Abstract
Many of the enzymes that catalyze reactions at nucleotide glycosidic linkages proceed through either a reactive oxocarbenium-ion intermediate or a transition state with considerable oxocarbenium character. To investigate how an RNA active site deals with the catalytic challenge of nucleotide synthesis, we probed the transition state of a ribozyme able to promote the formation of a pyrimidine nucleotide. Primary and secondary kinetic isotope effects indicate that this ribozyme stabilizes a highly dissociative reaction with considerable sp2 hybridization and negligible bond order between the departing pyrophosphate leaving group and the anomeric carbon. The small primary 13C isotope effect of 1.002 ± 0.003 indicates that the reaction is likely to be less concerted than that observed for protein nucleotide synthesis enzymes, which typically have primary 13C isotope effects of 1.02–1.03. The dissociative nature of the ribozyme reaction most resembles the reaction of some hydrolytic enzymes, such as uracil DNA glycosylase, which uses the negative charges found in the phosphodiester backbone of its DNA substrate to transiently stabilize an oxocarbenium ion during hydrolysis. The detectable hydrolysis observed in the ribozyme reaction indicates that shielding of this reactive intermediate from water is a significant challenge for RNA, which protein enzymes that synthesize nucleotides have managed to overcome during evolution, apparently by the utilization of more concerted chemistry.
We previously isolated from a large pool of random RNA sequences a ribozyme able to synthesize a pyrimidine nucleotide in a reaction modeled after pyrimidine synthesis of contemporary metabolism (1, 2). The ribozyme promotes the formation of 4-thiouridine at its 3′ terminus, using tethered 5-phosphoribosyl 1-pyrophosphate (PRPP) and free 4-thiouracil (4SUra). This ribozyme and its catalytic mechanism are of interest because pyrimidine nucleotide synthesis is a challenging reaction, central to the RNA world, that has not been demonstrated by prebiotic chemistry (3, 4).
Proteins that catalyze the formation or cleavage of nucleotide glycosidic linkages have been shown to exploit a number of mechanistically distinct reaction pathways (5–10). One extreme involves the stabilization of oxocarbenium-ion intermediates. Hydrolytic enzymes, such as uracil DNA glycosylase and ricin A-chain, have been shown through experimental and quantum mechanical studies to form an oxocarbenium ion that ultimately is quenched by water (9, 11, 12). In contrast, enzymes such as pertussis toxin and many glycosyl transfer enzymes have been shown to promote concerted reactions having low but appreciable amounts of bond order during the chemical step (9).
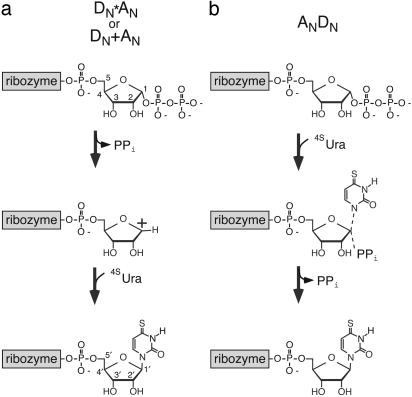
The ability of RNA, with its four bulky functional groups, to perform glycosidic chemistry might intrinsically be more limited than that of evolved protein catalysts. Thus an RNA catalyst might not be able to efficiently support the range of mechanisms used by protein enzymes, which possess a diverse set of functional groups and have been optimized through billions of years of evolution. A fundamental difficulty faced by all catalytic systems promoting nucleotide synthesis is that the uncatalyzed rate of PRPP hydrolysis greatly exceeds that of nucleotide synthesis, which occurs at an undetectably low rate (1). Because hydrolysis of PRPP is likely to take place through a very weakly concerted oxocarbenium-like transition state (5), a ribozyme, in analogy with protein enzymes, could employ at least two distinct catalytic strategies. One would be to promote the oxocarbenium-ion character of the uncatalyzed transition state while simultaneously limiting access of water to this reactive state (Fig. 1a). Alternatively, a ribozyme might promote a concerted mechanism in which the attack of the 4SUra and the departure of the pyrophosphate are made more synchronous (Fig. 1b). The relative accessibility of these catalytic strategies for emergent ribozymes is difficult to assess based on previous studies, because prior exploration of ribozyme reaction mechanisms and catalytic strategies has focused on ribozymes with concerted reactions (13, 14).
