Abstract
The mismatch repair proteins function upstream in the DNA damage signaling pathways induced by the DNA methylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). We report that MSH2 (MutS homolog 2) protein interacts with the ATR (ATM- and Rad3-related) kinase to form a signaling module and regulate the phosphorylation of Chk1 and SMC1 (structure maintenance of chromosome 1). We found that phosphorylation of Chk1 by ATR also requires checkpoint proteins Rad17 and replication protein A. In contrast, phosphorylation of SMC1 by ATR is independent of Rad17 and replication protein A, suggesting that the signaling pathway leading to SMC1 phosphorylation is distinct from that mediated by the checkpoint proteins. In addition, both MSH2 and Rad17 are required for the activation of the S-phase checkpoint to suppress DNA synthesis in response to MNNG, and phosphorylation of SMC1 is required for cellular survival. These data support a model in which MSH2 and ATR function upstream to regulate two branches of the response pathway to DNA damage caused by MNNG.
By maintaining genome stability, the DNA damage response network plays a significant role in preventing cancer development (1). Many tumor suppressors, such as p53, Chk2, and the breast cancer tumor suppressor BRCA1, are integral components of this network, which regulates cell cycle checkpoint activation and DNA repair. Genetic work from yeast has established the current conceptual framework of DNA damage response (2). In humans, the central checkpoint kinases ATM (ataxia telangiectasia mutated) and ATR (ATM- and Rad3-related), and the RFC (replication factor C)-like checkpoint protein Rad17/RFC2–5 complex have been demonstrated to function upstream of the DNA damage response pathway for the activation of the downstream checkpoint kinase Chk1 by phosphorylation (3). Because different DNA-damaging agents generate different DNA lesions, a key question in damage signaling is how the checkpoint proteins recognize different DNA lesions. One mechanism of DNA damage recognition has been elucidated recently, in which binding of replication protein A (RPA) to single-stranded DNA (ssDNA) results in recruitment of the ATR-interacting protein ATRIP to load the ATR/ATRIP complex to ssDNA, leading to activation of ATR (4). Because ssDNA is a common intermediate for many DNA repair pathways, this model explains nicely how ATR-dependent checkpoint pathways respond to different types of DNA damage.
In contrast to checkpoint pathways, DNA repair pathways use lesion-specific factors to recognize different lesions (5, 6). The mismatch repair (MMR) proteins play key roles in postreplicational mispair correction that is essential for genomic stability (7). Mutations in MMR genes, especially MSH2 and MLH1, are associated with predisposition to hereditary nonpolyposis colorectal cancer (8–10). The MMR reaction is initiated by binding of the MSH2 (MutS homolog 2)/MSH6 heterodimer to the mismatched DNA (11). The MMR proteins also function in signaling DNA damage (7, 11). Upon exposure to MNNG and crosslinking agents, cells deficient in MMR proteins exhibit impaired G2/M cell cycle arrest, reduced activation of the p53/p73 apoptosis pathway, and resistance to the cytotoxicity of these DNA-damaging agents (12–14). Recently, the MMR system has also been implicated in S-phase checkpoint activation in response to low-dosage IR (15). The ability of the MSH2/MSH6 heterodimer to bind a variety of modified DNA structure in vitro suggests a possibility that the MMR proteins may signal the DNA damage directly. Alternatively, the checkpoint response can be activated indirectly through recognition of repair intermediates that are generated by MMR. These possibilities have not been thoroughly tested.
In this study, we report a MSH2-dependent signaling pathway in response to DNA damage caused by MNNG. We found that MSH2 protein interacts with the ATR kinase constitutively to form a signaling module that regulates the phosphorylation of downstream effectors including Chk1 and SMC1 (structure maintenance of chromosome 1). Whereas phosphorylation of Chk1 also requires Rad17 and RPA, phosphorylation of SMC1 is independent of Rad17 and RPA. Thus, the pathway leading to SMC1 phosphorylation by ATR seems to branch out the checkpoint pathway mediated by Rad17 and RPA. In addition, both MSH2 and Rad17 are required for the activation of the S-phase checkpoint to suppress DNA synthesis in response to MNNG. Thus, MSH2 and ATR function upstream to regulate two branches of the response pathway to DNA damage caused by MNNG.
Materials and Methods
Cell Lines and Antibodies. TK6 and MT1 cells (provided by P. Modrich, Duke University, Durham, NC) were maintained in RPMI medium 1640 with 10% FBS. 293T and HeLa cells were cultured in DMEM with 10% FBS. Anti-RPA70 antibody was from Oncogene Research Products (La Jolla, CA). Anti-MSH2 (BL323), Rad17 (BL239G), Chk1 (BL234), SMC1 (BL308), MSH6 (BL903), SMC1-pS966 (BL311), Chk1-pS317 (BL229), and Chk1-pS345 (BL231) antibodies were from Bethyl Laboratories (Montgomery, TX). Rabbit anti-ATR was raised against GST-ATR (400–460) and affinity-purified. Anti-ATRIP antibody was provided by S. Elledge (Baylor College of Medicine).
