Abstract
Syringolin A (SylA) is a nonribosomal cyclic peptide produced by the bacterial pathogen Pseudomonas syringae pv syringae that can inhibit the eukaryotic proteasome. The proteasome is a multisubunit proteolytic complex that resides in the nucleus and cytoplasm and contains three subunits with different catalytic activities: β1, β2, and β5. Here, we studied how SylA targets the plant proteasome in living cells using activity-based profiling and imaging. We further developed this technology by introducing new, more selective probes and establishing procedures of noninvasive imaging in living Arabidopsis (Arabidopsis thaliana) cells. These studies showed that SylA preferentially targets β2 and β5 of the plant proteasome in vitro and in vivo. Structure-activity analysis revealed that the dipeptide tail of SylA contributes to β2 specificity and identified a nonreactive SylA derivative that proved essential for imaging experiments. Interestingly, subcellular imaging with probes based on epoxomicin and SylA showed that SylA accumulates in the nucleus of the plant cell and suggests that SylA targets the nuclear proteasome. Furthermore, subcellular fractionation studies showed that SylA labels nuclear and cytoplasmic proteasomes. The selectivity of SylA for the catalytic subunits and subcellular compartments is discussed, and the subunit selectivity is explained by crystallographic data.
The interaction between Arabidopsis (Arabidopsis thaliana) and the bacterial pathogen Pseudomonas syringae is an important model system to study plant-pathogen interactions. Besides Arabidopsis, strains of this bacterial pathogen cause various diseases on a wide range of plant species, including fruit trees, tomato (Solanum lycopersicum), and other crop plants (Hirano and Upper, 2000). P. syringae manipulates its host by injecting effector proteins through the type III secretion system into the host cell (Göhre and Robatzek, 2008; Boller and He, 2009; Büttner and He, 2009; Cunnac et al., 2009; Lewis et al., 2009). Many of these effectors suppress basal defense responses (Cunnac et al., 2009; Guo et al., 2009).
Besides type III effectors, P. syringae strains also produce different small molecule effectors to manipulate the host. Coronatine from P. syringae pv tomato DC3000, for example, induces the jasmonate signaling cascade, provoking the opening of stomata to overcome preinvasive immunity (Melotto et al., 2006). Other examples are tabtoxin from P. syringae pv tabaci, which inhibits Gln synthetase, and phaseolotoxin from P. syringae pv phaseolicola, which causes Arg deficiency by inhibiting Orn carbamoyl transferase (Bender et al., 1999). It is evident from these studies that small molecule effectors are equally important for bacterial diseases as type III effectors. Yet, studies on small molecule effectors and their targets are limited.
Here, we study the action of syringolin A (SylA), a small molecule effector produced by P. syringae pv syringae (Psy). SylA is a nonribosomal cyclic peptide that contributes to the development of disease symptoms (Groll et al., 2008). SylA is a 493-D molecule that consists of a 12-member macrocycle and a dipeptide tail (Wäspi et al., 1999). The ring contains an α,β-unsaturated amide and a second double bond. This second unsaturated bond is absent in SylB, a minor, additional metabolite produced by Psy (Wäspi et al., 1999). The dipeptide tail contains two l-Vals, linked through an ureido bond. SylA is produced by Psy by nonribosomal peptide and polyketide synthetases encoded by the sylC and sylD biosynthesis genes and presumably secreted from bacteria by the product of sylE, which encodes a transporter-like protein (Amrein et al., 2004; Imker et al., 2009; Ramel et al., 2009).
SylA inhibits the eukaryotic 26S proteasome (Groll et al., 2008). The 26S proteasome is a large, multicomponent protease residing in the cytosol and nucleus and consists of a 20S core protease and a 19S regulatory particle. The 19S regulatory particle accepts ubiquitinated substrates, unfolds them, and feeds them into the core protease (Kurepa and Smalle, 2008). The 20S core protease consists of four rings of seven subunits each that make a hollow cylinder. The inner two rings consist of β-subunits (Groll et al., 1997). The proteolytic activity is located in the inner cavity of the cylinder and resides in three β-subunits of the inner two rings. Each of these subunits has slightly different catalytic activities: β1 cleaves after acidic residues (caspase-like activity); β2 after basic residues (trypsin-like activity); and β5 after hydrophobic residues (chymotrypsin-like activity; Dick et al., 1998). Together, these subunits degrade proteins into three- to 20-amino-acid-long peptides that are released into the cytosol or nucleus (Kurepa and Smalle, 2008).
Crystallographic data revealed that the α,β-unsaturated amide of SylA is attacked by the N-terminal Thr of the catalytic β-subunits, resulting in an irreversible, covalent ether bond (Groll et al., 2008). Further studies showed that SylA has antiapoptotic properties in mammalian cells and therefore is a promising novel anticancer drug, having different properties when compared with bortezomib, a proteasome inhibitor that is currently used in the clinic as an anticancer drug (Coleman et al., 2006; Clerc et al., 2009a).
That SylA inhibits the plant proteasome in vivo has been demonstrated by its ability to promote the accumulation of cyclin-GUS fusion proteins in root tips (Groll et al., 2008). Beyond this, little is known of how this small molecule effector interacts with its natural host target and if the proteasome is the only target in plants. Here, we study the action of SylA in plants using activity-based probes, which are reporter-tagged inhibitors that react with active site residues of enzymes in a mechanism-dependent manner. The irreversible covalent bond facilitates the display of labeled enzymes on protein gels and/or the identification of labeled proteins by affinity capture and mass spectrometry (Cravatt et al., 2008). We recently introduced proteasome activity profiling in plants using MV151, a fluorescent vinyl sulfone probe that labels the β1-, β2-, and β5-subunits of the proteasome and several papain-like Cys proteases (PLCPs; Gu et al., 2010).
Activity-based protein profiling is a simple and robust approach that is now also used in plant science (Kolodziejek and van der Hoorn, 2010). In this study, we further developed the activity-based protein profiling technology to facilitate detailed studies of the selectivity of SylA on its natural host target, the plant proteasome. To this end, we introduced novel, selective proteasome probes and established procedures for in vivo proteasome activity profiling and imaging. With these tools, and using SylA derivatives, we examined the subunit selectivity and subcellular targeting of SylA in living plant cells.
RESULTS
Comparison of Three Proteasome Probes in Vitro
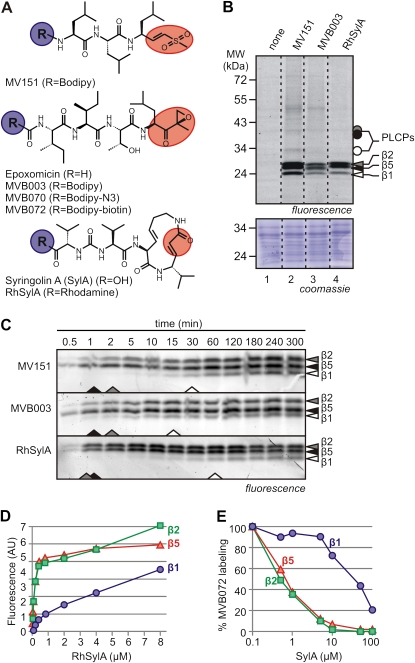
In this study, we used three different probes that target the proteasome (Fig. 1A; Supplemental Fig. S1). All three probes carry a fluorescent reporter tag but differ in their reactive group. MV151 contains a vinyl sulfone and has previously been used to investigate plant and animal proteasomes (Verdoes et al., 2006; Gu et al., 2010). MVB003 is based on the highly selective proteasome inhibitor epoxomicin and carries an epoxyketone reactive group (Meng et al., 1999). RhSylA is a fluorescent SylA derivative, previously used on animal extracts (Clerc et al., 2009a), carrying a reactive Michael system in the ring structure. Each of these reactive groups binds covalently and irreversibly with the N-terminal active site Thr of the catalytic subunits of the proteasome, but through distinct molecular mechanisms (Groll et al., 2008; Huang and Chen, 2009). In addition to MVB003, we used two more epoxomicin-based probes, which only differ from MVB003 in the reporter tags. MVB070 contains bodipy carrying an azide minitag for “click chemistry,” and MVB072 contains bodipy carrying a biotin for affinity purification (Supplemental Fig. S1). MVB003, MVB070, and MVB072 cause nearly identical labeling profiles (data not shown), but only MVB003 was used for in vivo imaging experiments.
