Abstract
Proteomics has become a critical tool in the functional understanding of plant processes at the molecular level. Proteomics-based studies have also contributed to the ever-expanding array of data in modern biology, with many generating Web portals and online resources that contain incrementally expanding and updated information. Many of these resources reflect specialist research areas with significant and novel information that is not currently captured by centralized repositories. The Arabidopsis (Arabidopsis thaliana) community is well served by a number of online proteomics resources that hold an abundance of functional information. These sites can be difficult to locate among a multitude of online resources. Furthermore, they can be difficult to navigate in order to identify specific features of interest without significant technical knowledge. Recently, members of the Arabidopsis proteomics community involved in developing many of these resources decided to develop a summary aggregation portal that is capable of retrieving proteomics data from a series of online resources on the fly. The Web portal is known as the MASCP Gator and can be accessed at the following address: http://gator.masc-proteomics.org/. Significantly, proteomics data displayed at this site retrieve information from the data repositories upon each request. This means that information is always up to date and displays the latest data sets. The site also provides hyperlinks back to the source information hosted at each of the curated databases to facilitate more in-depth analysis of the primary data.
The utilization of mass spectrometry for the characterization of proteins and biological systems has been widely embraced by plant researchers (Heazlewood and Millar, 2006; Weckwerth et al., 2008; Jorrín-Novo et al., 2009). The adoption of proteomics by the plant community can be attributed to the availability of plant genomes during the early phase of this technological development (Heazlewood and Millar, 2003). In recent years, a number of large-scale studies in the model plant Arabidopsis (Arabidopsis thaliana) have utilized proteomics and emerging technologies in mass spectrometry. These have included comparative proteomic studies (Niittylä et al., 2007; Wienkoop et al., 2008), characterization of subcellular structures within the plant cell (Heazlewood et al., 2004; Kleffmann et al., 2004; Eubel et al., 2008; Zybailov et al., 2008; Mitra et al., 2009), profiling of protein composition of plant tissues and organs (Wienkoop et al., 2004; Zou et al., 2009), examination of posttranslational modifications (Zybailov et al., 2009; Nakagami et al., 2010), and providing a genomic context to the proteome through proteogenomic mapping (Baerenfaller et al., 2008; Castellana et al., 2008). Many of these studies have resulted in large data sets comprising either protein identifications or interpreted mass spectral data. While these data sets are usually available as supplemental material or deposited into public repositories, many of these studies have led to the creation of specific online resources to facilitate further interaction with the data (Weckwerth et al., 2008). This has resulted in an increasing number of online resources where pieces of the Arabidopsis proteomic puzzle can be assembled by the informed researcher to create a picture of their protein of interest. Unfortunately, the vast majority of researchers are unaware of the presence of these resources, have limited time to expend on learning resource interfaces, or do not need to fully utilize the often overwhelming amount of information that can be provided by these sites.
Overcoming these issues of usability and awareness of resources can be rectified through the centralization and grouping of biological data at a single portal such as The Arabidopsis Information Resource (TAIR; Swarbreck et al., 2008). A major problem with a centralized repository or database is its inability to respond rapidly to data set updates and modifications. Such a situation is common in the fast-developing area of proteomics, where new tools for data analysis are continually evolving. A further issue that centralized resources must contend with is dealing with the volume of data currently produced by advanced analytic techniques such as mass spectrometry. The ability to successfully capture information beyond the very basic data found in publications is a major difficulty for many of these centralized resources, which often need to rely on a third party to provide data dumps of processed information. The concept of specialized curated databases and services developed by experts that interact through Web services has been discussed for a number of years (Wilkinson and Links, 2002). Such a process has been successfully implemented through BioMoby, a defined ontology designed to enable the exchange and processing of information from biological resources and services (Vandervalk et al., 2009). The advantages become apparent when you consider that research groups producing and analyzing specific data types have a vested interest in actively maintaining and updating the data structure as well as applying the latest analysis techniques. The distributed data resource model becomes even more apparent given the uncertainty associated with funding for many of these centralized resources (Editorial, 2009). Thus, an interlinked web of resources and services could provide stability given the vagaries of research funding and support. Such approaches employing distributed models for data curation, management, and analysis may represent the future direction for online biological resources.
Model plant systems such as rice (Oryza sativa) and Arabidopsis have been exceptionally well served by centralized databases (Lawrence et al., 2007; Ouyang et al., 2007; Swarbreck et al., 2008). These resources have provided community portals for gene annotations, gene and protein models, and links to resources such as seed stocks. Importantly, these resources have defined the framework for gene models and sequences as well as been involved in developing naming nomenclatures that have been widely adopted by the plant research community. Nonetheless, a significant issue with these resources has been their evolution from their respective genome sequencing programs. Consequently, these resources have had a tendency to become feature rich with information that pertains directly to the genome sequence. More recently, with the contextualization of proteomics to the genome through proteogenomic mapping of mass spectra, some proteomic information is interacting with the plant genomics information (Baerenfaller et al., 2008; Castellana et al., 2008). While this has provided some protein context to the genome, resources generally supply simplistic Web links for each and every gene/protein to an assortment of external resources, some of which contain proteomics data. In fact, the Web link appears to be the primary method for interfacing with the majority of online resources, but it provides a very restricted overview of the information present with no reference to the actual availability of any data in the linked site, a situation that is in complete contrast to the objectives of the resource.
The Multinational Arabidopsis Steering Committee (MASC) developed from the coordinated efforts involved in the international genome sequencing program. Its role has been to support and coordinate international Arabidopsis research programs, especially in the area of functional genomics. Several years ago, subcommittees within MASC were initiated to provide focus points in key areas of research in Arabidopsis. The MASC Proteomics Subcommittee (MASCP) was formed to coordinate international proteomics research in Arabidopsis, and its members have been active in establishing proteomics databases and resources (Weckwerth et al., 2008). As part of this effort, members of the MASCP have created a proteomics aggregator (MASCP Gator) that summarizes information about a given Arabidopsis gene model from a variety of international Arabidopsis proteomic databases. The portal provides an initial reference point for researchers to quickly view the extent of tandem mass spectral information, posttranslational modifications, subcellular localization, and organ profiles for a given Arabidopsis protein.