RNA Interference, in Vitro Kinase Assay, in Vitro Binding Assays, Immunoprecipitation (IP), and Mass Spectrometry. The small interfering RNA (siRNA) duplexes were synthesized by Dharmacon Research (Boulder, CO) and prepared by annealing two 21-ribonucleotide oligonucleotides according to the manufacturer's suggestions. The sequences targeting each gene were as follows: MSH2, 5′-AAU CUG CAG AGU GUU GUG CUU-3′; ATR, 5′-AAC GAG ACU UCU GCG GAU UGC-3′; Rad17, 5′-AAC AGA CUG GGU UGA CCC AUC-3′; and ATM, 5′-AAG CAC CAG UCC AGU AUU GGC-3′. The siRPA70 was described in ref. 4. (Throughout, “si” before a gene name indicates gene-specific siRNA.) The siGFP and siVimentin were purchased from Dharmacon Research and used as mock controls. HeLa cells were transfected with siRNA duplex by using Oligofectamine (Invitrogen) according to the manufacturer's protocols. Transfected cells were replated 48 h after transfection with a 1:3 ratio and treated with DNA-damaging agents 60–72 h after transfection (16).
The MSH2 and MSH6 proteins were overexpressed in sf9 cells transfected with baculovirus overexpressing MSH2 or MSH6 and purified by IP with antibodies. Flag-ATR was overexpressed in 293T cells, purified with anti-Flag agarose, and eluted with Flag peptide. MSH2 or MSH6 bound to protein A-Sepharose were incubated at 4°C with either purified Flag-ATR or 35S-labeled, in vitro translated SMC1 for 1 h and washed three times with NETN buffer (0.5% Nonidet P-40/1 mM EDTA, pH 8.0/20 mM Tris, pH 8.0/100 mM NaCl). The binding was analyzed with Western blotting or autoradiography.
Purification of MSH2 complex was carried out in 5 ml of HeLa nuclear extracts (NE) (≈10 mg/ml protein) with 50 μg of MSH2 antibody essentially as described (17). The protein components were separated on a SDS/PAGE and analyzed by mass spectrometry as described (17). An in vitro kinase assay using overexpressed Flag-ATR and GST-SMC1 was carried out as described (18).
Inhibition of DNA Synthesis and Clonogenic Survival Assays. Inhibition of DNA synthesis assay was carried out as described with some modifications (19). Briefly, 24 h after siRNA transfection, cells were labeled with 50 nCi (1 Ci = 37 GBq) of [14C]thymidine per ml for 24 h as a control for the total DNA content of different samples. Sixty to 72 h after transfection, cells were treated with the indicated MNNG concentration for 1 h and allowed to recover for indicated times. They were then pulse-labeled with 1 μCi/ml [3H]thymidine for 30 min. Labeled cells were harvested, washed twice with PBS, and fixed with 80% ethanol at –20°C overnight. The fixed cells were then pelleted by centrifugation, washed in 80% ethanol, and pelleted again twice. Finally, the washed cell pellets were dissolved in 0.5 ml of 0.25 N NaOH solution, and 10 ml of scintillation counting solution was added for radioactivity counting in a liquid scintillation counter. DNA synthesis was calculated by using the ratio of 3H/14C. Overlapping 3H and 14C emissions were corrected with quenched 3H and 14C standards. Three replicas were measured for each sample to derive the standard deviation.
Transient transfection of GFP-wt-SMC1 or GFP-S966A-SMC1 in HeLa cells was carried out with Lipofectamine (Invitrogen). Cells overexpressing wild-type or S966A GFP-SMC1 were plated in triplicate at limiting dilutions 30 h after transfection and treated with a range of MNNG 18 h later (48 h after transfection). After incubation in the presence of MNNG for 1 h, cells were replaced with fresh medium and recovered for 7 days. Colonies were then fixed in methanol and stained with Giemsa. A population of >50 cells was counted as one colony.