Figure 1.
Characterization of three probes for in vitro proteasome labeling. A, Inhibitors and probes used in this study. Probes and inhibitors are based on vinyl sulfone (top), epoxomicin (middle), or SylA (bottom). The reactive groups are indicated in red. Probes carry a reporter tag at position R (blue). B, Comparison of in vitro labeling with the three probes. Extracts from cell cultures were labeled for 2 h with 2 μm MV151, RhSylA, or MVB003. Proteins were detected from protein gels by fluorescence scanning and Coomassie Brilliant Blue staining. Dashed lines indicate lanes combined from the same gel. C, Time course of proteasome labeling. Arabidopsis leaf extracts were labeled with 2 μm MV151, MVB003, or RhSylA, and samples were taken at different time points and quenched in SDS sample buffer. Fluorescent proteins were detected by fluorescence scanning. Arrowheads at the bottom of the gels indicate the probe concentrations required for half-maximal labeling. D, Labeling by SylA at different probe concentrations. Arabidopsis leaf extract was labeled with various concentrations of RhSylA, and signals of the different subunits were quantified from protein gels and plotted against the RhSylA concentration. E, Suppression of proteasome labeling at different SylA concentrations. Arabidopsis leaf extracts were preincubated with various SylA concentrations and labeled with 2 μm MVB072. Signals of the different subunits were quantified from protein gels and plotted against SylA concentration. Fluorescence was normalized to the no-inhibitor control.
To determine in vitro labeling with the probes, extracts from Arabidopsis cell cultures were incubated with the probes for 2 h. Proteins were separated on protein gels and analyzed for fluorescently labeled proteins by fluorescence scanning. MV151 labeling of Arabidopsis extracts causes three strong signals in the 25-kD range, representing the catalytic subunits of the proteasome (Fig. 1B, lane 2; Gu et al., 2010). MV151 labeling also causes weak signals at 30 and 40 kD, representing PLCPs (Gu et al., 2010). MVB003 and RhSylA labeling of extracts causes strong signals in the 25-kD range, similar to those of MV151-labeled extracts (Fig. 1B, lanes 3 and 4). MVB003-labeled proteins run slightly lower on the gel, probably because MVB003 has a smaller molecular mass compared with MV151 and RhSylA.
To investigate the potential β-subunit selectivity of the different probes, we performed time course labeling experiments in vitro. Labeling in extracts occurs within minutes for all three probes (Fig. 1C). MV151 and MVB003 label β5 within 1 min, followed quickly by β2, whereas β1 becomes labeled within 15 min (for MVB003) or 30 min (for MV151; Fig. 1C). In contrast, RhSylA labels β2 and β5 simultaneously within minutes, but β1 labeling takes 1 h, which is longer when compared with MV151 and MVB003 labeling (Fig. 1C). These data show that MV151 and MVB003 preferentially target β5, whereas RhSylA preferentially targets β5 and β2.
To further validate the subunit specificity of RhSylA, Arabidopsis leaf extracts were labeled for 2 h with different concentrations of RhSylA. Both β2 and β5 reach maximum labeling at concentrations below 1 μm, whereas β1 labeling is not saturated even at 8 μm (Fig. 1D), consistent with a slow labeling of β1. To determine if SylA itself has low affinity for β1, Arabidopsis leaf extracts were preincubated with various SylA concentrations and then labeled with MVB072. This showed that labeling of β2 and β5 is prevented at low SylA concentrations (concentration for 50% inhibition = 0.36 and 0.31 μm, respectively), whereas inhibition of β1 requires high SylA concentrations (Fig. 1E). Taken together, these experiments demonstrate that SylA and RhSylA preferentially target β2- and β5-subunits of the proteasome.
Comparison of Three Proteasome Probes in Vivo
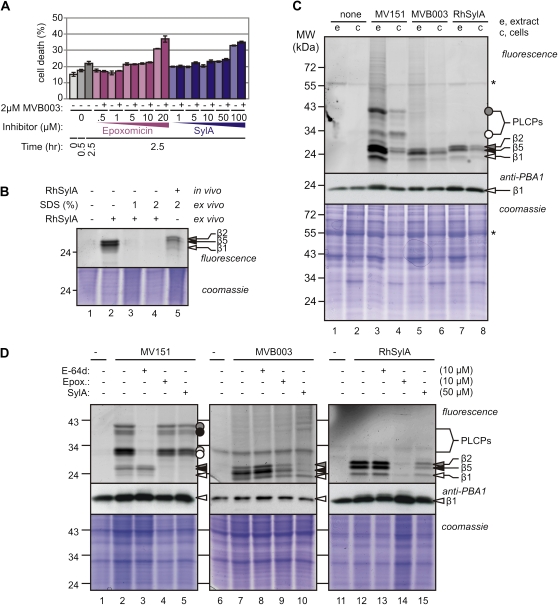
To establish in vivo labeling in Arabidopsis cell cultures, the toxicity of the probes and inhibitors was first determined using Evans blue staining (Kaffarnik et al., 2009). Concentrations of 100 μm SylA or 20 μm epoxomicin caused significant cell death when incubated for 2.5 h with cell culture (Fig. 2A). Lower concentrations of inhibitors (50 μm SylA or 10 μm epoxomicin) or addition of probes (2 μm) did not affect cell viability and were used for in vivo inhibition assays (Fig. 2A).
Figure 2.
Characterization of in vivo proteasome labeling. A, Viability of cell culture after incubations with probes and inhibitors, measured with Evans blue staining and compared with the heat shock control. Cell cultures were preincubated for 30 min with various inhibitor concentrations and labeled for 2 h with 2 μm MVB003. Cell death was quantified from Evans blue staining and compared with a heat shock control (=100% cell death). Error bars represent se of three measurements. This experiment was repeated twice with similar results. B, SDS during extraction prevents ex vivo labeling. Lanes 1 to 4, Cell cultures were preincubated with DMSO and washed, and proteins were extracted in 2 μm RhSylA containing 0%, 1%, or 2% SDS (lanes 2, 3, and 4, respectively); lane 5, cell cultures were preincubated with 2 μm RhSylA, and proteins were extracted in 2% SDS. C, Comparison of in vitro and in vivo labeling. Cell cultures (c; in vivo) or extracts of cell cultures (e; in vitro) were labeled for 2 h with 2 μm MV151, RhSylA, or MVB003. Proteins were extracted and detected from protein gels by fluorescence scanning and Coomassie Brilliant Blue staining. Differences in protein amounts are caused by differences in protein extraction efficiency from cell cultures. Asterisks indicate background signal. D, In vivo inhibition confirms target selectivity. Arabidopsis cell cultures were preincubated with 10 μm E-64d or epoxomicin or 50 μm SylA for 30 min and then labeled with 2 μm MV151, RhSylA, or MVB003 for 2 h. Proteins were extracted under denaturing conditions, separated on protein gels, and detected by fluorescence scanning and Coomassie Brilliant Blue staining.
