The realization that crop yields are reaching a plateau, while population increases continue at pace, has placed manipulation of photosynthesis in a central position to achieve increases in yield. Increasing flux through the C3 cycle will be a major focus of this effort. Through application of new technologies together with novel modeling approaches, increased yield through improved photosynthetic carbon fixation should be an attainable goal in the near to mid term. This Update article reviews the past approaches and successes in this area. An outline of some of the known targets for future work is given, and approaches to identify novel targets for exploitation are outlined.
Improving crop yield to meet the demands of an increasing world population for food and fuel is a central challenge for plant biology (Edgerton, 2009). This goal must be achieved in a sustainable manner (i.e. with minimal agricultural inputs and environmental impacts) in the face of elevated levels of CO2 and more extreme conditions of water availability and temperature. Agricultural yields have generally kept pace with demand in the recent past as a result of the gains made through breeding programs and farming practice, but crops yields are now reaching a plateau. One fundamental component of plant productivity that has not been used to select for increased yield is photosynthesis. There is now the opportunity to exploit our extensive knowledge of this fundamental process for the benefit of humankind (von Caemmerer and Evans, 2010; Zhu et al., 2010a).
In the plant kingdom, there are three pathways of photosynthetic, atmospheric CO2 fixation. However, the vast majority of plant species fix atmospheric CO2 using the enzyme Rubisco in the Calvin-Benson cycle. The first stable product of this cycle is a three-carbon compound, phosphoglycerate (3-PGA), and for this reason this process is referred to as the C3 cycle. Plants utilizing this pathway are often referred to as C3 species. A major problem with the C3 cycle is the enzyme Rubisco. This is because Rubisco is not only an inefficient enzyme with a low turnover number, but it also catalyzes two competing reactions: carboxylation and oxygenation (Portis and Parry, 2007). The oxygenation reaction directs the flow of carbon through the photorespiratory pathway (Fig. 1), and this can result in losses of between 25% and 30% of the carbon fixed. Environmental variables, such as high temperature and drought, can result in an increase in the oxygenase reaction. Therefore, reducing the Rubisco oxygenase reaction has the potential to increase carbon assimilation significantly and would represent a step change in photosynthesis (up to 100% depending on temperature; Long et al., 2006).
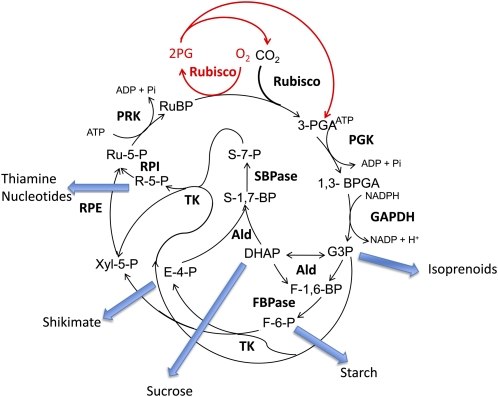
Figure 1.
The C3 cycle. The carboxylation reaction catalyzed by Rubisco fixes CO2 into the acceptor molecule RuBP, forming 3-PGA. The reductive phase of the cycle follows with two reactions catalyzed by 3-PGA kinase (PGK) and GAPDH, producing G-3-P. The G-3-P enters the regenerative phase catalyzed by aldolase (Ald) and either FBPase or SBPase, producing Fru-6-P (F-6-P) and sedoheptulose-7-P (S-7-P). Fru-6-P and sedoheptulose-7-P are then utilized in reactions catalyzed by TK, R-5-P isomerase (RPI), and ribulose-5-P (Ru-5-P) epimerase (RPE), producing Ru-5-P. The final step converts Ru-5-P to RuBP, catalyzed by PRK. The oxygenation reaction of Rubisco fixes O2 into the acceptor molecule RuBP, forming PGA and 2-phosphoglycolate (2PG), and the process of photorespiration (shown in red) releases CO2 and PGA. The five export points from the pathway are shown with blue arrows.
Two metabolic pathways have evolved to overcome this, the C4 (the first stable compound synthesized is a C4 acid, oxaloacetate) and crassulacean acid metabolism pathways. Both the C4 and crassulacean acid metabolism pathways are additional to the C3 cycle and increase the supply of CO2 to Rubisco, thereby reducing the oxygenation reaction and flux to the photorespiratory pathway. This review will focus on progress and future prospects to improve the C3 cycle.
