Abstract
Temperature variations at the nonextreme range modulate various processes of plant growth, development, and physiology, but how plants perceive and transduce these temperature signals is not well understood. Moderate cooling from 28°C to 22°C induces transcription of a number of genes in salicylic acid-dependent and -independent manners. Here, we report the study of the transcriptional control of the BON1-ASSOCIATED PROTEIN1 (BAP1) gene that is responsive to a moderate decrease of temperature as well as to many environmental stimuli. Using reporter genes under the control of series of regions of the BAP1 promoter, we identified a 35-bp fragment that is necessary and sufficient for the BAP1 transcript induction by a moderate cooling. This fragment also confers an induction of BAP1 by cold and reactive oxygen species-generating paraquat. Furthermore, the INDUCER OF CBF EXPRESSION1 (ICE1) protein that is involved in transcriptional control of cold responses is found to bind to a MYC element in this promoter and is required for the cooling induction of BAP1. The ice1 mutant has a low induction of BAP1 and enhanced resistance to a bacterial pathogen. Thus, responses to a moderate decrease in temperature may utilize components in the cold response as well as a potentiating signaling involving salicylic acid.
Plants, being sessile, have evolved to adapt to their environment to maximize their fitness and reproduction. One of the major environmental factors they monitor and respond to is temperature, which fluctuates daily and seasonally. Almost all processes of growth and development are modulated by temperature at the molecular, cellular, physiological, and ecological levels (Long and Woodward, 1988; Penfield, 2008). Transcriptional regulation is one of the major responses plants assume to achieve adaptation. Both cold acclimation and heat acclimation involve the up-regulation of transcription of genes that are important for adaptation to extreme conditions (Hua, 2009). For cold responses, one transcriptional cascade has been identified by molecular and genetic studies on a number of cold-induced genes named COLD REGULATED (COR) or LOW TEMPERATURE INDUCED (Thomashow, 1999). This cascade includes the A/GCCGAC motif named C-REPEAT (CRT)/DEHYDRATION RESPONSIVE ELEMENT (DRE) that is found in the promoter region of many COR genes (Thomashow, 1999; Yamaguchi-Shinozaki and Shinozaki, 2006). The CTR element is bound by AP2 domain-containing transcription factors CRT BINDING FACTOR (CBF)/DRE BINDING PROTEIN (Thomashow, 1999; Yamaguchi-Shinozaki and Shinozaki, 2006). The CBF3 gene is transcriptionally regulated by a MYC-type transcription factor INDUCER OF CBF EXPRESSION1 (ICE1) through ICEr1 and ICEr2 sequences in its promoter (Chinnusamy et al., 2003). The significance of this transcriptional cascade is demonstrated by the profound effect on cold/freezing tolerance with altered expression of CBFs and ICE1 (Chinnusamy et al., 2003; Sung et al., 2003). For heat shock responses, transcriptional cascade has also been identified to control the expression of HEAT SHOCK PROTEIN (HSP; Vierling, 1991). Heat shock factors are transcription factors that bind to the heat shock element consisting of AGAAnnTTCT found in promoters of many HSP genes (Kotak et al., 2007; von Koskull-Döring et al., 2007). Some of the heat shock factors have been demonstrated to be essential for thermotolerance (Sung et al., 2003; von Koskull-Döring et al., 2007).
Moderate temperature variations also greatly influence many aspects of growth and development such as growth rate (Cuadrado et al., 1989), flowering time (Blázquez et al., 2003), metabolism (Kaplan et al., 2004), hormonal responses (Larkindale and Huang, 2004), and circadian rhythms (Gardner et al., 2006). Additionally, they influence interaction between plants and other organisms, including plant disease resistance (Wang et al., 2009). Relatively less is known about the molecular mechanism underlying plants’ responses to these moderate temperature variations. Recently, it is shown that ARP6, a subunit of the SWR1 complex, represses expression of warm genes at low temperatures in Arabidopsis (Arabidopsis thaliana), likely through the temperature-sensitive occupancy of the alternative histone H2A.Z on promoter sequences (Kumar and Wigge, 2010). Our early studies with marker genes revealed shared and distinct mechanisms for responses to drastic and moderate decrease in temperature at the transcriptional level (Wang and Hua, 2009). Both a salicylic acid (SA)-dependent and an SA-independent pathway are found to function in cooling induction of genes. A small decrease of temperature from 28°C to 22°C induces COR15a expression in an SA-independent manner. The induction is mediated by the CBF genes and could contribute to the enhanced cold tolerance. It appears that some of the cooling responses may prepare plants to anticipate and prepare for extreme conditions.
We initiated an investigation on the SA-dependent transcriptional response to moderate temperature decrease in the BON1-ASSOCIATED PROTEIN1 (BAP1) gene. BAP1 is a membrane-associated C2 domain protein that negatively regulates defense responses (Hua et al., 2001; Yang et al., 2006; Yang et al., 2007). Its loss-of-function mutant has constitutive defense responses, and its overexpression in plants and yeast (Saccharomyces cerevisiae) suppresses programmed cell death induced by a number of reagents including pathogens, proapopotic genes, and reactive oxygen species (ROS; Yang et al., 2007). BAP1 is itself induced by multiple stimuli including temperature variations, mechanical stress, and biotic stresses (op den Camp et al., 2003; Yang et al., 2006). The BAP1 gene has a higher expression level at stable 22°C than at 28°C and is rapidly induced by a cooling from 28°C to 22°C. Interestingly, a number of genes involved in defense responses including BON1, BAP1, ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1; Wiermer et al., 2005), and PHYTOLEXIN DEFICIENT4 (PAD4; Jirage et al., 1999) have differential expression at 28°C and 22°C (Yang and Hua, 2004). It is possible that this temperature modulation reflects a more critical role of these genes to regulate defense responses at 22°C than 28°C as the loss of BON1 or BAP1 function has more detrimental effects at 22°C than 28°C.