We next established procedures to prevent ex vivo labeling. In vitro proteasome labeling with the probes is so quick (Fig. 1C) that a significant labeling occurs when probes are added during protein extraction (Fig. 2B, lane 2). However, extraction in the presence of 1% or 2% SDS completely prevented ex vivo labeling (Fig. 2B, lanes 3 and 4). Signals that appear after labeling in vivo, followed by extraction in 2% SDS, are therefore only caused by in vivo labeling (Fig. 2B, lane 5). For all subsequent in vivo labeling assays, we used 2% SDS in the extraction procedure.
In vivo labeling of Arabidopsis cell cultures with MV151, MVB003, and RhSylA causes profiles that are different from the in vitro labeling profiles (Fig. 2C). Most obvious are the relatively strong signals at 30 and 40 kD in the MV151 profile (Fig. 2C, lane 4). These data are consistent with the observation that PLCPs are more active in vivo when compared with in vitro (Kaschani et al., 2009). Second, the proteasome-derived signals are significantly weaker for all three probes when compared with in vitro labeling (Fig. 2C), but this correlates with lower protein levels. However, despite differences in intensities, the profiles of the proteasome subunits are not significantly changed when compared with in vitro labeling. MV151 and MVB003 still preferentially label β5 (Fig. 2C, lanes 4 and 6), whereas RhSylA preferentially labels both β2 and β5 (Fig. 2C, lane 8). Signals in the 30- and 40-kD range or any other molecular mass are absent with MVB003 or RhSylA, indicating that these probes do not label PLCPs in vivo and are specific for the catalytic subunits of proteasome.
In Vivo Inhibition Studies Confirm the Identities of Labeled Proteins
To verify the identity of the in vivo labeled signals, we preincubated cell cultures with various inhibitors and then labeled the cultures with the different probes. As inhibitors, we used sublethal concentrations of SylA, E-64d, and epoxomicin. Preincubation with E-64d prevents MV151 labeling of the 30- and 40-kD signals of MV151 (Fig. 2D, lane 3), consistent with the fact that these signals are derived from PLCPs (Kaschani et al., 2009; Gu et al., 2010). Pretreatment with E-64d has no influence on the labeling of MVB003 or RhSylA (Fig. 2D, lanes 8 and 13). Pretreatment with epoxomicin or SylA selectively prevents labeling of all three 25-kD signals in the MV151 profile (Fig. 2D, lanes 4 and 5). In contrast, epoxomicin significantly suppresses overall MVB003 labeling (Fig. 2D, lane 9) and prevents RhSylA labeling (Fig. 2D, lane 14). Interestingly, SylA pretreatment suppresses MVB003 labeling of β5 and β2 but not β1 (Fig. 2D, lane 10) and suppresses RhSylA labeling of all signals (Fig. 2D, lane 15). In summary, these data confirm that the signals at 30 and 40 kD are PLCPs and the signals at 25 kD are from the proteasome. These data also support the previous notion that MVB003 and MV151 preferentially target β5 but eventually label also β2 and β1, whereas RhSylA targets both β2 and β5 and labels β1 only to a low extent.
Epoxomicin-Based Probes Target β5 in Vivo
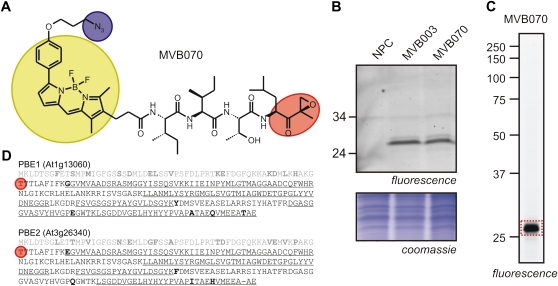
MVB003 preferentially labels a 25-kD protein that we assumed is β5. However, since this signal runs faster compared with MV151 and RhSylA signals, we determined the identity of the proteins labeled in vivo by epoxomicin-based probes. We used MVB070 (Fig. 3A), an azide (N3) minitagged version of MVB003, which causes identical labeling profiles when compared with MVB003 (Fig. 3B). Cell cultures were labeled with MVB070, and proteins were extracted under denaturing conditions. Azide-labeled proteins were biotinylated with biotin-alkyne using copper (I)-catalyzed click chemistry reaction (Speers and Cravatt, 2004; Kaschani et al., 2009). Biotinylated proteins were purified and separated on gels, and the 25-kD signal (Fig. 3C) was excised and digested with trypsin (Kaschani et al., 2009). Peptides were analyzed by mass spectrometry and matched to the Arabidopsis protein database. Proteasome subunits PBE1 (At1g13060) and PBE2 (At3g26340) were identified with high coverage and multiple unique peptides (Fig. 3D). Both PBE1 and PBE2 are β5 catalytic subunits that only differ in a few amino acids (Fig. 3D, boldface). These data confirm that the epoxomicin-based probes preferentially label the β5-subunit in vivo.
Figure 3.
Identification of epoxomicin-labeled proteins in vivo. A, Structure of the minitagged fluorescent epoxomicin derivative MVB070. Red, Reactive group; yellow, fluorescent bodipy group; blue, azide minitag. B, Comparison of labeling profiles using MVB003 and MVB070. Arabidopsis cell cultures were labeled in vivo with MVB003 and MVB070, and fluorescently labeled proteins were detected from protein gels using fluorescence scanning. NPC, No-probe control. C, Identification of in vivo labeled proteins. Cell cultures were labeled with MVB070. Proteins were extracted under denaturing conditions, and azide-labeled proteins were biotinylated using click chemistry. Biotinylated proteins were purified and separated on protein gels, and the fluorescent signal at approximately 25 kD was excised, digested with trypsin, and identified by mass spectrometry. D, Identified peptides are underlined in the full-length protein sequences of two different β5-subunits, PBE1 and PBE2. Differences between PBE1 and PBE2 are indicated in boldface; the prodomain (first line) is in gray, and the N-terminal catalytic Thr is circled in red.
