Abstract
Early forms of the genetic code likely generated “statistical” proteins, with similar side chains occupying the same sequence positions at different ratios. In this scenario, groups of related side chains were treated by aminoacyl-tRNA synthetases as a single molecular species until a discrimination mechanism developed that could separate them. The aromatic amino acids tryptophan, tyrosine, and phenylalanine likely constituted one of these groups. A crystal structure of human tryptophanyl-tRNA synthetase was solved at 2.1 Å with a tryptophanyl-adenylate bound at the active site. A cocrystal structure of an active fragment of human tyrosyl-tRNA synthetase with its cognate amino acid analog was also solved at 1.6 Å. The two structures enabled active site identifications and provided the information for structure-based sequence alignments of ≈45 orthologs of each enzyme. Two critical positions shared by all tyrosyl-tRNA synthetases and tryptophanyl-tRNA synthetases for amino acid discrimination were identified. The variations at these two positions and phylogenetic analyses based on the structural information suggest that, in contrast to many other amino acids, discrimination of tyrosine from tryptophan occurred late in the development of the genetic code.
Aminoacyl-tRNA synthetases are ancient components of the genetic code. These enzymes establish the algorithm of the code (matching specific amino acids to nucleotide triplets) by aminoacylation reactions whereby each amino acid is linked to the tRNA bearing the cognate anticodon triplet. The synthetases are divided into two classes of ten enzymes each. [Although most lysyl-tRNA synthetases (LysRS) are found in class II, examples of class I LysRS are known (1).] Each class is defined by a distinct active site architecture that is shared by all members of the same class (2). Significantly, each class can be broken down into three subclasses where synthetases of the same subclass are more similar to each other than to members of other subclasses (3, 4) (Fig. 1). The two distinct architectures enable synthetases of opposite subclasses to bind to a single tRNA acceptor stem without any steric clashes (5). These subclass-specific pairings provide a way to cover and protect the tRNA acceptor stem, possibly reflecting an early environment where RNAs were susceptible to degradation by high temperatures, metal ions, and nucleases (5). The subclass organization is also thought to reflect in part the existence of statistical proteins in the development of a primitive genetic code where amino acids that were roughly similar were treated as equivalent and inserted into the same position in a growing polypeptide. Indeed, statistical proteins have been generated in vivo by interference with the mechanism of discrimination of similar amino acids in the same subclass (6). Interestingly, the symmetry of the two classes is violated between subclass Ic and IIc where two enzymes tyrosyl- and tryptophanyl-tRNA synthetases (TyrRS and TrpRS) from class Ic are paired with a single enzyme, phenylalanyl-tRNA synthetase, from class IIc.
Fig. 1.
Classification of aminoacyl-tRNA synthetases adapted from ref. 5. The 20 synthetases are divided into 2 classes of 10 enzymes each. The exceptional class I LysRS is shown in gray. Highlighted with a yellow box, TyrRS and TrpRS from class Ic are paired with PheRS from class IIc.
The progenitor of each subclass is thought to be the primitive synthetase that activated some or all of the related amino acids of that subclass. To make the transition from statistical proteins to specific chemical entities required refinements to the primitive active sites. In some cases, specificity was gained by the acquisition of editing domains that cleared misactivated amino acids. In other cases, like those present herein, enzymes lacking editing functions have evolved highly specific amino acid discrimination motifs within their catalytic sites for aminoacylation (Table 2, which is published as supporting information on the PNAS web site).
In these regards, the discrimination of the aromatic amino acids tryptophan (class Ic), tyrosine (class Ic), and phenylalanine (class IIc) is of particular interest. TyrRS and TrpRS are close homologs, and yet no editing functions have been found in these two enzymes. The sequence identities between TyrRSs and TrpRSs (10–20%) are similar to that between eukaryotic and bacterial TyrRS orthologs, and between eukaryotic and bacterial TrpRS orthologs. Although exceptions like this are known, typically, bacterial and eukaryotic tRNA synthetase orthologs of a particular amino acid are more similar to each other than to synthetases of any other type (7, 8). Thus, Tyr vs. Trp discrimination may have developed late and may have been contemporaneous with the emergence of Tyr vs. Phe discrimination.