RESULTS
Construction of the MASCP Gator
In order to design an effective data-interchange scheme, the types of data being aggregated need to be accurately identified. For the services included in the aggregator, a number of different data types were identified: the PhosPhAt (Durek et al., 2010) and RIPP-DB (Nakagami et al., 2010) databases describe sets of phosphorylation sites (both experimental and theoretical); the SUBcellular Arabidopsis database SUBA (Heazlewood et al., 2007) lists the subcellular localizations for a given Arabidopsis Gene Identifier (AGI); AtProteome (Baerenfaller et al., 2008) returns tandem mass spectrometry (MS/MS) data with information regarding experimentally observed peptides for different plant organs; The Plant Proteomic Database (PPDB; Sun et al., 2009) returns experimentally derived spectra from various studies; and ProMEX (Hummel et al., 2007) returns a reference to the experimentally observed spectra for a particular locus. Both the AtPeptide data (Castellana et al., 2008) and gene model information from TAIR (Swarbreck et al., 2008) are hosted locally and provide experimentally derived spectra and sequence data, respectively (Table I). Given the wide variety of data being returned and the differing database schema employed, a single and simple data structure could not be readily employed. While the basis of the data exchange employs the AGI, we chose not to use an approach where returned data contained self-describing embedded meta-data, since the service implementation and consuming code would become lengthier and a burden for providers to implement. Instead, each data provider is free to format the data in a fashion that is appropriate for the data being returned, and the service provider must simply document their format.
Table I. Proteomics data sources served by the MASCP Gator.
Collectively, these data contain proteomics information on 21,415 Arabidopsis proteins and contain experimental evidence for approximately 64.1% of the potential proteome encoded by Arabidopsis (genome release 9 from TAIR). FP, Fluorescent protein.
| Data Source | Description | URL | Reference |
| SUBA | Subcellular localization (MS, FP) | http://suba.plantenergy.uwa.edu.au/ | Heazlewood et al. (2007) |
| AtProteome | MS/MS; organ profiles | http://fgcz-atproteome.unizh.ch/ | Baerenfaller et al. (2008) |
| ProMEX | MS/MS | http://www.promexdb.org/ | Hummel et al. (2007) |
| PhosPhAt | Phosphorylation; MS/MS | http://phosphat.mpimp-golm.mpg.de/ | Heazlewood et al. (2008) |
| PPDB | MS/MS; modifications | http://ppdb.tc.cornell.edu/ | Sun et al. (2009) |
| RIPP-DB | Phosphorylation; MS/MS | http://phosphoproteome.psc.database.riken.jp/ | Nakagami et al. (2010) |
| AT_CHLOROa | MS/MS | http://www.grenoble.prabi.fr/at_chloro/ | Ferro et al. (2010) |
| AtPeptide | MS/MS | MASCP (internally hosted) | Castellana et al. (2008) |
| TAIR | Genome annotation | MASCP (internally hosted) | Swarbreck et al. (2008) |
Web service currently under development.
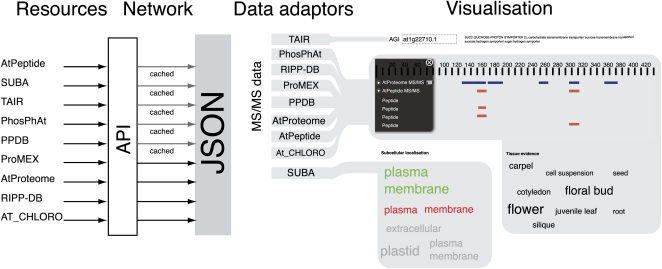
In general, the data types could be classified into three broader families: amino acid modification data (single value on protein sequence), tandem mass spectrometry peptide data (a value range on the protein sequence), and protein localization or expression data (multiple distinct values). These three distinct types of data have different requirements for efficient visualization. Sequence annotation data lend themselves to superposition upon the actual sequence, so that common regions across annotations can be identified. Localization lends itself to a map-based approach, but given that any representations of actual localization will only be illustrative, a cumulative method was employed to indicate occurrence, creating tag clouds for each set of subcellular localization and organ evidence. A tag cloud simply represents keywords presented visually in a weighted state (Sinclair and Cardew-Hall, 2008). For subcellular location information, the SUBA database employs AmiGO (Carbon et al., 2009) controlled vocabularies for subcellular locations and are found as descendents of the terms intracellular part (GO:0044424), membrane (GO:0016020), and cellular component (GO:0005575). For data pertaining to plant organ evidence, the controlled vocabularies available from the Plant Ontology Consortium are used wherever possible (Avraham et al., 2008). Currently the AtPeptide and AtProteome resources broadly employ the plant structure ontology, with only the PhosPhAt database currently utilizing an undocumented ontology. Controlled vocabularies allow for better manipulation of data through reducing the ambiguity found within free text fields. In this way, source information from one service can be compared with data from other resources (Fig. 1). It is anticipated that current and future resources will standardize their vocabularies to ensure integration.
Figure 1.
Schematic diagram of the retrieval processes for the MASCP Gator. Data are requested from the various proteomic resources via an API and passed to the data adaptors that can each understand the data and populate the appropriate parts of the MASCP Gator for visualization. A total of nine resources (AtPeptide, AtProteome, SUBA, TAIR, PhosPhAt, PPDB, ProMEX, RIPP-DB, and AT_CHLORO) are integrated into the MASCP Gator, with the AT_CHLORO service currently under development.
