability despite sharing the same catalytic chemistry (Fig. 4). Unfortunately, comprehensive catalytic studies have only been made for relatively few Rubiscos (Supplemental Table S1), limiting our capacity to fully appreciate the connections between catalytic and sequence diversity and the influence of temperature on the activity of evolutionarily diverse Rubiscos. As highlighted by Tcherkez et al. (2006), improvements in CO2 fixation rate for forms I and II Rubiscos generally come at the expense of affinity for CO2. Therefore, photosynthetic organisms that live under high CO2 and low O2 (e.g. proteobacteria such as Rhodospirillum rubrum) or that have evolved complex, energy-expensive, biochemical CO2-concentrating mechanisms that elevate CO2 levels around Rubisco, as seen in C4 plants, many algae, and cyanobacteria, have Rubiscos with lower CO2 affinities (i.e. higher Km for CO2) but higher carboxylation rates (vCO2; Fig. 4, A and B). This is usually accompanied by lower specificities for CO2 over O2 (SC/O; Fig. 4C). In contrast, C3 plants (which include most crops) and algae that lack a CO2-concentrating mechanism have higher CO2 affinities, better CO2/O2 specificities, but slower carboxylation rates.
Figure 4.
Comparative catalytic features of different Rubisco forms measured at 25°C. Individual dashes in each column represent separate catalytic measurements for each Rubisco form (detailed in Supplemental Table S1). Yellow circles indicate the catalytic measurements for green algal Rubisco. SC/O values are calculated as (vCO2/KmO2)/(vO2/KmCO2), where vCO2 and vO2 are the maximum rates of RuBP carboxylation and oxygenation and KmO2 and KmCO2 are the apparent Km values for O2 and CO2, respectively.
A growing realization is that the catalytic diversity of Rubiscos originates from residues distant from the active site and generally not conspicuous in the more than 20 different Rubisco x-ray structures currently available (Andersson and Backlund, 2008). The significant catalytic diversity observed in Rubiscos from diverse C4 and C3 species (Galmes et al., 2005; Ghannoum et al., 2005; Kubien et al., 2008; Carmo-Silva et al., 2010) has prompted the use of bioinformatic analyses to identify potential “catalytic switches,” key residues responsible for the faster vCO2 rates of C4 Rubiscos (Christin et al., 2008). This approach has yet to be applied to unveil residues responsible for the variation in CO2/O2 specificity of Rubiscos of related C3 species (Galmes et al., 2005).
Confidence that catalytic improvements of C3 Rubiscos are possible stems from the finding that some red algae have the most efficient Rubiscos (Tcherkez et al., 2006; Parry et al., 2007). Indeed, the successful transfer of the catalytic properties of the Rubisco enzyme from the red alga Griffithsia monilis into a C3 crop has the potential to raise yields by approximately 30% (Long et al., 2006) as a result of higher discrimination between CO2 and O2 and reduced photorespiration (Fig. 3). The carbon losses due to photorespiration are even larger at elevated temperatures, which favor increasing the relative oxygenation activity of Rubisco (Sage, 2002).