Structural information and sequence alignments are needed to understand the development of discrimination of the three aromatic amino acids. Substrate complex structures are available for bacterial and archaeal TyrRS and bacterial TrpRS (9–13). These structures reveal contacts critical for amino acid discrimination at the active site. However, because of the weak sequence similarities between eukaryotic and bacterial synthetases, alignments are not reliable without structural information on eukaryotic TyrRS and TrpRS. This situation contrasts with other tRNA synthetases where alignments of orthologs through evolution are mostly straightforward. In the absence of reliable alignments of sequences of TyrRS and TrpRS, little insight can be gained into the development of aromatic amino acid discrimination. To address this issue, we crystallized and determined the structures of an active fragment of human TyrRS (miniTyrRS) with bound cognate amino acid analog tyrosinol. In addition, we solved the crystal structure of human TrpRS with bound tryptophanyl-adenylate (Trp-AMP). These structures, together with our recently published structure of the unligand human mini-TyrRS (14), enabled us to achieve structure-based alignments that show a clear path for the development of Trp vs. Tyr discrimination.
Materials and Methods
Crystallization, Data Collection, and Structure Determination of Human TrpRS. Human TrpRS was prepared as described (15) and maintained in a stock solution of 10 mM Hepes (pH 7.5), 20 mM KCl, 0.02% NaN3, 2 mM 2-mercaptoethanol. Initial crystallization trials were conducted by using the proprietary high throughput protein crystallization platform developed at Syrrx, Inc. (La Jolla, CA) as described (14). Single crystals were obtained by vapor diffusion of sitting drops (2 μl of protein sample and 2 μl of reservoir solution) against a reservoir of 2% PEG 8000, 0.1 M Mes/Mes-Na (pH 6.32) at 4°C. Selenium-labeled crystals were obtained in the same way by using selenomethionine-containing protein prepared as described (16).
The structure of human TrpRS was determined by the single wavelength anomalous dispersion method using a selenium-labeled crystal. Data were collected with beamline 9-2 at the Stanford Synchrotron Radiation Laboratory. Data were integrated and scaled with hkl2000 (17). Twenty-two selenium sites were identified by using solve (18). After density modification in resolve (19), the overall figure of merit at 2.3 Å increased from 0.37 to 0.54. arp/warp (20) then successfully traced and built ≈75% of the final model. The remaining model was built manually in o (21). The refinement of the model was performed in cns (22) with a final Rwork = 20.80% and Rfree = 23.86% at 2.1 Å. Data collection and refinement statistics of human TrpRS are summarized in Table 1.
Table 1. Data collection and refinement statistics.
| TrpRS/Trp-AMP | Mini-TyrRS/tyrosinol | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P21212 |
| a, Å | 152.1 | 75.8 |
| b, Å | 95.7 | 163.0 |
| c, Å | 98.5 | 35.3 |
| β, ° | 91.6 | 90.0 |
| Wavelength, Å | 0.9794 (Sepeak) | 1.2800 |
| Resolution, Å | 2.1 | 1.6 |
| Unique reflections | 82479 | 55381 |
| Completeness, %* | 96.5 (87.3) | 93.8 (64.5) |
| Redundancy | 6.8 | 6.7 |
| Rmerge, %*† | 9.9 (61.4) | 6.2 (30.1) |
| 〈I/σ(I)〉* | 13.9 (1.0) | 47.0 (3.8) |
| Refinement statistics | ||
| Resolution range, Å | 20-2.1 | 50-1.6 |
| Rwork/Rfree, %‡ | 20.80/23.86 | 19.66/21.70 |
| rms deviation bond lengths, Å | 0.006 | 0.004 |
| rms deviation bond angle, ° | 1.2 | 1.1 |
| Ramachandran plot, % | ||
| Favored | 92.4 | 91.4 |
| Allowed | 7.0 | 7.6 |
| Generously allowed | 0.3 | 0.7 |
| Disallowed | 0.3 | 0.3 |
| Average B-factors for protein, Å2 | 40.6 | 22.0 |
| Average B-factors for substrate, Å2 | 26.6 | 13.7 |
| Average B-factors for waters, Å2 | 42.1 | 32.7 |
Numbers in parentheses refer to the highest resolution shell.