Results
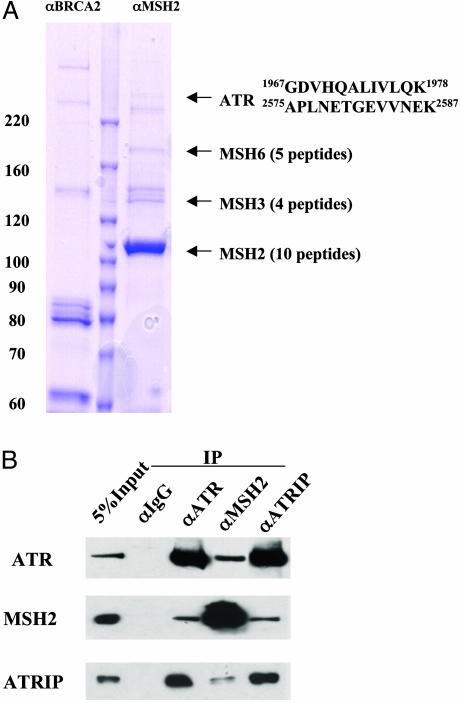
ATR Coimmunoprecipitates with MSH2. To identify signaling components in the MSH2 pathway, we carried out an IP from HeLa NE using an anti-MSH2 antibody. An IP using a BRCA2 antibody was used as a negative control. We analyzed the immunoprecipitates by mass spectrometry (Fig. 1A). Besides MSH6 and MSH3, which are known to be associated with MSH2, we identified a substoichiometrical component with an apparent molecular mass of ≈250 kDa as the checkpoint kinase ATR. The association between ATR and MSH2 was confirmed by reciprocal IP detected by Western blotting (Fig. 1B). The recently identified ATRIP (18), although not detected by mass spectrometry, was also found to be coimmunoprecipitated with MSH2 detected by Western blotting (Fig. 1B). Moreover, mass spectrometry analysis of immunoprecipitates of ATR from HeLa NE also identified ATRIP, MSH2, and MSH6 proteins (data not shown), consistent with the IP/Western blot results. This suggests that ATRIP in the MSH2 IP is below the detection limit of Coomassie blue staining because of its lower molecular mass (90 kDa). Elution profiles of MSH2 from ion exchange and gel filtration columns suggest that MSH2 exists in different complexes in the HeLa NE (data not shown). The small percentage of MSH2 and ATR that coimmunoprecipitates suggests that only a small proportion of MSH2 complexes contains ATR.
Fig. 1.
MSH2 associates with ATR in HeLa NE. (A) The MSH2 complex was isolated by anti-MSH2 IP and separated on a SDS/PAGE. A parallel IP using a BRCA2 antibody serves as a negative control. Protein bands stained by Coomassie blue were analyzed by mass spectrometry. The sequences of the two ATR peptides identified are shown. (B) Coimmunoprecipitations of MSH2, ATR, and ATRIP detected by Western blotting. IPs were carried out in HeLa NE by using indicated antibodies. Five percent of the total protein used in the IP was loaded in the input lane.
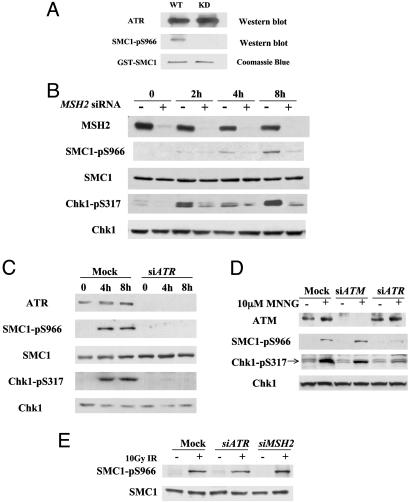
SMC1 and Chk1 Are Downstream Effectors in the MSH2/ATR Pathway in Response to MNNG. The association of ATR with MSH2 implicates ATR as a transducer kinase in the MMR protein-mediated response pathway. SMC1 was recently identified as a component of the ATM/NBS1 branch of the S-phase checkpoint pathway in response to IR (20, 21). SMC1 is phosphorylated at S966 and S957 by ATM in response to IR. To investigate whether SMC1 is a downstream effector in response to DNA methylation-induced damage, we treated the cells with MNNG. We found that S966 of SMC1 is phosphorylated in a dosage-dependent manner in HeLa and primary human fibroblast IMR90 cells (data not shown). To determine whether ATR directly phosphorylates SMC1 in vitro, we expressed a fragment of SMC1 containing the in vivo phosphorylation site (amino acids 890-1233) as a GST fusion protein. Flag-tagged wild-type ATR but not the kinase-inactive form of ATR (kd-ATR) expressed in 293T cells can directly phosphorylate GST-SMC1 at S966 in an in vitro kinase assay (Fig. 2A).
Fig. 2.