THE C3 CYCLE
The C3 cycle is the primary pathway of carbon assimilation in the majority of photosynthetic organisms. It is the single largest flux of organic carbon in the biosphere and assimilates about 100 billion tons of carbon a year (15% of the carbon in the atmosphere). Understanding the responses of the Calvin cycle to altered demands for photosynthate within the plant and to external environmental conditions is essential for attempts to increase yield and to redirect carbon into important products. The C3 cycle utilizes the products of the light reactions of photosynthesis, ATP and NADPH, to fix atmospheric CO2 into carbon skeletons that are used to fuel the rest of plant metabolism (Fig. 1; Stitt et al., 2010). The C3 cycle is initiated by the enzyme Rubisco that catalyzes the carboxylation of the CO2 acceptor molecule ribulose-1,5-bisP (RuBP). 3-PGA formed by this reaction is used to form the triose phosphates glyceraldehyde phosphate (G-3-P) and dihydroxyacetone phosphate via two reactions that consume ATP and NADPH. The regenerative phase of the cycle involves a series of reactions that convert triose phosphates into the CO2 acceptor molecule RuBP (Fig. 1). Carbon compounds produced in this cycle are essential for growth and development of the plant (Raines and Paul, 2006; Smith and Stitt, 2007). While the majority (five-sixths) of the triose phosphate produced in the Calvin cycle remain within the cycle to regenerate RuBP, one-sixth of the carbon exits the cycle for biosynthesis of a range of compounds. Triose and hexose phosphates are used to synthesize Suc and starch, and erythrose-4-P (E-4-P) goes directly to the shikimate pathway for the biosynthesis of amino acids and lignin, G-3-P to the isoprenoid pathway, and Rib-5-P (R-5-P) for nucleotide, thiamine metabolism, and cell wall biosynthesis (Fig. 1). Clearly, the Calvin cycle occupies a central position in carbon metabolism. Manipulation of this pathway has the potential to increase yield and production of carbon for the synthesis of products made in secondary metabolic pathways.
WHAT LIMITS C3 PHOTOSYNTHESIS?
It has been known for some time that a large proportion of the limitation to carbon assimilation in plants using the C3 cycle is due to the catalytic properties of the enzyme Rubisco (Portis and Parry, 2007). During the 1990s, metabolic control analysis was used to explore the possibility that enzymes other than Rubisco may also have a role in determining rates of carbon flux through the C3 cycle (Stitt and Schulze, 1994; Raines, 2003). To undertake metabolic control analysis of a pathway, it is necessary to be able to reduce specifically the amount of an individual enzyme in that pathway; the effect of this reduction on flux can then be compared to the control. The flux control coefficient can vary from 0, for an enzyme that makes no contribution to control, to 1, for an enzyme that exerts total control. One fundamental difference between this approach and that based on the kinetics of individual enzymes is that metabolic control analysis allows for all enzymes in a pathway to share control of flux in that pathway. The flux control value for any single enzyme is not a constant and can change depending on the conditions under which the analysis was carried out.
Transgenic plants with reductions in Rubisco protein levels produced using an antisense construct were used to assess the relative contribution that Rubisco imposed on carbon flux. This approach has demonstrated clearly that the limitation imposed by Rubisco on C3 carbon fixation is greatest in high light and temperature conditions (Stitt and Schulze, 1994). In contrast, as expected, this is reduced in plants grown in elevated CO2 (Masle et al., 1993). Importantly, these transgenic studies also showed that Rubisco does not limit the C3 cycle in all conditions and that enzymes of the regenerative phase of the cycle also play a role in determining the rate of photosynthesis (Raines, 2003, 2006; Stitt et al., 2010).
Regenerative Capacity
Regeneration of the CO2 acceptor molecule RuBP involves eight enzymes (Fig. 1). To identify enzymes that have significant control over carbon flux through the Calvin cycle, antisense plants with reduced levels of individual enzymes were created and analyzed (Stitt and Schulze, 1994; Raines, 2003). Under many conditions, reductions in the activities of enzymes catalyzing highly regulated, effectively irreversible reactions, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Fru bisphosphatase (FBPase), and phosphoribulokinase (PRK), had little impact on carbon assimilation (for review, see Raines, 2003; Stitt et al., 2010). In contrast, small reductions in the enzyme sedoheptulose-1,7-bisphosphatase (SBPase) resulted in a decrease in CO2 fixation and growth, identifying this enzyme as a major control point in the C3 cycle. The relative importance of SBPase in determining flux through this cycle varied with development and environmental conditions, confirming that no one enzyme limits the C3 cycle under all conditions (Harrison et al., 1998, 2001; Olçer et al., 2001). Somewhat surprisingly carbon flux through the C3 cycle was also reduced in plants with relatively small reductions in the activity of either transketolase (TK) or aldolase, enzymes that are not highly regulated and operate close to equilibrium (Haake et al., 1998, 1999; Henkes et al., 2001). This was unexpected, as it was thought that enzymes that were highly regulated would be most important in determining the rate of carbon flow through the cycle. These analyses of the antisense C3 cycle plants identified enzymes other than Rubisco that can limit CO2 fixation. This work also indicated that it may be possible to increase photosynthetic carbon fixation and growth by increasing the levels of SBPase or, to a lesser extent, TK. Experiments exploring this hypothesis are described below.
