Abstract
Plant mannose-binding lectins (MBLs) are crucial for plant defense signaling during pathogen attack by recognizing specific carbohydrates on pathogen surfaces. In this study, we isolated and functionally characterized a novel pepper (Capsicum annuum) MBL gene, CaMBL1, from pepper leaves infected with Xanthomonas campestris pv vesicatoria (Xcv). The CaMBL1 gene contains a predicted Galanthus nivalis agglutinin-related lectin domain responsible for the recognition of high-mannose N-glycans but lacks a middle S-locus glycoprotein domain and a carboxyl-terminal PAN-Apple domain. The CaMBL1 protein exhibits binding specificity for mannose and is mainly localized to the plasma membrane. Immunoblotting using a CaMBL1-specific antibody revealed that CaMBL1 is strongly expressed and accumulates in pepper leaves during avirulent Xcv infection. The transient expression of CaMBL1 induces the accumulation of salicylic acid (SA), the activation of defense-related genes, and the cell death phenotype in pepper. The G. nivalis agglutinin-related lectin domain of CaMBL1 is responsible for cell death induction. CaMBL1-silenced pepper plants are more susceptible to virulent or avirulent Xcv infection compared with unsilenced control plants, a phenotype that is accompanied by lowered reactive oxygen species accumulation, reduced expression of downstream SA target genes, and a concomitant decrease in SA accumulation. In contrast, CaMBL1 overexpression in Arabidopsis (Arabidopsis thaliana) confers enhanced resistance to Pseudomonas syringae pv tomato and Alternaria brassicicola infection. Together, these data suggest that CaMBL1 plays a key role in the regulation of plant cell death and defense responses through the induction of downstream defense-related genes and SA accumulation after the recognition of microbial pathogens.
Plants protect themselves against a wide range of pathogens, such as bacteria, fungi, and viruses, with numerous defense mechanisms, including a complicated innate immune system against pathogens (Chisholm et al., 2006; Jones and Dangl, 2006). Plants can distinguish between self and nonself or detect specific pathogens by recognizing pathogen-associated molecular patterns mostly generated or secreted from pathogens. Defense responses mediated via signal transduction pathways lead to the reinforcement of plant cell walls, the production of antimicrobial metabolites (phytoalexins) and pathogenesis-related (PR) proteins, and the hypersensitive response (HR), a form of programmed cell death at infection sites that limits pathogen development (Dangl and Jones, 2001; Mur et al., 2008). Signaling molecules, such as reactive oxygen species (ROS), ethylene, salicylic acid (SA), and jasmonic acid, play important roles in the complex signaling networks that activate defense mechanisms (Hammond-Kosack and Parker, 2003). Although the regulation and execution of cell death associated with the HR are not fully understood, the production of ROS, ion fluxes, defense-related gene activation, and the induction of signaling molecules have been proposed to be implicated in the cell death process (Torres and Dangl, 2005).
Plant cell walls are dynamic structures whose composition and architecture change during growth, development, and defense responses. Preformed physical and chemical barriers, such as cell walls, and constitutively produced antimicrobial compounds protect plants against invading pathogens. Proteins embedded in the cell wall and plasma membrane are involved in a monitoring system that is required for the recognition and transduction of environmental, developmental, and defense-associated signals (Garcia-Brugger et al., 2006). These proteins are released from plants or pathogens, and pathogens can trigger cell wall-mediated defense responses. Disruption of cell wall-plasma membrane adhesion during pathogen penetration into host cells may lead to a reduction of cell wall-associated defense responses, thereby making the plant more susceptible to disease (Mellersh and Heath, 2001).
Carbohydrate-binding proteins, commonly referred to as lectins or agglutinins, are ubiquitous in many plant species (Peumans and Van Damme, 1995) and function in defense responses to pathogen invaders. The most abundant structural proteins in plant cell walls, Hyp-rich glycoproteins, are also induced in disease resistance responses, especially during incompatible plant-pathogen interactions (Davies et al., 1997). There is convincing evidence that Hyp-rich glycoproteins act as impenetrable physical barriers against pathogen ingress (Deepak et al., 2007). While the function of lectins in plants is believed to be the binding of glycoproteins on cell surfaces, their role in animals also includes the binding of soluble extracellular and intercellular glycoproteins. Plant lectins are involved in specific protein-carbohydrate interactions within the cytoplasmic and/or nuclear compartments (Van Damme et al., 2004). The broad spectrum of the carbohydrate-binding specificity of lectins can be interpreted as the successful recognition by plant cells of different types of sugar-containing receptors. Lectins bind to the glycans of glycoproteins, glycolipids, or polysaccharides with high affinity, and they are the only plant proteins that recognize the glycoconjugates present on the surfaces of microorganisms such as bacteria and fungi. For example, chitin-binding plant lectins recognize a carbohydrate that is a typical constituent of fungal cell walls (Broekaert et al., 1989). Several plant lectins play a role in the defense against bacteria through an indirect mechanism, based on an interaction with cell wall carbohydrates or extracellular glycans. The potato (Solanum tuberosum) lectin immobilizes avirulent strains of Pseudomonas solanacearum on the plant cell wall; however, a virulent strain is not recognized by lectins (Sequeira and Graham, 1977).
Recent studies demonstrate that all known plant lectins can be classified into 12 lectin families of structurally and evolutionarily related proteins (Van Damme et al., 2008). There are the amaranthins, Cucurbitaceae phloem lectins (now called the Nictaba lectins), lectins with hevein domains, jacalin-related lectins, legume lectins, monocot Man-binding lectins (now called the GNA-related lectins [for Galanthus nivalis agglutinin]), and type II ribosome-inactivating proteins (also known as the Ricin-B family) in plants (Van Damme et al., 2007b). More recently, galectins (previously called the S-type lectins, having a strong affinity for β-galactosides) and the calnexin/calreticulin lectin families, Agaricus bisporus agglutinins, class V chitinase homologs with lectin activity, EEA, LysM family, and cyanovirins, are newly identified and/or regrouped (Shridhar et al., 2009). However, the physiological role of lectins is poorly understood (Sequeira and Graham, 1977; Van Damme et al., 2008; Michiels et al., 2010).
Man-binding proteins that constitute one or two domains equivalent to the GNA purified from the bulbs of snowdrop are believed to play an important role in defense against microbial pathogens by recognizing Man-type glycans of foreign microorganisms (Van Damme et al., 1987, 1998, 2007b; Barre et al., 2002). GNA was originally considered a Man-specific lectin that possesses three similar Man-binding sites per subunit (Van Damme et al., 2008). Many of them interact only weakly with Man but exhibit a strong affinity toward oligomannosides and high-Man N-glycans. A monomeric Man/Glc-binding lectin inhibits the germination of Aspergillus flavus and Fusarium moniliforme spores and hyphal growth in red cluster pepper (Capsicum frutescens) seed (Ngai and Ng, 2007). Some lectin-like receptor-like kinases (LecRLKs) have been implicated in plant defense, senescence, and wounding (Riou et al., 2002). RLKs containing an extracellular Man-binding lectin (MBL) have also been reported in rice (Oryza sativa; Chen et al., 2006). The MBL domain in a Ser/Thr RLK gene, Pi-d2, is required for R gene-mediated resistance to rice blast. However, the function of plant carbohydrate-binding proteins containing this domain is not fully understood. Although several R genes that encode RLKs have been cloned and characterized, none has been shown to have an extracellular lectin domain. There is convincing in vitro evidence that some plant MBLs exert a deleterious effect on fungal pathogens. Gastrodianin, which is a monomeric Man-binding protein homolog in Gastrodia elata and Epipactis helleborine, displays in vitro antifungal activity against Rhizoctonia solani and Phytophthora nicotianae (Wang et al., 2001). Gastrodianin-like proteins also inhibit the mycelial growth of Botrytis cinerea, Gibberella zeae, Ganoderma lucidum, R. solani, and Valsa ambients (Cox et al., 2006). Dendrobium findleyanum agglutinin inhibits the growth of Alternaria alternata (Sudmoon et al., 2008).
In this study, we isolated genes differentially expressed in pepper (Capsicum annuum ‘Nokwang’) during the incompatible interaction with Xanthomonas campestris pv vesicatoria (Xcv). Among them, the novel pepper pathogen-responsive gene CaMBL1 encodes a putative glycoprotein with a GNA-related lectin domain that localizes to the plasma membrane. To our knowledge, this is the first report suggesting that glycoproteins with the GNA-related lectin domain play a crucial role for cell death and defense responses and the expression of defense-related genes in pepper plants. Here, we show that the CaMBL1 gene is involved in suppressing disease development in pepper plants during Xcv infection. We also report that overexpression of CaMBL1 in Arabidopsis (Arabidopsis thaliana) enhances innate immunity against Pseudomonas syringae pv tomato DC3000 (Pst DC3000) and Alternaria brassicicola.
RESULTS
Isolation and Identification of CaMBL1 cDNA
In a previous study, we constructed a cDNA library using RNA isolated from pepper leaves inoculated with the Xcv avirulent strain BV5-4a (Jung and Hwang, 2000). The full-length cDNA clone, designated CaMBL1 and containing glycoprotein-homologous sequences, was isolated from this pepper cDNA library. The cloned CaMBL1 cDNA had an insert of 1,160 bp containing an 894-bp coding region encoding a protein of 298 amino acids (Supplemental Fig. S1). The CaMBL1 cDNA sequence was analyzed using the BLASTx program (http://www.ncbi.nlm.nih.gov/BLAST/), the ExPASy Proteomics Server (http://www.expasy.org), and the PLecDom program (http://www.nipgr.res.in/plecdom.html). CaMBL1 shares 47% to 55% amino acid sequence identities with an unknown grape (Vitis vinifera) protein (accession no. CAN75015), the sugar beet (Beta vulgaris) SIEP1L protein (accession no. CAA61158), the carrot (Daucus carota) cell attachment protein (accession no. BAD24818), the Arabidopsis curculin-like (Man-binding) lectin family protein (CLLFP; At1g78860; accession no. NP_178007), the flax (Linum usitatissimum) secreted glycoprotein (accession no. AAO15899), and another Arabidopsis CLLFP (At1g78830; accession no. NP_565191; Supplemental Fig. S2). A database search revealed that CaMBL1 and related plant proteins have the GNA-related lectin domain commonly present in the plant lectin family. After the identification of GNA, similar lectins were isolated and characterized from many other plant species (Van Damme et al., 2007a). Previously, GNA and related lectins were classified into the so called “monocot Man-binding lectins.” Therefore, the term bulb-type MBL domain can no longer be used and has been replaced by GNA domain (Van Damme et al., 2007a, 2008). Based on the BLAST search with CaMBL1, we revealed that CaMBL1 contains the putative GNA-related lectin domain but lacks a middle S-locus glycoprotein domain (SLP) and a C-terminal PAN-Apple domain that are widespread in plant lectin proteins (Van Damme et al., 2008). Moreover, the GNA-SLP-PAN-type protein itself corresponds to the N-terminal half of the so-called receptor kinases, which constitute a transmembrane helix and a C-terminal protein kinase domain next to the GNA-SLP-PAN module. Notably, some S-locus receptor kinases that constitute these four domains were demonstrated in Brassicaceae species (Naithani et al., 2007). Thus, CaMBL1 containing only the GNA-related lectin domain may be a truncated homolog of the GNA-SLP-PAN-type protein in pepper plants.