The MSH2/ATR complex is required for MNNG-induced phosphorylation of SMC1 and Chk1. (A) In vitro kinase assays of SMC1 by ATR. Transiently expressed Flag-tagged ATR (wild type or kinase dead) in 293T cells was used to phosphorylate a GST-SMC1 fragment (amino acids 890-1233). The phosphorylation product was immunoblotted with SMC1 S966 phosphospecific antibody. Equal amounts of GST-SMC1 proteins used in the kinase assays were monitored by Coomassie blue staining. (B and C) Dependence of SMC1 S966 and Chk1 S317 phosphorylations on MSH2 (B) and ATR (C) in response to MNNG. HeLa cells transfected with mock (siGFP), siATR, or siMSH2 were treated with 10 μM MNNG for 1 h and harvested at indicated times. Efficiency of RNA interference was monitored by Western blotting to each protein. (D) ATR, not ATM, is the major kinase involved in early response to MNNG-induced damage. Mock, siRNA to ATM-or ATR-transfected HeLa cells was treated with 10 μM MNNG for 1 h and harvested after 4 h. Cell lysates were analyzed with indicated antibodies. Chk1 blot also serves as a loading control. (E) Independence of SMC1 phosphorylation on ATR and MSH2 in response to γ irradiation. HeLa cells transfected with indicated siRNA were treated with 10 Gy of IR and harvested after 1 h.
To determine whether MSH2 and ATR are required in signaling MNNG-induced damage, we used siRNA to inhibit their expression (16). Transfections of siMSH2 effectively reduced MSH2 protein level and led to defective phosphorylation of both SMC1 at S966 and Chk1 at S317 in response to 10 μM MNNG treatment (Fig. 2B). Similarly, phosphorylation of SMC1 at S966 and Chk1 at S317 were largely abolished in cells depleted of ATR (Fig. 2C), consistent with its role as a checkpoint kinase. These results demonstrate that MSH2 and the ATR kinase function upstream in response to MNNG that leads to phosphorylation of SMC1 and Chk1. Defective phopshorylation of SMC1 and Chk1 was also observed in the MSH6-deficient MT1 cell line when compared with the parental TK6 cell line (data not shown), suggesting that a functional MSH2/MSH6 complex is required for SMC1 and Chk1 phosphorylation.
ATM and ATR are the central checkpoint kinases in signaling DNA damage, and they play distinct yet sometimes overlapping roles in signaling different damages (22). To differentiate the role of ATM plays from ATR in signaling MNNG-induced damage, we compared phosphorylation of checkpoint effectors in HeLa cells transfected with siATM or siATR 4 h after 10 μM MNNG treatment. Phosphorylation of SMC1 and Chk1 remains largely intact in cells lacking ATM and is more defective in cells transfected with siATR (Fig. 2D). HeLa cells transfected with siATM under the same conditions are defective in activation of S-phase checkpoint in response to 10 Gy of IR (data not shown), confirming that the ATM function has been compromised by siRNA transfection. Therefore, we conclude that ATR is the major kinase responsible for checkpoint activation in response to MNNG-induced damage.
Because the MSH2/MSH6 heterodimer is a lesion-specific factor that binds DNA mismatches and a variety of modified DNA structures including those caused by MNNG, it has been proposed as a putative damage sensor in the MMR pathway (7, 13). To test whether MSH2 and ATR function in a lesion-specific manner for SMC1 phosphorylation, we examined their roles in response to IR. In contrast to the requirement of MSH2 and ATR in MNNG-induced damage, phosphorylation of SMC1 in response to 10 Gy of IR is normal in siMSH2- or siATR-transfected cells (Fig. 2E). Thus, the MSH2/ATR pathway is required to phosphorylate SMC1 specifically in response to damage induced by DNA methylation but not for double-stranded break caused by IR.
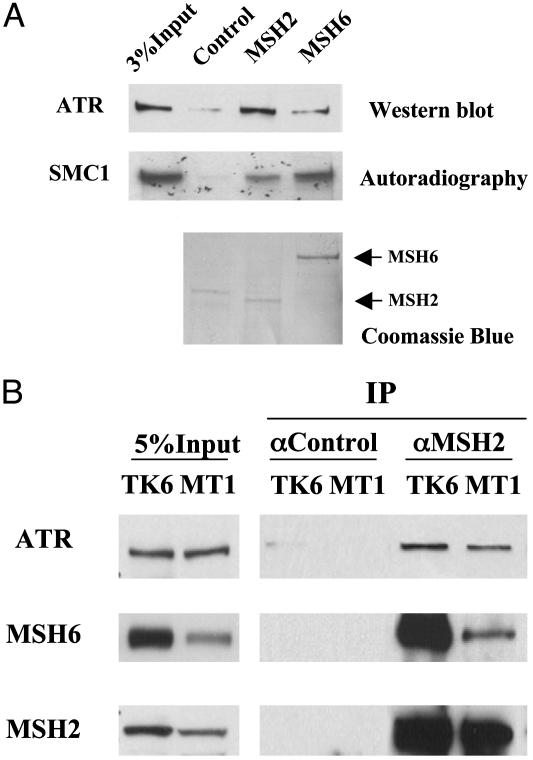
Interaction of MSH2 or MSH6 with ATR and SMC1. To understand the mechanism of MSH2/MSH6-dependent phosphorylation of SMC1 by ATR, we tested whether ATR and SMC1 interact directly with MSH2 or MSH6. Recombinant MSH2 and MSH6 were expressed in insect cells and purified by IP by using anti-MSH2 or MSH6 antibodies. Flag-ATR was expressed in 239T cells, purified with Flag beads, and eluted with the Flag peptide. As shown in Fig. 3A, MSH2 is able to pull down purified ATR in vitro, suggesting that they may interact directly. When the same amount of protein A-bound MSH2 or MSH6 was mixed with in vitro translated SMC1, more SMC1 was brought down by MSH6 than by MSH2. This suggests that MSH6 may interact directly with SMC1.