PAST SUCCESSES TO IMPROVE THE C3 CYCLE
1. Rubisco-Related Bottlenecks
Three approaches to improve the C3 cycle have targeted Rubisco-associated bottlenecks. Expression of the cyanobacterial ictB protein in higher plants to reduce the oxygenation reaction of Rubisco resulted in the production of transgenic tobacco (Nicotiana tabacum) with higher photosynthetic rates under limiting, but not saturating, CO2 levels. Similar results were obtained in Arabidopsis (Arabidopsis thaliana) plants expressing the ictB from Anabaena PCC7120; in low humidity conditions, the growth of transgenic Arabidopsis plants was faster and the final dry weight was higher than that of wild-type plants (Lieman-Hurwitz et al., 2003). A second approach introduced the enzymes of the bacterial glycolate pathway into plants, creating a photorespiratory bypass in the chloroplast (Peterhansel and Maurino, 2011). In Arabidopsis, this resulted in reduced photorespiration and increased CO2 assimilation and biomass yield (Kebeish et al., 2007; Leegood, 2007). These two strategies for overcoming the oxygenase reaction of Rubisco offer more immediate alternatives to improve crop photosynthesis in the near future, compared to the more ambitious CO2-concentrating strategies described below in the future approaches section.
A. Manipulation of Rubisco Activation
The activation state of Rubisco is dependent on the enzyme Rubisco activase, which at temperatures above 30°C has been shown to be unstable in vitro. This led to the suggestion that Rubisco activase is responsible for the reversible temperature-sensitive reduction in the activation state of Rubisco, leading to inhibition of photosynthesis when plants are subjected to mild heat stress (Salvucci and Crafts-Brandner, 2004). The introduction of a thermostable version of the Rubisco activase enzyme into an Arabidopsis activase deletion mutant resulted in transgenic lines with higher photosynthetic rates and increased biomass and seed yield when compared with control plants expressing the wild-type activase (Kurek et al., 2007). All three of these approaches have demonstrated the potential to stimulate photosynthesis and growth in model species, and the next step will be to test this in crop plants in natural environments.
2. Increasing Regenerative Capacity of the C3 Cycle
Transgenic antisense work described above identified SBPase, plastid aldolase, and TK as enzymes that might be targets for improving C3 cycle flux. Indeed, overexpression of either a bifunctional SBPase/FBPase or plant SBPase in tobacco plants resulted in increased photosynthetic CO2 fixation and growth (Miyagawa et al., 2001; Lefebvre et al., 2005). Analysis of CO2 response curves revealed that this increase in photosynthesis could be attributed to an increase in the capacity to regenerate the CO2 acceptor molecule RuBP. Measurements of ambient photosynthesis over the daily cycle revealed an increase in photosynthetic rates of between 6% and 12%. Of particular interest for crop improvement is that these plants also displayed an increase in leaf area and biomass of up to 30%. Young seedlings (4/5 leaves) with increased SBPase activity were shown to have an increase in leaf area compared to wild-type plants. Chlorophyll fluorescence imaging of these young plants revealed a higher photosynthetic capacity at the whole plant level. These plants also showed an increased growth rate during the early phase of development (Lefebvre et al., 2005). This result is of significance in an agricultural context as rapid, early seedling establishment is known to contribute to yield increases at maturity. In contrast to the results with tobacco, increasing SBPase activity in rice (Oryza sativa) did not lead to increases in photosynthesis or growth. However, if the plants were subjected to heat or salt stress conditions, photosynthesis rates in the transgenic plants with increased SBPase plant were higher than in wild-type controls (Feng et al., 2007a, 2007b). The rice studies highlight the fact that increasing photosynthesis and yield by manipulation of SBPase is likely to be dependent not only on species but also growth conditions.