The data retrieval components for the MASCP Gator encapsulate the methods needed to retrieve data for a single AGI, and functions are provided for extracting the data from the retrieval components. Generally speaking, each request for a new data set for an AGI results in a new separate request to the resource database. Written as a software package, the individual components have been organized so that retrieval of data and display of data are handled by separate components able to operate independently of each other. This division of responsibilities allows for the independent maintenance of Application Programming Interface (API) consumption for each service as well as increasing the number of possibilities for the use of the libraries in various situations, such as ad-hoc analysis tools. Furthermore, the libraries have been specifically structured so that third parties can integrate extra functionality. The data-retrieval components within the library retrieve data using asynchronous requests to the remote Web servers. By directly making requests on the original databases, it is not necessary to create data synchronization routines, as the data being returned will always be the latest data. Making these asynchronous requests is part of a technique known as Asynchronous JavaScript and XML. The technique has wide support across Web browsers due to its simplicity and ability to utilize a variety of Web technologies (Woychowsky, 2007).
The MASCP Gator Interface
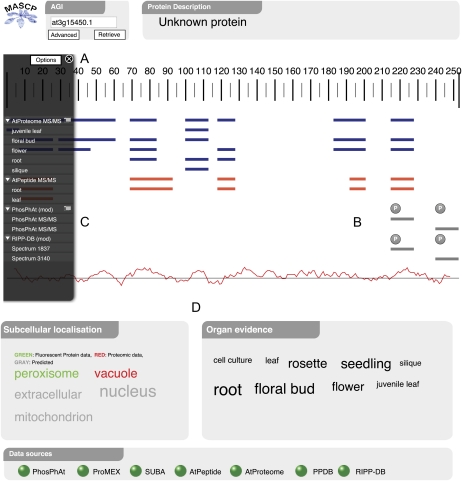
Data from the external databases are fetched and rendered live using a number of visualization techniques. Underpinning the whole interface is a sequence view, which allows for the examination of the entire amino acid sequence at varying levels of detail that range from the amino acid level to a high-level overview (Fig. 2A). Each peptide from the mass spectrometric data sources is overlaid onto the sequence information hosted locally, showing data relevant to the particular area of the sequence in the same region of the protein (Fig. 2B). The overlaid peptides from each data source can be unfurled using the triangle icon in the control panel, providing a mechanism to expand individual peptides that may constitute a region of the protein (Fig. 2C). Complete peptide context and modification location are attained through the zoom function on the toolbox (or by utilizing the wheel on the mouse), and regions can be specifically examined through the cursor-driven panning feature. The “Options” function in the control panel allows tracks to be rearranged and removed from the display. A hydropathy plot of the displayed protein is also available through the Options menu to visualize peptide coverage with regard to hydrophobic regions that could encode transmembrane domains.
Figure 2.
Screenshot of the MASCP Gator interface and result output. The interface was created to be visually intuitive with all necessary information available at a glance. A user simply enters an AGI into the field at the top of the page and clicks the “Retrieve” button (or ENTER). The multiple AGI retrieval facility is accessible through the “Advanced” button. A, Protein sequence is represented as a scale bar. B, Peptides from various data sources are displayed as colored lines with a hydropathy plot shown underneath. C, The control panel provides access to extra features. D, Subcellular information and mass spectral source organ evidence are shown as tag clouds to provide weighted abundance information. “Fluorescent protein” indicates localization by a fluorescent protein, “Proteomic” indicates localization by proteomics, and “Predicted” indicates the predicted subcellular localization. Green and red markers at the bottom of the page indicate whether communication with the external resource was successful.
Source organ information is associated with much of the spectral data and thus could be used to illustrate a protein’s presence in a particular organ. Consequently, we were able to combine this information in an “Organ evidence” tag cloud to convey the protein’s differential presence in plant organs (Fig. 2D). These data are displayed by relating the spectral count to the size of the font for an organ type. For example, “floral bud” is written in larger type relative to “root” if there are more spectra data derived from this organ for the AGI (Fig. 2D). Data for this section are compiled on page load from information associated with spectra in AtProteome, AtPeptide, and PhosPhAt resources. The data are presented as raw spectral counts with no normalization or statistical interpretation and thus should not be used to compare a protein’s relative organ abundance or protein-to-protein organ abundance. The information simply indicates whether a given protein has been identified in a particular plant organ.
Similarly for “Subcellular localization,” a tag cloud was created to visualize existing subcellular localization data comprising organelle proteomic studies, fluorescent protein localizations, and precomputed predictions housed at the SUBA database (Heazlewood et al., 2007). The font size represented by this tag cloud is proportional to a simple tally of data found in SUBA that reports a localization for a given published report (both proteomics and fluorescent protein). For “Fluorescent protein” and “Proteomic” tags, the font size is related to the number of references wherein a protein has been experimentally localized. For “Predicted” tags, the font size is a consolidation of 10 precomputed predictions of subcellular localization. This information comprises data from subcellular proteomic studies (red), fluorescent protein localizations (green), and subcellular prediction (gray).
To facilitate the examination of the underlying data and to provide the ability to obtain information on multiple AGIs, an advanced search feature is also available. Due to communication constraints, the input is limited to a total of 50 AGI codes. The output is arrayed in a tabular format and indicates sites of experimental phosphorylation, potential modulated phosphorylation sites, a “winner takes all” output for subcellular location (fluorescent protein and proteomic), subcellular predictions, and the actual number of spectra identified from each plant organ for each AGI. Thus, it is possible to obtain an overview of proteomics data from a subset of proteins (e.g. biochemical pathway). For convenience, these data can be exported as a comma-separated data sheet.
Using the MASCP Gator Utility
The MASCP Gator was designed to present Arabidopsis proteome data in a simple visual format. The primary use of this tool is to easily investigate protein data for a given Arabidopsis protein (AGI) of interest. By retrieving data and integrating them into a single interface, the ability to comparatively examine data from different sources is enabled. Through a consistent user interface, it is now much easier to see the differences in collected data between organ types, modification states, and subcellular localizations. The arrangement of identified peptides in a linear fashion from diverse sources provides a simple overview of current mass spectral information relevant to the protein of interest. Such collective information could be used to further assess the validity of gene models or be used to provide direct evidence for the actual expression of a protein in Arabidopsis.