Fig. 3.
Interaction of MSH2 or MSH6 with ATR and SMC1. (A) In vitro interactions of MSH2 or MSH6 with ATR and SMC1, respectively. Affinity-purified human MSH2 and MSH6 from sf9 cells were incubated with purified Flag-ATR or 35S-labeled, in vitro translated SMC1. An unrelated antibody incubated with sf9 cell lysate was used as a control. Coomassie blue staining of the membrane after Western blotting shows that human MSH2 and MSH6 used in the binding reactions do not copurify with the insect MSH6 or MSH2. (B) MSH6-independent association of MSH2 and ATR. IPs were carried out in NE prepared from lymphoblastoid cell line TK6, which expresses wild-type MSH6, and MT1, which expresses a low level of a mutant form of MSH6.
The MMR-deficient cell line MT1 was isolated from TK6 by its resistance to killing by MNNG. MT1 expresses a lower level of a mutant MSH6 than TK6 does. To determine whether MSH6 is required for MSH2/ATR association in vivo, we compared the coimmunoprecipitation of MSH2/ATR in TK6 and MT1 cells. We found that the MSH2/ATR association in MT1 cells appeared to be intact compared with that in the parental TK6 cells (Fig. 3B). The slightly lower amount of coprecipitated ATR in MT1 cells is likely a result of a lower level of MSH2 in the anti-MSH2 IP from MT1. The above result is in agreement with the in vitro binding results, suggesting that the MSH2 may directly interact with ATR in vivo.
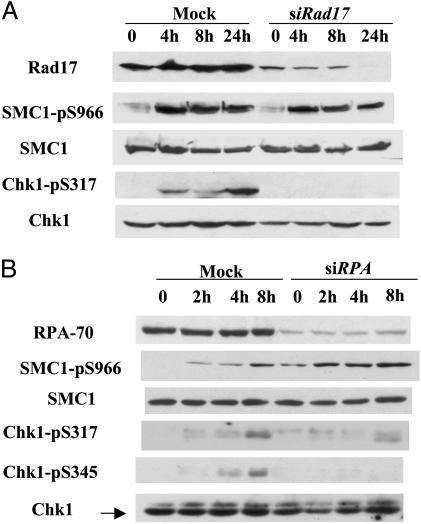
Phosphorylation of SMC1 Does Not Depend on Rad17or RPA. The replication factor C-like checkpoint protein Rad17 has been well established as an upstream element in the DNA damage response (2). To test whether phosphorylation of SMC1 and Chk1 requires Rad17, we depleted Rad17 by small RNA interference. Surprisingly, phosphorylation of SMC1 remains largely unaffected in the absence of Rad17 (Fig. 4A). In contrast, phosphorylation of Chk1 is greatly reduced, as expected. Thus, SMC1 as an ATR substrate can be phosphorylated through a mechanism that is distinct from the Rad17-mediated signaling pathway.
Fig. 4.
Rad17 and RPA are not required for the phosphorylation of SMC1. Shown is MNNG-induced phosphorylation of SMC1 and Chk1 in the absence of Rad17 (A) or RPA (B). HeLa cells transfected with indicated siRNAs were treated with 10 μM MNNG for 1 h and harvested at indicated times. Lysates were analyzed with indicated antibodies.
RPA has been shown recently to activate ATR by recruiting the ATRIP to the damaged sites (4). Likewise, we found that RPA is required for the phopshorylation of Chk1 but not for the phosphorylation of SMC1 (Fig. 4B). Taken together, these data suggest that, while MSH2 and ATR are required for the MNNG-induced response, the pathway leading to phosphorylation of SMC1 is distinct from that mediated by Rad17 and RPA. Thus, the SMC1 pathway seems to branch out from that mediated by the checkpoint protein Rad17 and RPA.