RUBISCO AND ITS INTERACTIONS WITH OTHER PROTEINS
Rubisco Expression and Assembly
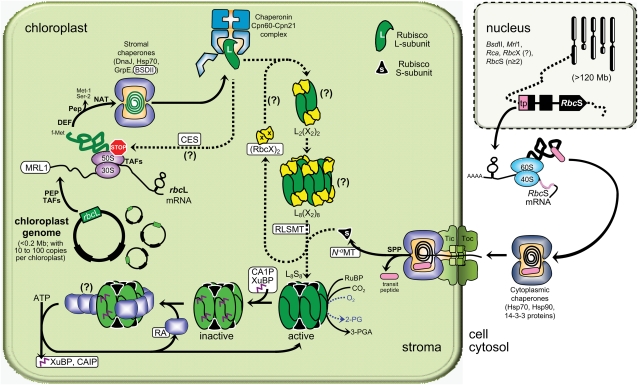
Key to developing strategies to engineer Rubisco is a better understanding of its biogenesis and regulation. The mechanism and requirements that coordinate the expression and assembly of L- and S-subunits into L8S8 Rubisco in chloroplasts are still rudimentary (Fig. 5; Nishimura et al., 2008). In prokaryotes and the plastid genome of nongreen algae, genes for the L- (rbcL) and S- (rbcS) subunits of form I Rubiscos are colocated in an operon. In higher plants and green algae, the single rbcL gene remains encoded by the plastid genome, while multiple copies of the RbcS gene are located in the nucleus (Fig. 5). The process(es) by which expression of the plastid- and nucleus-encoded L- and S-subunits, respectively, are coordinated remains unclear. Recent evidence suggests that L-subunit expression may be controlled by the epistasy of synthesis (CES) paradigm, wherein unassembled L-subunit motifs bind to the rbcL mRNA to autoregulate its translation (Wostrikoff and Stern, 2007). The importance of regulating Rubisco synthesis in plastids is paramount, as it is produced in high quantities to account for its slow and unspecific enzymatic activity. For example, in C3 plants, between 20% and 30% of the leaf protein (i.e. approximately 25% of the leaf nitrogen) is invested in Rubisco.
Figure 5.
The complexity of Rubisco biogenesis and its regulation by RA in vascular plant chloroplasts. Putative and known Rubisco-specific processes and interacting molecular partners (coded by genes in the nucleus as shown) are highlighted in white rectangles. See text for details of the processes, abbreviations, and the challenges faced in modifying the L8S8 enzyme. Figure details were adapted from Nishimura et al. (2008). Uncertainties in the biogenesis process are indicated by question marks. Hsp, Heat shock proteins; PEP, plastid-encoded RNA polymerase proteins; Pep, unknown Met-1, Ser-2 peptidase activity; Rca, nucleus gene coding RA; TAFs, nucleus-encoded trans-acting factors, Tic and Toc, chloroplast translocon inner and outer membrane complexes respectively; 30S/50S and 40S/60S, stromal and cytosolic ribosomal subunits.
Our understanding of Rubisco S-subunit biogenesis is limited. As with other plastid-localized proteins, synthesis of the S-subunit in the cytosol necessitates an appropriate N-terminal transit peptide for transfer (with the aid of molecular chaperones) to, and then passage through, the chloroplast envelope translocon complexes (Fig. 5; Jarvis, 2008). Within the stroma, the S-subunits undergo further posttranslational modification (transit peptide cleavage, Met-1 αN-methylation) prior to assembly into L8S8 complexes (Grimm et al., 1997).
Biogenesis of L-subunits in the chloroplast begins with the transcription of rbcL via a plastid-encoded RNA polymerase that requires a variety of nucleus-encoded factors (Shiina et al., 2005), including sequence-specific RNA-interacting regulatory proteins. A conserved pentatricopeptide repeat protein, MRL1, has recently been identified in Chlamydomonas and Arabidopsis (Arabidopsis thaliana) that specifically binds to the 5′ untranslated region of rbcL to stabilize the mRNA and/or ensure correct processing of the transcript (Johnson et al., 2010). The fundamental aspects of rbcL translation and posttranslational processing in chloroplasts also lack detail but appear to show similarities to the bacterial translational machinery (consistent with its prokaryotic ancestry), albeit reliant on nucleus-encoded factors for proper functioning (for review, see Nishimura et al., 2008). Nascent L-subunits are targeted for extensive N-terminal processing (for review, see Houtz et al., 2008) that begins with the deformylation of N-formyl-Met-1 by peptide deformylase and then Met-1 and Ser-2 removal via an uncertain peptidase process, leaving an N-terminal Pro-3 that is acetylated by an unknown αN-acetyltransferase. In some species, Lys-14 is also trimethylated by a Rubisco L-subunit εN-methyltransferase. Recent single-particle cryoelectron microscopy analysis revealed a large contact area between Rubisco L-subunit εN-methyltransferase and the C- and N-terminal domains of L-subunit pairs in L8S8 Rubisco but not the S-subunits (Raunser et al., 2009), suggesting that trimethylation of Lys-14 occurs after L2 assembly (see below) and prior to S-subunit assembly (Fig. 5). Although the functions of these posttranslational modifications remain unclear, it is assumed that they protect plant Rubisco from proteolytic degradation (Houtz et al., 2008).