Structure-Activity Relationships of SylA Derivatives
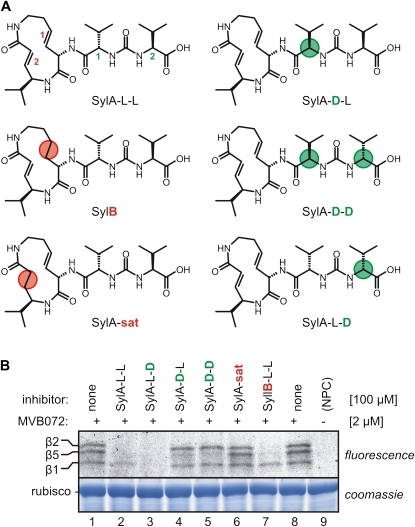
SylA contains a 12-member ring with two double bonds. A natural SylA variant, SylB, differs from SylA by having one of these bonds saturated (Fig. 4A). Preincubation of leaf extracts with SylB suppresses MVB072 labeling, but concentrations needed for inhibition are higher when compared with SylA (Fig. 4B, lane 7), similar to the weaker binding of SylB to the human proteasome (Clerc et al., 2009b). To verify the importance of the second unsaturated bond, we tested a SylA derivative where this bond is saturated (SylA-sat; Fig. 4A). Preincubation with SylA-sat does not prevent proteasome labeling (Fig. 4B, lane 6), indicating that the double bond is important for proteasome inhibition. The importance of this double bond is consistent with the proposed inhibition mechanism because this is the Michael system that is attacked by the catalytic Thr of the proteasome (Groll et al., 2008).
Figure 4.
Structure-activity relationship of SylA derivatives. A, Structures of SylA derivatives. Differences with the naturally occurring SylA-L-L are indicated with circles and concern double bonds 1 and 2 (red) and stereocenters in Val-1 and Val-2 (green). B, Inhibition of labeling by SylA derivatives. Arabidopsis leaf extracts were preincubated for 30 min with 100 μm SylA derivatives and then labeled for 2 h with 1.6 μm MVB072.
Besides the 12-member ring, SylA also contains two l-Val amino acids linked through an ureido bond. To test the importance of the conformation of these two Vals, stereoisomers were generated and tested. SylA-D-L and SylA-D-D (Fig. 4A), which both carry d-Val at the first position (position 1), are able to inhibit β5 but not β1 or β2 (Fig. 4B, lanes 4 and 5). In contrast, SylA-L-D, which carries a d-Val at position 2 (Fig. 4A), prevents proteasome labeling, similar to natural SylA (SylA-L-L; Fig. 4B, lane 3), indicating that the conformation of the Val at position 2 is not important for the selectivity of proteasome inhibition by SylA. Thus, the Michael system is essential for overall reactivity, whereas specificity for inhibition of the β2-subunit requires the l-configuration of the Val at position 1.
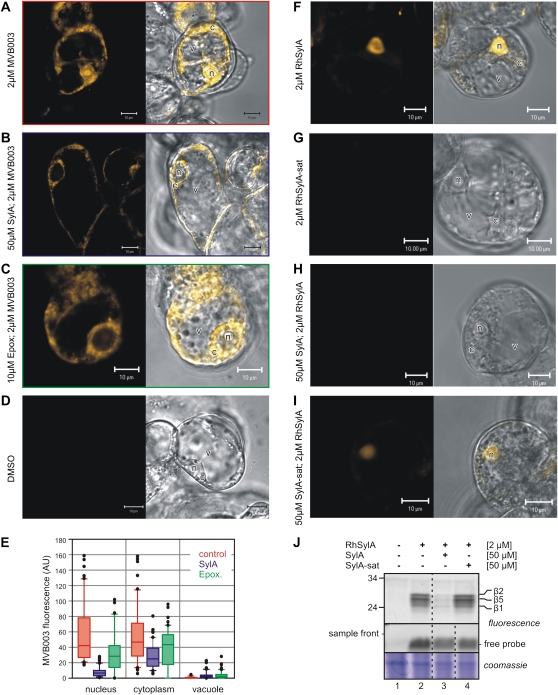
SylA Suppresses MVB003 Fluorescence Mostly in the Nucleus
We next used MVB003 to image proteasome labeling of Arabidopsis cell cultures by confocal microscopy. MV151 was not used, since this probe also labels PLCPs in vivo (Fig. 2). Cell cultures were incubated with 2 μm MVB003 and subsequently washed before imaging. MVB003 labeling causes fluorescence in the nucleus and cytoplasm, but not in the vacuole, consistent with the subcellular location of the proteasome (Fig. 5A). Preincubation with a sublethal dose of SylA and epoxomicin suppresses fluorescence in the nucleus and in the cytoplasm (Fig. 5, B and C, respectively). However, in contrast to epoxomicin, which suppresses overall fluorescence, SylA suppresses mostly nuclear fluorescence. Quantification of the fluorescence over multiple images confirms that MVB003 fluorescence is suppressed upon preincubation with SylA mostly in the nucleus, but also in the cytoplasm (Fig. 5E, blue bars). In contrast, preincubation with epoxomicin only moderately suppresses fluorescent signals in the nucleus and cytoplasm (Fig. 5E, green bars). Interestingly, these data suggest that SylA suppresses proteasome labeling mostly in the nucleus.
Figure 5.
SylA and RhSylA accumulate in the nucleus. A to D, Imaging of cells with MVB003 after preincubation with and without SylA or epoxomicin. Arabidopsis cell cultures were preincubated for 30 min with DMSO, 50 μm SylA, or 10 μm epoxomicin and labeled for 2 h with 2 μm MVB003. Cells were washed and imaged by confocal microscopy. E, Quantification of fluorescence images. Fluorescence intensities in the nucleus, vacuole, and cytoplasm were quantified at 10 positions per cell and for 10 cells and represented in a box plot. F to I, Imaging of cells with RhSylA and RhSylA-sat after preincubation with and without SylA or SylA-sat. Arabidopsis cell cultures were preincubated for 30 min with DMSO or 50 μm SylA or SylA-sat and labeled for 2 h with 2 μm RhSylA or RhSylA-sat. Cells were washed and imaged by confocal microscopy. J, Analysis of labeled proteins and unlabeled probe from cell cultures labeled as described in F to I. Proteins were extracted from labeled cells in the presence of 2% SDS and analyzed on protein gels. Detection was done using fluorescence scanning and Coomassie Brilliant Blue staining. The sample front is shown, since it contains unlabeled probe. c, Cytoplasm; n, nucleus; v, vacuole.
Accumulation of RhSylA in the Nucleus Requires a Reactive Ring System
To further investigate a potential subcellular targeting by SylA, we used RhSylA for in vivo imaging. Importantly, fluorescence caused by RhSylA accumulates in the nucleus, whereas only weak fluorescence resides in the cytoplasm (Fig. 5F). The RhSylA colocalizes with Hoechst33342, which stains the nuclei of living cells (Supplemental Fig. S2). To determine if the signal depends on the reactivity of SylA, we generated a rhodamine-tagged version of SylA-sat, which lacks the reactive Michael system and is unable to compete with proteasome labeling (Fig. 4). Imaging of cells incubated with RhSylA-sat did not show an accumulation of nuclear fluorescence (Fig. 5G), suggesting that the nuclear fluorescence observed with RhSylA represents RhSylA that is covalently bound to the proteasome. Furthermore, nuclear fluorescence of RhSylA could be suppressed by preincubation with 50 μm SylA but not with SylA-sat (Fig. 5, H and I, respectively). Addition of 50 μm SylA after preincubation with 2 μm RhSylA does not suppress proteasome labeling and nuclear fluorescence, indicating that nuclear accumulation of RhSylA is irreversible (data not shown). Analysis of the proteins extracted from the labeled cell cultures confirmed that RhSylA labels the proteasome and that RhSylA labeling can be suppressed by SylA but not by SylA-sat (Fig. 5J). Notably, significant amounts of free, unreacted probe were found in the frontier of the protein gel (Fig. 5J), indicating that some of the free probe stayed inside the cells after washing. However, since free probe is also present in the absence of nuclear fluorescence (Fig. 5, H and J, lane 3), the free probe is probably distributed throughout the cells. In conclusion, these data show that RhSylA accumulates in the nucleus. That nuclear accumulation can be prevented by adding excess SylA before, but not during, the labeling indicates that nuclear RhSylA accumulation is irreversible.