Fig. 1.
Two mechanistic extremes for ribozyme-promoted 4-thiouridine synthesis. (a) A stepwise dissociative mechanism (International Union of Pure and Applied Chemistry nomenclature: DN+AN or DN*AN) involving the displacement of pyrophosphate (PPi), and the formation of an oxocarbenium-ion intermediate, followed by nucleophilic addition of 4SUra and the production of 4-thiouridine. (b) A concerted reaction (ANdN) involving the attack of 4SUra and concerted displacement of pyrophosphate.
Examining the effects of isotopic substitution at or near the substrate reactive center has provided an invaluable tool for characterizing the transition states of proteinaceous enzymes (15, 16). Isotopic modification of an enzyme substrate leaves the static (geometric) arrangement of atoms unchanged but modifies the kinetic (vibrational and translational) properties of the transition state and thus provides an experimental avenue for examining transition-state structure. To extend the analysis of kinetic isotope effects (KIEs) to the study of a ribozyme transition state, we synthesized ribozyme-PRPP molecules with isotopic substitutions of key PRPP atoms and compared the ribozyme reaction rates of the substituted molecules with those of analogously prepared molecules with natural isotopic abundance.
Materials and Methods
Preparation of Radiolabeled Ribozyme-PRPP Pairs. In vitro evolution was used to generate a variant of isolate a15 from our initial selection with an efficiency suitable for characterization by using kinetic isotope methods (1, 2) (improved sequence: 5′-GGA UAA UGC GCA CAA GUA AGC GUG UUG CAA UCC UGC CUA UUG AAA CAC GAA AUC AGU CGU AAU GGA AAC GAG GGG GAU AGG UUG CUC GUU ACU CCC AUU UUG GCA CCA AAU ACG GCA GUA CAG UAA UCG ACA AGU ACU GGA UGG CCA UAG UGG CAC AAA AUA GGC CUC UAC CAG CGG UGU GGG GUA ACC CAC AUC GCA GCA ACC). This ribozyme performs the 4-thiouridine synthesis reaction with an apparent kcat/Km4SUra of 50 M–1·min–1, which is at least 108 times more efficient than the uncatalyzed reaction. 32P- and 33P-labeled ribozyme was produced by T7 transcription with a final specific activity ≈9,600 GBq/mmol (isotopes from New England Nuclear). Gel-purified RNA transcripts were attached to PRPP by using preadenylated PRPP (AppRpp) and T4 RNA ligase [1–5 μM RNA, 40–50 μM isotopically labeled AppRpp, 50 mM Hepes (pH 8.3), 5 mM MgCl2, 3.3 mM DTT, 10 μg/ml BSA in 8.3% glycerol, and 0.5 units/μl enzyme from Pharmacia in a 4-h incubation at room temperature]. Ligation was terminated by the addition of EDTA followed by phenol/chloroform extraction, ethanol precipitation, and resuspension in water. To ensure equal treatment of both species of a dual-labeled pair, the appropriate dual-labeled species were mixed together to form pairs no. 1 and no. 2 (Table 1) and then purified on a denaturing sequencing gel before being used for kinetic analysis.
Table 1. The four dual-labeled pairs used to measure either the secondary ([1-1H, 1-2H]) or the primary ([1-12C, 1-13C]) KIE.
| Determination | Dual label no. 1 | Dual label no. 2 | Measured KIE |
|---|---|---|---|
| [1-1H, 1-2H] | (32P, 1-1H)* + (33P, 1-2H) | (33P, 1-1H)* + (32P, 1-2H) | 1.15 ± 0.012 |
| [1-12C, 1-13C] | (32P, 1-12C)* + (33P, 1-13C) | (33P, 1-12C)* + (32P, 1-13C) | 1.002 ± 0.003 |
Six different RNA-PRPP constructs were generated. The label (X, 1-Y) refers to the RNA radiolabel (X = 32P or 33P) and the isotopic substitution (Y = 1H, 2H, 12C, or 13C) found at the 1-carbon of the tethered PRPP substrate.