In plastids, it is thought that newly translated L-subunits interact with the general Hsp70 chaperone system (DnaK/DnaJ/GrpE) and the Rubisco-specific chaperone BSDII (for review, see Nishimura et al., 2008). These chaperones prevent misfolding and convey the unfolded L-peptide to the folding cage of the chaperonin-60/21 complexes (plant homologous of the GroEL and GroES Escherichia coli proteins). Studies with cyanobacteria L8S8 Rubisco have shown that chaperonin-folded L-subunits interact with RbcX, a Rubisco-specific chaperone whose gene (rbcX) is often located between rbcL and rbcS in cyanobacteria (Saschenbrecker et al., 2007; Liu et al., 2010). RbcX dimers facilitate the assembly of L-subunits into (L2)4 complexes and are then displaced by the stable binding of S-subunits that produce the native L8S8 enzyme. In Synechococcus PCC7942, where rbcX is located separate from the rbcL-rbcS operon, deletion of rbcX has no effect on Rubisco synthesis (Emlyn-Jones et al., 2006). This begs the question of whether the nucleus-encoded RbcX homolog(s) in higher plants has a functional role in Rubisco assembly in plastids (Fig. 5).
Regulating Rubisco Activity
Attempts to manipulate Rubisco in higher plants and green algae may also need to consider the requirement for Rubisco’s chiropractic protein Rubisco activase (RA; Fig. 5). This nucleus-encoded protein uses the energy of ATP to remove active-site bound sugar-phosphate inhibitors, which are produced naturally to regulate Rubisco activity (such as CA1P) or as the result of misfired reactions (e.g. xyulose-1,5-bisphosphate; Pearce, 2006; Parry et al., 2008). The inability to obtain a crystal structure of RA continues to hamper our understanding of how oligomers of RA subunits interact with Rubisco. A current model proposes that amino acids between positions 89 and 94 of the L-subunits, located centrally on the surface of form I Rubiscos, interact with residues in the C-terminal sensor 2 domain of RA (Portis et al., 2008). Movement of the sensor 2 domain following ATP hydrolysis is thought to promote L-subunit conformational changes that result in the movement of loop 6 (a flexible loop that “closes” over the active site after RuBP binding), allowing inhibitor release (Fig. 5).
ADVANCES AND CHALLENGES IN ENGINEERING BETTER RUBISCOS
Conceptual breakthroughs in the general understanding of Rubisco biology and its pervasive influence on photosynthesis, along with technological advances in its genetic engineering, have provided pivotal insights into structure-function relationships and enabled its catalytic enhancement. These breakthroughs have rekindled the challenge of manipulating the catalytic properties of Rubisco and scrutinizing their effects on photosynthesis and plant growth.
Is the Evolution of Rubisco Naturally Constrained?
Tradeoffs between acquiring beneficial catalytic changes while maintaining catalytic chemistry and satisfying the many molecular interaction requirements may have limited the capacity of many Rubiscos to evolve improved catalytic prowess (Mueller-Cajar and Whitney, 2008). The survival dependency of photosynthetic organisms on Rubisco functionality, its high expression levels, and its necessity to interact with so many molecular partners (Fig. 5) may have constrained its mutational tolerance. Moreover, the evolution of Rubisco’s complex catalytic chemistry under high CO2 and no O2 (Fig. 1), and the fact CO2 binds directly to the RuBP enediol without forming a Michaelis complex with Rubisco, making it problematic to distinguish it from O2 (Kannappan and Gready, 2008), may encumber large catalytic improvements. However, the kinetic diversity of natural Rubiscos indicates that there is room for improvement (Fig. 4) and begs the question of how Rubiscos in nongreen algae have evolved further and faster. Possibly, beneficial sequence mutations are more accessible in nongreen algae because the genes for both Rubisco subunits and their molecular partners chaperonin 60 and DnaK are all located within the plastome. Additionally, “red” Rubiscos may not be encumbered with a requirement for regulation by RA (Pearce, 2006).