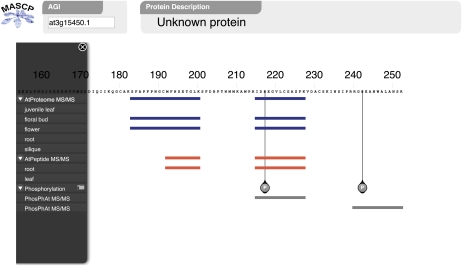
The integration of these data can quickly reveal interesting features that would otherwise be onerous to manually uncover. By presenting both peptides and phosphopeptides, the MASCP Gator can highlight potential phosphoregulated sites on a protein of interest. This is exemplified with the Gator entry for protein At3G15450.1 (Fig. 3). Of particular interest is the presence of the phosphorylation site (Ser-218) derived from phosphopeptide data retrieved from the phosphorylation databases PhosPhAt and RIPP-DB. Unmodified peptide information is also found for this region with data sourced from both AtProteome and AtPeptide. Both the phosphopeptide and unmodified peptide are tryptic and comprise residues 216 to 243. This indicates that this site of the protein At3G15450.1 is likely to be subject to phosphoregulation and is thus a significant functional feature easily observed by this utility. While the biological meaning of this modulation cannot be elucidated from the MASCP Gator, it does provide a starting point for further examination. Other functional conclusions can be drawn about this protein by examining the subcellular localization information and organ evidence. In this particular example (Fig. 2), there is the potential for dual subcellular localization of the protein (peroxisome and vacuole) and some organ evidence indicating its presence in the rosette, seedling, root, and floral bud. The actual “counts” that comprise the tag cloud can be observed by simply hovering the cursor over each subcellular location or organ type.
Figure 3.
A detailed view of the MASCP Gator interface. The zoom and pan features allow full peptide-to-protein context and can display the precise location of any known modifications in the amino acid sequence. This view also clearly shows overlapping peptide information and can be used to identify modulated modification sites and reliable and compatible peptide tags for mass spectrometry-based quantitative studies such as selected reaction monitoring in a triple quadrupole mass spectrometer. [See online article for color version of this figure.]
Further details for data presented at the MASCP Gator can be obtained from the parent databases where publication information for source material and raw data are housed. Relevant links to the precise data source are available in the control panel and in the Web page footer, which also displays whether data were successfully downloaded from the resource. A green marker indicates data retrieval, while a red marker indicates no communication with the resource and that the database may be currently inaccessible. A simple refresh or reload will attempt to retrieve missing data and may correct any communication issues. Further details on the use of the utility are available through a tutorial via the Help link.
Mining Arabidopsis Proteomic Resources
To assess whether further experimental data already exist within current Arabidopsis online resources for the modulation of phosphorylation, we analyzed the protein kinase family of Arabidopsis, which comprises nearly 1,000 members (Gribskov et al., 2001). Protein kinases are known to exhibit autophosphorylation (Harper et al., 2004) and were chosen as likely candidates to assess the efficacy of uncovering sites of phosphoregulation using the MASCP Gator. Employing the data-retrieval libraries developed as part of the utility or the advanced search feature (limited to 50 AGIs), it is possible to find sets of proteins that exhibit this feature. These libraries have been made publicly available at a code repository (http://gator.masc-proteomics.org/source) and can be readily employed through custom scripts to automate bulk data retrieval. The presence of mass spectrometric information was retrieved for a total of 989 Arabidopsis kinases obtained from the PlantsP resource (Gribskov et al., 2001). In total, there were 354 proteins that had data that included both phosphorylation and unmodified information. A further search was performed for the presence of the phosphopeptide (PhosPhAt) and the presence of a corresponding unmodified peptide (AtPeptide or AtProteome). A total of 65 proteins showed this putative pattern of phosphoregulation through complete modulation of phosphorylation. Furthermore, a number of these putative kinases contained multiple sites of modulation, with a total of 92 sites/regions identified (Table II).
Table II. Arabidopsis kinases with evidence for complete modulation of phosphorylation.
Utilizing the MASCP Gator libraries, a total of 65 Arabidopsis putative protein kinases (92 sites) currently contain experimentally determined sites of phosphorylation by mass spectrometry and corresponding mass spectral data, indicating that this site has also been identified in the unmodified form. Such information provides actual experimental evidence for sites of major phosphoregulation. Description (TAIR9) lists the gene descriptions available from TAIR; Residues Where Phosphorylation Resides lists the amino acid range that constitutes the identified peptide with the determined phosphorylation site.