Rmerge = (ΣhΣi|Ii(h) — 〈I(h)〉|/ΣhΣi Ii(h)) × 100, where 〈I(h)〉 is the average intensity of I symmetry-related observations of reflections with Bragg index h.
Rwork = (Σhkl|Fo — Fc|/Σhkl |Fo|) × 100, where Fo and Fc are the observed and the calculated structure factors, respectively, for 95% of the reflections uesd in the refinement. Rfree was calculated as for Rwork but on 5% of reflections excluded before refinement.
Crystallization, Data Collection, and Structure Determination of MiniTyrRS with Tyrosinol. An active fragment of human TyrRS (miniTyrRS) was expressed and purified as described (14). MiniTyrRS (18 mg/ml) was maintained in the same stock solution as for human TrpRS, with 5 mM of tyrosinol. Crystals of miniTyrRS/tyrosinol complex were grown by sitting drop vapor diffusion against the reservoir of 1.8 M (NH4)2SO4, 0.1 M NaH2PO4/K2HPO4 (pH 6.9), and 2% acetone at 4°C.
Data for the miniTyrRS/tyrosinol complex were collected with beamline X26C at the National Synchrotron Light Source at Brookhaven National Laboratory (Upton, NY). The crystal diffracted to 1.6 Å and had the same space group and similar lattice as for miniTyrRS alone (14). The asymmetric unit comprised one monomer of the dimeric complex. The miniTyrRS/tyrosinol complex structure was readily solved by molecular replacement in cns (22) using the structure of miniTyrRS alone as the search model. The initial model was improved in arp/warp (20), and then further refined by using cns. The final structure of miniTyrRS/tyrosinol complex had an Rwork of 19.66% and an Rfree of 21.70%. Data collection and refinement statistics are summarized in Table 1.
Sequences and Structures. The sequences used in our analyses are available in the GenBank database of protein sequences or in the aminoacyl-tRNA synthetase database (23). Except for the structures newly reported here, all of the protein coordinates used for the generation of alignments were obtained from the Protein Data Bank.
Generation of the Alignments. The structural coordinates of all available TyrRS and TrpRS were used for the generation of the structure-based alignments. The alignments were constructed according to a described method (24).
Phylogenetic Calculations. Maximum parsimony (MP), neighbor joining (NJ), and maximum likelihood (ML) analyses were done by using the software package phylip 3.6 (25). MP trees were calculated by using the program protpars from 100 replicate heuristic searches whereas confidence limits of branch points (for MP and NJ) were estimated by 1,000 bootstrap replications. NJ phylogenies were based on distances between amino acid sequences calculated with the programs neighbor and protdist, using the Dayhoff 120 substitution matrix.
Maximum likelihood calculations were done by using the program protml, using a JTT substitution matrix, and the gamma distribution model of rate variation, with five rate categories. Confidence limits of branch points for maximum likelihood trees were estimated by 100 bootstrap replications.
Estimation of ancestral sequences at internal nodes of the maximum likelihood trees was also calculated with the mlprot program. This was done by selecting three representative sequences among all available bacterial TyrRS (Thermotoga maritima, Aquifex aeolicus, and Rickettsia prowazecki), eukaryotic/archaeal TyrRS (Pyrococcus abyssi, Leishmania major, and Homo sapiens), bacterial TrpRS (Thermotoga maritima, Aquifex aeolicus, and Rickettsia prowazecki), and eukaryotic/archaeal TrpRS (P. abyssi, Giardia lamblia, and H. sapiens) (Fig. 5, which is published as supporting information on the PNAS web site). The resulting twelve sequences were analyzed with protml under varying numerical parameters and substitution models. Under all conditions, the ancestral sequences of the central nodes of the trees remained constant at the α8-D and β2-Y positions.