A phylogenetic tree was constructed with CaMBL1 and homologous proteins retrieved from the GenBank database and the PLecDom program (Supplemental Fig. S3), in which MBL family members are divided into two groups containing GNA-related lectin domain or kinase domain. MBL proteins in the first group contain the GNA-related lectin domain alone, and those in the second group contain GNA-related lectin and kinase domains. Pepper GNA-related lectins including CaMBL1 were constructed in the phylogenetic tree with the GNA-related lectin members from sugar beet, grape, carrot, flax, and Arabidopsis (Supplemental Fig. S3). Some GNA-related lectins were identified in pepper, although the role of the GNA-related lectins remains unknown. CaMBL1 was closely related to the pepper GNA-related lectin (TC7720) rather than other pepper GNA-related lectins.
Expression of the CaMBL1 Gene in Pepper
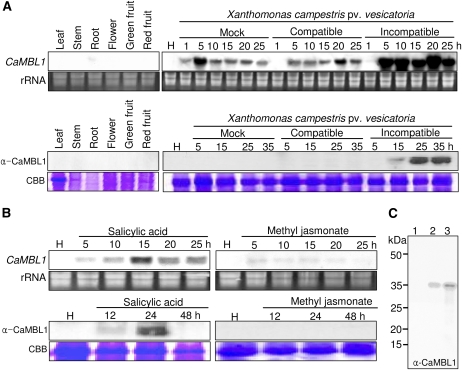
To determine the expression profiles of CaMBL1, we performed RNA gel-blot and immunoblot analyses of pepper plant tissues (Fig. 1). The expression of CaMBL1 was undetected or nearly so in leaves, stems, roots, flowers, and green and red fruits during normal growth and development (Fig. 1A). Northern analysis was also performed to determine whether CaMBL1 expression is induced by the virulent (compatible) Ds1 and avirulent (incompatible) Bv5-4a strains of Xcv. CaMBL1 transcript accumulation in pepper leaves began 5 h after inoculation with the avirulent Bv5-4a strain, and it reached maximal levels from 5 to 25 h (Fig. 1A). Mock inoculation and virulent Xcv infection induced CaMBL1 in pepper leaves in a similar manner. This indicates that infiltration by itself is sufficient to induce CaMBL1. Furthermore, infiltration may prime for enhanced expression of CaMBL1 upon inoculation with avirulent Xcv. However, avirulent Xcv infection led to high levels of CaMBL1 protein, as detected by western blotting. Treatments with SA, but not with methyl jasmonate, also significantly induced the CaMBL1 gene in pepper leaves 5 to 25 h after exposure (Fig. 1B). After SA treatment, transcripts were detected within 5 h, reached their highest levels between 10 and 15 h, and gradually decreased by 20 h. A high level of CaMBL1 protein expression was also detected 24 h after SA treatment. In contrast, treatment with methyl jasmonate did not induce CaMBL1 protein expression. We tested whether the anti-CaMBL1 antibody used in this study is specific to the corresponding CaMBL1 protein (Fig. 1C). The anti-CaMBL1 antibody raised in rabbits against a synthetic peptide that corresponds to C-terminal residues 284 to 297 of CaMBL1 specifically bound to His-tagged CaMBL1 proteins that were expressed in Escherichia coli.
Figure 1.
RNA gel- and western-blot analyses of the expression of CaMBL1 in pepper plants. A, Expression of the CaMBL1 gene and CaMBL1 protein in healthy organs of pepper plants and in pepper leaves at various times after inoculation with the virulent strain Ds1 and the avirulent strain Bv5-4a of Xcv. B, Expression of the CaMBL1 gene and CaMBL1 protein in pepper leaves at various times after treatment with SA (5 mm) and methyl jasmonate (100 μm). Equal loadings (10 μg) of RNA were verified by visualizing rRNA on gels stained with ethidium bromide. Coomassie Brilliant Blue (CBB) staining is shown for the 60-kD region of protein extracts. C, Immunoblot of His-tagged CaMBL1 protein using the anti-CaMBL1 antiserum raised in rabbits against a synthetic peptide corresponding to C-terminal residues 284 to 297 of CaMBL1. His-tagged CaMBL1 proteins expressed in E. coli BL21 was used. Lane 1, Uninduced E. coli BL21 cell extracts (1 μg of protein); lane 2, 0.1 μg of His-tagged CaMBL1 protein expressed in E. coli BL21 cells after isopropylthio-β-galactoside induction; lane 3, 1 μg of His-tagged CaMBL1 protein expressed in E. coli BL21 cells after isopropylthio-β-galactoside induction. [See online article for color version of this figure.]
Binding of CaMBL1 to d-Man
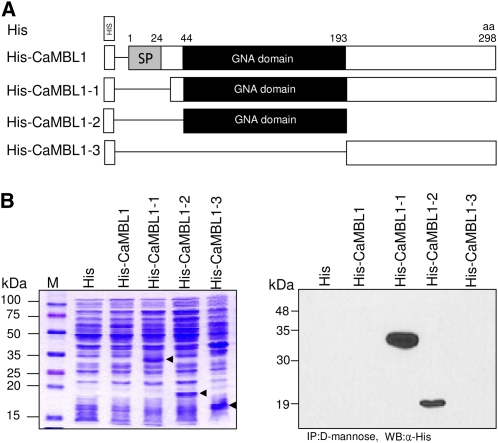
CaMBL1 was predicted to contain the GNA-related lectin domain. To determine whether it has an essential function as an MBL, we generated a series of CaMBL1 deletion mutants (Fig. 2A) and expressed them in E. coli as fusion proteins with a 6×His tag at the N terminus (Fig. 2B, left panel). As a result, His-CaMBL1-1 that lacks the putative signal peptide region, His-CaMBL1-2 that contains a GNA-related lectin domain, and His-CaMBL1-3 that contains only the C-terminal region were distinctly expressed in E. coli, followed by their purification using d-Man-agarose columns. However, full-length 6×His-tagged CaMBL1 was not expressed in E. coli (no arrowhead), even with the use of a GNA-related lectin domain and glutathione S-transferase-tagged ones (data not shown). This indicated that the presence of the signal peptide may block the expression of CaMBL1 in E. coli. Immunoblot analysis of the purified CaMBL1 proteins using the anti-His antibody showed that protein bands from CaMBL1 deletion mutants that lack the signal peptide region but contain the GNA-related lectin domain region could be immunodetected by this antibody (Fig. 2B, right panel). These immunoblotting data indicate that the GNA-related lectin domain of CaMBL1 is essential for its binding to d-Man. The glycan array screening of CaMBL1 was performed by the Consortium for Functional Glycomics at the School of Medicine, Emory University. Binding levels of CaMBL1 to the various glycans on the arrays were measured by fluorescence scanning (Supplemental Fig. S5; Supplemental Table S1). These glycan array data indicate that CaMBL1 has affinity toward Manα and/or Manβ and GalNAc residues.
Figure 2.
Binding of CaMBL1 to d-Man. A, Schematic representation of CaMBL1 and CaMBL1 deletion constructs. B, Immunoblot analysis of CaMBL1 using the anti-His antibody. His-tagged CaMBL1 and CaMBL1 deletion constructs were expressed in E. coli (left panel) and purified using d-Man-agarose columns. Arrowheads indicate expressed His-tagged recombinant proteins. aa, Amino acids; CaMBL1-1, signal peptide deleted; CaMBL1-2, GNA-related lectin domain only; CaMBL1-3, C-terminal region only; GNA domain, GNA-related lectin domain; His, His tag; SP, signal peptide region; α-His, anti-His antibody; IP, immunoprecipitation; M, molecular mass markers; WB, western blotting. [See online article for color version of this figure.]
Subcellular Localization of CaMBL1
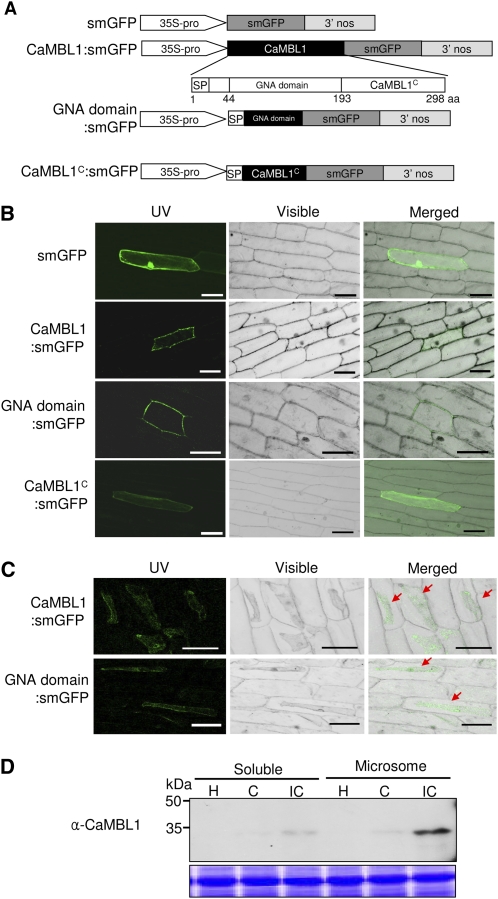
To identify the subcellular location of the CaMBL1 protein, and to determine whether it depends on the GNA-related lectin domain, soluble modified GFP (smGFP) was fused to the full-length CaMBL1 protein (CaMBL1:smGFP), the GNA-related lectin domain region (GNA domain:smGFP), and the C-terminal region (CaMBL1c:smGFP; Fig. 3A). The subcellular localization of these fusion proteins was visualized in onion (Allium cepa) epidermal cells by confocal microscopy. The merging of fluorescence and bright-field images revealed that both the full-length and GNA-related lectin domain fusion proteins localized to the plasma membrane, whereas the C-terminal region of CaMBL1 was expressed only in the cytosol (Fig. 3B). In contrast, the control smGFP was uniformly distributed throughout the cell. Onion epidermis was treated with 1 m NaCl to induce plasmolysis. The effect of plasmolysis on onion cells expressing full-length and GNA-related lectin domain fusion proteins also confirmed that CaMBL1 is localized to plasma membrane and that the GNA-related lectin domain is required for its localization (Fig. 3C). We also tested the subcellular localization of CaMBL1 in pepper leaves. Total proteins were extracted from the pepper leaves during Xcv infection. To separate soluble and microsomal fractions, total proteins were centrifuged at 92,000g for 90 min. Immunoblotting data with anti-CaMBL1 antibody indicated that CaMBL1 was enriched in the microsomal fraction including the plasma membrane or the endoplasmic reticulum membrane from the leaves infected by the avirulent (incompatible) strain Bv5-4a of Xcv. However, CaMBL1 was slightly detected in the soluble fraction including secretory proteins (Fig. 3D). Collectively, these results indicate that CaMBL1 is a membrane-associated protein and requires the GNA-related lectin domain to localize to the plasma membrane.
Figure 3.
Subcellular localization of CaMBL1. A, Schematic representation of CaMBL1 constructs. B, Bright-field images of subcellular localization of CaMBL1:smGFP, GNA domain:smGFP, and CaMBL1C:smGFP proteins in onion epidermal cells. GFP images were observed by confocal laser scanning fluorescence microscopy. Bars = 0.1 mm. C, Subcellular localization of CaMBL1:smGFP and GNA domain:smGFP in onion epidermal cells after plasmolysis with 1 m NaCl. Red arrows indicate plasmolyzed plasma membrane. Bars = 0.1 mm. D, Immunodetection of CaMBL1 with anti-CaMBL1 antibody in soluble and microsomal fractions from pepper leaves infected by Xcv. H, Healthy leaves; C, compatible; IC, incompatible.