| AGI | Description (TAIR9) | Residues Where Phosphorylation Resides |
| AT1G06840.1 | Leu-rich protein kinase | 758–778, 894–915, 915–939 |
| AT1G10940.1 | SNRK2.4; SNF1-related protein kinase 2.4 | 157–173 |
| AT1G11330.1 | S-locus lectin protein kinase | 487–503 |
| AT1G25320.1 | Leu-rich protein kinase | 372–394 |
| AT1G28440.1 | HAESA-like 1 Ser/Thr kinase | 957–972 |
| AT1G30570.1 | Protein kinase family protein | 682–694 |
| AT1G34210.1 | SERK2; somatic embryo receptor-like kinase 2 | 460–469 |
| AT1G34300.1 | Lectin protein kinase | 560–575 |
| AT1G35670.1 | ATCDPK2; calcium-dependent protein kinase 2 | 476–495 |
| AT1G50700.1 | CPK33; calmodulin-dependent protein kinase | 487–500 |
| AT1G51800.1 | Leu-rich protein kinase | 560–572, 816–828 |
| AT1G52540.1 | Protein kinase, putative | 317–347 |
| AT1G53430.1 | Leu-rich protein kinase | 810–822 |
| AT1G53730.1 | SRF6; strubbelig-receptor family 6 | 374–385, 376–385 |
| AT1G55610.1 | BRL1 (BRI 1 LIKE); kinase | 1,138–1,153 |
| AT1G56140.1 | Leu-rich protein kinase | 980–995; 995–1,010 |
| AT1G60940.1 | SNRK2.10; SNF1-related protein kinase 2.10 | 148–157 |
| AT1G70530.1 | Protein kinase family protein | 632–646 |
| AT1G72710.1 | CKL2; casein kinase 1-like protein 2 | 427–438 |
| AT1G73450.1 | Protein kinase, putative | 609–625 |
| AT2G16250.1 | Leu-rich protein kinase | 832–844 |
| AT2G17290.1 | CPK6; calcium-dependent protein kinase 6 | 26–46, 534–542 |
| AT2G19470.1 | ckl5; casein kinase I-like 5 | 383–400 |
| AT2G35050.1 | Protein kinase family protein | 554–566, 766–784 |
| AT2G36570.1 | Leu-rich protein kinase | 646–660 |
| AT3G08680.1 | Leu-rich protein kinase | 308–321 |
| AT3G13530.1 | MAPKKK7; Ser/Thr kinase | 480–510 |
| AT3G17420.1 | GPK1; protein Ser/Thr kinase | 67–89 |
| AT3G17750.1 | Protein kinase family protein | 653–669 |
| AT3G17840.1 | RLK902; Ser/Thr kinase | 324–336, 505–524 |
| AT3G17850.1 | Protein kinase, putative | 655–668, 668–684 |
| AT3G20410.1 | CPK9; calmodulin-domain protein kinase 9 | 22–41, 60–71, 75–88, 250–265, 462–471, 505–518 |
| AT3G21630.1 | CERK1; chitin elicitor receptor kinase 1 | 276–306 |
| AT3G23310.1 | Protein kinase, putative | 301–312 |
| AT3G24550.1 | ATPERK1; Pro extensin-like receptor kinase 1 | 427–439 |
| AT3G24660.1 | TMKL1; transmembrane kinase-like 1 | 328–350, 329–350 |
| AT3G28450.1 | Leu-rich protein kinase | 262–276 |
| AT3G50500.1 | SNRK2.2; SNF1-related protein kinase 2.2 | 27–38, 167–176 |
| AT3G51550.1 | FER (FERONIA); kinase/ protein kinase | 505–522, 870–893 |
| AT3G51740.1 | IMK2; inflorescence meristem receptor-like kinase 2 | 768–782 |
| AT3G51850.1 | CPK13; calmodulin-dependent protein kinase | 211–228 |
| AT3G53030.1 | SRPK4; Ser/Arg-rich protein kinase 4 | 262–282 |
| AT3G56370.1 | Leu-rich protein kinase | 817–825 |
| AT3G58640.1 | Protein kinase family protein | 415–429 |
| AT3G63260.1 | ATMRK1; Ser/Thr/Tyr kinase | 38–46, 110–129 |
| AT4G08850.1 | Kinase | 975–998 |
| AT4G18950.1 | Ankyrin protein kinase, putative | 22–46, 184–198 |
| AT4G24400.1 | CIPK8; CBL-interacting protein kinase 8 | 165–182 |
| AT4G29810.1 | ATMKK2; Arabidopsis MAP kinase kinase 2 | 45–74 |
| AT4G35230.1 | BSK1; BR-signaling kinase 1 | 227–242, 383–401 |
| AT4G38470.1 | Protein kinase family protein | 244–262 |
| AT5G10290.1 | Leu-rich protein kinase | 319–334 |
| AT5G14720.1 | Protein kinase family protein | 343–365, 415–433, 478–493 |
| AT5G16590.1 | LRR1; Ser/Thr kinase | 552–573, 615–625 |
| AT5G18500.1 | Protein kinase family protein | 72–91 |
| AT5G18610.1 | Protein kinase family protein | 391–406, 428–447 |
| AT5G19450.1 | CDPK19; calcium-dependent protein kinase 19 | 523–533 |
| AT5G24010.1 | Protein kinase family protein | 476–491 |
| AT5G38560.1 | Protein kinase family protein | 649–664 |
| AT5G44290.1 | Protein kinase family protein | 65–91 |
| AT5G49760.1 | Leu-rich protein kinase | 899–921, 928–949 |
| AT5G51350.1 | Leu-rich protein kinase | 639–656 |
| AT5G54380.1 | THE1; theseus 1 kinase | 657–672, 824-853 |
| AT5G65700.1 | BAM1; barely any meristem 1 kinase | 33–43, 975–991 |
| AT5G66880.1 | SNRK2.3; (SNF1)-related protein kinase 2.3 | 175–191 |
To determine the validity of this list, we wanted to verify sites that had been characterized using methods other than mass spectrometry. In recent years, a number of early events in the brassinosteroid signaling pathway have been characterized in Arabidopsis. Two protein kinases known to be involved in these early events are presented in the list of 65, namely the BRASSINOSTEROID INSENSITIVE1-LIKE RECEPTOR KINASE (BRL1; At1G55610.1) and the BRASSINOSTEROID SIGNALING KINASE1 (BSK1; At4G35230.1). It was recently demonstrated through in vitro assays that the BRL1 receptor kinase paralog BRASSINOSTEROID INSENSITIVE1 phosphorylates BSK1 at Ser-230 (Tang et al., 2008). This is precisely the region and site identified by the MASCP Gator as a potentially active phosphoregulated residue. While there is no non-mass-spectrometry evidence for phosphorylation of BRL1, the protein has been shown to exhibit autophosphorylation activity, and the potential phosphoregulated residues outlined in Table I coincide with the intracellular Ser/Thr kinase domain of this protein (Zhou et al., 2004). The accuracy of a given phosphorylation site can be further assessed through hyperlinks back to the relevant resource (e.g. PhosPhAt) to view, download, and analyze the spectra for candidates such as the BSK1 phosphopeptide. Such a process allows users to directly assess a phosphorylation claim using external analysis tools such as the PhosCalc utility (MacLean et al., 2008).