The cooling induction of BAP1 provides an entry point to dissect the transcriptional response to moderate decrease in temperature in the SA-dependent manner. Here, we report the identification of a 35-bp fragment in the BAP1 promoter as a cis-acting region to confer response to a cooling from 28°C to 22°C. This temperature-sensitive region also mediates responses to cold and ROS but not to wounding and pathogen infection. Furthermore, ICE1 is found to bind to this element and mediate the induction of BAP1 by cooling, cold, and ROS. Thus, this study reveals a cooling induction utilizing the ICE1 protein, suggesting a common mechanism for responding to extreme and nonextreme temperatures.
RESULTS
Induction of BAP1 Expression by a 28°C to 22°C Shift Requires the SA Pathway
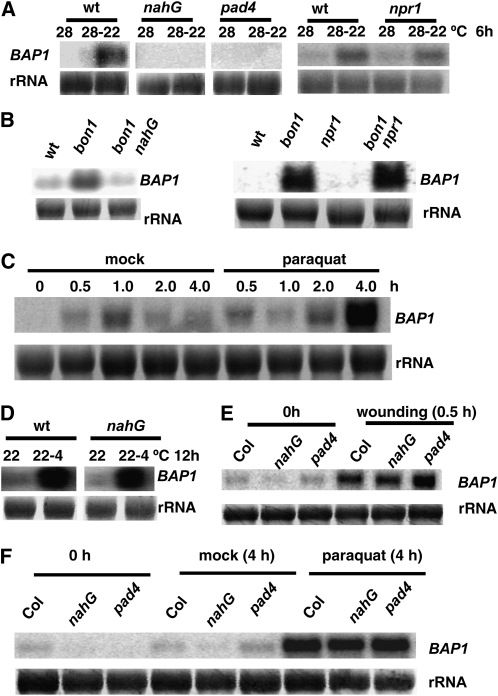
Earlier studies revealed both SA-dependent and -independent pathways for cooling induction from 28°C to 22°C (Wang and Hua, 2009). As BAP1 expression is induced by SA (Yang et al., 2006), we determined whether the cooling induction of BAP1 from 28°C to 22°C requires SA and the function of PAD4, a regulator of cell death and resistance. To this end, we used Arabidopsis plants either containing nahG coding for a bacterial enzyme degrading SA (Clarke et al., 2000) or with a loss-of-function pad4 mutation (Jirage et al., 1999). The wild-type Columbia-0 (Col-0), nahG, and pad4 plants were subjected to the 28°C to 22°C cooling treatment, and the BAP1 induction was analyzed by RNA blots. In contrast to the wild type, neither nahG nor pad4 had up-regulated BAP1 expression 6 h after shift (Fig. 1A), indicating a requirement of SA and PAD4 for this rapid induction.
Figure 1.
Induction of BAP1 by multiple stimuli. Shown are BAP1 expressions analyzed by RNA blot. Two-week-old plants grown at 28°C were used for cooling treatment (28°C–22°C). Three-week-old plants grown at 22°C were used for paraquat, SA, and cold (22°C–4°C) treatments. Total RNAs were prepared from tissues collected at indicated time points, and rRNAs were used as the loading controls. A, BAP1 induction by cooling is SA and PAD4 dependent but not NPR1 dependent. BAP1 induction is abolished in nahG and pad4 plants but is present in the npr1 mutant. B, BAP1 induction in bon1 is SA dependent but not NPR1 dependent. The up-regulation of BAP1 in the bon1 mutant is abolished in nahG but is retained in npr1. C, BAP1 is induced by paraquat. Plants were sprayed with a 20 μm paraquat solution containing 0.1% Tween 20 or a mock solution of 0.1% Tween 20. BAP1 expression was greatly induced at 4 h after paraquat treatment. An apparent increase in the 1.0-h mock sample is largely due to an overloading of total RNA. D, BAP1 induction by cold is not SA dependent. BAP1 induction by this cold treatment is present in nahG and pad4. E, BAP1 induction by wounding is not SA or PAD4 dependent. Plants were wounded by a needleless syringe. BAP1 was rapidly induced at 0.5 h after wounding. F, BAP1 induction by paraquat is not SA or PAD4 dependent. BAP1 induction by paraquat is retained in nahG or pad4 mutant. wt, Wild type.
NPR1 mediates many aspects of SA responses and is a positive regulator of systemic acquired resistance (Durrant and Dong, 2004). RNA-blot analysis shows that the npr1 mutant had the same induction of BAP1 as the wild type, indicating that NPR1 is not involved in this induction (Fig. 1A). A similar genetic requirement was observed in the up-regulation of the BAP1 in the bon1 mutant. BAP1 is induced in the bon1-1 mutant compared with the wild type and this induction is abolished in bon1-1nahG but not in bon1-1npr1-1 (Fig. 1B). Thus, BAP1 induction by cooling and the loss of BON1 function are both mediated by a NPR1-independent SA function.
Requirement of SA for BAP1 Induction by Multiple Stimuli
The BAP1 gene is induced by multiple stimuli including temperature variations, mechanical stresses, and biotic stresses (Yang et al., 2006). One common event following these stimuli might be the generation of ROS. Indeed, it has been shown that singlet oxygen species induces BAP1 rapidly (op den Camp et al., 2003), and hydrogen peroxide also induces BAP1 according the public array data (The Arabidopsis Information Resource array no. 185). To analyze the induction of BAP1 by the other ROS superoxide, we treated plants with paraquat to induce the generation of superoxide in chloroplasts (Babbs et al., 1989). The BAP1 transcript was greatly induced at 4 h after the spray with paraquat compared with the mock treatment (Fig. 1C). Thus, BAP1 could be induced by various ROS.
We tested whether SA and PAD4 are required for BAP1 induction by other stimuli by comparing responses in nahG and pad4 to those of the wild type. RNA-blot analysis showed that BAP1 induction by cold, wounding, or paraquat was similar in nahG and pad4 as in the wild type (Fig. 1, D–F). Thus, the 28°C to 22°C induction of BAP1 appears to be unique among other stimuli in that it requires SA and PAD4.