Induction of Cell Death by Transient Expression of CaMBL1
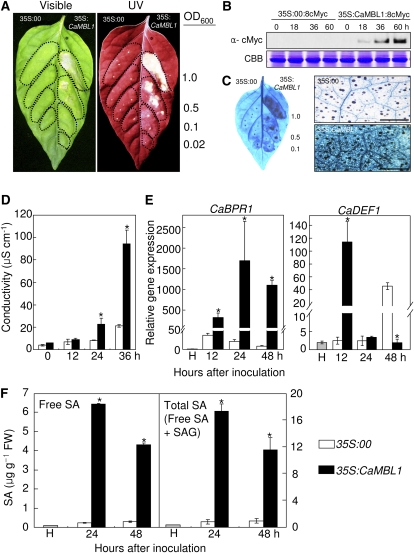
To further test whether the CaMBL1 gene and the CaMBL1-mediated signaling pathway affect the induction of cell death, we transiently expressed CaMBL1 in pepper leaves by infiltration with Agrobacterium tumefaciens carrying 35S:00 (empty vector) or 35S:CaMBL1 constructs. As shown in Figure 4A, a distinct necrotic phenotype was observed in pepper leaves 4 d after infiltration with different concentrations of Agrobacterium carrying 35S:CaMBL1. Whitish necrotic symptoms appeared in pepper leaves 48 h after agroinfiltration. In contrast, infiltration with the Agrobacterium empty vector control did not induce a cell death response. Plant cells undergoing HR-like cell death accumulated autofluorescent compounds, which may contain cross-linked phenolics that could serve to strengthen the cell wall (Dixon and Paiva, 1995). As observed under UV illumination, autofluorescence was detected in pepper leaves as early as 2 d after infiltration with Agrobacterium (35S:CaMBL1). This result indicates that CaMBL1 transient expression triggers the accumulation of fluorescent phenolic compounds as a cell death-related defense reaction. An increased expression of c-Myc-tagged CaMBL1 was detected in leaf tissues 18, 36, and 60 h after infiltration with Agrobacterium (35S:CaMBL1), as shown by western-blot analysis (Fig. 4B). However, the protein expressed by 35S:c-Myc was not transiently induced in empty vector control leaves. Large clusters of dead cells that stained with trypan blue were seen in the Agrobacterium (35S:CaMBL1)-infiltrated leaf areas but not in leaves transfected with the empty vector (Fig. 4C). The severity of cell necrosis caused by membrane damage in leaves was estimated by measuring electrolyte leakage from leaf tissues. Pepper leaves that transiently expressed CaMBL1 exhibited drastic electrolyte leakage compared with empty vector control leaves 36 h after infiltration, when the cell death phenotype appeared (Fig. 4D). We used real-time reverse transcription (RT)-PCR to determine whether the HR-like cell death in leaves transiently expressing CaMBL1 is accompanied by the expression of defense-related genes (Fig. 4E). CaBPR1 (for basic pathogenesis-related gene) was selected as a marker gene responsible for the activation of plant defense responses. The transcript level of CaBPR1 increased significantly during the transient expression of CaMBL1, indicating that induction by CaMBL1 contributes to the defense response in plants. However, the transcript level of CaDEF1 (for defensin) was not affected by CaMBL1 transient expression, except for that at the 12-h time point. As shown in Figure 4F, increased CaMBL1 expression led to the accumulation of free SA and total SA (free SA plus Glc-conjugated SA) in pepper leaves. In particular, SA levels 24 and 48 h after infiltration were significantly higher in leaves transiently expressing CaMBL1 than those in leaves transfected with the empty vector control. Increased SA levels in CaMBL1 transiently expressed leaves 24 h after agroinfiltration were higher than those 48 h after agroinfiltration. These increased SA levels correlated with the increased CaBPR1 expression levels during the transient expression of CaMBL1. Overall, these results indicate that the transient expression of CaMBL1 induces cell death in pepper leaves, which is supported by the expression of the defense-related gene CaBPR1 and SA accumulation.
Figure 4.
Transient expression of cell death in pepper leaves infiltrated with Agrobacterium GV3101 carrying the 35S:00 (empty vector) or 35S:CaMBL1 construct. A, Visible and UV light-illuminated phenotypes of CaMBL1-transiently expressing leaves 4 d after infiltration with different bacterial concentrations (OD600). B, Expression of CaMBL1 protein in empty vector control leaves and 35S:CaMBL1 leaves as detected by immunoblotting. Coomassie Brilliant Blue (CBB) staining is shown for the 60-kD regions of protein extracts. C, Staining with trypan blue of leaf tissues 1 d (right) and 2 d (left) after infiltration. Bars = 0.5 mm. D, Electrolyte leakage assay of CaMBL1-transiently expressing leaves at different times after agroinfiltration. E, Real-time quantitative PCR analysis of CaBPR1 and CaDEF1 in CaMBL1-transiently expressing leaves. Transcript levels were normalized to the expression of pepper 18S ribosomal RNA measured in the same samples. F, Levels of SA in CaMBL1-transiently expressing leaves. All experiments were repeated three times with similar results. Values are presented as means ± sd. Asterisks indicate significant differences between the means, as determined by Student’s t test (P < 0.05). FW, Fresh weight; H, healthy plants. [See online article for color version of this figure.]
The GNA-Related Lectin Domain of CaMBL1 Is Responsible for Cell Death Induction
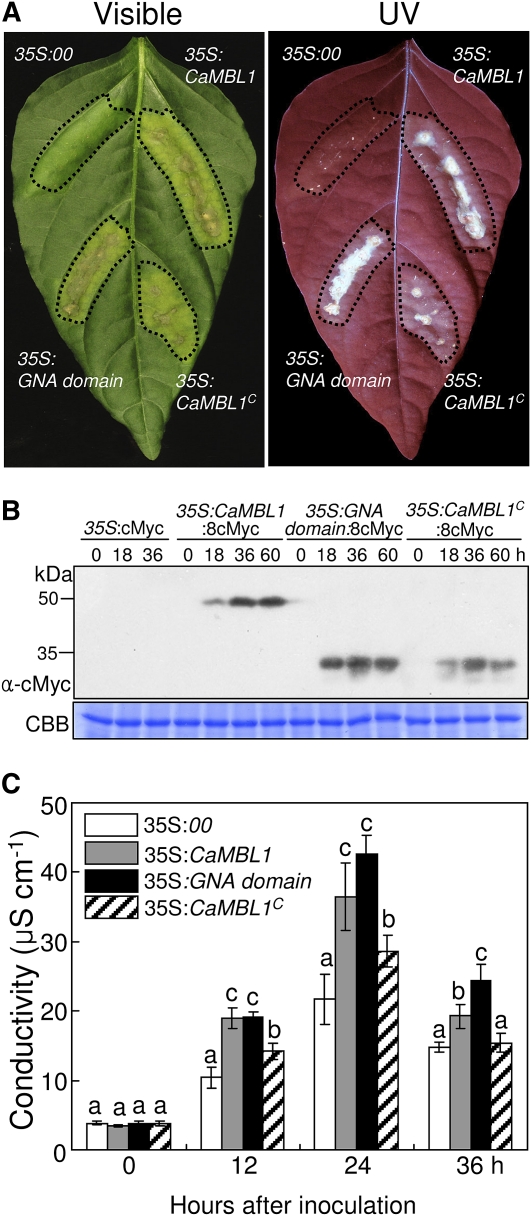
Induction of CaMBL1 by Agrobacterium-mediated transient expression led to cell death, which was accompanied by defense responses, including the induction of defense-related genes and SA accumulation (Fig. 4). Within 36 to 48 h after agroinfiltration, hypersensitive cell death was seen in the infiltrated area of pepper leaves, which collapsed 3 to 4 d later (data not shown). To determine which region of the CaMBL1 gene is responsible for the induction of cell death in pepper leaves, we transformed the constructs pBIN:CaMBL1, pBIN:GNA domain, and pBIN:CaMBL1c, which contains only the C-terminal region, into Agrobacterium strain GV3101. As expected, the transient expression of 35S:CaMBL1 and of 35S:GNA domain induced much more hypersensitive cell death 4 d after agroinfiltration compared with that induced by 35S:CaMBL1c (Fig. 5A). All three proteins from these constructs were detected in leaf protein extracts by immunoblotting using an anti-cMyc antibody (Fig. 5B), showing that the constructs are transiently expressed in pepper leaves. The hypersensitive cell death responses were also well supported by ion conductivity data (Fig. 5C). Distinct electrolyte leakage, a measure of membrane damage, began within 12 h after agroinfiltration. Leaves transiently expressing 35S:CaMBL1 and 35S:GNA domain exhibited significantly enhanced electrolyte leakage compared with empty vector and CaMBL1c-expressing leaves 12, 24, and 36 h after agroinfiltration (Fig. 5C). Collectively, these data indicate that the GNA-related lectin domain of the CaMBL1 gene contributes to CaMBL1-specific cell death induction.
Figure 5.
Agrobacterium-mediated transient expression of 35S:00 (empty vector), 35S:CaMBL1, 35S:GNA domain, and 35S:CaMBL1c in pepper leaves. A, Visible and UV light-illuminated phenotypes of the 35S:00 (empty vector), 35S:CaMBL1, 35S:GNA domain, and 35S:CaMBL1c-transiently expressing pepper leaves 4 d after agroinfiltration. B, Immunoblot analysis of 35S:8cMyc, 35S:CaMBL1:8cMyc, 35S:GNA domain:8cMyc, and 35S:CaMBL1c:8cMyc protein expression in pepper leaves using an anti-cMyc antibody. Coomassie Brilliant Blue (CBB) staining is shown for the 60-kD regions of protein extracts. C, Electrolyte leakage assay of 35S:00 (empty vector), 35S:CaMBL1, 35S:GNA domain, and 35S:CaMBL1c-transiently expressing pepper leaves at different times after agroinfiltration. All experiments were repeated three times with similar results. Values are presented as means ± sd. Different letters indicate significant differences as determined by Fisher’s lsd test (P < 0.05).
Enhanced Susceptibility of CaMBL1-Silenced Pepper to Xcv Infection
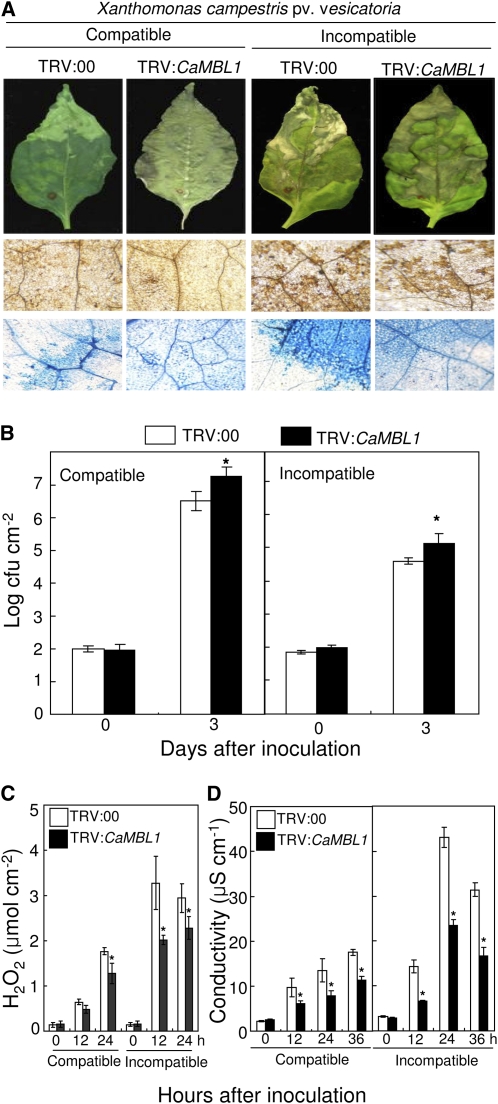
To characterize the loss of function of CaMBL1 in pepper plants, we used the virus-induced gene silencing (VIGS) system with tobacco rattle virus (TRV)-based vectors to knock down the expression of CaMBL1. CaMBL1-silenced pepper plants (TRV:CaMBL1) were morphologically comparable to empty vector control plants (TRV:00). We examined the levels of resistance to Xcv infection in CaMBL1-silenced plants. Three and 4 weeks after VIGS, empty vector control and silenced plants were challenged with the virulent strain Ds1 (compatible) and the avirulent strain Bv5-4a (incompatible) of Xcv. Disease symptoms on pepper plants were monitored 7 d after inoculation (Fig. 6A). In leaves infected with the virulent strain Ds1, silencing of CaMBL1 resulted in severe disease symptoms accompanied by increased bacterial growth (Fig. 6, A and B). The virulent Xcv strain Ds1 grew less in empty vector control (TRV:00) leaves than in CaMBL1-silenced leaves during infection, especially 3 d after inoculation with 5 × 104 colony-forming units (cfu) mL−1 (Fig. 6A). In the compatible interaction with Xcv Ds1, neither the empty vector control leaves nor CaMBL1-silenced leaves exhibited high levels of hydrogen peroxide (H2O2; Fig. 6C) or hypersensitive cell death. In the incompatible interaction with the Xcv avirulent strain Bv5-4a, silencing of the CaMBL1 gene enhanced bacterial growth in pepper leaves (Fig. 6B), which was accompanied by susceptible disease symptoms. We further measured H2O2 formation using the xylenol orange assay and also hypersensitive cell death by monitoring electrolyte leakage. Silencing of CaMBL1 compromised the induction of H2O2 and electrolyte leakage in pepper leaves infected by either virulent or avirulent Xcv strains (Fig. 6, C and D). These data indicate that the CaMBL1 gene is involved in the early events of cell death and defense responses of pepper plants.