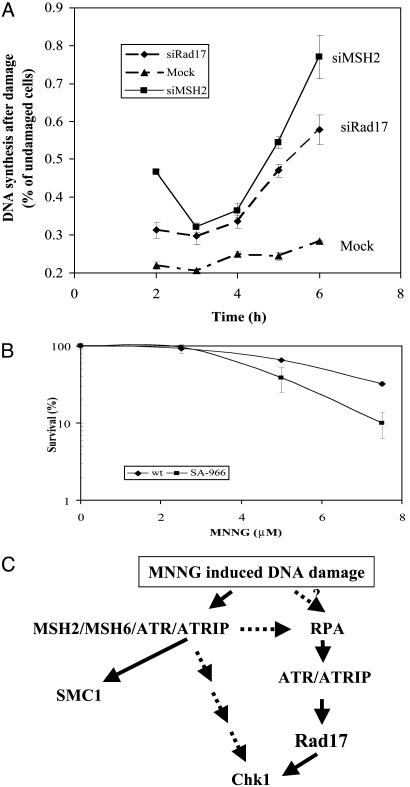
MSH2 and Rad17 Both Are Required for Inhibition of DNA Synthesis, and Phosphorylation of SMC1 Is Important for Cellular Survival in Response to MNNG. In response to DNA damage, eukaryotic cells activate the S-phase checkpoint to slow down DNA synthesis. To investigate whether MSH2 and Rad17 are required for S-phase checkpoint activation in response to MNNG, we monitored DNA synthesis in HeLa cells transfected with siMSH2, siRad17, or a control siRNA through a 6-h time course after 10 μM MNNG treatment. Similar to slowed DNA synthesis in response to IR, DNA synthesis measured by thymidine incorporation in control siRNA-transfected cells was reduced to 20% of that in cycling cells 3 h after MNNG treatment and was 25% at 6 h. Such slowing down of DNA synthesis is characteristic of the activation of the S-phase checkpoint (Fig. 5A). In contrast, cells transfected with siMSH2 or siRad17 consistently exhibit a higher level of DNA synthesis after MNNG treatment, indicating that the ability to inhibit DNA synthesis is compromised. We therefore conclude that inactivation of MSH2 or Rad17 leads to defective inhibition of DNA synthesis in response to MNNG.
Fig. 5.
(A) Both MSH2 and Rad17 are required for activation of MNNG-induced S-phase checkpoint. HeLa cells transfected with mock, MSH2, or Rad17 siRNA were treated with 10 μM MNNG for 1 h. DNA synthesis measured by [3H]thymidine incorporation was evaluated at time indicated after the treatment and normalized to that in cycling cells. The measurements were performed in triplicate. The error bar represents the standard deviation. (B) SMC1 phosphorylation is required for cellular survival after MNNG treatment. HeLa cells expressing either GFP-wt-SMC1 or GFP-SA966-SMC1 were treated with MNNG of indicated concentrations for 1 h and recovered for 1 week before assaying for colony formation. The assay was performed in triplicate. The error bar represents the standard deviation. (C) A schematic model of MNNG-induced DNA damage response. Upon exposure to MNNG, binding of the MSH2/MSH6 heterodimer to O6-methyl-G·C activates the checkpoint kinase ATR, leading to the phosphorylation of SMC1 and Chk1. The RPA is also required to recruit the ATR/ATRIP complex to the damaged sites, possibly through binding to MMR intermediates, resulting in Rad17-dependent phosphorylation of Chk1. However, SMC1 can be phosphorylated independent of RPA and Rad17, suggesting the existence of a response pathway through direct interaction between ATR/MSH2 and MSH6/SMC1.
To determine the long-term effect of damage-induced SMC1 phosphorylation, we evaluated the clonogenic survival of HeLa cells transiently transfected with a wild-type SMC1 or a phosphorylation site mutant of SMC1 after MNNG treatment. HeLa cells expressing S966A of SMC1 (Ser-966 is mutated to Ala-966) display increased sensitivity to MNNG-induced killing compared with those transfected with the wild-type SMC1 (Fig. 5B), suggesting that phosphorylation of SMC1 is important for cellular survival. This finding is consistent with the previous report that SMC1 phosphorylation is important for cellular survival in response to IR (21).
Discussion
The MSH2/ATR Signaling Module. In this study, we show that the MSH2 protein physically interacts with ATR to form a signaling module. They are required for the phosphorylation of SMC1 and Chk1. This MSH2-dependent response is lesion-specific, because it responds primarily to MNNG, not IR. The MMR proteins have been implicated as upstream elements in response to MNNG- and cisplatin-induced damage. Our findings further strengthen this notion and establish that ATR is the transducer kinase that participates in the MSH2-dependent DNA damage response pathway. The MSH2/ATR signaling module is analogous to the established signaling module of the double-stranded break repair complex Mre11/Rad50/NBS1 (M/R/N) with ATM, which responds primarily to double-stranded break (23–25). Such arrangement is intriguing, in that the repair proteins physically associate with transducer kinases constitutively. Because the M/R/N complex binds double-stranded break and MSH2/MSH6 binds the O6-methyl-G·C generated by MNNG, the physical association of these repair proteins with transducer kinases puts them in close proximity for the possibility of direct damage signaling.