Laboratory Evolution of Better Rubiscos
During the last decade, the practice of laboratory-directed protein evolution has become a versatile tool for artificially mimicking and accelerating natural evolution (Bershtein and Tawfik, 2008). Directed evolution typically comprises the generation of a library of mutants followed by a selection process to identify mutant proteins with the desired phenotype. For Rubisco, a variety of selection systems using photosynthetic (Rhodobacter capsulatus, C. reinhardtii) and nonphotosynthetic (E. coli) hosts have been successfully developed (for review, see Mueller-Cajar and Whitney, 2008), with most successfully identifying changes that improve Rubisco folding and assembly (“solubility”). However, there are reports where mutants of C. reinhardtii and cyanobacterial Rubisco that show improvements in catalytic prowess (i.e. vCO2, KmCO2, and SC/O) have been selected (Greene et al., 2007; Zhu et al., 2010). In one case, catalytic improvements have been found to be transposable to Rubisco in tobacco (Nicotiana tabacum; Zhu et al., 2010), although how these improvements translate to changes in photosynthesis and plant growth remain unknown.
Advances in Manipulating Rubisco in Planta
The propensity of plant Rubisco L-subunits to form misfolded, insoluble aggregates in E. coli continues to restrict such studies to in planta genetic manipulation. Commonly, nucleus or chloroplast genome (plastome) transformation techniques are applied to model organisms where both genetic engineering tools are most developed, such as the unicellular alga Chlamydomonas and the C3 plant tobacco. Chlamydomonas is a particularly elegant engineering host, as changes can be made to both rbcL and rbcS genes. It has been successfully used to show the importance of the S-subunit in establishing Rubisco catalytic efficiency via complementarily directed mutagenesis of L- and S- subunits and recently for the creation of hybrid Rubiscos comprising higher plant S-subunits with Chlamydomonas L-subunits (Spreitzer et al., 2005; Genkov et al., 2010). Phylogenetic and structural comparisons of variant Rubiscos have identified candidate L- and S-subunit residues that benefit catalysis. Several of these sites and regions have been explored experimentally using Chlamydomonas, identifying important roles for loop 6 and Asp-473 in the L-subunit and the loop between β-strands A and B of the S-subunit (Satagopan and Spreitzer, 2004; Karkehabadi et al., 2005, 2007). Undeniably, future endeavors to generate and test better Rubiscos will be heavily reliant on the Chlamydomonas model system.
Genetically engineering the entire L8S8 Rubisco in tobacco is hindered by the disparate location of rbcL and RbcS in different genomes (Fig. 5). Recent modifications of Rubisco in tobacco have focused on manipulating rbcL by plastome transformation and have successfully demonstrated the feasibility of replacing higher plant Rubisco with phylogenetically distinct bacterial R. rubrum (L2) and archeal M. burtonii (L10) Rubiscos (Whitney and Andrews, 2001a; Alonso et al., 2009). This technology has also demonstrated the feasibility of assembling hybrid L8S8 Rubiscos comprising sunflower (Helianthus annuus) L-subunits and tobacco S-subunits, which show no catalytic demise (Sharwood et al., 2008). In each case, the growth and photosynthetic properties of the transplastomic plants have corresponded with the content and catalytic properties of the recombinant Rubisco, confirming the accuracy of the models used to predict photosynthetic carbon assimilation.