Figure 6.
Enhanced susceptibility of CaMBL1-silenced pepper plants to Xcv infection with the virulent (compatible) strain Ds1 and the avirulent (incompatible) strain Bv5-4a. A, Disease symptoms developed on the empty vector control (TRV:00) and silenced (TRV:CaMBL1) leaves infected by Xcv strains. Infected leaves were stained with DAB and trypan blue. Bars = 0.2 mm. B, Bacterial growth in leaves of the empty vector control (TRV:00) and CaMBL1-silenced (TRV:CaMBL1) plants 0 and 3 d after inoculation with Xcv strains (5 × 104 cfu mL−1). C, Quantification of H2O2 in leaves of empty vector control plants and CaMBL1-silenced plants after inoculation with Xcv strains using the xylenol orange assay. D, Measurement of electrolyte leakage from the leaf tissues at different time points after inoculation with Xcv strains. All experiments were repeated three times with similar results. Values are presented as means ± sd. Asterisks indicate significant differences between the means as determined by Student’s t test (P < 0.05).
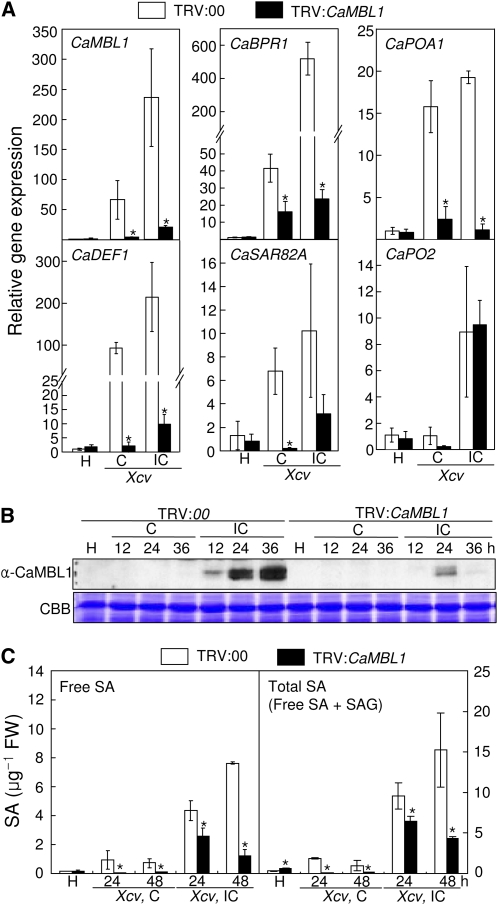
We next examined the expression profiles of CaMBL1 and defense-related genes in empty vector control (TRV:00) and silenced (TRV:CaMBL1) pepper leaves in compatible and incompatible interactions with Xcv. The CaMBL1 gene was almost uninduced in CaMBL1-silenced leaves during Xcv infection, indicating that CaMBL1 silencing was successfully achieved. More importantly, expression of the CaBPR1 (a SA molecular marker), CaDEF1, CaSAR82A (for systemic acquired resistance [SAR] 8.2), and CaPOA1 (for ascorbic peroxidase) genes was significantly reduced in CaMBL1-silenced plants during Xcv infection (Fig. 7A). Immunoblot analysis using the anti-CaMBL1 antibody identified a strong expression of the CaMBL1 protein during incompatible interactions, peaking at 36 h after Xcv inoculation in empty vector control plants (Fig. 7B). However, CaMBL1 proteins were not induced in empty vector control leaves during Xcv virulent (compatible) infection. In contrast, induction of CaMBL1 was compromised in silenced pepper leaves during avirulent Xcv infection. To test whether defense responses were compromised in CaMBL1-silenced plants, SA levels in empty vector control and CaMBL1-silenced leaves were measured using HPLC (Fig. 7C). CaMBL1-silenced leaves exhibited significantly lower levels of free SA and total SA (free SA plus Glc-conjugated SA) than empty vector control leaves during compatible and incompatible interactions with Xcv.
Figure 7.
Effects of CaMBL1 silencing on gene expression, protein expression, and SA accumulation in pepper leaves infected by the virulent (compatible) strain Ds1 and the avirulent (incompatible) strain Bv5-4a of Xcv. A, Real-time quantitative PCR analysis of the expression of CaMBL1 and pepper defense-related genes in empty vector control leaves (TRV:00) and CaMBL1-silenced leaves (TRV:CaMBL1) at 18 h after inoculation with Xcv. The experiments were carried out three times. Transcript levels were normalized to the expression of pepper 18S ribosomal RNA measured in the same samples. H, Healthy leaves; C, compatible; IC, incompatible. B, Western-blot analysis of CaMBL1 proteins in empty vector control and CaMBL1-silenced leaves. Coomassie Brilliant Blue (CBB) staining is shown for the 60-kD regions of protein extracts. C, Levels of SA in empty vector control and CaMBL1-silenced leaves. Values are presented as means ± sd. Asterisks indicate significant differences with respect to empty vector control plants (Student’s t test, P < 0.05). FW, Fresh weight. [See online article for color version of this figure.]
Enhanced Resistance of CaMBL1-Overexpression Arabidopsis to P. syringae pv tomato Infection
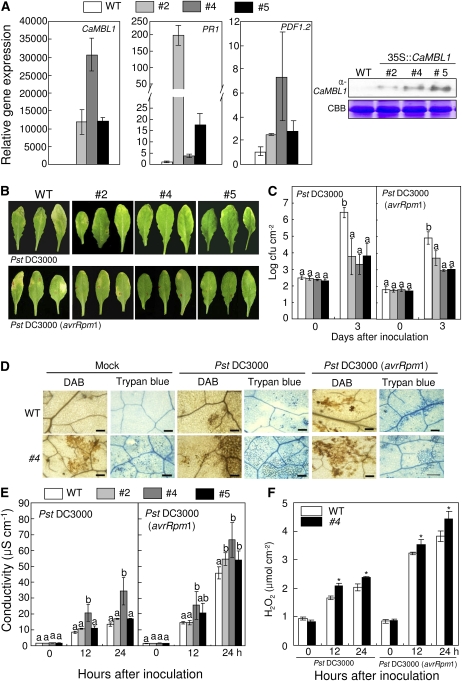
Because stable transformation of pepper plants is difficult, we constructed transgenic Arabidopsis plants constitutively expressing CaMBL1 under the control of the cauliflower mosaic virus 35S constitutive promoter to study the gain-of-function phenotype of CaMBL1 in heterologous plants during pathogen infection. When detected with the CaMBL1-specific antibody by western blotting, at least 10 independent transgenic lines were found to constitutively express the CaMBL1 protein, which was not detected in wild-type plants. Three transgenic lines with a single insertion of the CaMBL1 transgene, lines 2, 4, and 5, were selected for this study (Fig. 8). Expression of the pathogenesis-related genes PR1 and PDF was constitutively up-regulated in the three CaMBL1-overexpression (OX) transgenic lines compared with that in wild-type plants, although the levels of expression differed among these lines (Fig. 8A). CaMBL1-OX transgenic plants did not exhibit any apparent phenotypic abnormality compared with wild-type plants (data not shown). The three CaMBL1 transgenic Arabidopsis lines were tested for resistance against Pst DC3000 and the Pst DC3000 strain carrying avrRpm1 (Pst DC3000 avrRpm1). Five days after inoculation, the leaves of CaMBL1-OX plants exhibited mild chlorotic lesions compared with wild-type leaves (Fig. 8B). Overexpression of CaMBL1 significantly inhibited the growth of Pst DC3000 and Pst DC3000 avrRpm1 in Arabidopsis leaves (Fig. 8C). Reduction of disease symptoms was intimately correlated with the bacterial growth suppressed in the leaves of CaMBL1-OX plants.
Figure 8.
Enhanced resistance of CaMBL1-OX transgenic Arabidopsis plants to P. syringae pv tomato infection. A, Real-time quantitative PCR and western-blot analyses of the expression of CaMBL1 and Arabidopsis defense-related genes in wild-type (WT) and transgenic plants (lines 2, 4, and 5). Transcript levels were normalized to the expression of pepper 18S ribosomal RNA measured in the same samples. B, Disease symptoms of wild-type and transgenic plants inoculated with Pst DC3000 strains. Leaves of 4-week-old Arabidopsis plants were infiltrated with a suspension (105 cfu mL−1) of Pst DC3000 or Pst DC3000 avrRpm1. Disease symptoms were photographed 5 d after inoculation. C, Bacterial growth in leaves of wild-type and transgenic lines inoculated with Pst DC3000 and Pst DC3000 avrRpm1. D, DAB and trypan blue staining of leaf tissues of wild-type and transgenic plants 24 h after inoculation with Pst DC3000 or Pst DC3000 avrRpm1. Bars = 0.1 mm. E, Quantification of electrolyte leakage from leaf tissues inoculated with Pst DC3000 or Pst DC3000 avrRpm1. Samples were taken 0, 12, and 24 h after inoculation. F, Quantification of H2O2 in wild-type and transgenic plants after inoculation with Pst DC3000 or Pst DC3000 avrRpm1 using the xylenol orange assay. Values are presented as means ± sd. Different letters indicate significant differences from three independent experiments based on the lsd test (P < 0.05). Asterisks indicate significant differences with respect to empty vector control plants (Student’s t test, P < 0.05). [See online article for color version of this figure.]
We next examined the necrotizing process at the microscopic level by assessing H2O2 production after 3,3′-diaminobenzidine (DAB) staining (dark brown) and HR-like cell death after trypan blue staining (dark blue) of leaves infected by Pst DC3000 and Pst DC3000 avrRpm1 (Fig. 8D). DAB polymerizes instantly and locally upon contact with H2O2 to form reddish brown polymers. CaMBL1-OX plants, which were either uninoculated or inoculated with Pst DC3000 or Pst DC3000 avrRPM1, exhibited DAB-stained spots, indicating H2O2 accumulation to high levels. CaMBL1-OX plants exhibited significantly increased dark blue zones (HR-like cell death) compared with wild-type plants after inoculation with Pst DC3000 and Pst DC3000 avrRPM1. Cell death associated with the HR was quantified using the electrolyte leakage assay (Fig. 8E). CaMBL1-OX leaves exhibited higher levels of electrolyte leakage compared with wild-type leaves during Pst DC3000 and Pst DC3000 avrRpm1 infection. We next quantified H2O2 levels at different times after infection using the xylenol orange assay (Fig. 8F). H2O2 production was significantly promoted in CaMBL1-OX leaves during Pst DC3000 and Pst DC3000 avrRPM1 infection, as observed by DAB staining. Together, these results suggest that CaMBL1 overexpression in Arabidopsis may contribute to both basal defense and HR-mediated resistance to hemibiotrophic Pst infection.