Results
Overall Structure of Human TrpRS. The crystal structure of the human TrpRS was solved at 2.1 Å by the single wavelength anomalous dispersion method using selenium-labeled protein. As observed for its bacterial ortholog (10), human TrpRS was a dimer, and the whole dimer was accommodated in the crystallographic asymmetric unit (Fig. 2). In one monomer of the dimer, all three domains of human TrpRS [N-terminal appended domain (Ala-7-Ala-60), Rossmann fold catalytic domain (Glu-82-Ser-353), and anticodon recognition domain (Asp-354-Ala-467] were resolved. A disordered region of 21 residues (Asp-61-Glu-81) is likely to be a flexible linker between the N-domain and the Rossmann fold. However, in the other monomer, the first 96 residues, which include the N-terminal domain, the flexible linker, and part of the Rossmann fold catalytic unit, were completely disordered. A bound Trp-AMP was found only in the monomer with the resolved N-domain, suggesting that the ligand helped to hold the three domains together. Indeed, the anticodon recognition domain of the other monomer was in a slightly more opened conformation, and part of the “KMSKS” loop (Ala-346-Asp-354) of that monomer was disordered probably because it lacked bound ligand.
Fig. 2.
Structure of the dimeric human TrpRS with one monomer shown in color. The circled CP1 insertion of the Rossmann fold domain forms the dimerization interface. All three domains [N-terminal appended domain (blue), Rossmann fold catalytic domain (yellow), and anticodon recognition domain (green)] were resolved in one monomer of the dimer with a disordered linker of 21 residues connecting the N-domain and the Rossmann fold domain. However, in the other monomer, the first 96 residues, which include the N-terminal domain, the linker region, and part of the Rossmann fold catalytic domain, were completely disordered. A bound Trp-AMP was found only in the monomer with the resolved N-domain.
Structural Alignments and Active Site Identifications Enabled by These Structures. To locate definitively the amino acid-binding pocket, a cocrystal structure of human miniTyrRS with the amino acid analog tyrosinol was also obtained. Except for the active site residues involved in amino acid binding, miniTyrRS in the complex has essentially the same conformation as in the unligand form (14). Superpositions of TrpRS and miniTyrRS structures from human and the published TyrRS (9, 11, 12) and TrpRS (10) structures from bacteria together enabled accurate alignment of over 90 sequences in their common core elements. Five examples of the 93 structure-based alignments, each with 173 residues that cover ≈50% of the total sequences, are shown in Fig. 3A. Elements of secondary structure [β-strands, α-helices, and 310-helices (η-helices)] were brought into register in the structure-based alignments. These alignments, together with the structures of the active sites seen in the human TyrRS/tyrosinol complex and human TrpRS/Trp-AMP complex reported here, and in published substrate complexes of Thermus thermophilus TyrRS (12) and Bacillus stearothermophilus TrpRS (10), enabled us to see clearly the path of amino acid replacements through evolution that facilitate binding and differentiation of Tyr and Trp, on the one hand, and the rejection of Phe on the other. Unlike other synthetases that charge amino acids with hydrophilic side chain groups, where specific amino acid contact residues are essentially fixed through evolution (Table 2), particular changes are seen at the active sites of TyrRS and TrpRS that suggest a late development of discriminatory mechanisms.
Fig. 3.
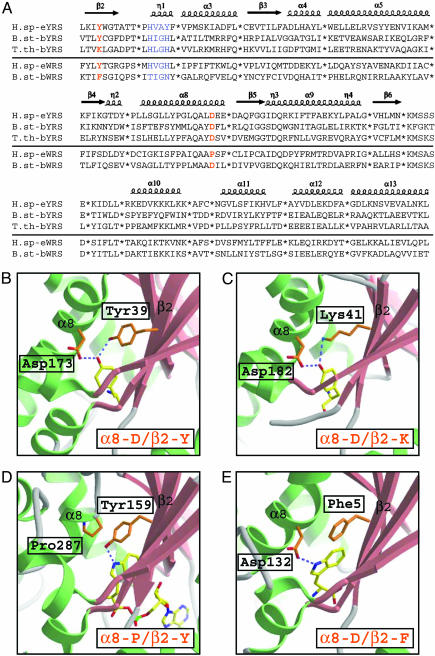
(A) Structure-based alignment of human TyrRS, B. stearothermophilus TyrRS, Thermus thermophilus TyrRS, human TrpRS, and B. stearothermophilus TrpRS. Variable regions, which usually correspond to terminal or loop regions, were removed to give a total of 173 aa in the alignment of all sequences. The secondary structure elements of human TyrRS were superimposed on top of the alignment (α,α-helices; η,310 helices; β,β-strands). “HIGH” and “KMSKS” signature sequences were colored in blue, and the two positions of amino acid recognition residues (inα8 andβ2) were colored in orange. (B) Active site of human TyrRS for the recognition of the tyrosinol side chain, which represent the active site of all eukaryotic, archaeal, and one group of bacterial TyrRSs including B. stearothermophilus TyrRS. The tyrosinol