DISCUSSION
The MASCP Gator is a unique resource in the Arabidopsis community in that it provides an aggregating portal for protein information from a number of independently curated Arabidopsis proteomics resources. The creation of an aggregating portal is a highly collaborative endeavor, as it summarizes multiple data sources in a single location. Its development requires coordination between the portal developers and the data providers to ensure that services to retrieve data are available and that the data are well understood. While the developmental processes and coordination can be onerous, the advantages are clear. The data being viewed will always remain up to date, and rather than a centralized repository, specialist curation will be maintained by parties with a vested interest in maintaining data integrity. With the creation of the MASCP Gator, it was constructive that the investigators who developed many of the online proteomics data resources were all members of the MASCP. Thus, a staged integration of resources took place with initial prototyping of interfaces and finally the formalization of the data access. The resultant utility provides a visual overview of protein-based information in Arabidopsis and can provide a source of functional information to the researcher.
Formalized Data Retrieval
The use of a data API is preferable to using a Web-scraping technique, as the latter methodology is extremely fragile. Since Web scraping does not enshrine an agreement between data provider and consumer (since no collaboration is required between the two), Web pages that rely on this technique must maintain the same format for the HTML source. Since the provider does not know that they must maintain the format, there is a high likelihood that the page-parsing algorithm will not be able to accept any changes. Thus, the utilization of an agreed-upon interface provides a more robust structure. Development of public APIs requires careful thought, as any functionality exposed in an API is generally expected to be supported for long periods of time. Moreover, since each database contains unique data, the sets of functionality between databases are generally disjointed. For these reasons, each source database must be examined individually to better understand the needs for the API. The MASCP Gator interfaces with the proteomics resources AtProteome, SUBA, PhosPhAt, PPDB, AtPeptide, ProMEX, RIPP-DB, and AT_CHLORO (under development) through a series of APIs to achieve the aggregated result for a given AGI. The MASCP Gator infrastructure has been designed as a series of modular components that can be included in other resources and allows for the rapid adoption of new data sources. By consuming data APIs provided by databases and online resources, combining them with interfaces for client-side interaction, and presenting them as simple libraries, integration can be more easily achieved.
The communication protocols provide the mechanism by which the APIs transmit data, and since the structures are loosely defined for the return data, a flexible encoding was chosen for the protocol. The MASCP Gator principally employs JSON, a text-based format for communication. JSON provides a number of advantages, the most significant of which is that it is easily parsed in modern Web browsers. This fact again reduces the burden on service providers, since no requirements for service descriptors are prescribed. Furthermore, requests to the data provider services are simple, in this case with the use of a single AGI sent to the service as a parameter, which then returns the data. Simplified and transparent structures thus provide uncomplicated accessibility as issues can be more readily isolated and resolved.
Functional Proteomics
The increased development in proteomic technologies and the increased data production have resulted in an explosion in resources (Vizcaíno et al., 2010). Many researchers have complemented published material with online resources, allowing for expanded and future interpretations of data (Ferro et al., 2010). Unfortunately, much of these data are served from a variety of online data sources that can be difficult to identify and can be complicated to navigate for the casual user. Thus, the strength of the MASCP Gator lies in both community coordination and the ability to compare and contrast data from a variety of repositories. The utility has been specifically designed to summarize disparate data in the area of Arabidopsis proteomics in an intuitive visual manner to quickly get an overview of relevant information. Undertaking this type of comparative display has previously been impractical due to the differences in interfaces provided by the various providers. The ability to see the data on the whole protein level, as well as down to the individual amino acid level, allows great scope for the exploration of the available data by a greater cross-section of the plant research community. By using a Web services model for data retrieval, the tool also will remain up to date, always retrieving the latest versions of the data.
The clear advantages of this aggregation process can be readily observed when phosphopeptides and unmodified peptides are brought together for a given AGI. It is possible to easily identify functionally significant regions of a protein, such as the modulation of a phosphorylation site in the protein. To assess how convenient this approach was at identifying such modifications, we analyzed the list of 65 protein kinases for evidence of phosphoregulation in the literature. Over 50 of these 65 protein kinases (Table II) have been previously identified by mass spectrometry in a variety of proteomic surveys. Many of these studies do not contain any detailed mass spectral information; therefore, it is impossible to assess the phosphorylation state for a number of the protein kinases on the list (Elortza et al., 2003; Fukao et al., 2003; Alexandersson et al., 2004; Carter et al., 2004; Bayer et al., 2006; Dunkley et al., 2006; Qi and Katagiri, 2009). Nonetheless a number of Arabidopsis proteomic analyses have identified several of these kinases and have included peptide information as supplemental material, indicating unmodified phosphorylation states (Nelson et al., 2006; Marmagne et al., 2007; Mitra et al., 2009). While these studies present evidence for kinase autophosphorylation for 10 of the 65 kinases on the list, the data are only found in the large lists of supplemental material associated with each paper. Such an arrangement is not particularly useful for data mining. The evidence for phosphorylation of these 65 kinases is derived from a collection of studies collated at the PhosPhAt database. Unfortunately, the majority of these studies only report the presence of phosphorylation sites on these protein kinases, and no unmodified peptide information is supplied (Nühse et al., 2003, 2004, 2007; Hem et al., 2007; Niittylä et al., 2007; de la Fuente van Bentem et al., 2008; Sugiyama et al., 2008; Whiteman et al., 2008; Jones et al., 2009; Li et al., 2009; Reiland et al., 2009; Chen et al., 2010). Finally, a number of studies have utilized phosphoproteomics techniques and presented both modified and unmodified peptide information as supplemental material. A total of 18 of the 65 protein kinases have been previously identified with evidence for both modified and unmodified peptides (Benschop et al., 2007). Unfortunately, again, this information is only found in large supplemental tables and is not straightforward to readily extract information from. While, collectively, these data provide some of the information outlined in Table I, they represent nearly 15 independent studies and identify less than half of the kinases. Such an approach is no match for the advantages associated with the portal and resources developed as part of the MASCP Gator.