Plastid TK was also identified as a potential target to increase the regenerative capacity of the C3 cycle (Henkes et al., 2001). To explore this possibility, transgenic plants have been produced overexpressing TK, either on its own or together with overexpression of SBPase. Analysis of these transgenic plants has shown that photosynthetic rates were similar to wild type but that the plants have reduced growth and display a chlorotic phenotype. The basis of this phenotype has not been fully resolved, but preliminary analysis would suggest that increasing TK activity has perturbed the balance of export of carbon from the C3 cycle (M. Khozaei, S.C. Lefebvre, T. Lawson, and C.A. Raines, unpublished data). This is in keeping with data from antisense studies that revealed a reduction in carbon flow from the C3 cycle to the shikimate pathway that correlated with reductions in TK activity (Henkes et al., 2001). This work highlights the need for modeling approaches that not only allow identification of potential targets for improving CO2 assimilation but that will also allow flux from the C3 cycle to be determined. This is discussed in the modeling section below.
FUTURE PROSPECTS FOR IMPROVING THE C3 CYCLE
1. Improving Known Bottlenecks
The oxygenase reaction of Rubisco and subsequent photorespiratory pathway are clear targets for improvement of the C3 cycle (Whitney et al., 2011). Over the last 20 years, considerable effort has been focused on reengineering the Rubisco protein, using site-directed mutagenesis to increase the carboxylase relative to the oxygenase reaction. However, this approach has yet to produce an improved Rubisco (Parry et al., 2007). Recently, it has been shown that it is possible to form an active Rubisco in vitro using unfolded large subunits with the chaperone proteins GroEL/ES, RbcX, and ATP (Liu et al., 2010). This work paves the way for in vitro analysis of mutants and has the potential to allow screening for improvements in the Rubisco catalytic parameters of genetically engineered mutants. A new opportunity has also arisen from interspecies comparison of Rubisco catalytic properties that has revealed variation in the specificity, kcat, and temperature response of Rubisco from natural vegetation in the Mediterranean (Sage, 2002; Galmes et al., 2005). The variation in the properties would indicate that transferring Rubisco from these species into crop plants could substantially increase the rate of CO2 assimilation. The recent demonstration of the successful expression of Rubisco large and small subunits as a single fusion protein in the chloroplast that assembled into an active holoenzyme provides the opportunity to use expression of foreign Rubisco as a strategy to improve the C3 cycle (Whitney and Sharwood, 2007; Whitney et al., 2009).
CO2-concentrating mechanisms (CCMs) have been found in all plant groups, in algal species, and in the cyanobacteria. These adaptations present the possibility to exploit existing solutions that have evolved in these organisms to improve C3 plants (Gowik and Westhoff, 2011; Price et al., 2011). To realize the potential of these strategies will likely be in the long term; nevertheless, it is essential to begin the research now to determine the feasibility as a realistic option for future improvements. Transferring C4 photosynthesis to C3 species is one strategy to reduce photorespiration. But engineering C3 plants to carry out C4 metabolism will be a challenge as not only is there a requirement for additional metabolism but there are also changes in anatomy needed (Hibberd et al., 2008; Wang et al., 2009; Zhu et al., 2010b). Fundamental research aimed at understanding the molecular basis of the anatomical specialization and the regulatory processes determining the location and the level of expression of the C4-specific enzymes is under way (Hibberd and Covshoff, 2010). Although this has not yet led to any major breakthroughs, the combination of new sequencing and proteomic technologies together with a large international program give optimism for future success in this area.
The prokaryotic carboxysome provides a microcompartment in the cell that transports and fixes CO2, acting as a CCM (Badger et al., 2006; Moroney and Ynalvez, 2007; Price et al., 2008). Eukaryotic algae (and some primitive land plants) possess a structure, the pyrenoid, that may act to concentrate CO2 at Rubisco (Moroney and Ynalvez, 2007). Both of these CCMs provide components that could be transferred to higher plants to reduce photorespiration and increase water and nitrogen use efficiency. However, it is likely that engineering the complete CCM from either of these groups of organisms into higher plants will be a complex feat of gene engineering, and given that the components of the pyrenoid remain to be elucidated, these approaches are likely to be successful only in the longer term. However, both the carboxysome and pyrenoid CCMs involve more than just the microcompartmentation of Rubisco. It might be more attainable in the shorter term to exploit the CO2 transport systems in these organisms to boost CO2 levels in C3 plants and thereby have a positive effect on atmosphere CO2 assimilation by Rubisco (Price et al., 2011).