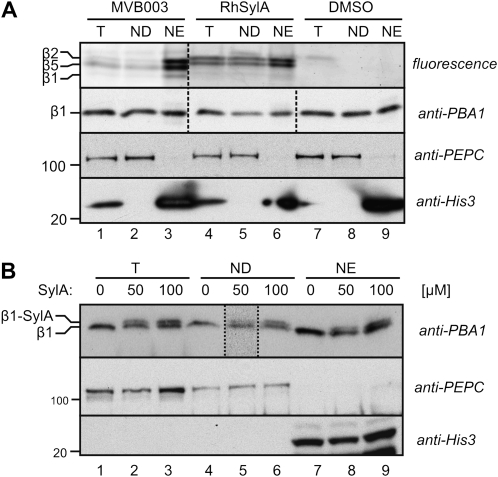
SylA Labels Both the Nuclear and Cytoplasmic Proteasomes
To verify the subcellular targeting biochemically, subcellular fractionation experiments were performed. We labeled cell cultures with and without MVB003 and RhSylA and generated nuclei-enriched (NE) and nuclei-depleted (ND) fractions. Subcellular markers PEPC for cytoplasmic proteins and histone H3 for nuclear proteins confirmed that the fractions were not cross-contaminated (Fig. 6A). The proteasome subunit PBA1 was detected in both the ND and NE fractions, and the relative signals indicate that only 5% of the cellular proteasomes were localized in the nucleus, given the fact that the NE fraction was 10× concentrated compared with the ND fraction (Fig. 6A). Importantly, treatment with MVB003 or RhSylA does not affect the PBA1 levels in the different compartments, indicating that these probes do not influence the subcellular distribution of the proteasome. Analysis of fluorescently labeled proteins reveals that RhSylA labels both β2- and β5-subunits in the nucleus and the cytoplasm, but the signals in the nucleus are relatively stronger (Fig. 6A, lanes 5 and 6). Surprisingly, MVB003 also causes relatively strong labeling of the nuclear proteasome (Fig. 6A, lane 3), which is in contrast to the strong fluorescence of MVB003 in the cytoplasm (Fig. 5).
Figure 6.
RhSylA and SylA label the proteasome in both the nucleus and cytoplasm. A, Nuclear fractionation of MVB003- and RhSylA-labeled cell cultures. Arabidopsis cell cultures were incubated with 2 μm MVB003 or RhSylA or DMSO for 2 h. Cells were homogenized, and total extracts (T) were separated into ND and NE fractions. Fluorescent proteins were detected by in-gel fluorescence scanning, and proteasome subunit PBA1, cytoplasmic marker PEPC, and nuclear marker histone-3 were detected by western blotting on the same samples. B, Nuclear fractionation of SylA-labeled cell cultures. Arabidopsis cell cultures were incubated with 50 and 100 μm SylA or DMSO for 2 h. Cells were homogenized, and T were separated into ND and NE fractions. Labeling of the β1-subunit was visualized as a shift on the western blot probed with anti-PBA1 antibody. The image of this blot was stretched vertically. A longer exposure is shown for the lane bordered by dashed lines. Antibodies against PEPC and histone-3 were used as markers for the cytoplasmic and nuclear proteins, respectively. The NE fractions correspond to 20× more tissue when compared with the T and ND samples.
To investigate if SylA itself targets the nuclear proteasome, we took advantage of the fact that covalent labeling of the β1-subunit causes a shift on the western blot with anti-PBA1 antibody (Gu et al., 2010). To maximize β1 labeling with SylA, we incubated cell cultures with 50 and 100 μm SylA. Even though the difference in molecular mass is small, the SylA-β1 conjugate is clearly separated from the unreacted β1, since a signal with a slightly higher molecular mass appears in total extracts of SylA-treated cells (Fig. 6B, lanes 1–3). Subcellular fractionation of this sample into ND and NE fractions showed that the SylA-β1 conjugates occur in both the cytoplasmic and nuclear fractions (Fig. 6B, lanes 4–9). The ratio of the SylA-β1 when compared with the unreacted β1 is similar between the nuclear and cytoplasmic fractions, indicating that SylA labels the proteasome in both the cytoplasm and nucleus.
DISCUSSION
Through a thorough characterization of in vivo and in vitro profiling and imaging with three unrelated proteasome probes, we have established new procedures for proteasome studies and determined the subunit and subcellular specificity of the proteasome inhibitor SylA, a bacterial small molecule effector released by some P. syringae strains during infection.
SylA Targets β2 and β5 Catalytic Subunits of the Plant Proteasome
We found that SylA preferentially targets only two of the three catalytic subunits of the plant proteasome. RhSylA preferentially labels the β2- and β5-subunits in vitro during short labeling times and at low RhSylA concentrations (Fig. 1, C and D). The same subunit selectivity by RhSylA was observed in vivo (Figs. 2, C and D, and 6A). The subunit selectivity does not reside in the reporter tag, since we observed that SylA itself preferentially competes with labeling on β2 and β5, both in vitro (Fig. 1E) and in vivo (Fig. 2D). The subunit selectivity is different from that of MV151 and MVB003, which preferentially label β5. β1 labeling is slow for all probes, although β1 is best labeled by MVB003 or MV151 (Fig. 1C).
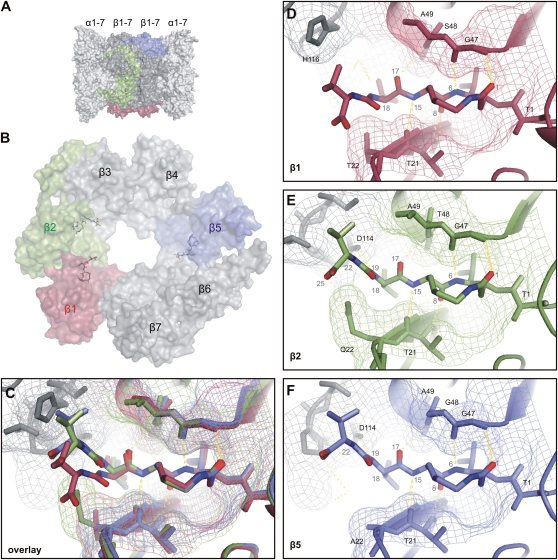
The subunit selectivity of SylA was also observed with studies on the yeast proteasome (Groll et al., 2008) and can be explained using the crystal structure of the yeast proteasome inhibited by SylA (Groll et al., 2008; Protein Data Bank code 2ZCY). The crystal structure of the 20S yeast proteasome contains six SylA molecules, three on each of the two middle rings of β-subunits (Fig. 7A). SylA is covalently bound to the N-terminal Thr of β1, β2, and β5, and the dipeptide tail of SylA also interacts with the adjacent subunit (Fig. 7B). The structure of the adjacent subunits has important implications for how SylA can bind to each of the three binding pockets. Overlay of the SylA structures shows that the dipeptide tail of SylA is pushed downward when bound to the β1-subunits but not when bound to the β2- and β5-subunits (Fig. 7C). This is caused by a bulky His-116 side chain in the subunit adjacent to the β1-subunit that makes the β1 binding pocket smaller when compared with that of the β2- and β5-subunits (Fig. 7, D–F). Consequently, SylA bound to the β1 binding pocket is unable to make a hydrogen bond with Asp-114 of the adjacent subunit, which is an important interaction of SylA bound to the binding pocket of β2 and β5 (Fig. 7, D–F). The presence of the Asp-114 interaction in β2 and β5 binding pockets explains why SylA preferentially targets the β2- and β5-subunits. Since many properties including His-116 and Asp-114 are conserved in the proteasome subunits of Arabidopsis, it seems likely that this interpretation from the yeast crystal structure might also apply for the Arabidopsis proteasome.