(1-1H)PRPP and (1-12C)PRPP are equivalent and indicate PRPP with natural abundance of hydrogen and carbon.
Isotopic AppRpp Synthesis. PRPP was prepared from isotopically derivatized d-ribose, either 1-2H (98%) or 1-13C (99%) substituted from Cambridge Isotope Laboratories (Cambridge, MA). Ribose 5-phosphate was synthesized from ATP and ribose by using ribokinase (EC 2.7.1.15), and pyrophosphate was transferred from ATP by using phosphoribosyltranferase (EC 2.7.6.1) to generate PRPP (17, 18). Ribose 5-phosphate and PRPP were purified on a DEAE A-25 column, with a linear gradient from 50 to 900 mM triethylammonium bicarbonate (TEAB), pH 7.0. Ribose 5-phosphate and PRPP eluted at ≈250 mM and ≈600 mM TEAB, respectively. Six hundred millimolar PRPP was incubated with 300 mM adenosine 5′-phosphorimidazole (19) in the presence of 200 mM MgCl2 for 2 h at 50°C. The reaction was quenched with EDTA, and AppRpp was purified on a reversephase C18 HPLC column. An analogous synthesis of AppRpp starting from isotopically unenriched PRPP (Sigma) was also performed to generate the AppRpp used to synthesize the ribozyme-[1-1H]PRPP/ribozyme-[1-12C]PRPP species.
Analysis of Isotopic Purity. A 2.6-nmol sample of each isotopically labeled AppRpp synthesis was mixed with GpC and ApApC oligoribonucleotide mass standards in a matrix of 22.5 g/liter 3-hydroxypicolinic acid, 2.5 g/liter l-tartaric acid in 50% acetonitrile for matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) analysis (Voyager instrument, PerSeptive Biosystems, Framingham, MA). Calibration was performed by using the oligoribonucleotide mass standards, which bracket the mass of AppRpp. As expected, the 13C- and 2H-labeled AppRpp samples were 1 atomic mass unit heavier than the unenriched AppRpp (predicted m/z for lightest isotopic peak was 718.00 atomic mass units, observed was 718.08 atomic mass units; predicted m/z for lightest isotopic peak of 13C- and 2H-substituted molecules was 719.00 atomic mass units, observed were 719.00 and 719.19 atomic mass units, respectively). To evaluate purity, MALDI-TOF spectra for each isotopic derivative of AppRpp were first baseline subtracted and normalized, then aligned along the m/z axis until the highest channel from each derivative was in register with underivatized AppRpp. The peak shapes were virtually identical and showed <5% variability from sample to sample in the regions diagnostic of potential isotopic contamination. From this analysis we infer that the isotopic purity of each AppRpp synthesis was >95%.
Kinetic Analysis. RNA pairs were heated to 80°C in water and cooled to room temperature before addition of buffer. Ribozyme reactions were performed with 1 mM 4SUra in 50 mM Tris·HCl (pH 7.6)/25 mM MgCl2/150 mM KCl at 23°C. Reacted ribozyme was purified away from unreacted ribozyme by using [(N-acryloylamino)phenyl]mercuric chloride (APM)/polyacrylamide gels (1). To achieve approximately equal yield of reacted ribozyme product for each time point, smaller aliquots were loaded for the later time points. Scintillation counting was then used to determine R(t), the relative ratios of 32P to 33P in the product at each time point, correcting for the radioactive decay of the samples during counting. The amount of 4-thiouridine generated by one isotopically modified ribozyme relative to another is given by the equation R(t)* = (1 – e–k2t)/(1 – e–k1t), where t is time, k1 and k2 are the respective rates for the 32P- and 33P-tagged isotopic substitutions, and R(t)* is R(t) normalized by the asymptotic ratio. This asymptotic ratio reflects the relative amounts of 33P and 32P radioactive label used in a particular experiment and was determined from time points taken after the reaction was more than 99.8% complete (t > 128 min). After directly measuring k1 from PhosphorImager analysis of the gel used to purify the ribozyme product, k2 was fitted as a function of R(t)* by using the combined data from dual-label pair no. 2 together with the inverse of the data from dual-label pair no. 1.