2. Identifying Novel Targets
A. Modeling-Based Strategies
Redistribution of Enzymes of Carbon Fixation. Although antisense studies described above revealed enzymes in the C3 cycle as targets for manipulation, it would be impossible to analyze every potential manipulation in this way. It is even less attainable to select multiple targets that may impact synergistically to improve the cycle. In order that significant advances can be made in a sensible time frame, it will be essential to have a dynamic kinetic model that is predictive. A model of photosynthetic carbon metabolism has been developed, and its application, in conjunction with an evolutionary algorithm, indicated that the distribution of resources between enzymes of carbon metabolism (that is the amount of each) is not optimal (Zhu et al., 2007). An overinvestment in enzymes of the photorespiratory pathway, namely Gly decarboxylase, and an underinvestment in ADP-Glc pyrophosphorylase, SBPase, and plastid aldolase enzymes were identified. This suggests that the rate of light-saturated photosynthesis in existing C3 plants could be increased significantly by redistribution of the amounts of these enzymes in carbon assimilation, photorespiration, and starch biosynthesis. The hypotheses generated from this modeling can easily be tested using current transgenic technology using a model plant system. Importantly, the results from this work have the potential to provide clear strategies for improvement of photosynthesis in a time frame of 2 to 3 years.
A neglected aspect of the C3 cycle is the control of flux from the cycle to the output pathways of starch, Suc, isoprenoids, shikimate, and nucleotides (Fig. 1). Although detailed analysis of the regulation of Suc, starch, and shikimate biosynthetic pathways have been undertaken, there is no complete model that can simulate the way the relative flux to these pathways changes during development, in response to stress or availability of light. This lack of knowledge places limitations on the ability to exploit the enormous potential for manipulation of the cycle for the synthesis of high value chemicals such as pharmaceuticals, plant protectants, nutraceuticals, and flavor/color compounds that are products of the shikimate and isoprenoid output pathways of the C3 cycle.
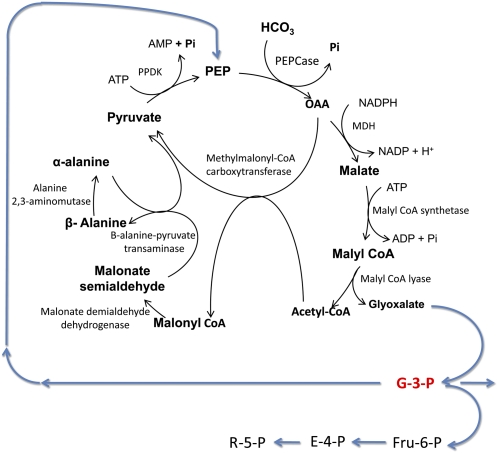
Design and Introduction of a Synthetic Carbon Fixation Pathway. Although the majority of photosynthetic organisms fix atmospheric CO2 through the C3 cycle, in nature there are five other CO2 fixation pathways that are found in the Archaea (organisms that have structure and metabolism similar to bacteria and transcriptional regulation more closely aligned with the eukaryotes; Berg et al., 2010). Although none of these organisms use either Rubisco or phosphoenolpyruvate carboxylase as the carboxylation enzyme, they do reveal a diversity of metabolism that has evolved in response to a range of environmental niches. This raises the possibility of employing a combination of modeling together with synthetic biology, to construct completely novel CO2 fixation pathways that may be more efficient than the C3 cycle. In addition, this approach may offer a solution to the problem of Rubisco without having to redesign plant development, required for introduction of the C4 pathway. Using a constraint-based modeling approach that included approximately 5,000 enzymes known to occur naturally, a number of hypothetical alternative carbon fixation pathways were identified (Bar-Even et al., 2010). This study showed that some of these hypothetical pathways may be more efficient than the naturally occurring C3 cycle. Interestingly, all of the pathways predicted to be 2 to 3 times faster than the C3 cycle included the enzyme phosphoenolpyruvate carboxylase, which is the carboxylating enzyme found in C4 species, and not any of the carboxylating enzymes found in the Archaea. These model-generated, hypothetical pathways also share a core set of reactions and have been termed the malonyl-CoA-oxaloacetate-glyoxylate (MOG) pathways (Fig. 2). Although the glyoxylate produced could be converted to G-3-P by introducing enzymes from Escherichia coli, there is still a requirement to synthesize hexose phosphates, Fru-6-P, E-4-P, and R-5-P, needed to supply the essential carbon compounds for growth and development of the plant. If a MOG-type synthetic pathway was to be engineered into it may require coupling to a partial Calvin cycle, creating Rubisco-independent carbon assimilation. To maintain the MOG pathway, it would also be necessary to use some of the G-3-P to regenerate the phosphoenolpyruvate needed for CO2 assimilation (Fig. 2). The technical challenges to engineer such large changes in the core CO2 pathway(s) in higher plants using synthetic biology will be significant. But redesigning metabolism based on the modeling strategy of Bar-Even et al. (2010) offers a novel and exciting approach that warrants experimental study.