hydroxyl group is a hydrogen bond donor to the carboxylate oxygen of Asp in α8 and an acceptor for the hydroxyl of a Tyr in β2. (C) Active site of Thermus thermophilus TyrRS, which represent the other group of bacterial TyrRS. Here, the hydroxyl of the Tyr is replaced by the ε-amino of a Lys at the same position in β2 to donate a hydrogen bond to the tyrosinol hydroxyl group. (D) Active site of human TrpRS for the recognition of the tryptophan side chain, which represents the active site of all of the eukaryotic and one archaeal TrpRS from P. abyssi. The indole nitrogen of the tryptophan side chain accepts just one hydrogen bond from the hydroxyl of a Tyr in β2. (E) Active site of B. stearothermophilus TrpRS, which represent the active site of all of the bacterial and, except P. abyssi, all other archaeal TrpRSs. The indole nitrogen of the tryptophan side chain here accepts the one hydrogen bond from the carboxylate group of Asp in α8.
Key Residues for Amino Acid Recognition. The key residues for amino acid recognition are close in space but well separated in the primary structure in strand β2 and helix α8. In the human TyrRS complex with tyrosinol reported here, the tyrosinol hydroxyl group is a hydrogen bond donor to the carboxylate oxygen of Asp-173 in α8 and an acceptor for the hydroxyl of Tyr-39 in β2 (Fig. 3B). From the alignments, this combination of α8-D/β2-Y is fixed in all eukaryotic and archaeal sequences of TyrRS. However, bacteria split into two groups: one that is α8-D/β2-Y (the same as eukaryotes and archaea) and another where the tyrosine of β2 is replaced with a lysine to give α8-D/β2-K as in Thermus thermophilus TyrRS (Fig. 3C).
Remarkably, in the active site of TrpRS, as revealed by our crystal structure of the human enzyme with bound Trp-AMP and by the structure of the B. stearothermophilus TrpRS/tryptophan complex (10), the same two positions in the α8-helix and β2-strand of TyrRS are used by TrpRS to recognize the indole nitrogen of the tryptophan side chain. However, unlike the hydroxyl of tyrosine that can both accept and donate hydrogen bonds simultaneously, the indole nitrogen accepts only one H-bond, either from the β2-tyrosine as in the human enzyme (Fig. 3D), or from the α8-aspartate as in the B. stearothermophilus enzyme (Fig. 3E). Based on the alignments made possible by the new structure, the H-bond donor in TrpRS from all bacteria and most archaea is α8-D, that is, the same as seen in all TyrRSs through evolution. However, another key residue in β2 for TyrRSs is invariably replaced by one of several hydrophobic amino acids in TrpRSs (Fig. 3E). In eukaryotes, on the other hand, the aspartate of α8 is replaced by proline, and the H-bond donor comes from strand β2 and is a tyrosine (Fig. 3D), the same amino acid seen in all eukaryotic, archaeal, and one group of bacterial TyrRSs.
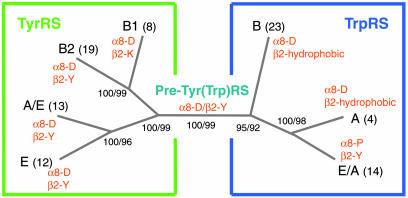
Phylogenetic Analysis. Structure-based alignments of over 90 sequences of TyrRSs and TrpRSs (that exclude idiosyncratic regions) were used to carry out a phylogenetic analysis (Fig. 4; and Fig. 6, which is published as supporting information on the PNAS web site). The generated phylogenetic tree is closer to the early one of Brown et al. (26) than to that of Ribas de Pouplana et al. (27), both of which lacked the structural information on eukaryotic TyrRS and TrpRS, and the many sequences now available. The bootstrap frequencies are exceptionally robust and consistent through the different phylogenetic methods used, thereby giving high confidence to the tree that was generated. For TyrRS, the α8-D/β2-Y combination is fixed in two branches (eukaryotes and archaea) and present in one branch of bacteria that is split into two groups. Although the first group (including B. stearothermophilus, Escherichia coli, Staphylococcus aureus, etc.) has the “canonical” α8-D/β2-Y, the second has α8-D/β2-K and includes among others Thermus thermophilus, T. maritima, and A. aeolicus. (The same phylogeny tree was produced, even with the β2-Y/K residue removed from the alignment.) For TrpRS, the same α8-D of TyrRS is present in all bacteria and most archaea, but with one exception is replaced by α8-P in all eukaryotes (α8-T is found in Saccharomyces cerevisiae). This D → P replacement is coordinated through all eukaryotes with a hydrophobic → Y replacement in β2. As stated above, the β2-Y for recognition of tryptophan is the same β2-Y providing tyrosine discrimination by TyrRS.