Two Branches of the Damage Response That Are Regulated by MSH2 and ATR. The participation of Rad17 and RPA as upstream elements for the phopshorylation of Chk1 in the response to MNNG is expected. It is surprising that they are not required for the phosphorylation of SMC1. Although we cannot rule out the possibility that SMC1 lies upstream of Rad17, the current understanding of the mechanism by which Rad17 is recruited to the damaged sites suggests that the pathway that governs SMC1 phosphorylation is likely branched out from the well established checkpoint pathway that is mediated by Rad17 and RPA (Fig. 5C). Similar dependence of SMC1 phosphorylation in response to IR was also observed, in which SMC1 phosphorylation depends on the repair protein NBS1 (20, 21) but not the checkpoint protein Rad17 (26).
Although both SMC1 and Chk1 are bone fide ATR substrates, the molecular mechanism by which they are phosphorylated can be different. On the SMC1 branch, the phosphorylation of SMC1 by ATR may be through a mechanism of direct damage signaling. Given that MSH2/MSH6 heterodimer is able to recognize DNA lesions directly, the MSH2/ATR association suggests that ATR kinase can be activated at the site of damaged DNA, leading to SMC1 phopshorylation. The in vitro binding of MSH6 with SMC1 raised the possibility that the MSH6 may function as an “adaptor.” This is also consistent with the requirement for a functional MSH6 in the MT1 cell line for SMC1 phopshorylation but not for the MSH2/ATR association.
On the Chk1 branch, phosphorylation of Chk1 by ATR may be through an indict mechanism of damage recognition. The requirement for RPA suggests that ssDNA is generated either as a direct result of MNNG damage or, more likely, as a repair intermediate that is processed by the MMR system. Thus, ATR/ATRIP is loaded by RPA-ssDNA, which in turn phosphorylates Chk1 in a Rad17-dependent manner. It is possible that the pool of ATR that phosphorylates Chk1 is different from the pool of ATR that phosphorylates SMC1. The former does not need to associate with MSH2, but the latter does, consistent with our finding that only a small percentage of ATR associates with MSH2 in the cell.
The existence of two branches in damage response to MNNG that are governed by MSH2 suggests that the MMR proteins may be involved in two aspects of damage signaling. (i) They generate repair intermediates during MMR, which are recognized by the checkpoint proteins, leading to activation of the Chk1 branch. (ii) They signal damage directly to phosphorylate a unique set of substrates, including SMC1, thus are directly involved in checkpoint signaling. A reconstituted in vitro system to demonstrate directly the MMR protein-dependent activation of the ATR kinase to phosphorylate SMC1 remains to be established.
SMC1 Phosphorylation Is Important for Cellular Survival. Consistent with its role for survival in response to IR (21), we show here that phosphorylation of SMC1 is also involved in the modulation of cellular sensitivity to MNNG damage. In Schizosaccharomyces pombe, phosphorylation of Chk1 is also important for cellular survival in response to UV damage (27). Thus, the ATR-dependent pathways are important for cellular survival in response to DNA damage. Paradoxically, a hallmark of MMR-deficient cells is their resistance to killing by DNA-alkylating agents, including MNNG. Clearly, the inability to phosphorylate SMC1 or Chk1 in MMR-deficient cells is not the cause for such MNNG resistance. Because the MMR proteins function upstream in the DNA damage response, they must also signal to other pathways, which may include the apoptotic pathway, whose components remain to be identified. Elucidation of components in this pathway will provide insights into mechanisms involved in MNNG resistance in MMR-deficient tumors.
Acknowledgments
We thank P. Modrich, J. Jiricny, and S. Elledge for reagents and cell lines and P. Yazdi for critical reading of the manuscript. This work was supported by National Institutes of Health Grant CA84199 (to J.Q.). J.Q. is the recipient of Career Development Award DAMD17-00-1-0146 from the Department of Defense Breast Cancer Research Program.
Abbreviations: MNNG, N-methyl-N′-nitro-N-nitrosoguanidine; MMR, mismatch repair; ATM, ataxia telangiectasia mutated; ATR, ATM- and Rad3-related; MSH, MutS homolog; ATRIP, ATR-interacting protein; SMC, structure maintenance of chromosome; RPA, replication protein A; ssDNA, single-stranded DNA; siRNA, small interfering RNA; NE, nuclear extracts; IP, immunoprecipitation.