Finally, displaying data from multiple data sources builds further confidence in the presence of a given peptide. The multiple presence of a peptide that is compatible with mass spectrometry also provides a specific peptide tag for further quantitative experiments involving mass spectrometry (Wienkoop and Weckwerth, 2006; Lehmann et al., 2008). Such approaches involving targeted analyses like selected reaction monitoring are becoming more prevalent due to the ability to accurately and simultaneously monitor multiple tags from a given protein or biochemical pathway in highly complex mixtures (Lange et al., 2008b). While the selection of such tags can be accomplished through software packages (Mallick et al., 2007; Lange et al., 2008a), the ability to cross-correlate potential candidates with experimentally identified peptides that are compatible with mass spectrometry is extremely advantageous. The potential power of the MASCP Gator for functional proteomics is thus the ability to fast track the development of experiments and to provide new directions. By incorporating both subcellular information and organ evidence in the context of this functional information, a user can more intuitively develop further experiments on any protein of interest.
CONCLUSION
The abundance of data now being produced in the biological sciences has fueled a massive increase in online resources and databases. While there are clear advantages in having a centralized repository for research focus areas, these resources can often be exposed to the unpredictability of funding. The development of an Arabidopsis proteomics aggregator (MASCP Gator) provides a portal where relevant data are summarized from a variety of databases and data types. With the establishment of the MASCP Gator and the associated component libraries, it is now also possible to integrate these data with complementary resources. This could include expression-based resources such as eFP Browser at the Bio-Array Resource (Winter et al., 2007) and protein-protein interaction resources such as the Arabidopsis Membrane Interactome Project (Lalonde et al., 2010). In addition, we are currently examining the feasibility of connecting the MASCP Gator with community-based databases that archive raw proteomics data from Arabidopsis (Vizcaíno et al., 2010). Integration of information pertaining to the identification of a protein, its presence in a plant organ, its subcellular location, and the presence of posttranslational modifications with both expression and protein interactions could significantly enhance our understanding of biochemical processes. Finally, as future Arabidopsis (and/or plant) proteomics resources are developed, a framework is now in place to integrate these initiatives to create a network of information.
MATERIALS AND METHODS
Languages
The aggregator was implemented using basic Web technology languages: HTML (http://www.w3.org/TR/html401/), JavaScript, and SVG (http://www.w3.org/Graphics/SVG/). Scripts to cache Web requests to provide extra stability were written in Perl. The aggregator page itself was implemented using a combination of HTML and SVG.
Software Libraries
In order to develop this tool, numerous libraries were used to deliver the desired functionality within the Web browser. A major library used is the JQuery (http://jquery.com) JavaScript library, used to provide a consistent document model for the various Web browsers supported by the tool. Since SVG support is lacking in some browsers, a SVG compatibility layer named SVGWeb is optionally supported.
Web Services
Web services were provided from the data sources in JSON format (http://www.json.org/). JSON data are provided by each of the services, responding to a query based upon a given AGI. The format of the JSON response varies between services, depending on the data that the database contain. Clients for the services were written in JavaScript and used to populate the data in the aggregator.
Source Code
The source code for the full aggregator is available online at http://gator.masc-proteomics.org/source. Documentation, unit tests, and examples are provided so that individuals can utilize the libraries developed for the aggregator.
Bioinformatics
To provide further information about the protein of interest, a hydropathy plot is calculated by deriving the mean hydrophobic index of residues given a six-residue window using a Kyte-Doolittle scale (Kyte and Doolittle, 1982). To derive the list of kinases containing potential regions of phosphoregulation, a list of kinases was obtained from the PlantsP database (Gribskov et al., 2001), and the aggregator software was used to retrieve data from AtProteome (Baerenfaller et al., 2008), AtPeptide (Castellana et al., 2008), and PhosPhAt (Durek et al., 2010). This list of kinases was filtered to only accept proteins that contained peptides and phosphopeptides in the same region.
References
- Alexandersson E, Saalbach G, Larsson C, Kjellbom P. (2004) Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol 45: 1543–1556 [DOI] [PubMed] [Google Scholar]
- Avraham S, Tung CW, Ilic K, Jaiswal P, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, et al. (2008) The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res 36: D449–D454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Bayer EM, Bottrill AR, Walshaw J, Vigouroux M, Naldrett MJ, Thomas CL, Maule AJ. (2006) Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6: 301–311 [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci USA 105: 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Hoehenwarter W, Weckwerth W. (2010) Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J 63: 1–17 [DOI] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Anrather D, Dohnal I, Roitinger E, Csaszar E, Joore J, Buijnink J, Carreri A, Forzani C, Lorkovic ZJ, et al. (2008) Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J Proteome Res 7: 2458–2470 [DOI] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX. (2010) PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res 38: D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial (2009) Access denied? Nature 462: 252. [DOI] [PubMed] [Google Scholar]
- Elortza F, Nühse TS, Foster LJ, Stensballe A, Peck SC, Jensen ON. (2003) Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol Cell Proteomics 2: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Eubel H, Meyer EH, Taylor NL, Bussell JD, O’Toole N, Heazlewood JL, Castleden I, Small ID, Smith SM, Millar AH. (2008) Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Hara-Nishimura I, Nishimura M. (2003) Novel glyoxysomal protein kinase, GPK1, identified by proteomic analysis of glyoxysomes in etiolated cotyledons of Arabidopsis thaliana. Plant Cell Physiol 44: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Gribskov M, Fana F, Harper J, Hope DA, Harmon AC, Smith DW, Tax FE, Zhang G. (2001) PlantsP: a functional genomics database for plant phosphorylation. Nucleic Acids Res 29: 111–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A. (2004) Decoding Ca(2+) signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX. (2008) PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res 36: D1015–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Millar AH. (2003) Integrated plant proteomics: putting the green genomes to work. Funct Plant Biol 30: 471–482 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Millar AH. (2006) Plant proteomics: challenges and resources. Finnie C, , Plant Proteomics, Vol 28. Blackwell Publishing, Oxford, pp 1–31 [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis subcellular database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hem S, Rofidal V, Sommerer N, Rossignol M. (2007) Novel subsets of the Arabidopsis plasmalemma phosphoproteome identify phosphorylation sites in secondary active transporters. Biochem Biophys Res Commun 363: 375–380 [DOI] [PubMed] [Google Scholar]