Isolation of a Temperature-Responsive Element in the BAP1 Promoter
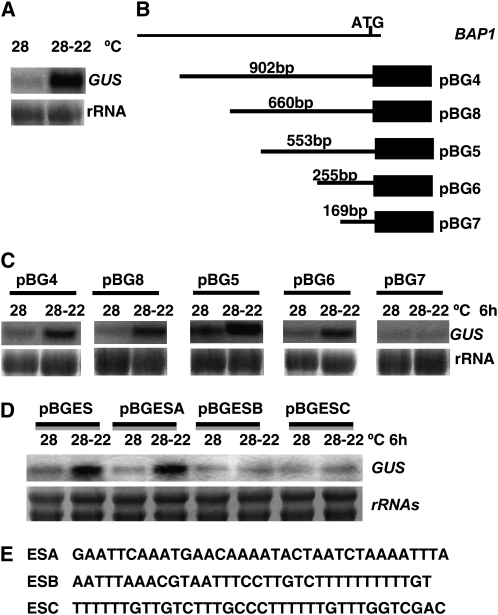
We determined that the induction of BAP1 by cooling is at the transcriptional level. A 2.1-kb fragment of the BAP1 promoter was fused to the reporter gene GUS and the pBAP1::GUS transgene (pBG) was transformed into wild-type Arabidopsis Col-0 plants. More than 50 transgenic plants were generated and 10 were tested for the induction of GUS by cooling treatment. Eight of the 10 lines showed an induction of GUS transcripts by a temperature shift from 28°C to 22°C for 6 h (Fig. 2A). Thus, BAP1 induction by cooling is at the transcriptional level and its promoter contains a temperature-responsive (TR) element(s).
Figure 2.
Isolation of a TR fragment in the BAP1 promoter. Cooling induction was carried out similar as in Figure 1. Total RNAs were isolated, and expression of the GUS reporter gene was analyzed by RNA blots. The rRNAs were used as loading controls. A, Temperature regulation of BAP1 expression is at the transcriptional level. GUS was induced by cooling treatment in the eight of the 10 pBG2 transgenic lines. A representative line is shown. B, Diagram of pBG fusion series. The GUS gene is represented by the black bar and specific fragments of the BAP1 promoter used in the promoter fusions are indicated. C, Analysis of cooling induction of GUS in pBG transgenic plants. The pBG4, pBG8, pBG5, and pBG6 lines, but not the pBG7 line, showed a cooling induction of GUS. D, The ESA fragment confers a cooling induction to the GUS reporter gene. GUS induction was seen in the pBGES and pBGESA transgenic lines, but not the pBGESB and pBGESC lines. E, Nucleotide sequences of three overlapping subfragments of ES: ESA, ESB, and ESC.
To define this TR element in the BAP1 promoter, we generated a series of GUS promoter fusions with various lengths of the truncated BAP1 promoter (Fig. 2B). The five promoter fragments started from −902, −660, −553, −255, and −169 bp, respectively, and ended at −1 bp relative to the translation start site and the transcriptional initiation site of BAP1 is 27-bp upstream of the translation start site. These fusions are named pBG4, pBG8, pBG5 pBG6, and pBG7, respectively.
All of these fusion constructs were transformed into wild-type Col-0 plants. More than 10 independent transgenic lines for each promoter deletion fusion were subject to cooling treatment, and the expression of both the endogenous BAP1 gene and the reporter GUS genes before and after the treatment were compared. We found that GUS staining was not sensitive and accurate enough to reflect the transcriptional regulation by the BAP1 promoter, so we used RNA-blot hybridizataion for all promoter analyses. The majority of each of the pBG4, pBG8, pBG5, and pBG6 lines (10 of 10, 10 of 10, five of 10, and eight of 10) showed GUS induction by the 28°C to 22°C shift (Fig. 2C). In contrast, none of the pBG7 lines exhibited induction by cooling (Fig. 2C) while the endogenous BAP1 exhibited induction. Thus, the TR elements reside in between −255 and −169 bp upstream to the translational start site of BAP1.
Refining the TR Element to a 35-bp Fragment
To determine whether this 87-bp fragment is sufficient to confer temperature response, we fused this fragment with a minimal 35S promoter to the GUS reporter gene and named it pBGES. Five independent transgenic lines were tested and four of them exhibited GUS induction after a 28°C to 22°C shift analyzed by RNA blots (Fig. 2D), indicating that this fragment contains the TR element. In general, a shorter promoter confers a relatively weaker induction than a longer promoter, suggesting that there are additional activating elements in the BAP1 promoter although this 87-bp fragment is the most prominent one responsive to cooling.
To refine the TR element, we divided this fragment into three overlapping subfragments named A, B, and C (Fig. 2E). Three repeats of A, four repeats of B, and four repeats of C were fused, respectively, to the GUS reporter gene with a minimal 35S promoter, and they were named pBGESA, pBGESB, and pBGESC (Fig. 2E). None of the pBGESB or pBGESC transgenic lines had any induction of GUS by a 28°C to 22°C shift, while four of the 13 pBGESA lines exhibited an induction of GUS similarly to the endogenous BAP1 gene (Fig. 2D). Therefore, the fragment ESA is responsive to a cooling of 28°C to 22°C downshift and contains the TR element. However, there was a lower proportion of transgenic lines of pBGESA (four of 13) exhibiting induction compared with that of pBGES (four of five), indicating that ESB and ESC contain additional element(s) enhancing cooling induction by ESA.