Figure 7.
Affinities of SylA derivatives explained by crystallographic data. A, Structure of the 20S core protease of the yeast proteasome. The core protease of the proteasome consists of four rings of α- and β-subunits: the outer rings of seven α-subunits each (light gray) and the inner rings of seven β-subunits each (gray and colors). B, One ring of seven β-subunits shown from the side of the inner cavity, with three SylA molecules bound to the catalytic subunits: β1 (red), β2 (green), and β5 (blue). Please note that the tail of SylA interacts with the adjacent subunit. C, Overlay of SylA bound to the three catalytic subunits. Please note the distinct conformation of the tail of SylA bound to β1 (red). D, SylA bound to β1. SylA (middle, ball and stick) is bound to Thr-1 (T1) of the β1-subunit (red). Hydrogen bonding between SylA and Thr-21, Gly-47, and Ser-48 exist, but His-116 of the adjacent β2-subunit (gray) pushes the tail of SylA downward, preventing hydrogen bonding with Asp-114 of the adjacent subunit. E, SylA bound to β2. SylA (middle, ball and stick) is bound to Thr-1 (T1) of the β2-subunit (green). Hydrogen bonding between SylA and Thr-21, Gly-47, and Thr-48 of β2 exist, as well as with Asp-114 of the adjacent β3-subunit. However, the substrate-binding cleft is relatively narrow compared with that of β1 and β5 because of the Gln-22 side chain. F, SylA bound to β5. SylA (middle, ball and stick) is bound to Thr-1 (T1) of the β5-subunit (blue). Hydrogen bonding between SylA and Thr-21, Gly-47, and Gly-48 of β5 exist, as well as with Asp-114 of the adjacent β6-subunit. The substrate-binding cleft is relatively wide compared with that of β2 because of the small Ala-22 side chain.
We found that the conformation of the Val at position 1 of the dipeptide tail of SylA contributes to the specificity for the β2-subunit, since the SylA derivatives carrying a d-Val at this position have a reduced affinity for β2 (Fig. 4B). Also, this observation can be explained using the crystal structure of the yeast proteasome bound to SylA (Fig. 7). The binding cleft of β2 is narrower compared with β5 because it carries Gln at position 22 (Q22) compared with Ala (A22) in β5. The side chain of d-Val at position 1 (atom 18 in SylA) would clash with the narrow cleft of β2 (Fig. 7E) but not with the wider cleft of β5 (Fig. 7F).
Subunit selectivity might be an important aspect of SylA function. Hatsugai et al. (2009) demonstrated that the proteasome mediates the discharge of the vacuolar content into the apoplast during the hypersensitive response, triggered by avirulent P. syringae bacteria. Inhibition studies indicated that this is regulated through the caspase activity of the β1 proteasome subunit (Hatsugai et al., 2009). Consistent with this observation, we found that SylA-producing bacteria are able to prevent early host cell death (J.C. Misas-Villamil, I. Kolodziejek, and R.A.L. van der Hoorn, unpublished data), but our data indicate that this is more likely mediated by inhibition of the β2- and β5-subunits of the host proteasome. Subunit selectivity is also an important aspect of drug development, since proteasome inhibitors are important anticancer drugs. Interestingly, SylA was found to induce apoptosis and inhibit cancer proliferation (Coleman et al., 2006) and selectively targets the proteasome in cancer cells that have adapted to the proteasome-targeting drug bortezomib (Clerc et al., 2009a).
Nuclear Accumulation of SylA
Unexpectedly, imaging experiments show that SylA and RhSylA accumulate in the nucleus. MVB003 causes fluorescence in both the nucleus and cytoplasm, but preincubation with SylA suppresses mostly the nuclear fluorescence, in contrast to preincubation with epoxomicin, which suppresses fluorescence in both compartments (Fig. 5, A–E). Furthermore, RhSylA causes mostly nuclear fluorescence (Fig. 5F). The fact that RhSylA-sat does not accumulate in the nucleus (Fig. 5G) and that nuclear fluorescence by RhSylA labeling is not washed out (Fig. 5F) or suppressed by adding an excess SylA after RhSylA labeling indicates that RhSylA is immobilized in the nucleus. However, subcellular fractionation studies could not confirm that SylA preferentially labels the nuclear proteasome (Fig. 6). There can be many explanations for this apparent discrepancy. One explanation might be that the protocol for subcellular fractionation does not exclude ex vivo labeling because SDS would disintegrate the nuclear compartment before separation. Free, unreacted probes that accumulate in the cells and remain after washing (Fig. 5J) might react ex vivo with the proteasome in both the cytoplasmic and nuclear fractions, hiding signals caused by in vivo labeling. However, nuclear fractionations in the presence of the proteasome inhibitor MG132 did not reduce labeling of the nuclear proteasome by MVB003 (Supplemental Fig. S3). Another explanation might be that when used at high SylA concentrations, proteasome labeling occurs also in the cytoplasm.
Several mechanistic explanations for potential nuclear targeting of SylA can be excluded by the current data set. One possibility is that SylA is transported to the nucleus and reacts more efficiently with the nuclear proteasome because the SylA concentration is higher in the nucleus. This would imply that SylA is a cargo for the nuclear import machinery. However, we can rule out the possibility that free SylA concentrates in the nucleus, since nuclear fluorescence by MVB003 or RhSylA is competable by preincubation with SylA but not the inactive SylA-sat derivative (Fig. 5). It might be that SylA blocks nuclear import at high concentrations, preventing probes from entering the nucleus. However, SylA was found to react with the nuclear proteasome using subcellular fractionation experiments. A third explanation is that SylA-labeled proteasomes move from the cytoplasm to the nucleus. However, although nuclear import of proteasomes from the cytoplasm is regulated and proteasome inhibition may increase proteasome import into the nucleus (Takeda and Yanagida, 2005), we did not see an increased PBA1 concentration in the nucleus upon SylA treatment during fractionation experiments (Fig. 6). A fourth explanation is that the different reporter tags in MVB003 (bodipy) and RhSylA (rhodamine) have different fluorescence in the nucleus and cytoplasm. However, such a difference in subcellular fluorescence has not been described for these fluorophores before, and we also found that untagged SylA suppresses nuclear fluorescence of MVB003 labeling. A fifth possibility is that the proteasome in the nucleus is more active when compared with the proteasome in the cytoplasm. This would be consistent with the strong labeling of the nuclear proteasome by both MVB003 and RhSylA (Fig. 6B). A sixth mechanism for nuclear targeting may be that SylA has a higher affinity for the nuclear proteasome (e.g. mediated by a different composition of the proteasome). Although a different composition of the nuclear proteasome has not yet been described, compositions and functions of proteasomes may differ. The immunoproteasome described in animals, for example, appears during immune responses and contains different catalytic subunits that are responsible for the release of more hydrophobic peptides that are used for antigen display (Rock et al., 1994; Goldberg et al., 2002). Differences in proteasome compositions and possible subcellular targeting by SylA are interesting topics for future studies.