References
- 1.Myung, K., Datta, A. & Kolodner, R. D. (2001) Cell 104, 397–408. [DOI] [PubMed] [Google Scholar]
- 2.Lydall, D. & Weinert, T. (1995) Science 270, 1488–1491. [DOI] [PubMed] [Google Scholar]
- 3.Zou, L., Cortez, D. & Elledge, S. J. (2002) Genes Dev. 16, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou, L. & Elledge, S. J. (2003) Science 300, 1542–1548. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg, E. C., Walker, G. C. & Sinclair, D. A. (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC.).
- 6.Wood, R. D., Mitchell, M., Sgouros, J. & Lindahl, T. (2001) Science 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- 7.Modrich, P. (1997) J. Biol. Chem. 272, 24727–24730. [DOI] [PubMed] [Google Scholar]
- 8.Fishel, R., Lescoe, M. K., Rao, M. R., Copeland, N. G., Jenkins, N. A., Garber, J., Kane, M. & Kolodner, R. (1993) Cell 75, 1027–1038. [DOI] [PubMed] [Google Scholar]
- 9.Drummond, J. T., Li, G. M., Longley, M. J. & Modrich, P. (1995) Science 268, 1909–1912. [DOI] [PubMed] [Google Scholar]
- 10.Papadopoulos, N., Nicolaides, N. C., Wei, Y. F., Ruben, S. M., Carter, K. C., Rosen, C. A., Haseltine, W. A., Fleischmann, R. D., Fraser, C. M., Adams, M. D., et al. (1994) Science 263, 1625–1629. [DOI] [PubMed] [Google Scholar]
- 11.Duckett, D. R., Drummond, J. T., Murchie, A. I., Reardon, J. T., Sancar, A., Lilley, D. M. & Modrich, P. (1996) Proc. Natl. Acad. Sci. USA 93, 6443–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickman, M. J. & Samson, L. D. (1999) Proc. Natl. Acad. Sci. USA 96, 10764–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duckett, D. R., Bronstein, S. M., Taya, Y. & Modrich, P. (1999) Proc. Natl. Acad. Sci. USA 96, 12384–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong, J. G., Costanzo, A., Yang, H. Q., Melino, G., Kaelin, W. G. J., Levrero, M. & Wang, J. Y. (1999) Nature 399, 806–809. [DOI] [PubMed] [Google Scholar]
- 15.Brown, K. D., Rathi, A., Kamath, R., Beardsley, D. I., Zhan, Q., Mannino, J. L. & Baskaran, R. (2003) Nat. Genet. 33, 80–84. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 17.Wang, Y., Cortez, D., Yazdi, P., Neff, N., Elledge, S. J. & Qin, J. (2000) Genes Dev. 14, 927–939. [PMC free article] [PubMed] [Google Scholar]
- 18.Cortez, D., Guntuku, S., Qin, J. & Elledge, S. J. (2001) Science 294, 1713–1716. [DOI] [PubMed] [Google Scholar]
- 19.Morgan, S. E., Lovly, C., Pandita, T. K., Shiloh, Y. & Kastan, M. B. (1997) Mol. Cell. Biol. 17, 2020–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yazdi, P. T., Wang, Y., Zhao, S., Patel, N., Lee, E. Y. & Qin, J. (2002) Genes Dev. 16, 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. T., Xu, B. & Kastan, M. B. (2002) Genes Dev. 16, 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abraham, R. T. (2001) Genes Dev. 15, 2177–2196. [DOI] [PubMed] [Google Scholar]
- 23.Lim, D. S., Kim, S. T., Xu, B., Maser, R. S., Lin, J., Petrini, J. H. & Kastan, M. B. (2000) Nature 404, 613–617. [DOI] [PubMed] [Google Scholar]
- 24.Wu, X., Ranganathan, V., Weisman, D. S., Heine, W. F., Ciccone, D. N., O'Neill, T. B., Crick, K. E., Pierce, K. A., Lane, W. S., Rathbun, G., et al. (2000) Nature 405, 477–482. [DOI] [PubMed] [Google Scholar]
- 25.Zhao, S., Weng, Y. C., Yuan, S. S., Lin, Y. T., Hsu, H. C., Lin, S. C., Gerbino, E., Song, M. H., Zdzienicka, M. Z., Gatti, R. A., et al. (2000) Nature 405, 473–477. [DOI] [PubMed] [Google Scholar]
- 26.Wang, X., Zou, L., Zheng, H., Wei, Q., Elledge, S. J. & Li, L. (2003) Genes Dev. 17, 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capasso, H., Palermo, C., Wan, S., Rao, H., John, U. P., O'Connell, M. J. & Walworth, N. C. (2002) J. Cell Sci. 115, 4555–4564. [DOI] [PubMed] [Google Scholar]