Plastome transformation and ethyl methane sulfonate mutant studies in tobacco have also highlighted limitations to Rubisco engineering. More efficient Rubiscos from red algae cannot be produced in plant plastids due to evolutionary divergence in their folding and assembly requirements (Whitney et al., 2001). Similar compatibility problems with translation, folding, and/or assembly of sunflower L-subunits with tobacco S-subunits also limit their assembly into hybrid L8S8 complexes (Sharwood et al., 2008). Point mutations Gly-332-Ser and Ser-112-Phe in the tobacco L-subunit were also found to hamper L8S8 synthesis (Avni et al., 1989; Shikanai et al., 1996), reaffirming the notion that critical interactions with molecular partners during Rubisco biogenesis limit the accessible mutational sequence space. Challenges associated with engineering the S-subunit in tobacco have also become evident with the apparent propensity for nucleus-encoded S-subunits to preferentially assemble ahead of plastid-synthesized recombinant S-subunits (Whitney and Andrews, 2001b). Some success has been obtained by reducing the cytosolic availability of the S-subunit using anti-RbcS plant lines (Dhingra et al., 2004). The feasibility of entirely excluding cytosolic S-subunit assembly by tethering the S- and L-subunits together with flexible linker peptides has also been demonstrated (Whitney et al., 2009), although the versatility of this S-L fusion strategy for engineering altered or foreign Rubiscos in plastids has yet to be demonstrated.
Prospects for Increasing Yield and Resource Use by Speeding up Rubisco
Recent work showing that CO2 enrichment can increase crop yield provides support for the idea that increases in photosynthesis can improve yield potential (Long et al., 2006; Zhu et al., 2007). Similarly, greater photosynthetic rates in C4 plants lead to more biomass being produced for a given amount of sunlight relative to C3 crops. Such findings are key drivers behind strategies to “supercharge” photosynthesis in C3 plants and improve crop yield potential. The focal point of these strategies is to overcome the catalytic inefficiencies of Rubisco by emulating the carbon-concentrating process found in C4 plants, which elevates CO2 around Rubisco to minimize photorespiration and its associated energy costs and carbon loss (Fig. 3).
Over the last 60 million years, C4 plants have evolved a variety of CO2-concentrating strategies that enabled their Rubiscos to persevere with lower CO2 affinities while retaining enhanced CO2 fixation rates (Fig. 4). As a result, C4 plants maintain high photosynthetic rates with less Rubisco (increasing nitrogen use efficiency) and can operate efficiently under low CO2 levels, alleviating the need for wide stomata apertures, thereby reducing leaf water loss (Ghannoum et al., 2005). A variety of strategies for introducing CO2-concentrating approaches into C3 plants to minimize photorespiration are under way (Peterhansel et al., 2008; Maurino and Peterhansel, 2010). These aim to introduce C4-like features into rice, improve productivity by introducing CO2/HCO3− transporter proteins from cyanobacteria into chloroplast membranes, or engineer new pathways into plastids that bypass photorespiration and release CO2 in the stroma. While each strategy faces challenges in their fine-tuning and integration into crops, further improvement in yields and in water and nitrogen use efficiencies will likely follow the lead of C4 plants by increasing the vCO2 of the inherent C3 Rubisco.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Catalytic properties for different Rubisco forms determined at 25°C.
Supplementary Material
References
- Alonso H, Blayney MJ, Beck JL, Whitney SM. (2009) Substrate-induced assembly of Methanococcoides burtonii D-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers. J Biol Chem 284: 33876–33882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson I, Backlund A. (2008) Structure and function of Rubisco. Plant Physiol Biochem 46: 275–291 [DOI] [PubMed] [Google Scholar]
- Avni A, Edelman M, Rachailovich I, Aviv D, Fluhr R. (1989) A point mutation in the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase affects holoenzyme assembly in Nicotiana tabacum. EMBO J 8: 1915–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Hanson D, Price GD. (2002) Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct Plant Biol 29: 161–173 [DOI] [PubMed] [Google Scholar]
- Bershtein S, Tawfik DS. (2008) Advances in laboratory evolution of enzymes. Curr Opin Chem Biol 12: 151–158 [DOI] [PubMed] [Google Scholar]