Role of CaMBL1 and the Arabidopsis Ortholog At1g78830 in Resistance to A. brassicicola Infection
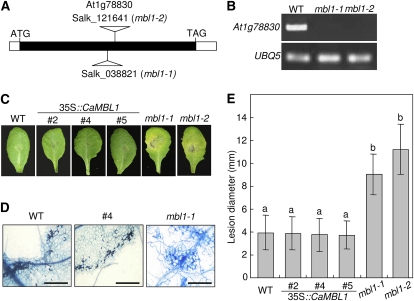
To investigate whether the expression of CaMBL1 and the Arabidopsis ortholog At1g78830 affects fungal infection, a spore suspension of A. brassicicola was drop inoculated on leaves of wild-type (ecotype Columbia [Col-0]), CaMBL1-OX, and At1g78830-defective Arabidopsis plants (Fig. 9). The predicted amino acid sequence of At1g78830, which contains an MBL domain, is 48% identical to that of CaMBL1 (Supplemental Figs. S2 and S3). Two At1g78830 T-DNA insertion mutants were investigated, mbl1-1 (SALK_003821) and mbl1-2 (SALK_121641; Fig. 9A). RT-PCR analysis did not detect At1g78830 transcripts in mbl1-1 or mbl1-2 plants during Pst DC3000 avrRpm1 infection (Fig. 9B), indicating that the At1g78830 function is completely lost due to the T-DNA insertion. Typical spreading of susceptible lesions was observed in the leaves of mbl1-1 and mbl1-2 plants 5 d after inoculation with A. brassicicola (Fig. 9, C and E). However, wild-type and CaMBL1-OX leaves exhibited small necrotic lesions no larger than the initial inoculation droplet during the A. brassicicola infection. Trypan blue staining of infected leaf tissues showed restricted hyphal growth and cell death in wild-type and CaMBL1-OX leaves but extensive hyphal growth in At1g78830 mutant leaves (Fig. 9D). These data indicate that the Arabidopsis ortholog At1g78830 is required for resistance to A. brassicicola infection of Arabidopsis plants.
Figure 9.
Disease development on the leaves of wild-type (WT), CaMBL1-OX, and Arabidopsis ortholog mutant (mbl1-1 and mbl1-2) plants inoculated with A. brassicicola. Leaves of 4-week-old Arabidopsis plants were inoculated with 10-μL droplets of 5 × 104 spores mL–1. A, Schematic representation of T-DNA insertion sites in mbl1-1 (SALK_038821) and mbl1-2 (SALK_121641) plants and the genomic structure of the At1g78830 gene. The At1g78830 exon is represented by the black box. B, RT-PCR analysis of expression of the mbl1-1 and mbl1-2 genes 15 h after inoculation with Pst DC3000 avrRpm1. C, Disease symptoms on leaves 5 d after inoculation. D, Microscopic images of fungal structures and damaged cells of infected leaves stained with trypan blue. Bars = 0.1 mm. E, Quantification of disease development based on the average diameter of lesions caused by A. brassicicola. The size of lesions is an average of 30 lesions per line. Values are presented as means ± sd. Different letters indicate significant differences from three independent experiments based on the lsd test (P < 0.05). [See online article for color version of this figure.]
Distinct Responses of CaMBL1-OX and CaMBL1 Arabidopsis Ortholog, mbl1, Mutants to Hyaloperonospora arabidopsidis and P. syringae pv tomato Infection
To determine whether the Arabidopsis defense response to biotrophic oomycete infection is induced by CaMBL1 and the CaMBL1 Arabidopsis ortholog, we tested the responses of the wild type (Col-0), CaMBL1-OX Arabidopsis, and the CaMBL1 Arabidopsis ortholog At1g78830 mutant (mbl1-1 and mbl1-2) plants to the biotrophic oomycete H. arabidopsidis. Over 100 wild-type and CaMBL1-OX plants were inoculated with an asexual spore suspension of H. arabidopsidis isolate Noco2 (5 × 104 conidiospores mL−1), which is virulent to Arabidopsis Col-0. As shown in Supplemental Figure S4A, the infected cotyledons of wild-type plants exhibited high levels of mycelial growth, sporulation, and sporangiophores. CaMBL1-OX plants were also highly susceptible to H. arabidopsidis isolate Noco2. No differences were observed between wild-type and CaMBL1-OX plants in mycelial growth, sporulation, and sporangiophores, as observed by trypan blue staining. The formation of conidiospores was scored for 7 d (Supplemental Fig. S4B). Extensive sporulation occurred in wild-type and CaMBL1-OX plants. Five days after inoculation, H. arabidopsidis infection was evaluated by measuring asexual sporangiophores on Arabidopsis cotyledons (Supplemental Fig. S4C). CaMBL1-OX plants exhibited the formation of sporangiophores similar to that on the cotyledons of wild-type plants. These results indicate that the CaMBL1 gene does not confer enhanced defense response against the biotrophic oomycete H. arabidopsidis isolate Noco2. No significant defense responses to H. arabidopsidis isolate Noco2 and Pst DC3000 were observed in CaMBL1 Arabidopsis ortholog At1g78830 mutant plants (mbl1-1 and mbl1-2), as assayed by measuring sporangiophores and bacterial growth (Supplemental Fig. S4, D and E).
DISCUSSION
MBLs (recently called the GNA-related lectins) are involved in many biological processes, including cell-to-cell and host-pathogen interactions, by specifically binding to carbohydrates (Peumans and Van Damme, 1995; Lis and Sharon, 1998; Van Damme et al., 2008). Notably, MBLs play a crucial role in the innate immune response through binding to carbohydrates on the surface of a wide range of microbial pathogens (Vijayan and Chandra, 1999; Barre et al., 2001). In this study, we isolated and functionally characterized the pepper CaMBL1 gene, which was rapidly and strongly induced in pepper leaves during incompatible interactions with Xcv. The CaMBL1 protein contains the GNA-related lectin domain that is conserved in rice (Barre et al., 2001). Lectins known to be carbohydrate-binding proteins are ubiquitous in plants, and MBLs that contain the GNA-related lectin domain are representative of a new plant lectin family (Van Damme et al., 2008). Among the MBL genes, there are also LecRLK genes encoding receptor-like kinases with predicted extracellular bulb-type Man-specific B-lectin and intracellular Ser/Thr kinase domains (Riou et al., 2002). Some LecRLKs have been shown to be involved in Rhizobium symbiosis and in other cellular processes (Navarro-Gochicoa et al., 2003; Shiu et al., 2004). More recently, the rice LecRLK gene Pi-d2 with these domains was suggested to confer resistance to the Chinese blast strain ZB15 of Magnaporthe oryzae (Chen et al., 2006). Intriguingly, a specific feature of the CaMBL1 gene is that it contains only the GNA-related lectin domain, unlike these LecRLK genes, and phylogenetic analysis suggests that CaMBL1 is more closely related to the group having only the GNA-related lectin domain. Notably, some pepper GNA-related lectins fall into similar clusters with CaMBL1 in the phylogenetic tree.
It has been demonstrated that the consensus sequence motif (QxDxNxVxY) of GNA-related lectin domain is involved in α-d-Man recognition (Van Damme et al., 2008). However, CaMBL1 containing the GNA-related lectin domain lacks the putative Man-binding site sequence (QxDxNxVxY). Thus, we tested whether CaMBL1 has the Man-binding ability. Using d-Man-agarose (Sigma), we found that His-tagged CaMBL1 could bind to d-Man-agarose, suggesting that CaMBL1 may have Man-binding ability without the intact consensus sequence motif (QxDxNxVxY). The immunoblot analysis of purified CaMBL1 deletion mutants strongly supports the notion that its GNA-related lectin domain is responsible for binding d-Man, suggesting that pepper GNA-related lectin may recognize Man on the Xcv cell surface. However, we could not find any direct interaction (agglutination) between Xcv and purified CaMBP1 fusion protein (data not shown). Notably, the glycan array screening of CaMBL1 performed by the Consortium for Functional Glycomics also indicates that CaMBL1 has affinity toward Manα and/or Manβ and GalNAc residues. Tsutsui et al. (2006) revealed that only the first of the two consensus sequence motifs (QxDxNxVxY) is required for Man-binding activity of pufflectins identified in the skin mucus of the fugu (Takifugu rubripes). In that study, three mutants of the second motif (QxDxNxVxY) could bind to Man. These findings supports the possibility that the consensus sequence motif (QxDxNxVxY) of the GNA-related lectin domain is not necessarily essential for Man-binding activity.
Animal MBLs bind Man-based carbohydrates on bacterial plasma membranes (Fujita et al., 2004). The finding that CaMBL1 encodes a GNA-related lectin domain is consistent with the observation that GFP-tagged CaMBL1 localizes to the plasma membrane of onion epidermal cells, suggesting that CaMBL1 does the same in its native pepper cells. We also tested the subcellular localization of CaMBL1 in pepper leaves. Indeed, CaMBL1 was mainly detected in the microsomal fraction including plasma membrane from pepper leaves during Xcv infection. In a subcellular localization experiment with a GNA domain:smGFP construct, we also confirmed that the GNA-related lectin domain is responsible for the localization of CaMBL1 to the plasma membrane. Together, these data suggest that the plasma membrane targeting of the CaMBL1 protein may be involved in the specific recognition of plant pathogens by GNA-related lectins. This proposal is well supported by the finding of Chen et al. (2006) that a B-lectin receptor kinase protein that triggers rice blast resistance is plasma membrane localized.
Most plant lectins may be involved in defense responses during pathogen infection by recognizing a wide range of pathogens (Peumans and Van Damme, 1995). Lectins are generally expressed at relatively low levels in a tissue- or organ-specific manner. In contrast, some lectins are induced to higher levels in plants following attack by other organisms or by stress. Plant lectins, including the GNA-related lectins, are believed to play a role in recognizing and binding high-Man glycans of foreign microorganisms or plant pathogens (Barre et al., 2001). GNA-related lectins are associated with specific resistance responses to herbivorous higher animals or phytophagous invertebrates (Peumans et al., 2002). In wheat (Triticum aestivum) resistant to the Hessian fly, induced GNA-related lectins recognize Man-rich glycans of microorganisms or plant predators, suggesting that this early recognition may play a significant role in defense responses (Subramanyam et al., 2008). However, with the exception of LecRLKs, the functions of plant lectins, such as those of GNA-related lectins in plant-pathogen interactions, are poorly understood. We found CaMBL1 expression to be pathogen specific; however, the role of CaMBL1 as a defense protein in plants remains to be elucidated. In incompatible interactions with Xcv, CaMBL1 was strongly and rapidly induced compared with its response in compatible interactions. These results suggest that CaMBL1 is crucial in pepper plants for overall defense responses to pathogen invasion. Similarly, an MBL gene is expressed in maize (Zea mays) plants early during Colletotrichum graminicola infection (Sugui and Deising, 2002).