Requirement of the ESA for BAP1 Induction by Other Stimuli
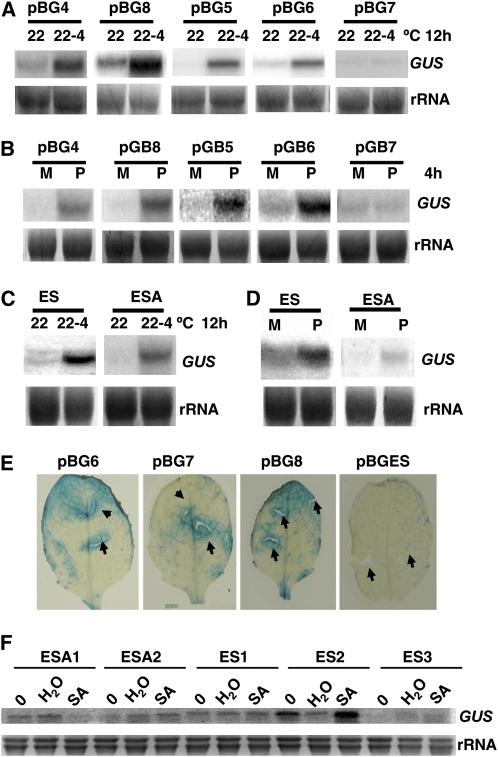
To determine whether BAP1 uses the same or different cis-element for induction by various stimuli, we assayed responses of pBG lines to other stimuli. A cold (4°C) treatment induced GUS expression in pBG4, pBG8, pBG5, and pBG6 lines but not pBG7 lines (Fig. 3A). Similarly, paraquat treatment induced GUS expression in pBG4, pBG8, pBG5, and pBG6 lines but not in the pBG7 lines (Fig. 3B), suggesting that cold and paraquat responses are mediated by the same ES fragment as cooling.
Figure 3.
The ESA fragment is required for cold and parquet induction of BAP1. Cold and paraquat treatments were carried as described in Figure 1. SA treatment was carried out by spraying 22°C-grown 3-week-old plants with 1 mm SA containing 0.1% Tween 20 or water with 0.1% Tween 20. Total RNAs were prepared from tissues collected at indicated time points. Expression of the GUS reporter gene was analyzed by RNA blots except for 3E. The rRNAs were used as loading controls. A, Cold induction of the GUS reporter gene in the pGBs lines. The pBG4, pBG8, pBG5, and pBG6 lines, but not the pBG7 line, showed a cold induction of GUS. B, Paraquat induction of GUS in pGB lines. The pBG4, pBG8, pBG5, and pBG6 lines, but not the pBG7 line, showed an induction of GUS by paraquat. C, Cold induction of GUS in pBGES and pBGESA lines. GUS induction was seen in both lines. D, Paraquat induction of GUS in pBGES and pBGESA lines. GUS induction was seen in both lines. E, GUS induction by wounding in pBG lines. Shown are GUS staining of treated tissues. The pBG6, pBG7, and pBG8, but not the pBGESA line showed GUS signals at the wounding sites. F, SA induction of GUS in pBGES and pBGESA lines. Two independent pBGESA lines (pBGESA1 and pBGESA2) and three pBGES lines (pBGES1, pBGES2, and pBGES3) were treated with water or SA. Only pBGES2 showed GUS induction by SA. P, Paraquat treatment; M, mock treatment. [See online article for color version of this figure.]
We found that the ESA fragment is sufficient for cold and paraquat induction. The GUS transcript was induced by cold and paraquat in the pBGESA lines (Fig. 3, C and D) but not pBGESB or pBGESC lines (data not shown). Thus, the same ESA is responsible for cold and paraquat induction.
In contrast, wounding induction of BAP1 is not mediated by ESA. As this wounding response is rapid and robust, and we could directly utilize tissue staining to assay GUS activity. Transgenic lines of pBG6, pBG7, pBG8, and pBGES were damaged with needless syringe and GUS activities were assayed half an hour after wounding. All of the lines except for pBGES gave a strong GUS staining (Fig. 3E). Thus, wounding response is mediated by the 169-bp fragment 5′ to the ESA fragment.
SA induction of BAP1 does not appear to be mediated by the ESA element either. We treated three pBGES (1, 2, and 3) lines and two pBGESA (1 and 2) lines with SA or water. Only the pBGES2 line exhibited a very weak GUS up-regulation and none of the others showed GUS induction upon SA treatment (Fig. 3F). Thus, the ESA fragment mediates BAP1 induction by some but not all stimuli.
Potential cis-Elements in the ESA Sequences
No obvious cold- or heat-inducible element is recognized in the ESA fragment. Therefore, we subjected it to plant cis-acting elements search through online softwares. Four motifs were found using PLACE (http://www.dna.affrc.go.jp/PLACE/index.html). The first one is A(A/T)TTCAAA starting from nucleotide position 2, an ethylene-responsive element initially found in the promoter of GLUTATHIONE S-TRANSFERASE1 (GST1; Itzhaki et al., 1994). The second is CAnnTG at nucleotide 6, a MYC binding site, conferring binding sites for bHLH transcription factors (Meshi and Iwabuchi, 1995). The third is (C/T)ACT at nucleotide 19, an element for mesophyll-specific expression in the promoter of phosphoenolpyruvate carboxylase gene (Gowik et al., 2004). The last one is AATACTAAT, a Suc-responsive element conserved among Suc-regulated genes (Grierson et al., 1994). Two other elements were identified in ESA with PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). One is a CAAT box starting at position 5, a common cis-acting element in the promoter or enhancer regions. The other is similar to Gap-box CAAATGAA(A/G)A, part of a light-responsive element (Conley et al., 1994).