Nuclear accumulation of SylA may have important biological implications. The proteasome degrades a series of nucleus-specific proteins, such as transcriptional regulators. For example, the proteasome degrades nuclear proteins NPR1, EIN2, and JAZ, which are important components of the signaling cascades of the stress hormones salicylic acid, ethylene, and jasmonate, respectively (Chini et al., 2007; Thines et al., 2007; Qiao et al., 2009; Spoel et al., 2009). SylA might be produced to interfere in these pathways. Indeed, we found that SylA promotes bacterial growth during SA-induced immunity (J.C. Misas-Villamil, I. Kolodziejek, and R.A.L. van der Hoorn, unpublished data). Interfering with nuclear but not cytoplasmic proteasome activities, therefore, might be beneficial for P. syringae producing SylA.
New Tools for in Vivo Proteasome Activity Profiling and Imaging
During our studies on SylA targeting, we made important technological advances to study the proteasome in vivo. Toxicity of the probes during in vivo labeling was found to be at acceptably low levels when low probe concentrations and relatively short labeling times were used, probably because these probes only label a fraction of the active proteasomes. We showed that the epoxyketone-based MVB003 and the syrbactin-based RhSylA are highly specific proteasome probes, whereas the vinyl sulfone-based MV151 also labels papain-like Cys proteases, especially in vivo (Gu et al., 2010). These characteristics have important implications for the use of these probes in vivo. MV151 has the advantage that it displays both proteasome and protease activities in activity profiles, allowing simultaneous monitoring of both proteolytic machinery. This revealed, for example, that the frequently used proteasome inhibitor MG132 preferentially targets PLCPs when used in vivo (Kaschani et al., 2009), which has important implications for the conclusions from studies where protein degradation was studied using MG132. In contrast to in vivo proteasome activity profiling, imaging of proteasome labeling should preferentially be done using MVB003 or RhSylA, since these probes do not label PLCPs. In addition, we found that the labeling of the different subunits depends on the probes used and on timing and probe concentration. Furthermore, we found that proteasome labeling occurs within minutes in vitro and in vivo. Therefore, special care had to be taken to prevent ex vivo labeling during extraction of in vivo labeled materials. Quick in vivo labeling indicates that there is an efficient uptake of these probes through the cell membrane. Thus, our study introduces the parameters and probes to study the proteasome in vivo.
MATERIALS AND METHODS
Chemicals
E-64d and epoxomicin were from Sigma and BioMol, respectively. Synthesis of MV151, SylA, RhSylA, and other SylA derivatives has been described previously (Verdoes et al., 2006; Clerc et al., 2009a, 2009b, 2010a, 2010b). Synthesis of MVB003, MVB070, and MVB072 is described in Supplemental File S1. Aliquots of the probes and inhibitors are available upon request.
Plant Materials
Arabidopsis (Arabidopsis thaliana ecotype Columbia) plants were grown in a growth chamber at 24°C (day)/20°C (night) under a 12-h light regime. Rosette leaves of 4- to 6-week-old plants were used for protein extraction. Cell cultures (ecotype Landsberg) were subcultured weekly in medium (3% [w/v] Suc, 0.5 mg L−1 naphthalene acetic acid, 0.05 mg L−1 6-benzylaminopurine, and 4.4 g of Murashige and Skoog Gamborg B5 vitamins [Duchefa], pH 5.7).
In Vitro Labeling of Arabidopsis Leaf Extracts
Proteins were extracted by grinding seven rosette leaves into 700 μL of water. The extract was cleared by centrifugation (2 min at 16,000g). Labeling was usually done by incubating approximately 100 μg of protein in 50 μL containing 125 mm Tris buffer (pH 7.0) in the presence of 1 or 2 μm probe for 2 h at room temperature (22°C–25°C) in the dark under gentle agitation. Equal volumes of dimethyl sulfoxide (DMSO) were added to the no-probe controls. The extract was mixed with 4× SDS-PAGE loading buffer containing β-mercaptoethanol and separated on 12% SDS gels (approximately 10 μg of protein per lane). Labeled proteins were visualized by in-gel fluorescence scanning using a Typhoon 8600 scanner (Molecular Dynamics) with excitation and emission at 532 and 580 nm, respectively. Alternatively, the Fuij FLA6000 fluorescence scanner was used. Fluorescent signals were quantified with ImageQuant 5.2 (Molecular Dynamics). After SDS-PAGE, proteins were transferred onto polyvinylidene fluoride membranes (Immobilon-P; Millipore) and detected using anti-PBA1 antibodies (1:5,000; BioMol), followed by horseradish peroxidase-conjugated anti-rabbit secondary antibodies (1:5,000; Amersham). Competition or inhibition assays were done by preincubating the protein extracts with competitor or inhibitor molecules for 30 min before labeling with activity-based probes. Time course experiments were done by taking 50-μL aliquots from a 1-mL reaction volume at various time points.
In Vivo Labeling and Competition
Cell cultures were diluted by adding 20 μL of culture medium to 80 μL of cell culture and kept at room temperature under gentle shaking in the presence or absence of inhibitors for 30 min. Probes were added and labeling continued in the dark for 120 min. The cell culture was washed twice with culture medium and three times with water and ground in the presence of 2% SDS. The extract was cleared by centrifugation (2 min at 16,000g), and the 80-μL supernatant was taken, mixed with 4× gel-loading buffer, and prepared for protein gel electrophoresis. Time course experiments were done by taking 100-μL aliquots from the reaction volume at various time points.
Nuclear Fractionation
Cell cultures were labeled with 2 μm RhSylA for 2 h. Cells were washed with Honda buffer (25 g L−1 Ficoll 400 [Sigma], 50 g L−1 Dextran T40 [Pharmacia Biotech], 0.4 m Suc, 25 mm Tris, pH 7.4, 10 mm MgCl2, protease inhibitor cocktail [Sigma], and 5 mm dithiothreitol) and homogenized in a mortar in 5 mL of Honda buffer. The extract was centrifuged gently at 201g through a 62-μm nylon mesh, and 0.5% Triton X-100 was added to the filtrate. After 15 min of incubation on ice, a 200-μL sample was taken as total extract and mixed with 80 μL of gel-loading buffer. The extract was centrifuged (5 min at 1,500g), and a 200-μL sample was taken from the supernatant as ND extract and mixed with 80 μL of gel-loading buffer. The pellet was washed twice with 3 mL of Honda buffer containing 0.1% Triton X-100 and dissolved in 200 μL of gel-loading buffer, resulting in a NE fraction that is 20-fold more concentrated than the ND and total extract fractions. Proteins were separated on protein gels and detected by fluorescence scanning and using antibodies for PBA1, PEPC, and histone, as described previously (Noël et al., 2007; Cheng et al., 2009).