Scintillation Counting. The maximum endpoint energies for 33P and 32P β decay are separated by a factor of 7, allowing the clean separation of 33P and 32P channels by using a conventional scintillation counter. Radiolabeled samples were eluted from gel slices, diluted in 15 vol of scintillation fluid (Formula 989, Packard) in glass scintillation vials, and counted (Beckman LS 6000SC scintillation counter) along with pure 32P- and 33P-radiolabeled RNA references, which allowed the determination of channel separation for each counting run. High (H) and low (L) windows were set by using channels 0–750 and 750–1,000, respectively. At these settings, L captured nearly 100% of the 33P signal and ≈50% of the 32P signal, whereas H captured 0.1% of the 33P signal and ≈50% of the 32P signal). The count rates in the L and H channels were background subtracted, and the amounts of 32P and 33P signal were determined by: C(33P) = (F22L – F21H)/(F11F22 – F21F12), C(32P) = (F11H – F12L)/(F11F22 – F21F12), with F11 = L/(L + H), F12 = H/(L + H), counting with pure 33P, and F21 = L/(L + H), F22 = H/(L + H), counting with pure 32P. A 30-min count time was used to minimize the effect of radioactive decay over the course of an individual count (effect <0.05%). However, as the time required to count an entire experiment was significant relative to the half-lives of the isotopes, the count rates, C(32P) and C(33P), observed for a particular vial were corrected for the effects of radioactive decay. Each sample vial was counted at least three times to improve counting statistics.
Forward Commitment. A ribozyme reaction initiated with buffer containing 1 mM 4SUra was diluted 10-fold into buffer with or without 1 mM 4SUra, resulting in 4SUra concentrations of 1.0 and 0.1 mM, respectively. Rates were compared with initial reaction rates in 0.1 and 1.0 mM 4SUra.
Hydrolysis Rate Determination. Ribozyme-PRPP complex was incubated in reaction buffer lacking 4SUra. At appropriate time points, aliquots were removed and supplemented with 4SUra to a final concentration of 1 mM. After reacting to completion, samples were analyzed on an APM gel. The total fraction of RNA able to react fit well to an exponential function fe–kt, where k is the PRPP hydrolysis rate, f is the fraction of active ribozyme, and t is the incubation time in buffer lacking 4SUra. These results agreed well with experiments monitoring the ribozyme active fraction as a function of 4SUra substrate concentration, which is sensitive to PRPP hydrolysis rate (1).
Results
The spectrum of transition states that describe nucleotide synthesis can be parameterized by degrees of freedom that include substrate and leaving group orientation, hybridization state, and sugar pucker. KIE measurements generate experimental constraints that help define the actual transition-state structure. Studies of enzyme-catalyzed nucleotide synthesis or hydrolysis have shown that the two most mechanistically informative sites of isotopic substitution are the primary substitution, in which carbon at the reaction center (1-C) of PRPP is replaced with a heavier isotope of carbon, and the α secondary substitution, in which the hydrogen proximal to the reaction center (1-H) is replaced with deuterium or tritium (16). Accordingly, we focused on these two substitutions to investigate the mechanism of ribozyme-mediated nucleotide synthesis.
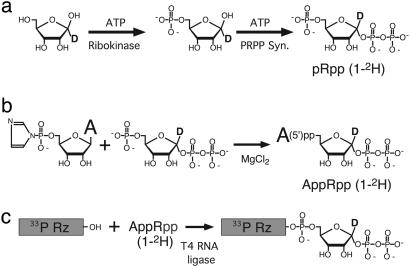
Accurate measurement of the small rate differences typical of KIEs is facilitated by using mixtures of dual-label substrates (17). To employ this strategy for our nucleotide synthase ribozyme, PRPP with natural isotope abundance or PRPP isotopically derivatized with either 1-2H or 1-13C was ligated to ribozyme RNA and body-labeled with either 32P or 33P to generate six different dual-labeled species, as illustrated for the dual-labeled species (33P, 1-2H) in Fig. 2. Isotopic purity of the adenylylated PRPP intermediate used to make each species was monitored by mass spectrometry and found to exceed 95%. For each isotopic substitution experiment, the appropriate ribozyme-PRPP constructs were mixed to generate two dual-label pairs (Table 1).