ICE1 Binds to the ESA Fragment
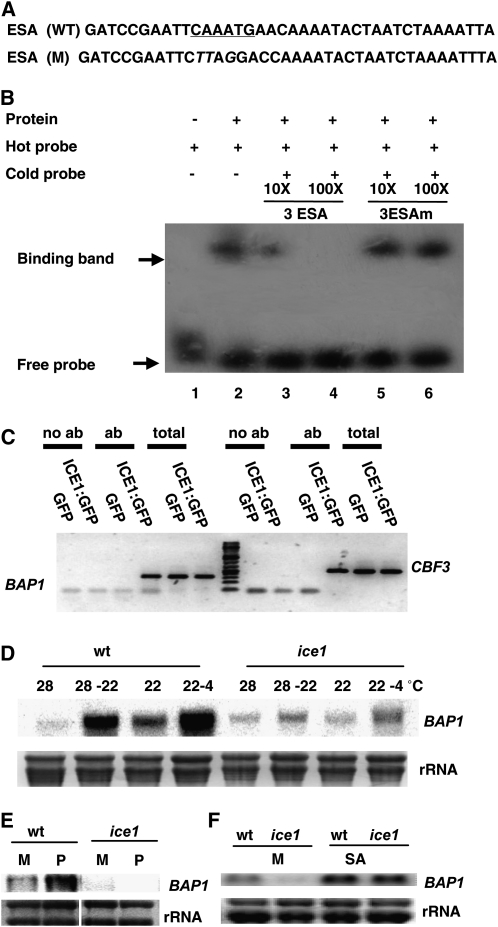
ICE1 is a MYC-type bHLH transcriptional activator that regulates the transcription of CBF3 in the cold response by recognizing the consensus DNA sequence CAnnTG in its promoter. To test whether ICE1 binds to the potential MYC recognition site AaaTG in the ESA element, we carried out an electrophoresis mobility shift assay (EMSA). The ICE1-GST fusion protein (Miura et al., 2007) expressed and purified from Escherichia coli was incubated with a labeled DNA probe containing three copies of the ESA fragment (3ESA; Fig. 4A). The ESA probe was shifted to a large Mr in the presence of ICE1:GST (Fig. 4B). The shifted band was abolished by addition of increasing amount of cold unlabeled DNA of 3ESA (Fig. 4B), indicating that ICE1 is able to specifically bind to the ESA element. To examine if the binding requires to the MYC binding site CAAATG, we mutated this motif into CTTAGG in ESA (Fig. 4A) and used this 3ESAm as a competitor for the wild-type 3ESA probe in EMSA. In contrast to the wild-type 3ESA, competition was not observed with the unlabeled DNA of 3ESAm (Fig. 4B), indicating that ICE1 binds specifically to the MYC recognition site in the ESA element.
Figure 4.
ICE1 binds to the ESA element of the BAP1 promoter and mediates the BAP1 induction. A, Oligonucleotide sequences of ESA used in EMSA. Letters underlined indicate the MYC-recognition sequences and letters in italic indicate the mutated nucleotides in the ESA element. B, In vitro binding of ICE1 to ESA element. EMSAs were preformed by using no protein (lane 1) or 0.2 μg of purified ICE1:GST protein (lane 2) and [32P]labeled wild-type ESA hot probe. Competition experiments were preformed by using increasing amounts (10× and 100×) of unlabeled cold probe of ESA (lane 3 and 4) or mutant ESA (lane 5 and 6). C, Enrichment of the BAP1 promoter DNA in the IP. Shown are PCR products from IPed samples. p35S::GFP and p35S::ICE1:GFP transgenic plants were cold treated (22°C–4°C) for 3 h, followed by formaldehyde cross linking and IP with anti-GFP antibodies. Products IPed with the GFP antibody (ab) were amplified with BAP1 promoter primers by PCR for 40 cycles. Products IPed without the GFP antibodies (no ab) were used as negative control. Total genomic DNA (total) was used as a PCR control. The presence of the CBF3 promoter was used as a positive control. D to F, RNA-blot analysis of BAP1 induction in wild type and ice1-1 by cooling and cold treatment (D), paraquat (E), and SA (F). BAP1 expression was analyzed by RNA-blot analysis and rRNAs were used as loading controls. BAP1 inductions were compromised in ice1-1 for cold, cooling, and paraquat treatments but not SA treatment. M, Mock; P, paraquat.
ICE1 Is Associated with the Promoter Region of BAP1 in Vivo
We carried out chromatin immunoprecipitation (ChIP) assay to determine whether ICE1 is associated with the promoter region of BAP1 in vivo under cooling or cold treatment. To this end, we utilized transgenic lines of p35S::ICE1:GFP (Kanaoka et al., 2008) to facilitate the detection of ICE1. Cold and cooling induction was carried out by shifting 3-week-old plants grown at 22°C to 4°C or 2-week-old plants grown at 28°C to 22°C, respectively. Anti-GFP antibodies were used to immunoprecipitate (IP) ICE1:GFP and its associated chromatin from the treated tissues; and the presence of the BAP1 promoter fragment in the IP was detected by PCR using a primer pair amplifying the promoter containing the ESA element. Strong amplification was observed in IPs of the cold-treated p35S::ICE1:GFP from BAP1 primers as well as from primers of CBF3 that are known to be bound by ICE1 (Fig. 4C). No amplification was obtained from IPs of cold-treated p35S::GFP plants or the IPs of p35S::ICE1:GFP without GFP antibody (Fig. 4C), nor was it observed from the tissues without cold treatment (data not shown). Therefore, ICE1 is specifically associated with the promoter region of BAP1 during the cold stress. We did not detect amplification of the BAP1 promoter from IPs of cooling shift from 28°C to 22°C. This suggests a weaker and more transient association between ICE1 and the BAP1 promoter region in cooling than cold induction, as BAP1 induction by cold is much stronger than by cooling.
Cooling and Cold Induction of BAP1 Is Compromised in the ice1 Mutant
We further tested the model that ICE1 is directly involved in the transcriptional regulation of BAP1 expression in cold and cooling responses by analyzing BAP1 induction in the ice1-1 mutant (Chinnusamy et al., 2003). For cold treatment, ice1-1 and the wild-type plants were grown at 22°C for 3 weeks before being shifted to 4°C for 6 h. For cooling induction, plants were grown at 28°C for 2 weeks before being shifted to 22°C for 9 h. RNA blots show that BAP1 inductions by cooling and cold treatments were both greatly reduced in ice1-1 (Fig. 4D), indicating that ICE1 is directly involved in BAP1 induction by cooling and cold.