Figure 2.
Rubisco-independent carbon assimilation. C4-glyoxalate (MOG) cycle—a model-developed hypothetical carbon fixation pathway. This pathway uses the enzyme phosphoenolpyruvate carboxylase (PEPCase) to fix atmospheric CO2 into oxaloacetate and the only output product of this cycle is glyoxalate. PPDK, Pyruvate phosphate dikinase; MDH, malate dehydrogenase. The arrows in blue are speculative routes for carbon that may be need to be engineered to maintain the MOG cycle in addition to linking with a partial C3 cycle to provide the intermediates for biosynthesis of isoprenoids (G-3-P), shikimate (E-4-P), Suc (G-3-P), starch (Fru-6-P), and nucleotides (R-5-P). Adapted from Bar-Even et al. (2010).
B. Elucidation of Mechanisms Underlying Natural Variation in Photosynthesis
There is considerable variation in photosynthetic rates per unit leaf area between C3 species. This is an overlooked and untapped resource yet has huge potential to identify natural mechanisms that have evolved to allow plants to survive in different environments. Systematic analysis of plants from different geographic regions, using high-throughput in vivo photosynthesis tools, has the potential to identify novel targets for manipulation in crop plants.
Development of photosynthetic capacity also differs in the same species when grown under different environmental conditions; this can result in changes in morphology and anatomy and is termed developmental acclimation. In addition, some plant species are able to alter photosynthetic capacity in fully mature leaves, displaying a dynamic acclimatory response. A recent study revealed that developmental and dynamic acclimation are distinct processes and that dynamic acclimation may have a role in increasing the fitness of plants in natural environments (Athanasiou et al., 2010). However, most studies focus on plants grown in constant conditions or after a shift to different conditions, e.g. low to high light. There have been few studies where plants have been exposed to fluctuating conditions. Elucidation of the mechanisms that are employed by plants during environmental fluctuations can provide novel targets for improving crop yield. This is particularly relevant given that many crop species lack any dynamic range to respond to this variation (Murchie et al., 2009).
Another aspect we know very little about is the relative importance of regulatory mechanisms in determining carbon flux in the C3 cycle. It has been known for several decades that a number of enzymes of the C3 cycle, FBPase, SBPase, PRKase, and GAPDH, are subjected to light activation via the ferredoxin/thioredoxin system. Importantly, there is a lag in this response when plants experience a dark to light shift, resulting in a delay of minutes in photosynthetic rates reaching a steady-state maximum (Sassenrath-Cole and Pearcy, 1994; Sassenrath-Cole et al., 1994). It has also been shown recently that the enzymes PRK and GAPDH are subject to rapid inactivation/activation in response to changes in light availability, mediated by a small protein, CP12, together with Trxf (Howard et al., 2008; Marri et al., 2009). These regulatory mechanisms are believed to match the rate of carbon fixation with the availability of energy from the light reactions. However, the relative importance of this regulation in determining photosynthetic capacity is unknown but may be important in the acclamatory response of plants dealing with fluctuating conditions. Furthermore, the extent to which this regulation varies between species is also relatively unexplored.
PERSPECTIVES
Manipulation of the C3 cycle offers an opportunity to increase photosynthesis and yield. A number of clear targets have been shown to have the potential to impact yield in the 3- to 5-year period but to date have been tested only in model species. It is now important that this knowledge is fully exploited in crops. Clearly, the reduction of the Rubisco oxygenase reaction remains a target for future improvement of photosynthesis. However, although the current strategies that might be exploited to achieve this goal are conceptually straightforward, all of these approaches will be technically demanding, requiring fundamental research to identify the genes involved. Improvement of the C3 cycle is not just about increasing CO2 fixation but should also aim to increase both nitrogen use efficiency and water use efficiency while maintaining high productivity. Therefore manipulation of the C3 cycle to improve these parameters is also an important goal. The range of genetic and molecular techniques that are now available, together with the development and application of rapid in vivo techniques to allow in-field analysis of a wider range of species in their natural environments, will facilitate the wider analysis of natural variation in photosynthetic carbon assimilation. This approach has enormous and unexplored potential for future exploitation to improve yield through manipulation of the C3 cycle.