- Carmo-Silva AE, Keys AJ, Andralojc PJ, Powers SJ, Arrabaça MC, Parry MAJ. (2010) Rubisco activities, properties, and regulation in three different C4 grasses under drought. J Exp Bot 61: 2355–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Muasya AM, Roalson EH, Russier F, Besnard G. (2008) Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Mol Biol Evol 25: 2361–2368 [DOI] [PubMed] [Google Scholar]
- Dhingra A, Portis AR, Jr, Daniell H. (2004) Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc Natl Acad Sci USA 101: 6315–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlyn-Jones D, Woodger FJ, Price GD, Whitney SM. (2006) RbcX can function as a Rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant Cell Physiol 47: 1630–1640 [DOI] [PubMed] [Google Scholar]
- Galmes J, Flexas J, Keys AJ, Cifre J, Mitchell RAC, Madgwick PJ, Haslam RP, Medrano H, Parry MAJ. (2005) Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant Cell Environ 28: 571–579 [Google Scholar]
- Genkov T, Meyer M, Griffiths H, Spreitzer RJ. (2010) Functional hybrid Rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas. J Biol Chem 285: 19833–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum O, Evans JR, Chow WS, Andrews TJ, Conroy JP, von Caemmerer S. (2005) Faster Rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiol 137: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene DN, Whitney SM, Matsumura I. (2007) Artificially evolved Synechococcus PCC6301 Rubisco variants exhibit improvements in folding and catalytic efficiency. Biochem J 404: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm R, Grimm M, Eckerskorn C, Pohlmeyer K, Röhl T, Soll J. (1997) Postimport methylation of the small subunit of ribulose-1,5-bisphosphate carboxylase in chloroplasts. FEBS Lett 408: 350–354 [DOI] [PubMed] [Google Scholar]
- Houtz RL, Magnani R, Nayak NR, Dirk LMA. (2008) Co- and post-translational modifications in Rubisco: unanswered questions. J Exp Bot 59: 1635–1645 [DOI] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. (2010) MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannappan B, Gready JE. (2008) Redefinition of Rubisco carboxylase reaction reveals origin of water for hydration and new roles for active-site residues. J Am Chem Soc 130: 15063–15080 [DOI] [PubMed] [Google Scholar]
- Karkehabadi S, Peddi SR, Anwaruzzaman M, Taylor TC, Cederlund A, Genkov T, Andersson I, Spreitzer RJ. (2005) Chimeric small subunits influence catalysis without causing global conformational changes in the crystal structure of ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry 44: 9851–9861 [DOI] [PubMed] [Google Scholar]
- Karkehabadi S, Satagopan S, Taylor TC, Spreitzer RJ, Andersson I. (2007) Structural analysis of altered large-subunit loop-6/carboxy-terminus interactions that influence catalytic efficiency and CO2/O2 specificity of ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry 46: 11080–11089 [DOI] [PubMed] [Google Scholar]
- Kubien DS, Whitney SM, Moore PV, Jesson LK. (2008) The biochemistry of Rubisco in Flaveria. J Exp Bot 59: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Liu CM, Young AL, Starling-Windhof A, Bracher A, Saschenbrecker S, Rao BV, Rao KV, Berninghausen O, Mielke T, Hartl FU, et al. (2010) Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463: 197–202 [DOI] [PubMed] [Google Scholar]
- Long SP, Zhu XG, Naidu SL, Ort DR. (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ 29: 315–330 [DOI] [PubMed] [Google Scholar]
- Maurino VG, Peterhansel C. (2010) Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol 13: 249–256 [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Whitney SM. (2008) Directing the evolution of Rubisco and Rubisco activase: first impressions of a new tool for photosynthesis research. Photosynth Res 98: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Ogawa T, Ashida H, Yokota A. (2008) Molecular mechanisms of Rubisco biosynthesis in higher plants. Plant Biotechnol 25: 285–290 [Google Scholar]
- Parry MAJ, Andralojc PJ, Mitchell RAC, Madgwick PJ, Keys AJ. (2003) Manipulation of Rubisco: the amount, activity, function and regulation. J Exp Bot 54: 1321–1333 [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Keys AJ, Madgwick PJ, Carmo-Silva AE, Andralojc PJ. (2008) Rubisco regulation: a role for inhibitors. J Exp Bot 59: 1569–1580 [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Madgwick PJ, Carvalho JFC, Andralojc PJ. (2007) Prospects for increasing photosynthesis by overcoming the limitations of Rubisco. J Agric Sci 145: 31–43 [Google Scholar]