We further tested whether ICE1 protein is required for BAP1 induction by other stimuli. The ice1-1 mutant and the wild type were sprayed with either 20 μm paraquat or 2 mm SA. RNA blots show that BAP1 induction by paraquat was totally abolished but its induction by SA was not altered in ice1-1 (Fig. 4, E and F). These results were consistent with the finding that the ESA element is required for BAP1 induction by ROS but not SA treatments (Fig. 3, D and F). Therefore, ICE1 might be involved in transcriptional regulation of BAP1 by temperature and ROS.
The ice1 Mutant Is More Resistant to Virulent Bacterial Pathogen Than the Wild Type
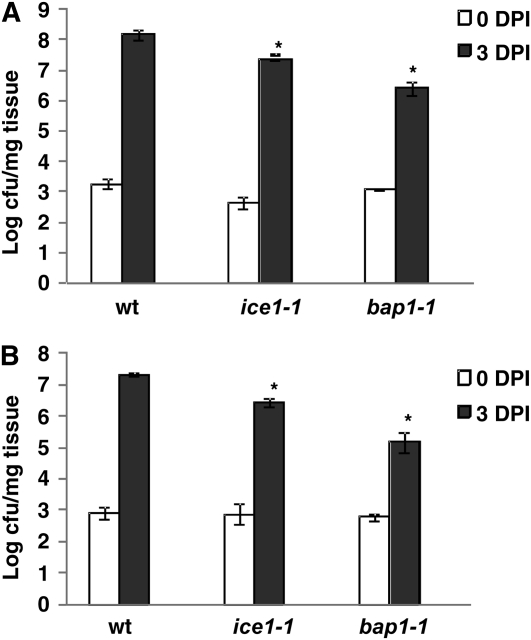
BAP1 is a negative regulator of cell death and SA-mediated disease resistance, and the bap1-1 mutant is more resistant to virulent bacterial pathogens (Yang et al., 2006). To determine if the low induction of BAP1 in the ice1-1 mutant renders the ice1-1 mutant more resistance to virulent pathogens, we assayed growth of Pseudomonas syringae pv tomato DC3000 in the ice1-1 mutant. As the ice1-1 mutant has more stomata that might facilitate bacterial invasion, we inoculated plants by dipping as well as vacuum infiltration to minimize the possible effects of easier entry of bacteria in ice1. In both methods, we saw an enhanced resistance to the bacterial pathogen in ice1. Compared with the wild type at 3 d after inoculation, there was a 7- and 8-fold decrease of bacterial growth in ice1-1 in dipping and infiltration, respectively, while there was, respectively, a 75- and 118-fold decrease in bap1-1 (Fig. 5). Thus, the ice1-1 mutant is more resistant to the bacterial pathogen than the wild type.
Figure 5.
The ice1-1 mutant is more resistant to a virulent P. syringae than the wild type. The virulent pathogen P. syringae pv. tomato DC3000 was inoculated either by vacuum infiltration (A) or dipping (B) at 106 colony-forming units/mL on the wild-type Col-0, ice1-1, and bap1-1 18-d-old plants. The amount of bacteria in plants 0 d postinoculation (DPI) and 3 d postinoculation were measured. Both ice1-1 and bap1-1 exhibited significant reduction of bacterial growth compared with the wild type in both inoculation methods. An asterisk indicates significant differences by the t test (P < 0.05).
DISCUSSION
Earlier studies suggest multiple transcriptional pathways in plant responses to a moderate decrease in temperature. In this study, we analyzed the transcriptional induction of the BAP1 gene by temperature as well as other stimuli in Arabidopsis. BAP1 is a negative regulator of programmed cell death and defense responses and it is induced by multiple environmental stimuli. Cooling from 28°C to 22°C and cold from 22°C to 4°C both induce BAP1, but the former not the latter induction is dependent on SA and PAD4. In addition, BAP1 is induced by other environmental stimuli including wounding, pathogen invasion, and ROS, which offers an opportunity to investigate temperature responses at the transcriptional level and potential interactions among different environmental stimuli.
With a series of promoter deletions, we identified a 35-bp TR element that is necessary and sufficient to confer induction by a moderate temperature cooling. We further showed genetically and biochemically that ICE1 binds to this fragment and regulates the induction of BAP1 by cold and cooling. A previous study revealed that CBFs, that bind the CRT/DRE elements, are directly involved in cooling induction of COR15a (Wang and Hua, 2009). In both cases, transcription factors formerly known to regulate transcription in cold responses are found to regulate transcriptional induction in response to a moderate decrease in temperature. Although two different transcription factors are utilized in cooling induction of COR15a and BAP1, respectively, they are in the same pathway in cold responses with ICE1 acting upstream of CBF3 by binding to its promoter directly. Thus, it appears that signaling cascade for cold responses is operating in cooling responses with more than one output into transcriptional regulation.
Transcriptional induction by a 28°C to 22°C cooling can be SA independent and SA dependent. Cooling induction of COR15a and a few other COR genes does not require SA while that of EDS1 and a few other defense-related genes requires SA (Wang and Hua, 2009). This study finds the SA-dependent induction of BAP1 utilizes ICE1, which raises the possibility that other SA-dependent cooling induction also requires ICE1. Although BAP1 can be induced by SA, this induction is not mediated by the same TR element for cooling. Therefore, SA and ICE likely operate independently on the promoter of BAP1, and they work together to promote its cooling induction. Intriguingly, cold induction of BAP1 utilizes the same TR element as cooling but does not require SA. Although we cannot exclude the possibility that a different motif in the same TR fragment mediates cold induction, it is likely that the same motif mediates both cold and cooling induction. In the latter case, induction is likely weaker by cooling than by cold and an additional factor mediating SA signals could work synergistically with ICE1 to fully activate cooling induction.