In plants, GNA-related lectins may be involved in pathogen recognition at an early stage of infection. The expression of CaMBL1 was induced rapidly and strongly during incompatible interactions with Xcv. Thus, its expression may lead to the activation of defense-related genes. Silencing of CaMBL1 resulted in enhanced disease susceptibility, increased bacterial growth, lowered accumulation of ROS, as well as reduced expression of PR genes in pepper leaves during virulent or avirulent Xcv infection. These findings support the notion that CaMBL1 expression during Xcv infection activates basal resistance in pepper plants, which is accompanied by the induction of PR genes. Reduced SA levels in CaMBL1-silenced plants impaired their ability to mount responses such as defense-related gene expression and HR-like cell death. Taken together, these data suggest that the CaMBL1 gene is a positive regulator of SA-dependent cell death and defense responses during Xcv infection.
Interestingly, CaMBL1 induced cell death responses in pepper leaves when it was transiently expressed by agroinfiltration, as evidenced by remarkably high levels of SA with concomitant PR1 activation in CaMBL1-overexpressing pepper leaves. More importantly, domain analyses of CaMBL1-induced cell death in pepper leaves revealed that the GNA-related lectin domain is responsible for this phenomenon. Collectively, these results suggest that induction of CaMBL1, as a positive regulator of cell death and defense responses, causes SA to accumulate in pepper leaf tissues and trigger the expression of downstream defense-related genes, such as CaBPR1 and CaSAR82A.
MBLs or GNA-related lectins may contribute to an innate immunity as a first line in plant defense (De Hoff et al., 2009). To determine whether overexpression of CaMBL1 confers defense response to heterologous plants, we generated CaMBL1-OX transgenic lines in Arabidopsis, which were resistant to virulent and avirulent P. syringae pv tomato infection. Resistance responses in the transgenic plants included ROS accumulation, cellular membrane damage, and necrotic disease symptoms. These findings suggest that overexpression of the CaMBL1 gene in Arabidopsis enhances basal or R gene-mediated resistance to bacterial infection. Inhibition of P. syringae pv tomato infection by overexpression of CaMBL1, which localizes to the plasma membrane, may be strongly supported by the activation of PR genes, induction of ROS accumulation, and cell death in Arabidopsis leaves following recognition of bacterial pathogens. However, there may be no correlation between CaMBL1 transcript and protein levels induced in the CaMBL1-OX Arabidopsis leaves by Pst DC 3000 infection. Furthermore, constitutively ectopic expression of the transgene CaMBL1 distinctly primes the induction of other defense-related genes, such as PR1, which may be dependent on the transgenic Arabidopsis lines. The susceptible response of Arabidopsis ortholog At1g78830 mutant plants to the fungal pathogen A. brassicicola also suggests that CaMBL1 is required for enhanced resistance to fungal pathogens in pepper plants.
GNA-related lectins constitute an extended superfamily of structurally and evolutionarily related proteins that play crucial roles for plant defenses. We have shown that CaMBL1 is required for enhanced resistance to the bacterial pathogen Xcv as well as to the fungal pathogen A. brassicicola. These results support the notion that CaMBL1 is effective for the recognition of bacterial and fungal pathogens on plant cell surfaces. Upon establishing contact with a host surface, pathogens release extracellular matrix-containing glycoproteins with Man residues (Sugui et al., 1998; Barre et al., 2001). Once GNA-related lectins recognize pathogens, their lectin domain could bind to Man or lipooligosaccharide residues on their surfaces. Therefore, expression of CaMBL1 may provide cues for the plant to specifically recognize pathogens and to promote further defense responses.
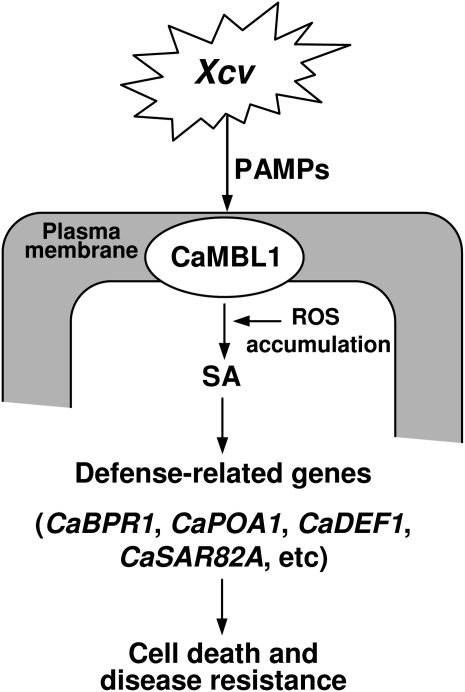
Taking these results together, we propose a working model for CaMBL1-mediated defense responses in plants (Fig. 10). Plants recognize a diversity of attacking pathogens, including virulent and avirulent Xcv, most likely by interactions of GNA-related lectins with a subset of pathogen-associated molecular patterns in the host plasma membrane. In this study, we showed that pathogen recognition by CaMBL1 triggers the accumulation of ROS and of the defense signal molecule SA in pepper plants, which in turn induces the expression of defense-related genes such as CaBPR1, CaPOA1, CaDEF1, and CaSAR82A. The CaMBL1 transcript does not accumulate in the absence of pathogens, but as a result of rapid CaMBL1 gene activation in response to their presence, pepper plants exhibit enhanced resistance to pathogens. In particular, plasma membrane-localized CaMBL1 is strongly expressed at an early stage of infection. However, it remains unclear how the recognition by CaMBL1 can lead to hypersensitive cell death, which is inevitably associated with ROS and SA accumulation, the expression of defense-related genes, and enhanced resistance. The presence of the GNA-related lectin domain supports a direct role for CaMBL1 in pathogen recognition, despite the absence of intracellular Ser/Thr kinase domains. Furthermore, experimental data using CaMBL1-silenced pepper and CaMBL1-OX Arabidopsis plants also support the notion that recognition by CaMBL1 may play a significant role in specific bacterial pathogen-plant interactions. Pathogen-induced CaMBL1 gene expression may alert the plant to invading pathogens, ultimately leading to cell death and disease resistance responses. We propose that CaMBL1 expression also triggers regulatory mechanisms to control defense responses in plants. Further studies of the GNA-related lectin protein CaMBL1 in pepper plants are required to reveal how this is achieved.
Figure 10.
A working model for CaMBL1 function in pepper leaf cells during Xcv infection. PAMPs, Pathogen-associated molecular patterns.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Pepper plants (Capsicum annuum ‘Nockwang’) were used in this study. Pepper seeds were germinated at 27°C. Plants were raised in a plastic tray (55 × 35 × 15 cm) and were transplanted to pots at the two-leaf stage. All plants were cultivated in a soil mix (peat moss:vermiculite:perlite, 3:3:1, v/v/v) at 26°C ± 2°C under a 16-h-light/8-h-dark cycle.
Wild-type ecotype Col-0 and transgenic lines of Arabidopsis (Arabidopsis thaliana) were also studied. CaMBL1 Arabidopsis ortholog At1g78830 mutant seeds (mbl1-1 and mbl1-2) were obtained from the Salk Institute T-DNA insertion library database (http://signal.salk.edu/cgi-bin/tdnaexpress). Prior to sowing on soil (peat moss:vermiculite:perlite, 1:1:0.5, v/v/v), seeds were vernalized at 4°C under low light for 2 d. Arabidopsis plants were raised in a growth chamber at 24°C with a 14-h-light/10-h-dark cycle.
Pathogen Inoculation and Disease Rating
The virulent strain Ds1 and avirulent strain Bv5-4a of Xanthomonas campestris pv vesicatoria were used to inoculate pepper plants. To prepare bacterial inocula, Xcv was grown on yeast-nutrient broth (5 g of yeast extract, 8 g of nutrient broth, and 1 L of water) at 28°C for 18 h. Pepper plants at the six-leaf stage were inoculated by infiltrating with a suspension (5 × 108 cfu mL−1) of the virulent strain Ds1 or the avirulent strain Bv5-4a or were mock infiltrated with 10 mm MgCl2. The inoculated plants were incubated for 18 h in a moist chamber at 28°C and then transferred to a growth room. Infected leaves were sampled at various times after inoculation.
Pseudomonas syringae pv tomato DC3000 and DC3000 expressing avrRpm1 were grown overnight at 28°C on yeast-nutrient broth containing rifampicin (50 μg mL−1). Arabidopsis plants were infiltrated with a 105 cfu mL−1 bacterial suspension in 10 mm MgCl2. To assess bacterial growth, infected leaves were collected 0 and 3 d after inoculation.
Hyaloperonospora arabidopsidis isolate Noco2 was maintained on Arabidopsis Col-0 by weekly subculturing. To produce a large quantity of inocula, 7- to 10-d-old seedlings were inoculated with H. arabidopsidis, and the spores produced on cotyledons were collected in water. The seedlings were spray inoculated with a suspension of asexual inoculum (5 × 104 conidiosporangia mL−1), covered with a transparent dome to maintain high humidity (80%–100%), and incubated for 7 d at 17°C. Asexual sporulation of H. arabidopsidis was assessed by using a light stereomicroscope to count the number of sporangiophores on both sides of the cotyledons 7 d after inoculation. The extent of disease was assigned to five classes based on the number of sporangiophores per cotyledon: zero to five, six to 10, 11 to 15, 16 to 20, and over 20. For the spore-count assay, the infected cotyledons of each line were excised and shaken vigorously. Spores harvested from 20 cotyledons per replicate were counted using a hemacytometer.
Alternaria brassicicola was cultured on potato dextrose agar medium at 24°C for 10 d. Conidial concentration was determined using a hemacytometer and adjusted to 5 × 104 conidia mL−1. A. brassicicola was inoculated by placing 10-μL droplets of suspension on leaves of 5-week-old plants. For mock treatment, 10-μL droplets of water were placed onto the leaves. Inoculated plants were kept at 100% relative humidity at 24°C. Four days after inoculation, lesion diameters were measured.
Treatment with SA and Methyl Jasmonate
The leaves of pepper plants at the six-leaf stage were treated with SA (5 mm) and methyl jasmonate (100 μm). SA and methyl jasmonate were sprayed onto pepper plants at the six-leaf stage. The pepper plants treated with methyl jasmonate were incubated in a vinyl bag. Control plants were sprayed with water. Pepper plants were sampled at various times after treatment, frozen in liquid nitrogen, and stored at −70°C for RNA isolation.
Isolation of the CaMBL1 Gene from a Pepper cDNA Library
A pathogen-induced cDNA library was prepared from total RNA extracted from pepper leaves 18 h after inoculation with Xcv using a λZAPII-cDNA library synthesis kit (Jung and Hwang, 2000). Differential hybridization was performed to isolate pathogen-induced cDNAs from the library using cDNA probes from uninoculated pepper leaves or leaves inoculated with the Xcv avirulent strain Bv5-4a.
Gene-specific RT-PCR was performed to confirm the differential expression of genes identified on an expression array. Digoxigenin-labeled single-stranded cDNAs were synthesized using total RNA from healthy and bacteria-infected pepper leaves. Total RNA (20 μg) from pepper was mixed with 5× reaction buffer (5× avian myeloblastosis virus [AMV] RT buffer), AMV reverse transcriptase (Roche), and oligo(dT)15 as a primer in diethyl pyrocarbonate water. To produce probes, RT-PCR was performed using RNA and primer mixes at 25°C for 10 min, 42°C for 60 min, and 99°C for 5 min. Equal amounts of the single-stranded cDNA probes from uninoculated control leaves and leaves inoculated with Xcv were hybridized to the two pepper cDNA expression arrays in a separate bottle for 18 h at 65°C in a mixture of 5× SSC, 0.1% sodium lauroylsarcosine, 0.02% SDS, and 10% blocking reagent. The hybridization membranes were washed twice in solution 1 (2× SSC and 0.1% SDS) for 10 min each at room temperature and then twice at 65°C for 5 min each in solution 2 (0.1× SSC and 0.1% SDS). The blotted membranes were exposed to x-ray film. The hybridization patterns produced by 96 samples each of uninoculated and inoculated samples were compared.