- Hummel J, Niemann M, Wienkoop S, Schulze W, Steinhauser D, Selbig J, Walther D, Weckwerth W. (2007) ProMEX: a mass spectral reference database for proteins and protein phosphorylation sites. BMC Bioinformatics 8: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, MacLean D, Studholme DJ, Serna-Sanz A, Andreasson E, Rathjen JP, Peck SC. (2009) Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. J Proteomics 72: 439–451 [DOI] [PubMed] [Google Scholar]
- Jorrín-Novo JV, Maldonado AM, Echevarría-Zomeño S, Valledor L, Castillejo MA, Curto M, Valero J, Sghaier B, Donoso G, Redondo I. (2009) Plant proteomics update (2007-2008): second-generation proteomic techniques, an appropriate experimental design, and data analysis to fulfill MIAPE standards, increase plant proteome coverage and expand biological knowledge. J Proteomics 72: 285–314 [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjölander K, Gruissem W, Baginsky S. (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol 14: 354–362 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Sero A, Pratelli RJ, Pilot G, Chen J, Sardi MI, Parsa SA, Kim DY, Acharya BR, Stein EV, et al. (2010) A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front Physiol 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Malmström JA, Didion J, King NL, Johansson BP, Schäfer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, et al. (2008a) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics 7: 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. (2008b) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Schaeffer ML, Seigfried TE, Campbell DA, Harper LC. (2007) MaizeGDB’s new data types, resources and activities. Nucleic Acids Res 35: D895–D900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann U, Wienkoop S, Tschoep H, Weckwerth W. (2008) If the antibody fails: a mass western approach. Plant J 55: 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wong WS, Zhu L, Guo HW, Ecker J, Li N. (2009) Phosphoproteomic analysis of ethylene-regulated protein phosphorylation in etiolated seedlings of Arabidopsis mutant ein2 using two-dimensional separations coupled with a hybrid quadrupole time-of-flight mass spectrometer. Proteomics 9: 1646–1661 [DOI] [PubMed] [Google Scholar]
- MacLean D, Burrell MA, Studholme DJ, Jones AM. (2008) PhosCalc: a tool for evaluating the sites of peptide phosphorylation from mass spectrometer data. BMC Res Notes 1: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, et al. (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol 25: 125–131 [DOI] [PubMed] [Google Scholar]
- Marmagne A, Ferro M, Meinnel T, Bruley C, Kuhn L, Garin J, Barbier-Brygoo H, Ephritikhine G. (2007) A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol Cell Proteomics 6: 1980–1996 [DOI] [PubMed] [Google Scholar]
- Mitra SK, Walters BT, Clouse SD, Goshe MB. (2009) An efficient organic solvent based extraction method for the proteomic analysis of Arabidopsis plasma membranes. J Proteome Res 8: 2752–2767 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K. (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol 153: 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CJ, Hegeman AD, Harms AC, Sussman MR. (2006) A quantitative analysis of Arabidopsis plasma membrane using trypsin-catalyzed (18)O labeling. Mol Cell Proteomics 5: 1382–1395 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Fuglsang AT, Palmgren MG, Frommer WB, Schulze WX. (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol Cell Proteomics 6: 1711–1726 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Bottrill AR, Jones AM, Peck SC. (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J 51: 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC. (2003) Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics 2: 1234–1243 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35: D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Katagiri F. (2009) Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J 57: 932–944 [DOI] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J, Cardew-Hall M. (2008) The folksonomy tag cloud: when is it useful? J Inf Sci 34: 15–29 [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y. (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the plant proteomics database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandervalk BP, McCarthy EL, Wilkinson MD. (2009) Moby and Moby 2: creatures of the deep (web). Brief Bioinform 10: 114–128 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Foster JM, Martens L. (2010) Proteomics data repositories: providing a safe haven for your data and acting as a springboard for further research. J Proteomics 73: 2136–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W, Baginsky S, van Wijk K, Heazlewood JL, Millar H. (2008) The multinational Arabidopsis steering subcommittee for proteomics assembles the largest proteome database resource for plant systems biology. J Proteome Res 7: 4209–4210 [DOI] [PubMed] [Google Scholar]
- Whiteman SA, Serazetdinova L, Jones AM, Sanders D, Rathjen J, Peck SC, Maathuis FJ. (2008) Identification of novel proteins and phosphorylation sites in a tonoplast enriched membrane fraction of Arabidopsis thaliana. Proteomics 8: 3536–3547 [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Glinski M, Tanaka N, Tolstikov V, Fiehn O, Weckwerth W. (2004) Linking protein fractionation with multidimensional monolithic reversed-phase peptide chromatography/mass spectrometry enhances protein identification from complex mixtures even in the presence of abundant proteins. Rapid Commun Mass Spectrom 18: 643–650 [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Morgenthal K, Wolschin F, Scholz M, Selbig J, Weckwerth W. (2008) Integration of metabolomic and proteomic phenotypes: analysis of data covariance dissects starch and RFO metabolism from low and high temperature compensation response in Arabidopsis thaliana. Mol Cell Proteomics 7: 1725–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienkoop S, Weckwerth W. (2006) Relative and absolute quantitative shotgun proteomics: targeting low-abundance proteins in Arabidopsis thaliana. J Exp Bot 57: 1529–1535 [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Links M. (2002) BioMOBY: an open source biological web services proposal. Brief Bioinform 3: 331–341 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychowsky E. (2007) Introducing AJAX. AJAX: Creating Web Pages with Asynchronous JavaScript and XML. Prentice Hall, Upper Saddle River, NJ, pp 19–40 [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J. (2004) BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J 40: 399–409 [DOI] [PubMed] [Google Scholar]
- Zou J, Song L, Zhang W, Wang Y, Ruan S, Wu WH. (2009) Comparative proteomic analysis of Arabidopsis mature pollen and germinated pollen. J Integr Plant Biol 51: 438–455 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Sun Q, van Wijk KJ. (2009) Workflow for large scale detection and validation of peptide modifications by RPLC-LTQ-Orbitrap: application to the Arabidopsis thaliana leaf proteome and an online modified peptide library. Anal Chem 81: 8015–8024 [DOI] [PubMed] [Google Scholar]