ICE1 is identified in this study as a transcription factor that mediates cooling induction of BAP1 possibly through directly binding to the MYC motif in the TR fragment. Originally isolated as a regulator of cold-induced transcription and freezing tolerance, ICE1 was later found to play additional roles in stomata differentiation (Kanaoka et al., 2008). Transcriptome analysis revealed that the ice1 mutation affects gene expression even when plants are not cold treated (Lee et al., 2005), suggesting a broader role of ICE1 in environmental response and development. Previous study of siz1 and ice1 also revealed an up-regulation of SA-related defense response genes such as PR1 (Miura and Ohta, 2010). In this study, we found that the ice1-1 mutant has enhanced resistance to a virulent bacterial pathogen, similarly to the bap1 loss-of-function mutant although at a lesser degree. This phenotype is most likely due to postinvasion resistance as the ice1-1 mutant has more stomata than the wild type that presumably would make it more susceptible to pathogens. It raises a possibility that up-regulation of SA-related defense responses is due to down-regulation of the negative regulator of BAP1 or other regulators in ice1-1.
The cooling induction of BAP1 is likely carried out by ICE1 and its homologs such as SCRM2/ICE2 (Chinnusamy et al., 2003; Kanaoka et al., 2008). The ice1-1 mutation is dominant, possibly interfering with the function of itself and its homologs. We found that cooling induction of BAP1 was still present in the loss-of-function mutants ice1-2 and scrm2-1, suggesting that ICE1 has overlapping functions with SCRM2 or other homologs in regulating BAP1 induction. The lethality of the ice1-2 scrm2-1 double mutant prevented us to vigorously test this hypothesis. Therefore, it has yet to be determined how much ICE1 and its homolog(s) each contribute to this induction.
The TR fragment also mediates BAP1 induction by paraquat and this induction appears to be dependent on ICE1 as well. There are at least three known cis-elements for response to various ROS. One is the ocs-element that is partly required for the induction of GST gene by hydrogen peroxide (Chen and Singh, 1999). The second is the as-1 element in the tobacco (Nicotiana tabacum) GST gene responsible for its induction by ROS (Garretón et al., 2002). The third one is a CORE element responsive to oxidative stress in the promoters of three antioxidant defense genes (Tsukamoto et al., 2005). Although the ROS-responsive element needs to be further defined in the TR fragment, there is an interesting possibility that G box might be a new cis-element responsive to ROS. Intriguingly, ICE1 and its homolog(s) appear to be involved in ROS regulation of BAP1 transcription. How the ROS and temperature signals interact and integrate in transcriptional regulation awaits further investigation.
In sum, we have revealed shared regulators for plants to respond to both moderate and drastic decrease in temperature, suggesting a common mechanism in temperature perception and signaling. In addition, the combinatory regulation by a decrease of temperature and SA on plant defense-related genes might enable plants to acquire enhanced resistance in cooling environment. Further study on the cooling induction of BAP1 might facilitate the understanding of temperature sensing in plants and how temperature signal interacts with other environmental stimuli to regulate gene expression.
MATERIALS AND METHODS
Plant Material and Plant Treatment
Arabidopsis (Arabidopsis thaliana) plants were grown at 22°C or 28°C under 24 h (for growth) or 12 h (for pathogen test) of fluorescent light (100 μmol m−2 s−1) per day with 50% to 70% relative humidity. Arabidopsis seeds were either directly sowed on soil or selected on plates before being transferred to soil.
Arabidopsis plants were exposed to different stimuli. Wounding treatment was done by squeezing leaves with 1-mL needless syringe or cut with a pair of scissors. Temperature-shift assays were performed by moving plants from one growth temperature to another while maintaining other parameters constant in growth chambers. For paraquat induction, a solution of 20 μm paraquat and 0.1% Tween 20 was sprayed onto Arabidopsis plants as previously described (op den Camp et al., 2003). For SA treatment, a solution of 2 mm SA and 0.1% Tween 20 was sprayed on the Arabidopsis plants grown at 22°C. Mock treatment used a solution with 0.1% Tween 20. Seedlings were collected for RNA-blot analysis at 4 h (paraquat) or 1 d (SA) after spray.
Plasmid Construction
Standard molecular techniques were used (Sambrook et al., 1989). A BAP1 promoter fragment of 2.1 kb was amplified from the bacterial artificial chromosome clone F22O13 (from Arabidopsis Biological Resource Center) and fused to the GUS reporter gene in the binary vector pPZPGUS2 (Diener et al., 2000) to generate the pBG2 plasmid. A fragment of 1,662 bp in this promoter was amplified with a BamHI site introduced right before the start codon ATG. The 909-bp BamHI and BglII fragment, the 563-bp BamHI and SpeI fragment, the 255-bp BamHI and EcoRI fragment, and the 169-bp BamHI and SalI fragment were cloned, respectively, into the binary vector pPZPGUS2. Recombinant plasmids were designated as pBG4, pBG5, pBG6, and pBG7, respectively. The pBG8 plasmid contains a 762-bp fragment of the BAP1 promoter (directly amplified from the bacterial artificial chromosome clone) in pZPGUS2.
A 107-bp fragment (−106 to +1) of the cauliflower mosaic virus 35S promoter was amplified by PCR and inserted into the EcoRI site of the pBluescript vector to generate a p35S-mini construct. The 87-bp EcoRI and SalI fragment of the BAP1 promoter was placed in front of the 35S mini promoter in p35S mini to generate the pES construct. Complementary strands of ESA, ESB, and ESC were annealed and ligated to the vector pBluescirpt. Three tandem copies of ESA and four tandem copies of ESB and ESC were obtained in pBluescript and then placed in front of the 35S mini promoter in p35S-mini to generate pESA, pESB, and pESC, respectively. Cassettes of the ES, ESA, ESB, and ESC-35S-mini promoters were each moved into the binary vector pPZPGUS2 to generate pGES, pGESA, pGESB, and pGESC constructs, respectively. These constructs were transformed into Agrobacterium GV3101 (Koncz and Schell, 1986) and transformed into wild-type Col-0 by the Agrobacterium-mediated floral-dip method (Clough and Bent, 1998).