References
- Athanasiou K, Dyson BC, Webster RE, Johnson GN. (2010) Dynamic acclimation of photosynthesis increases plant fitness in changing environments. Plant Physiol 152: 366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Price GD, Long BM, Woodger FJ. (2006) The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J Exp Bot 57: 249–265 [DOI] [PubMed] [Google Scholar]
- Bar-Even A, Noor E, Lewis NE, Milo R. (2010) Design and analysis of synthetic carbon fixation pathways. Proc Natl Acad Sci USA 107: 8889–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber BE, Fuchs G. (2010) Autotrophic carbon fixation in archaea. Nat Rev Microbiol 8: 447–460 [DOI] [PubMed] [Google Scholar]
- Edgerton MD. (2009) Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol 149: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng LL, Han YJ, An BG, Yang J, Yang GH, Li YS, Zhu YG. (2007a) Overexpression of sedoheptulose-1,7-bisphosphatase enhances photosynthesis and growth under salt stress in transgenic rice plants. Funct Plant Biol 34: 822–834 [DOI] [PubMed] [Google Scholar]
- Feng LL, Wang K, Li Y, Tan YP, Kong J, Li H, Li Y, Zhu YG. (2007b) Overexpression of SBPase enhances photosynthesis against high temperature stress in transgenic rice plants. Plant Cell Rep 26: 1635–1646 [DOI] [PubMed] [Google Scholar]
- Galmes J, Flexas J, Keys AJ, Cifre J, Mitchell RAC, Madgwick PJ, Haslam RP, Medrano H, Parry MAJ. (2005) Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant Cell Environ 28: 571–579 [Google Scholar]
- Gowik U, Westhoff P. (2011) The path from C3 to C4 photosynthesis. Plant Physiol 155: 56–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Zrenner R, Sonnewald U, Stitt M. (1998) A moderate decrease of plastid aldolase activity inhibits photosynthesis, alters the levels of sugars and starch, and inhibits growth of potato plants. Plant J 14: 147–157 [DOI] [PubMed] [Google Scholar]
- Haake VM, Geiger M, Walch-Liu P, Engels C, Zrenner R, Stitt M. (1999) Changes in aldolase activity in wild-type potato plants are important for acclimation to growth irradiance and carbon dioxide concentration, because plastid aldolase exerts control over the ambient rate of photosynthesis across a range of growth conditions. Plant J 17: 479–489 [Google Scholar]
- Harrison EP, Olcer H, Lloyd JC, Long SP, Raines CA. (2001) Small decreases in SBPase cause a linear decline in the apparent RuBP regeneration rate, but do not affect Rubisco carboxylation capacity. J Exp Bot 52: 1779–1784 [DOI] [PubMed] [Google Scholar]
- Harrison EP, Willingham NM, Lloyd JC, Raines CA. (1998) Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation. Planta 204: 27–36 [Google Scholar]
- Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M. (2001) A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13: 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. (2010) The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Biol 61: 181–207 [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Sheehy JE, Langdale JA. (2008) Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr Opin Plant Biol 11: 228–231 [DOI] [PubMed] [Google Scholar]
- Howard TP, Metodiev M, Lloyd JC, Raines CA. (2008) Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proc Natl Acad Sci USA 105: 4056–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebeish RM, Niessen M, Thiruveedhi K, Bari R, Hirsch HJ, Rosenkranz R, Stäbler N, Schönfeld B, Kreuzaler F, Peterhänsel C. (2007) Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol 25: 593–599 [DOI] [PubMed] [Google Scholar]
- Kurek I, Chang TK, Bertain SM, Madrigal A, Liu L, Lassner MW, Zhu GH. (2007) Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 19: 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC. (2007) A welcome diversion from photorespiration. Nat Biotechnol 25: 539–540 [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Lawson T, Fryer M, Zakhleniuk OV, Lloyd JC, Raines CA. (2005) Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol 138: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieman-Hurwitz J, Rachmilevitch S, Mittler R, Marcus Y, Kaplan A. (2003) Enhanced photosynthesis and growth of transgenic plants that express ictB, a gene involved in HCO3- accumulation in cyanobacteria. Plant Biotechnol J 1: 43–50 [DOI] [PubMed] [Google Scholar]
- Liu CM, Young AL, Starling-Windhof A, Bracher A, Saschenbrecker S, Rao BV, Rao KV, Berninghausen O, Mielke T, Hartl FU, et al. (2010) Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463: 197–202 [DOI] [PubMed] [Google Scholar]
- Long SP, Zhu XG, Naidu SL, Ort DR. (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ 29: 315–330 [DOI] [PubMed] [Google Scholar]
- Marri L, Zaffagnini M, Collin V, Issakidis-Bourguet E, Lemaire SD, Pupillo P, Sparla F, Miginiac-Maslow M, Trost P. (2009) Prompt and easy activation by specific thioredoxins of calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol Plant 2: 259–269 [DOI] [PubMed] [Google Scholar]