Fig. 2.
The synthesis of the (33P, 1-2H) dual-labeled ribozyme-PRPP species. (a) Ribose isotopically substituted with 1-2H was converted first to ribose 5-phosphate and then PRPP by using ribokinase and PRPP synthase successively in the presence of ATP. (b) PRPP purified by using a DEAE column was reacted with adenosine-5′-phosphorimidazole (19) to synthesize adenylylated PRPP (AppRpp), which was purified by reverse-phase HPLC. (c) Ribozyme (Rz) radiolabeled during T7 transcription with 33P was covalently attached at its 3′ end to the isotopically modified PRPP by using T4 RNA ligase.
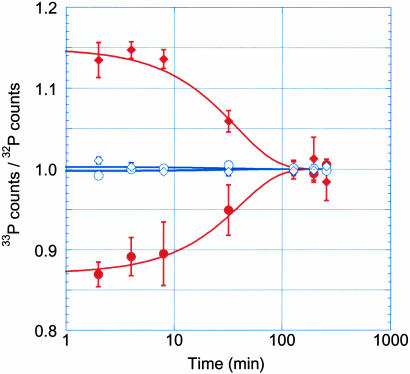
The large difference between β decay endpoint energies of 32P and 33P allowed the accurate determination of rate differences between the two species comprising each dual-labeled pair. The ratios of 33P to 32P counts observed for the secondary [1-1H, 1-2H] KIE determination are shown in Fig. 3 (filled symbols). The differences between the two species of each pair were greatest at early time points, then diminished at later times as both species reacted to completion. As expected, the ratios for the (32P, 1-1H) + (33P, 1-2H) pair were inverse to those of the (33P, 1-1H) + (32P, 1-2H) pair, indicating that the dual-label and counting procedures had little intrinsic bias. The data correspond to a KIE of 1.15 ± 0.012 (reported as KIE = k(1-1H)/k(1-2H) ± the standard error for the fit). Stated differently, the reaction is 15% more rapid with a hydrogen rather than deuterium proximal to the reaction center. Although secondary KIE values are not easily related directly to transitionstate structure, the large size of this secondary KIE is consistent with a transition state or intermediate possessing considerable sp2 hybridization at the anomeric carbon (8, 12). This effect is attributed to the large change in out-of-plane vibrational frequency that occurs when the mass of the vibrating proton is changed.
Fig. 3.
The effect of isotopic substitution on nucleotide synthesis rate. • and ♦ indicate average values for four independent experiments using the secondary dual-label pairs (32P, 1-1H) + (33P, 1-2H) and (33P, 1-1H) + (32P, 1-2H), respectively. ○ and ⋄ indicate average values for three independent experiments using the primary dual-label pairs (32P, 1-12C) + (33P, 1-13C) and (33P, 1-12C) + (32P, 1-13C), respectively. The bars report standard errors.
The secondary KIE is near the maximum predicted from theory. Such a large KIE shows that no steps along the reaction pathway were slower than the chemical step. Independent evidence of a rate-limiting chemical step was provided by an experiment in which no forward commitment to chemistry was detected when the ribozyme and substrate were incubated in standard conditions, then abruptly diluted 10-fold. The relative rate difference observed immediately after dilution was 6.5-fold, consistent with the rate difference observed for incubations that started with 0.1 and 1.0 mM 4SUra substrate concentrations. We also measured the rate of PRPP hydrolysis, which could lead to unanticipated isotopic fractionation. The rate of PRPP hydrolysis (0.005 min–1) was 1/10th the rate of 4-thiouridine synthesis (0.05 min–1 in the presence of 1 mM 4SUra) and therefore could not significantly effect the interpretation of the observed KIE values.