Activity-Based Imaging
The fluorescence of RhSylA (rhodamine; excitation, 543 nm/emission, 570 nm), MVB003 (bodipy; excitation, 532 nm/emission, 580 nm), and Hoechst33342 (excitation, 350 nm/emission, 450 nm) was detected by a Carl Zeiss LSM 510 confocal microscope. Confocal microscopy was performed with a HeNe1 (excitation, 534 nm) laser and a UV laser. The Zeiss LSM Image Examiner was used for confocal image processing. MVB003 signals were quantified by photometric measurements in situ within the 550- to 600-nm range. All experiments were done with identical acquisition settings for each probe.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structures of probes and inhibitors used in this study.
Supplemental Figure S2. Dual labeling of cell cultures with Hoechst33342 and RhSylA.
Supplemental Figure S3. Nuclear fractionation in the presence of the proteasome inhibitor MG132.
Supplemental File S1. Synthesis of MVB003, MVB070, and MVB072.
Supplementary Material
Acknowledgments
We thank Dr. Ana Garcia for her help with nuclear fractionation experiments and Dr. Elmon Schmelzer for sharing his expertise on imaging.
References
- Amrein H, Makart S, Granado J, Shakya R, Schneider-Pokorny J, Dudler R. (2004) Functional analysis of genes involved in the synthesis of syringolin A by Pseudomonas syringae pv. syringae B301 D-R. Mol Plant Microbe Interact 17: 90–97 [DOI] [PubMed] [Google Scholar]
- Bender CL, Alarcón-Chaidez F, Gross DC. (1999) Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63: 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, He SY. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D, He SY. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol 150: 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Germain H, Wiermer M, Bi D, Xu F, García AV, Wirthmueller L, Després C, Parker JE, Zhang Y, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Clerc J, Florea BI, Kraus M, Groll M, Huber R, Bachmann AS, Dudler R, Driessen C, Overkleeft HS, Kaiser M. (2009a) Syringolin A selectively labels the 20 S proteasome in murine EL4 and wild-type and bortezomib-adapted leukaemic cell lines. ChemBioChem 10: 2638–2643 [DOI] [PubMed] [Google Scholar]
- Clerc J, Groll M, Illich DJ, Bachmann AS, Huber R, Schellenberg B, Dudler R, Kaiser M. (2009b) Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition. Proc Natl Acad Sci USA 106: 6507–6512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc J, Nan L, Krahn D, Groll M, Bachmann AS, Florea BI, Overkleeft HS, Kaiser M. (September 7, 2010a) The natural product hybrid of syringolin A and glidobactin A synergizes proteasome inhibition potency with subsite selectivity. Chem Commun http://dx.doi.org/10.1039/C0CC02238A [DOI] [PubMed]
- Clerc J, Schellenberg B, Groll M, Bachmann AS, Huber R, Dudler R, Kaiser M. (2010b) Convergent synthesis and biological evaluation of syringolin A and derivatives as eukaryotic 20S proteasome inhibitors. Eur J Org Chem 3991–4003 [Google Scholar]
- Coleman CS, Rocetes JP, Park DJ, Wallick CJ, Warn-Cramer BJ, Michel K, Dudler R, Bachmann AS. (2006) Syringolin A, a new plant elicitor from the phytopathogenic bacterium Pseudomonas syringae pv. syringae, inhibits the proliferation of neuroblastoma and ovarian cancer cells and induces apoptosis. Cell Prolif 39: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. (2008) Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem 77: 383–414 [DOI] [PubMed] [Google Scholar]
- Cunnac S, Lindeberg M, Collmer A. (2009) Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr Opin Microbiol 12: 53–60 [DOI] [PubMed] [Google Scholar]
- Dick TP, Nussbaum AK, Deeg M, Heinemeyer W, Groll M, Schirle M, Keilholz W, Stevanović S, Wolf DH, Huber R, et al. (1998) Contribution of proteasomal β-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J Biol Chem 273: 25637–25646 [DOI] [PubMed] [Google Scholar]
- Göhre V, Robatzek S. (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46: 189–215 [DOI] [PubMed] [Google Scholar]
- Goldberg AL, Cascio P, Saric T, Rock KL. (2002) The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol Immunol 39: 147–164 [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. (1997) Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386: 463–471 [DOI] [PubMed] [Google Scholar]
- Groll M, Schellenberg B, Bachmann AS, Archer CR, Huber R, Powell TK, Lindow S, Kaiser M, Dudler R. (2008) A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism. Nature 452: 755–758 [DOI] [PubMed] [Google Scholar]
- Gu C, Kolodziejek I, Misas-Villamil J, Shindo T, Colby T, Verdoes M, Richau KH, Schmidt J, Overkleeft HS, van der Hoorn RAL. (2010) Proteasome activity profiling: a simple, robust and versatile method revealing subunit-selective inhibitors and cytoplasmic, defense-induced proteasome activities. Plant J 62: 160–170 [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR. (2009) The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I. (2009) A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev 23: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Upper CD. (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae: a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64: 624–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Chen CH. (2009) Proteasome regulators: activators and inhibitors. Curr Med Chem 16: 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imker HJ, Walsh CT, Wuest WM. (2009) SylC catalyzes ureido-bond formation during biosynthesis of the proteasome inhibitor syringolin A. J Am Chem Soc 131: 18263–18265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffarnik FA, Jones AM, Rathjen JP, Peck SC. (2009) Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol Cell Proteomics 8: 145–156 [DOI] [PubMed] [Google Scholar]
- Kaschani F, Verhelst SHL, van Swieten PF, Verdoes M, Wong CS, Wang Z, Kaiser M, Overkleeft HS, Bogyo M, van der Hoorn RAL. (2009) Minitags for small molecules: detecting targets of reactive small molecules in living plant tissues using ‘click chemistry’. Plant J 57: 373–385 [DOI] [PubMed] [Google Scholar]
- Kolodziejek I, van der Hoorn RAL. (2010) Mining the active proteome in plant science and biotechnology. Curr Opin Biotechnol 21: 225–233 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Smalle JA. (2008) Structure, function and regulation of plant proteasomes. Biochimie 90: 324–335 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Guttman DS, Desveaux D. (2009) The targeting of plant cellular systems by injected type III effector proteins. Semin Cell Dev Biol 20: 1055–1063 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BHB, Elofsson M, Sin N, Crews CM. (1999) Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA 96: 10403–10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S, Witte CP, Bhat R, Pochon N, Colby T, Parker JE. (2007) Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19: 4061–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR. (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel C, Tobler M, Meyer M, Bigler L, Ebert MO, Schellenberg B, Dudler R. (2009) Biosynthesis of the proteasome inhibitor syringolin A: the ureido group joining two amino acids originates from bicarbonate. BMC Biochem 10: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78: 761–771 [DOI] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF. (2004) Profiling enzyme activities in vivo using click chemistry methods. Chem Biol 11: 535–546 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Yanagida M. (2005) Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell 122: 393–405 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Verdoes M, Florea BI, Menendez-Benito V, Maynard CJ, Witte MD, van der Linden WA, van den Nieuwendijk AMCH, Hofmann T, Berkers CR, van Leeuwen FWB, et al. (2006) A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem Biol 13: 1217–1226 [DOI] [PubMed] [Google Scholar]
- Wäspi U, Hassa P, Staempfli A, Molleyres LP, Winkler T, Dudler R. (1999) Identification and structure of a family of syringolin variants: unusual cyclic peptides from Pseudomonas syringae pv. syringae that elicit defence responses in rice. Microbiol Res 154: 1–5 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.