Fig. 4.
Simplified phylogenetic tree calculated from the alignment of 93 sequences of TyrRSs and TrpRSs by using maximum parsimony, neighbor joining, and maximum likelihood methods. The numbers in the branch nodes correspond to bootstrap frequencies after using maximum parsimony (1,000 cycles) and maximum likelihood (100 cycles) analyses. The numbers in parentheses of each group indicate the total number of sequences associated with that group. The two key residues for amino acid recognition of each group are listed on the tree in orange. The ancestral sequence estimated by maximum likelihood method for the central node of the tree has α8-D/β2-Y.
Discussion
A polar environment for rejection (from the binding pocket for tyrosine) of the highly hydrophobic benzene ring of phenylalanine is provided by the universal presence of charged aspartate in α8 of all TyrRS and the presence of either tyrosine or lysine in β2. At the same time, TrpRS uses the same residues (either α8-D or β2-Y, but not both) to hydrogen bond to the indole nitrogen of tryptophan. Thus, substrate discrimination by these two enzymes uses small variations of just two residues with wide adoption of one charged residue (aspartate in α8) and the polar tyrosine in β2.
The widespread use of the α8-D/β2-Y is consistent with this amino acid pair being the most parsimonious candidate for the ancestral sequence of TyrRS and TrpRS. This finding was confirmed by the estimation by maximum likelihood methods of the ancestral sequences of a selected set of TyrRS and TrpRS sequences (Fig. 5). Although no TrpRS has this combination of two residues, one or the other (α8-D or β2-Y) is found in all TrpRSs through evolution. [Thus, a single Tyr(Trp)RS could have been present in the last common ancestor, perhaps contemporaneous with either or both of the emerging TyrRS and TrpRS.] Additionally, the splitting of TyrRSs into two bacterial groups (α8-D/β2-Y and α8-D/β2-K), against a backdrop of all archaeal and eukaryotic TyrRSs being fixed with the α8-D/β2-Y combination, is consistent with a later evolution of this system. Remarkably, the bacterial group that utilizes the α8-D/β2-K sequence contains the oldest bacterial lineages known (Thermotoga, Aquifex, and Thermus). This observation suggests that the establishment of the final active site architectures of TyrRS and TrpRS took place in the early stages of bacterial evolution. Thus, the fixation of tyrosine and tryptophan in the modern genetic code might have been contemporaneous with the first split of the tree of life and the initial speciation events among bacteria.
This conclusion is supported by the observation that, in a number of other class I and class II synthetases (such as, among others ArgRS, MetRS, LysRS II, and ProRS) active site residues for specific hydrogen-bonding interactions with amino acid substrates are fixed throughout evolution (Table 2), thus suggesting that these substrate interactions were already present at the time of the last common ancestor.
Supplementary Material
Acknowledgments
We thank Dr. Howard Robinson at National Synchrotron Light Source for collecting the x-ray data for human TyrRS with tyrosinol, Dr. Karla Ewalt for help with collecting eukaryotic TyrRS and TrpRS sequences, and Prof. Tamara Hendrickson for comments on the manuscript. This work was supported by Grant GM15539 from the National Institutes of Health and a fellowship from the National Foundation for Cancer Research.
Abbreviations: TyrRS, tyrosyl-tRNA synthetase; TrpRS, tryptophanyl-tRNA synthetase; Trp-AMP, tryptophanyl-adenylate; LysRS, lysyl-tRNA synthetases.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1R6T for human TrpRS with Trp-AMP and 1Q11 for human mini-TyrRS with tyrosinol).