- Pearce FG. (2006) Catalytic by-product formation and ligand binding by ribulose bisphosphate carboxylases from different phylogenies. Biochem J 399: 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhansel C, Niessen M, Kebeish RM. (2008) Metabolic engineering towards the enhancement of photosynthesis. Photochem Photobiol 84: 1317–1323 [DOI] [PubMed] [Google Scholar]
- Portis AR, Jr, Li C, Wang D, Salvucci ME. (2008) Regulation of Rubisco activase and its interaction with Rubisco. J Exp Bot 59: 1597–1604 [DOI] [PubMed] [Google Scholar]
- Raines CA. (2006) Transgenic approaches to manipulate the environmental responses of the C3 carbon fixation cycle. Plant Cell Environ 29: 331–339 [DOI] [PubMed] [Google Scholar]
- Raunser S, Magnani R, Huang Z, Houtz RL, Trievel RC, Penczek PA, Walz T. (2009) Rubisco in complex with Rubisco large subunit methyltransferase. Proc Natl Acad Sci USA 106: 3160–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. (2002) Variation in the kcat of Rubisco in C3 and C4 plants and some implications for photosynthetic performance at high and low temperature. J Exp Bot 53: 609–620 [DOI] [PubMed] [Google Scholar]
- Saito Y, Ashida H, Sakiyama T, de Marsac NT, Danchin A, Sekowska A, Yokota A. (2009) Structural and functional similarities between a ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco)-like protein from Bacillus subtilis and photosynthetic Rubisco. J Biol Chem 284: 13256–13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saschenbrecker S, Bracher A, Rao KV, Rao BV, Hartl FU, Hayer-Hartl M. (2007) Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell 129: 1189–1200 [DOI] [PubMed] [Google Scholar]
- Satagopan S, Spreitzer RJ. (2004) Substitutions at the Asp-473 latch residue of Chlamydomonas ribulosebisphosphate carboxylase/oxygenase cause decreases in carboxylation efficiency and CO2/O2 specificity. J Biol Chem 279: 14240–14244 [DOI] [PubMed] [Google Scholar]
- Sato T, Atomi H, Imanaka T. (2007) Archaeal type III Rubiscos function in a pathway for AMP metabolism. Science 315: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Sharwood RE, von Caemmerer S, Maliga P, Whitney SM. (2008) The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol 146: 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. (2005) Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol 244: 1–68 [DOI] [PubMed] [Google Scholar]
- Shikanai T, Foyer CH, Dulieu H, Parry MAJ, Yokota A. (1996) A point mutation in the gene encoding the Rubisco large subunit interferes with holoenzyme assembly. Plant Mol Biol 31: 399–403 [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ, Peddi SR, Satagopan S. (2005) Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc Natl Acad Sci USA 102: 17225–17230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. (2008) Phylogenetic and evolutionary relationships of Rubisco and the Rubisco-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc Lond B Biol Sci 363: 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez GGB, Farquhar GD, Andrews TJ. (2006) Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA 103: 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Andrews TJ. (2001a) Plastome-encoded bacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) supports photosynthesis and growth in tobacco. Proc Natl Acad Sci USA 98: 14738–14743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Andrews TJ. (2001b) The gene for the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell 13: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Baldet P, Hudson GS, Andrews TJ. (2001) Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J 26: 535–547 [DOI] [PubMed] [Google Scholar]
- Whitney SM, Kane HJ, Houtz RL, Sharwood RE. (2009) Rubisco oligomers composed of linked small and large subunits assemble in tobacco plastids and have higher affinities for CO2 and O2. Plant Physiol 149: 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Stern D. (2007) Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc Natl Acad Sci USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Kurek I, Liu L. (2010) Engineering photosynthetic enzymes involved in CO2-assimilation by gene shuffling. Govindjee, , The Chloroplast: Advances in Photosynthesis and Respiration, Vol 31 Springer, Amsterdam, pp 307–322 [Google Scholar]
- Zhu XG, de Sturler E, Long SP. (2007) Optimizing the distribution of resources between enzymes of carbon metabolism can dramatically increase photosynthetic rate: a numerical simulation using an evolutionary algorithm. Plant Physiol 145: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.