Several cDNA clones that were expressed in pepper leaves inoculated with the avirulent strain Bv5-4a of Xcv were isolated and sequenced with an ABI 310 DNA sequencer (PE Biosystems) using PRISM BigDYE Terminator sequencing-ready reaction kits. The DNA sequences were verified and analyzed using BLAST network services at the National Center for Biotechnology Information and the ExPASy Proteomics Server. A full-length cDNA clone encoding a pepper glycoprotein, designated CaMBL1, was isolated and analyzed.
Expression of the CaMBL1 Protein and Its Binding Assay with Man-Agarose
The CaMBL1 gene was cloned into the vector pET28a to generate pET28a::CaMBL1, which created a translational fusion of CaMBL1 to a His tag at the N terminus. The CaMBL1 protein construct was expressed in Escherichia coli and purified on a Man-agarose column. The CaMBL1 deletion series was made by PCR with specific primers CaMBL1 forward (5′-GGATCCATGTCTCCTTCATGGACTACTAC-3′) and CaMBL1 reverse (5′-AAGCTTTTAAATCCTGCTGTAAGTTACC-3′); CaMBL1-1 forward (5′-GGATCCATGCAAGTTCCAGCTGAAAACA-3′) and CaMBL1-1 reverse (5′-AAGCTTTTAAATCCTGCTGTAAGTTACC-3′); CaMBL1-2 forward (5′-GGATCCATGATTGTCGAATACAGCGCTGA-3′) and CaMBL1-2 reverse (5′-TTAAATCCTGCTGTAAGTTACC-3′); and CaMBL1-3 forward (5′-GGATCCATGTATAGCTTGGTGGTGC-3′) and CaMBL1-3 reverse (5′-AAGCTTTTAAATCCTGCTGTAAGTTAC-3′). To express the CaMBL1 gene, pET28a::CaMBL1 was transformed into E. coli BL21 (DE3). Recombinant strains were grown in 50 mL of Luria-Bertani medium at 37°C to an optical density at 600 nm (OD600) = 0.4 to 0.5, induced with 1 mm isopropyl-d-thiogalactopyranoside for 3 h at 18°C, and harvested. Cells were collected by centrifugation, resuspended in 10 mL of Man-binding buffer (50 mm Tris, pH 7.5, 100 mm NaCl, and 5 mm CaCl2), and disrupted by sonication. Cell debris was removed by centrifugation, and the supernatant was applied to a d-Man-agarose column (Sigma). The column was incubated in the presence of the protein extract for 30 min. After unbound proteins were washed out with Man-binding buffer, bound proteins were eluted from the column using Man-binding buffer containing 100 mm d-Man. Purified CaMBL1 was subjected to 15% SDS-PAGE. Protein bands were electrotransferred from an unstained gel onto Hybond-P membranes (GE Healthcare Bioscience). For detection of proteins, an anti-His antibody (Sigma) was used at 1:5,000 dilution.
Subcellular Localization
Plasmids harboring smGFP, CaMBL1:smGFP, GNA domain:smGFP, and CaMBL1c:smGFP were transformed into onion (Allium cepa) epidermal cells by particle bombardment (Bio-Rad) using 5 μg of each plasmid DNA, which was precipitated onto gold particles. After bombardment, the onion epidermal cells on Murashige and Skoog agar plates were incubated for 18 h at 25°C in the dark. Subcellular localization of fused proteins was visualized in onion epidermal cells by confocal laser scanning fluorescence microscopy (Carl Zeiss LSM 5 Exciter).
Agrobacterium tumefaciens-Mediated Transient Expression
Constructs containing pBIN35S:CaMBL1 or 35S:CaMBL1:8Myc were introduced into the Agrobacterium strain GV3101 by direct transformation. Recombinant Agrobacterium was grown overnight at 28°C, collected by centrifugation, and resuspended to a final concentration of OD600 = 1.0 in induction medium (10 mm ethanesulfonic acid, pH 5.7, and 10 mm MgCl2). In experiments to test the efficacy of additives, acetosyringone was added to the liquid culture to a final concentration of 200 μm at pH 5.6. The cell suspensions were incubated at 28°C for 3 h before infiltration. Agrobacterium suspensions expressing CaMBL1 were injected at different concentrations into pepper leaves.
VIGS
TRV-based vectors (pTRV1/2) were used for VIGS of the CaMBL1 gene (Liu et al., 2002). An 861-bp fragment of the CaMBL1 cDNA was amplified by PCR to specifically silence CaMBL1. The resulting PCR product was cloned into pTRV2 to generate pTRV2:CaMBL1. The TRV2 derivative pTRV:CaMBL1 was transformed into Agrobacterium strain GV3101, and transformed cells were selected on yeast extract peptone medium with 50 μg mL−1 kanamycin and 100 μg mL−1 rifampicin. Agrobacterium GV3101 containing pTRV1 and pTRV2:CaMBL1 VIGS vectors was coinfiltrated into fully expanded pepper cotyledons using a syringe (OD600 = 0.4). A positive control was infiltrated with a VIGS clone containing the Phytoene Desaturase gene. A negative control was infiltrated with an empty vector clone (TRV:00). The inoculated plants were transferred to a growth chamber maintained at 17°C for 2 d with 60% relative humidity and then placed in a growth room at 26°C ± 2°C with a 16-h-light/8-h-dark cycle. Unsilenced (TRV:00) and CaMBL1-silenced (TRV:CaMBL1) plants were inoculated with the virulent Ds1 and avirulent Bv5-4a strains of Xcv to examine gene expression (106 cfu mL−1) and to assay bacterial growth (5 × 104 cfu mL−1). Bacterial cells were counted 0 and 3 d after inoculation.
Generation of Arabidopsis Plants Overexpressing CaMBL1
To generate the CaMBL1-OE construct, the CaMBL1 coding sequence obtained from a pBluescript SK2 (Stratagene) construct by XbaI and BamHI restriction was subcloned into the vector pCR2.1-TOPO (Invitrogen). This fragment was ligated into the binary expression vector pBIN35S. The primers used in the PCR to generate XbaI and BamHI sites were 5′-TCTAGAATGTCTCCTTCATGGACTACTAG-3′ (forward) and 5′-GGATCCTTAAATCCTGCTTAAGTTACC-3′ (reverse). The vector pBIN35S containing the cauliflower mosaic virus 35S promoter-CaMBL1 construct was transformed into Agrobacterium strain EHA105 via electroporation.
To generate transgenic Arabidopsis, 4-week-old Arabidopsis plants were infected with Agrobacterium strain EHA105 by the floral dip method (Clough and Bent, 1998). Seeds were collected from transformed Arabidopsis, and plants were selected on Murashige and Skoog agar plates with 50 μg mL−1 kanamycin to obtain independent transgenic lines. The presence of the transgene was confirmed by PCR.
Identification of T-DNA Insertion Mutant Lines
The T-DNA insertion lines (SALK_121641 and SALK_038821) of At1g78830 were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/abrc/) at Ohio State University. Homozygous mutant plants were identified by PCR using T-DNA and gene-specific primer sets as described on the T-DNA Express Web site (http://signal.salk.edu/tdnaprimers.html). Two sets of PCR were carried out using the three primers (LP, left gene-specific primer; RP, right gene-specific primer; LB, left border primer of the T-DNA insertion) for each Salk line: LP primer (5′-GAGCGGTATTCCAAGTGGAG-3′) and RP primer (5′-TACATCTGATTCGAACGGCTC-3′) for SALK_121641; LP primer (5′-ACGGTGGATCATTACAAGCAC-3′) and RP primer (5′-ATTCCAAATCTTACCCAACGG-3′) for SALK_038821; and LBa1 primer (5′-TGGTTCACGTAGTGGGCCATCG-3′).
Northern, RT-PCR, and Quantitative Real-Time PCR Analyses
Total RNA was isolated from pepper leaves, stems, roots, flowers, and fruits and from Arabidopsis plants using TRIzol (Invitrogen) according to the manufacturer’s instructions. Leaf samples were harvested and immersed in liquid nitrogen at different times after inoculation with Xcv. Total RNA was extracted from the samples by the guanidinium thiocyanate-phenol-chloroform extraction method (Chomczynski and Sacchi, 1987). The total RNA concentration and purity were estimated by spectrophotometry and by ribosomal RNA staining with ethidium bromide, respectively. Total RNAs (10 μg) were fractionated on 1.2% agarose gels containing 7.4% formaldehyde and transferred onto nylon membranes (Hybond+; Amersham) for 18 h followed by UV cross-linking. A biotin probe (14-dCTP-biotin) was prepared from the corresponding pepper cDNA, and CaMBL1 and CaBPR1 were amplified with the following specific primer sets: CaMBL1, 5′-AAAATCAACAACGAGAAGTTCACATGTTAT-3′ (forward) and 5′-AACTAATTTATTGGACTAGCTGTGGACCCA-3′ (reverse); CaBPR1, 5′-ATGGGACACTCTAATATTGCC-3′ (forward) and 5′-GACATCAGTTGGAAGTTCCAA-3′ (reverse). After probe generation, membranes were prehybridized for 3 h at 65°C and then hybridized in 5% dextran sulfate, 0.25 m disodium phosphate (pH 7.2), 7% SDS, and 1 mm EDTA at 65°C overnight. Membranes were washed twice with 2× SSC, 0.1% SDS for 10 min each at room temperature and twice in 0.1× SSC, 0.1% SDS for 15 min each at 65°C. Membranes were exposed to x-ray film.
For RT-PCR analysis, first-strand cDNA synthesis was carried out in a reaction volume of 20 μL with 1 μg of RNA, 10 units μL−1 AMV reverse transcriptase (BIO BASIC), 2 μL of oligo(dT), 2 μL of deoxyribonucleotide triphosphates, and 2× AMV-RT buffer at 42°C for 60 min. PCR was performed in a final volume of 50 μL per reaction containing 0.5 μg of cDNA, 250 nm forward and reverse primers, 10× Taq reaction buffer (Takara), 0.2 mm deoxyribonucleotide triphosphates (Takara), and 0.5 units of Taq polymerase (Takara). Thirty cycles of amplification were preceded by an initial denaturation at 95°C for 5 min, with each amplification cycle including a 30-s denaturation at 95°C, a 30-s annealing at 55°C to 58°C (primer pair specific), and a 1-min extension at 72°C. After the amplification cycles, the samples were subjected to a 10-min extension at 72°C.
Quantitative real-time PCR using the Bio-Rad iCycler System was performed to monitor levels of gene expression in plants. Each reaction (20 μL) mix contained 10 μL of SYBR Green super mix (Bio-Rad) and 0.2 μm gene-specific primers. Thermal cycling conditions consisted of 2 min at 50°C, 10 min at 95°C, and 50 cycles of 15 s at 95°C and 1 min at 55°C. Data acquisition and analysis were performed by using iCycler Real-Time PCR Detection System software (Bio-Rad). Transcript levels were normalized to the expression of pepper 18S ribosomal RNA measured in the same samples.