References
- 1.Ibba, M., Morgan, S., Curnow, A. W., Pridmore, D. R., Vothknecht, U. C., Gardner, W., Lin, W., Woese, C. R. & Söll, D. (1997) Science 278, 1119–1122. [DOI] [PubMed] [Google Scholar]
- 2.Carter, C. W., Jr. (1993) Annu. Rev. Biochem. 62, 715–748. [DOI] [PubMed] [Google Scholar]
- 3.Cavarelli, J. & Moras, D. (1993) FASEB J. 7, 79–86. [DOI] [PubMed] [Google Scholar]
- 4.Cusack, S. (1997) Curr. Opin. Struct. Biol. 7, 881–889. [DOI] [PubMed] [Google Scholar]
- 5.Ribas de Pouplana, L. & Schimmel, P. (2001) Cell 104, 191–193. [DOI] [PubMed] [Google Scholar]
- 6.Doring, V., Mootz, H. D., Nangle, L. A., Hendrickson, T. L., de Crecy-Lagard, V., Schimmel, P. & Marliere, P. (2001) Science 292, 501–504. [DOI] [PubMed] [Google Scholar]
- 7.Nagel, G. M. & Doolittle, R. F. (1991) Proc. Natl. Acad. Sci. USA 88, 8121–8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagel, G. M. & Doolittle, R. F. (1995) J. Mol. Evol. 40, 487–498. [DOI] [PubMed] [Google Scholar]
- 9.Brick, P., Bhat, T. N. & Blow, D. M. (1988) J. Mol. Biol. 208, 83–98. [DOI] [PubMed] [Google Scholar]
- 10.Doublie, S., Bricogne, G., Gilmore, C. & Carter, C. W., Jr. (1995) Structure 3, 17–31. [DOI] [PubMed] [Google Scholar]
- 11.Qiu, X., Janson, C. A., Smith, W. W., Green, S. M., McDevitt, P., Johanson, K., Carter, P., Hibbs, M., Lewis, C., Chalker, A., et al. (2001) Protein Sci. 10, 2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yaremchuk, A., Kriklivyi, I., Tukalo, M. & Cusack, S. (2002) EMBO J. 21, 3829–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, T., Nureki, O., Ishitani, R., Yaremchuk, A., Tukalo, M., Cusack, S., Sakamoto, K. & Yokoyama, S. (2003) Nat. Struct. Biol. 10, 425–432. [DOI] [PubMed] [Google Scholar]
- 14.Yang, X.-L., Skene, R. J., McRee, D. E. & Schimmel, P. (2002) Proc. Natl. Acad. Sci. USA 99, 15369–15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakasugi, K., Slike, B. M., Hood, J., Otani, A., Ewalt, K. L., Friedlander, M., Cheresh, D. A. & Schimmel, P. (2002) Proc. Natl. Acad. Sci. USA 99, 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. (1990) EMBO J. 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski, Z. & Minor, W. (1997) in Methods in Enzymology, eds. Carter, C. W., Jr., & Sweet, R. W. (Academic, San Diego), Vol. 276, pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrakis, A., Morris, R. M. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6, 458–463. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 22.Adams, P. D., Rice, L. M. & Brunger, A. T. (1998) Curr. Opin. Struct. Biol. 8, 606–611. [DOI] [PubMed] [Google Scholar]
- 23.Szymanski, M. & Barciszewski, J. (2000) Nucleic Acids Res. 28, 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribas de Pouplana, L., Brown, J. R. & Schimmel, P. (2001) J. Mol. Evol. 53, 261–268. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein, J. (1988) Annu. Rev. Genet. 22, 521–565. [DOI] [PubMed] [Google Scholar]
- 26.Brown, J. R., Robb, F. T., Weiss, R. & Doolittle, W. F. (1997) J. Mol. Evol. 45, 9–16. [DOI] [PubMed] [Google Scholar]
- 27.Ribas de Pouplana, L., Frugier, M., Quinn, C. L. & Schimmel, P. (1996) Proc. Natl. Acad. Sci. USA 93, 166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.