RNA-Blot Analysis
Total RNAs were extracted from 3-week-old plants using TriReagents (Molecular Research) according to the manufacturer’s protocol. Twenty micrograms of RNA for each sample was resolved on 1.2% agarose gels containing 1.8% formaldehyde. Ethidium bromide was used to visualize the rRNA bands to ensure equal loading. RNA gel blots were hybridized with gene-specific, 32P-labeled, single-stranded DNA probes.
GUS Activity Analysis
Plant tissues were lightly fixed half an hour later after wounding and incubated overnight at 37°C in staining solution as described previously (Hua et al., 2001). Tissues were then cleared through ethanol series.
EMSA
The ICE1-GST fusion protein was expressed and purified as previously described (Miura et al., 2007). EMSA was carried out similarly to previously described (Zhu et al., 2003). The DNA fragment containing the ESA element was labeled by the Klenow fragment (New England Biolabs) with 32P-dCTP. Labeled DNA probes of 10 fmol were incubated with 0.2 μg of purified ICE1:GST fusion protein at 25°C for 20 min and the resulting DNA-protein complexes were resolved by electrophoresis on a 4% polyacrylamide gel and visualized by autoradiograph. For the competition experiment, unlabeled competitors were incubated with purified ICE1:GST fusion proteins on ice for 15 min prior to the addition of labeled probe.
ChIP Assay
ChIP experiment was carried out following protocol previously described (Kwon et al., 2005; Sridhar et al., 2006). A BAP1-specific fragment containing the ESA element was amplified by using the following primers: 5′-ATGAACTAACACAGCAAAGAG-3′ and 5′-TCACTTAGGAAGGTGACAAGG-3′. CBF3-specific fragment containing the ICE1 binding site was amplified by using the following primers: 5′-GCACTAAAATGTTACATTTGATC-3′ and 5′-TGGTAATGCCACGTAAACTATAC-3′. PCR reaction was carried out for 40 cycles of 50 s at 94°C, 50 s at 55°C, and 1 min at 72°C.
Acknowledgments
We thank Dr. J. Zhu, Dr. Torri, Dr. K. Miura, and Dr. P. Hasegawa for strains and constructs.
References
- Babbs CF, Pham JA, Coolbaugh RC. (1989) Lethal hydroxyl radical production in paraquat-treated plants. Plant Physiol 90: 1267–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Ahn JH, Weigel D. (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33: 168–171 [DOI] [PubMed] [Google Scholar]
- Chen W, Singh KB. (1999) The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J 19: 667–677 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conley TR, Park SC, Kwon HB, Peng HP, Shih MC. (1994) Characterization of cis-acting elements in light regulation of the nuclear gene encoding the A subunit of chloroplast isozymes of glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. Mol Cell Biol 14: 2525–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Navarrete MH, Canovas JL. (1989) Cell size of proliferating plant cells increases with temperature: implications in the control of cell division. Exp Cell Res 185: 277–282 [DOI] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR. (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X. (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hubbard KE, Hotta CT, Dodd AN, Webb AA. (2006) How plants tell the time. Biochem J 397: 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garretón V, Carpinelli J, Jordana X, Holuigue L. (2002) The as-1 promoter element is an oxidative stress-responsive element and salicylic acid activates it via oxidative species. Plant Physiol 130: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P. (2004) cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Du JS, de Torres Zabala M, Beggs K, Smith C, Holdsworth M, Bevan M. (1994) Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J 5: 815–826 [DOI] [PubMed] [Google Scholar]
- Hua J. (2009) From freezing to scorching, transcriptional responses to temperature variations in plants. Curr Opin Plant Biol 12: 568–573 [DOI] [PubMed] [Google Scholar]
- Hua J, Grisafi P, Cheng SH, Fink GR. (2001) Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev 15: 2263–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki H, Maxson JM, Woodson WR. (1994) An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc Natl Acad Sci USA 91: 8925–8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU. (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD. (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Kwon CS, Chen C, Wagner D. (2005) WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev 19: 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Huang B. (2004) Thermotolerance and antioxidant systems in Agrostis stolonifera: involvement of salicylic acid, abscisic acid, calcium, hydrogen peroxide, and ethylene. J Plant Physiol 161: 405–413 [DOI] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK. (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Woodward FI. (1988) Plants and Temperature. Society for Experimental Biology, Cambridge, England [Google Scholar]
- Meshi T, Iwabuchi M. (1995) Plant transcription factors. Plant Cell Physiol 36: 1405–1420 [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ohta M. (2010) SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J Plant Physiol 167: 555–560 [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. (2008) Temperature perception and signal transduction in plants. New Phytol 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. (1989) Molecular Cloning, A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z. (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166 [DOI] [PubMed] [Google Scholar]
- Sung DY, Kaplan F, Lee KJ, Guy CL. (2003) Acquired tolerance to temperature extremes. Trends Plant Sci 8: 179–187 [DOI] [PubMed] [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Morita S, Hirano E, Yokoi H, Masumura T, Tanaka K. (2005) A novel cis-element that is responsive to oxidative stress regulates three antioxidant defense genes in rice. Plant Physiol 137: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E. (1991) The roles of heat-shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620 [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L. (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wang Y, Bao Z, Zhu Y, Hua J. (2009) Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact 22: 498–506 [DOI] [PubMed] [Google Scholar]
- Wang Y, Hua J. (2009) A moderate decrease in temperature induces COR15a expression through the CBF signaling cascade and enhances freezing tolerance. Plant J 60: 340–349 [DOI] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yang H, Li Y, Hua J. (2006) The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J 48: 238–248 [DOI] [PubMed] [Google Scholar]
- Yang H, Yang S, Li Y, Hua J. (2007) The Arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol 145: 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Hua J. (2004) A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16: 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Cai XL, Wang ZY, Hong MM. (2003) An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J Biol Chem 278: 47803–47811 [DOI] [PubMed] [Google Scholar]