Protein Extraction and Immunoblotting
For immunoblotting with the anti-cMyc (Sigma) and anti-CaMBL1 antibodies, total protein was extracted from 0.5 g of leaf tissues homogenized in 1 mL of extraction buffer (50 mm HEPES, pH 7.4, 50 mm NaCl, 10 mm EDTA, 0.2% Triton X-100, and 1× proteinase inhibitor cocktail [Roche]). Crude protein extracts were immunoprecipitated on an anti-c-Myc agarose affinity gel (Sigma) overnight at 4°C. Protein extracts were separated by 12% SDS-PAGE and blotted onto Hybond-P membranes (GE Healthcare Bioscience). Fusion proteins were detected using a mouse anti-c-Myc antibody (Sigma) at 1:2,000 dilution. An anti-CaMBL1 antibody was raised in a rabbit against a synthetic peptide corresponding to C-terminal residues 284 to 297 of CaMBL1 (AbFrontier). Leaf protein extracts were immunoblotted using the anti-CaMBL1 antibody at 1:5,000 dilution, which was able to detect only CaMBL1 from the leaf extracts.
Measurement of SA
SA and SA glycoside were extracted and quantified according to the method described by Verberne et al. (2002). Leaf tissue samples (0.5 g) were frozen in liquid nitrogen, ground to a fine power, and sequentially extracted with 90% and 100% methanol. As an internal standard for SA, 3-hydroxybenzoic acid (Sigma) was added at a mass ratio of 50 mg g−1 fresh weight. SA was determined by fluorescence (excitation, 305 nm; emission, 405 nm) after separation on a C18 reverse-phase HPLC column (Waters).
Measurement of Electrolyte Leakage
Pepper leaves were infiltrated with Agrobacterium strain GV3101 carrying the pBIN:CaMBL1 construct to determine electrolyte leakage. Leaf discs (1 cm in diameter) were washed in 20 mL of double distilled water for 30 min and transferred into 20 mL of double distilled water. Ion leakage was measured 0, 12, 24, and 36 h after infiltration. Conductance was measured with a conductivity meter (model sensION7; Hach). Electrical conductivities of the medium were expressed in μS cm–1.
Quantification of H2O2
H2O2 production in plants was measured by ferrous oxidation using the xylenol orange assay (Choi et al., 2007). One milliliter of freshly prepared assay reagent (25 mm FeSO4 and 25 mm [NH4]2SO4 dissolved in 2.5 m H2SO4) was added to 100 mL of 125 μm xylenol orange and 100 mm sorbitol. Leaf discs (0.5 cm2) were floated for 10 min in 1 mL of distilled water, followed by centrifugation at 5,000g for 1 min. The supernatant (100 μL) was incubated for 30 min in 1 mL of xylenol orange reagent. H2O2 was determined at 560 nm using a standard H2O2 curve.
Staining with DAB and Trypan Blue
Infection and development of pathogens were assessed by staining inoculated plants with 1 mg mL−1 DAB (Sigma D8001) and lactophenol-trypan blue (10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, and 10 mg of trypan blue, dissolved in 10 mL of distilled water). After overnight treatment with DAB, the stained leaves were cleared by boiling for 10 min in absolute ethanol and then destained overnight in absolute ethanol. The inoculated leaves were also boiled in trypan blue staining solution for 5 min and destained overnight in chloral hydrate (2.5 g of chloral hydrate dissolved in 1 mL of distilled water). The destained plant tissues were mounted in 70% glycerol for observation with a microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GQ265892 (CaMBL1), AF053343 (CaBPR1), AF442388 (CaDEF1), AF442387 (CaPOA1), AF313766 (CaSAR82A), DQ489711 (CaPO2), PR1 (At2g14610), PDF1.2 (At5g44420), NP_565191 (At1g78830), and UBQ5 (At3g62250).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide and deduced amino acid sequences of pepper CaMBL1 cDNA encoding a GNA-related lectin protein.
Supplemental Figure S2. Alignment of pepper CaMBL1 with other GNA-related lectins.
Supplemental Figure S3. Phylogenetic tree analysis of CaMBL1 with other GNA-related lectins.
Supplemental Figure S4. Disease responses of CaMBL1-OX and Arabidopsis ortholog mutants mbl1-1 and mbl1-2 to H. arabidopsidis isolate Noco2 and Pst DC3000 infection.
Supplemental Figure S5. Glycan-binding profile of CaMBL1 as analyzed by the Consortium for Functional Glycomics.
Supplemental Table S1. The glycan binding data of CaMBL1 as analyzed by the Consortium for Functional Glycomics.
Acknowledgments
We thank Dr. S.P. Dinesh-Kumar (Yale University) for the pTRV1 and pTRV2 vectors and Dr. U. Bonas (Martin-Luther-Universitaet) for Agrobacterium strain GV3101.
References
- Barre A, Bourne Y, Van Damme EJM, Peumans WJ, Rougé P. (2001) Mannose-binding plant lectins: different structural scaffolds for a common sugar-recognition process. Biochimie 83: 645–651 [DOI] [PubMed] [Google Scholar]
- Barre A, Herve C, Lescure B, Rougé P. (2002) Lectin receptor kinases in plants. Crit Rev Plant Sci 21: 379–399 [Google Scholar]
- Broekaert WF, van Parijs J, Leyns F, Joos H, Peumans WJ. (1989) A chitin-binding lectin from stinging nettle rhizomes with antifungal properties. Science 245: 1100–1102 [DOI] [PubMed] [Google Scholar]
- Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, et al. (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46: 794–804 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Lee SC, Hong JK, Hwang BK. (2007) Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol 145: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cox KD, Layne DR, Scorza R, Schnabel G. (2006) Gastrodia anti-fungal protein from the orchid Gastrodia elata confers disease resistance to root pathogens in transgenic tobacco. Planta 224: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Davies HA, Daniels MJ, Dow JM. (1997) Induction of extracellular matrix glycoproteins in Brassica petioles by wounding and in response to Xanthomonas campestris. Mol Plant Microbe Interact 10: 812–820 [DOI] [PubMed] [Google Scholar]
- Deepak S, Shailasree S, Kini RK, Hause B, Shetty SH, Mithöfer A. (2007) Role of hydroxyproline-rich glycoproteins in resistance of pearl millet against downy mildew pathogen Sclerospora graminicola. Planta 226: 323–333 [DOI] [PubMed] [Google Scholar]
- De Hoff PL, Brill LM, Hirsch AM. (2009) Plant lectins: the ties that bind in root symbiosis and plant defense. Mol Genet Genomics 282: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Matsushita M, Endo Y. (2004) The lectin-complement pathway: its role in innate immunity and evolution. Immunol Rev 198: 185–202 [DOI] [PubMed] [Google Scholar]
- Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Poinssot B, Wendehenne D, Pugin A. (2006) Early signaling events induced by elicitors of plant defenses. Mol Plant Microbe Interact 19: 711–724 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE. (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14: 177–193 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jung HW, Hwang BK. (2000) Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Lis H, Sharon N. (1998) Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem Rev 98: 637–674 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Mellersh DG, Heath MC. (2001) Plasma membrane-cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 13: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels K, Van Damme EJ, Smagghe G. (2010) Plant-insect interactions: what can we learn from plant lectins? Arch Insect Biochem Physiol 73: 193–212 [DOI] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E. (2008) The hypersensitive response: the centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Naithani S, Chookajorn T, Ripoll DR, Nasrallah JB. (2007) Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc Natl Acad Sci USA 104: 12211–12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Gochicoa MT, Camut S, Timmers AC, Niebel A, Hervé C, Boutet E, Bono JJ, Imberty A, Cullimore JV. (2003) Characterization of four lectin-like receptor kinases expressed in roots of Medicago truncatula: structure, location, regulation of expression, and potential role in the symbiosis with Sinorhizobium meliloti. Plant Physiol 133: 1893–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngai PHK, Ng TB. (2007) A lectin with antifungal and mitogenic activities from red cluster pepper (Capsicum frutescens) seeds. Appl Microbiol Biotechnol 74: 366–371 [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Barre A, Bras J, Rougé P, Proost P, Van Damme EJM. (2002) The liverwort contains a lectin that is structurally and evolutionary related to the monocot mannose-binding lectins. Plant Physiol 129: 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJM. (1995) Lectins as plant defense proteins. Plant Physiol 109: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, Herve C, Pacquit V, Dabos P, Lescure B. (2002) Expression of an Arabidopsis lectin kinase receptor gene, lecRK-a1, is induced during senescence, wounding and in response to oligogalacturonic acid. Plant Physiol Biochem 40: 431–438 [Google Scholar]
- Sequeira L, Graham TL. (1977) Agglutination of avirulent strains of Pseudomonas solanacearum by potato lectin. Physiol Plant Pathol 11: 43–54 [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shridhar S, Chattopadhyay D, Yadav G. (2009) PLecDom: a program for identification and analysis of plant lectin domains. Nucleic Acids Res 37: W452–W458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanyam S, Smith DF, Clemens JC, Webb MA, Sardesai N, Williams CE. (2008) Functional characterization of HFR1, a high-mannose N-glycan-specific wheat lectin induced by Hessian fly larvae. Plant Physiol 147: 1412–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmoon R, Sattayasai N, Bunyatratchata W, Chaveerach A, Nuchadomrong S. (2008) Thermostable mannose-binding lectin from Dendrobium findleyanum with activities dependent on sulfhydryl content. Acta Biochim Biophys Sin (Shanghai) 40: 811–818 [PubMed] [Google Scholar]
- Sugui JA, Deising HB. (2002) Isolation of infection-specific sequence tags expressed during early stages of maize anthracnose disease development. Mol Plant Pathol 3: 197–203 [DOI] [PubMed] [Google Scholar]
- Sugui JA, Leite B, Nicholson RL. (1998) Partial characterization of the extracellular matrix released onto hydrophobic surfaces by conidia and conidial germlings of Colletotrichum graminicola. Physiol Mol Plant Pathol 52: 411–425 [Google Scholar]
- Torres MA, Dangl JL. (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Tsutsui S, Tasumi S, Suetake H, Kikuchi K, Suzuki Y. (2006) Carbohydrate-binding site of a novel mannose-specific lectin from fugu (Takifugu rubripes) skin mucus. Comp Biochem Physiol B Biochem Mol Biol 143: 514–519 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Allen AK, Peumans WJ. (1987) Isolation and characterization of lectin with exclusive specificity towards mannose from snow drop (Galanthus nivalis) bulbs. FEBS Lett 215: 140–144 [Google Scholar]
- Van Damme EJM, Barre A, Rougé P, Peumans WJ. (2004) Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Culerrier R, Barre A, Alvarez R, Rougé P, Peumans WJ. (2007a) A novel family of lectins evolutionarily related to class V chitinases: an example of neofunctionalization in legumes. Plant Physiol 144: 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Lannoo N, Peumans WJ. (2008) Plant lectins. Adv Bot Res 48: 108–209 [Google Scholar]
- Van Damme EJM, Nakamura-Tsuruta S, Smith DF, Ongenaert M, Winter HC, Rougé P, Goldstein IJ, Mo H, Kominami J, Culerrier R, et al. (2007b) Phylogenetic and specificity studies of two-domain GNA-related lectins: generation of multispecificity through domain duplication and divergent evolution. Biochem J 404: 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Peumans WJ, Barre A, Rougé P. (1998) Plant lectins: a composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. Crit Rev Plant Sci 17: 575–692 [Google Scholar]
- Verberne MC, Brouwer N, Delbianco F, Linthorst HJ, Bol JF, Verpoorte R. (2002) Method for the extraction of the volatile compound salicylic acid from tobacco leaf material. Phytochem Anal 13: 45–50 [DOI] [PubMed] [Google Scholar]
- Vijayan M, Chandra N. (1999) Lectins. Curr Opin Struct Biol 9: 707–714 [DOI] [PubMed] [Google Scholar]
- Wang XC, Bauw G, Van Damme EJM, Peumans WJ, Chen ZL, Van Montagu M, Angenon G, Dillen W. (2001) Gastrodianin-like mannose-binding proteins: a novel class of plant proteins with antifungal properties. Plant J 25: 651–661 [DOI] [PubMed] [Google Scholar]