- Masle J, Hudson GS, Badger MR. (1993) Effects of ambient CO2 concentration on growth and nitrogen use in tobacco (Nicotiana tabacum) plants transformed with an antisense gene to the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Physiol 103: 1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S. (2001) Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat Biotechnol 19: 965–969 [DOI] [PubMed] [Google Scholar]
- Moroney JV, Ynalvez RA. (2007) Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot Cell 6: 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Pinto M, Horton P. (2009) Agriculture and the new challenges for photosynthesis research. New Phytol 181: 532–552 [DOI] [PubMed] [Google Scholar]
- Olçer H, Lloyd JC, Raines CA. (2001) Photosynthetic capacity is differentially affected by reductions in sedoheptulose-1,7-bisphosphatase activity during leaf development in transgenic tobacco plants. Plant Physiol 125: 982–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MAJ, Madgwick PJ, Carvalho JFC, Andralojc PJ. (2007) Prospects for increasing photosynthesis by overcoming the limitations of Rubisco. J Agric Sci 145: 31–43 [Google Scholar]
- Peterhansel C, Maurino VG. (2011) Photorespiration redesigned. Plant Physiol 155: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis AR, Jr, Parry MAJ. (2007) Discoveries in Rubisco (ribulose 1,5-bisphosphate carboxylase/oxygenase): a historical perspective. Photosynth Res 94: 121–143 [DOI] [PubMed] [Google Scholar]
- Price GD, Badger MR, von Caemmerer S. (2011) The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiol 155: 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM. (2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461 [DOI] [PubMed] [Google Scholar]
- Raines CA. (2003) The Calvin cycle revisited. Photosynth Res 75: 1–10 [DOI] [PubMed] [Google Scholar]
- Raines CA. (2006) Transgenic approaches to manipulate the environmental responses of the C3 carbon fixation cycle. Plant Cell Environ 29: 331–339 [DOI] [PubMed] [Google Scholar]
- Raines CA, Paul MJ. (2006) Products of leaf primary carbon metabolism modulate the developmental programme determining plant morphology. J Exp Bot 57: 1857–1862 [DOI] [PubMed] [Google Scholar]
- Sage RF. (2002) Variation in the k(cat) of Rubisco in C(3) and C(4) plants and some implications for photosynthetic performance at high and low temperature. J Exp Bot 53: 609–620 [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ. (2004) Mechanism for deactivation of Rubisco under moderate heat stress. Physiol Plant 122: 513–519 [Google Scholar]
- Sassenrath-Cole GF, Pearcy RW. (1994) Regulation of photosynthetic induction state by the magnitude and duration of low-light exposure. Plant Physiol 105: 1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassenrath-Cole GF, Pearcy RW, Steinmaus S. (1994) The role of enzyme activation state in limiting carbon assimilation under variable light conditions. Photosynth Res 41: 295–302 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M. (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Stitt M, Lunn J, Usadel B. (2010) Arabidopsis and primary photosynthetic metabolism—more than the icing on the cake. Plant J 61: 1067–1091 [DOI] [PubMed] [Google Scholar]
- Stitt M, Schulze D. (1994) Does rubisco control the rate of photosynthesis and plant-growth—an exercise in molecular ecophysiology. Plant Cell Environ 17: 465–487 [Google Scholar]
- von Caemmerer S, Evans JR. (2010) Enhancing C3 photosynthesis. Plant Physiol 154: 589–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XY, Gowik U, Tang HB, Bowers JE, Westhoff P, Paterson AH. (2009) Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol 10: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H. (2011) Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol 155: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Kane HJ, Houtz RL, Sharwood RE. (2009) Rubisco oligomers composed of linked small and large subunits assemble in tobacco plastids and have higher affinities for CO2 and O2. Plant Physiol 149: 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE. (2007) Linked Rubisco subunits can assemble into functional oligomers without impeding catalytic performance. J Biol Chem 282: 3809–3818 [DOI] [PubMed] [Google Scholar]
- Zhu XG, de Sturler E, Long SP. (2007) Optimizing the distribution of resources between enzymes of carbon metabolism can dramatically increase photosynthetic rate: a numerical simulation using an evolutionary algorithm. Plant Physiol 145: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR. (2010a) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61: 235–261 [DOI] [PubMed] [Google Scholar]
- Zhu XG, Shan LL, Wang Y, Quick WP. (2010b) C4 rice—an ideal arena for systems biology research. J Integr Plant Biol 52: 762–770 [DOI] [PubMed] [Google Scholar]