The finding that the ribozyme chemical step is mostly, if not totally, rate-limiting is important when considering the very low primary KIE of the reaction. The KIE value of the [1-12C, 1-13C] primary substitution was 1.002 ± 0.003, with no data point being further than 1% away from the mean of the total data set (Fig. 3, open symbols). A negligible KIE at the 1-C primary position, indicating a transition state with very low bond order, would be predicted for a stepwise dissociative reaction (17). The very low observed primary KIE is significantly smaller than the lower bound thought possible for concerted mechanisms. Geometric effects within the ribose ring distort this simple picture, but quantum mechanical calculations have not been able to generate primary 13C KIE values <1.013–1.015, even for highly dissociative concerted reactions (8, 12, 15, 20).
Discussion
Our primary and secondary KIE measurements combined with previous computational studies point to a sequential ribozyme reaction that exploits an oxocarbenium-ion intermediate (Fig. 1a). The negligible primary KIE indicates that very low or no bond order exists between the departing pyrophosphate leaving group and the anomeric carbon, whereas the high secondary KIE value indicates considerable sp2 hybridization at the anomeric carbon during the reaction (8). Although sufficient to characterize the reaction mechanism, the KIE values from primary and secondary isotopic substitutions do not provide complete details of the reaction geometry (16). These await further KIE measurements at additional atomic sites.
The KIE measurements observed for ribozyme-promoted 4-thiouridine synthesis can be compared with protein-catalyzed hydrolysis reactions, nucleotide synthesis enzymes, and several uncatalyzed reactions (Table 2). Our results are most similar to those of Schramm and colleagues, who conclude that the small inverse primary KIE observed during the depurination of 28S ribosomal RNA by ricin A-chain is consistent with a stepwise dissociative mechanism involving an oxocarbenium-ion intermediate with finite lifetime (8). Uracil-DNA glycosylase, which has a slightly larger primary KIE value than our ribozyme, is also proposed to exploit an oxocarbenium intermediate (20, 21). These hydrolytic enzymes differ from the nucleotide synthase ribozyme in that they promote the departure of a base initially in a β configuration, whereas the ribozyme acts initially on PRPP having pyrophosphate in the α configuration. We note that this difference might be expected to subtly affect the direct comparison of observed KIE values in a manner that is difficult to assess at this time (22).
Table 2. A comparison of primary and secondary KIE values for a variety of glycosidic bond formation and cleavage reactions.
| Reaction | Mediator | Primary isotope effect [1-12C, 1-13C] | Secondary isotope effect [1-1H, 1-2H] | Initial orientation of leaving group* |
|---|---|---|---|---|
| 4-Thiouridine synthesis | 4-Thiouridine synthase (RNA) | 1.002 ± 0.003 | 1.15 ± 0.012 | α |
| RNA depurination (8)† | Ricin A-chain (protein) | 0.996 ± 0.004 | 1.111 ± 0.009 | β |
| Deoxyuridine hydrolysis (20) | Uracil-DNA glycosylase (protein) | 1.010 | 1.201 ± 0.021 | β |
| OMP synthesis (6)†‡ | OPRTase (protein) | 1.021 ± 0.004 | 1.135 ± 0.007 | (β) |
| IMP synthesis (5)† | HGPRTase (protein) | 1.026 | 1.20 | α |
| AMP hydrolysis (24)† | Acid | 1.026 ± 0.009 | 1.159 ± 0.01 | β |
| Uridine hydrolysis (25) | Acid | ND | 1.11 | β |
| PRPP hydrolysis (5)† | Acid | 1.01 | 1.27 | α |
The KIE effects observed for our ribozyme-promoted nucleotide synthesis reaction are highlighted in bold. OMP, orotidine 5′-monophosphate; OPRTase, orotate phosphoribosyltransferase; HGPRTase, hypoxanthine/guanine phosphoribosyltransferase; ND, not done.
The orientation of the leaving group (α or β) might have a subtle effect on chemistry. For example, 1-glucopyranosyl fluorides have differing KIE values depending on the orientation of the fluoride leaving group (22).
Tritium and 14C isotope experiments that have been converted to the corresponding 2H and 13C values by using the approximation of Swain and Schaad (28).
KIE effects for OPRTase were studied by using the reverse reaction and phosphonoacetate (6).
KIE data for both purine and pyrimidine nucleotide synthesis enzymes (hypoxanthine/guanine phosphoribosyltransferase and orotate phosphoribosyltransferase) indicate that these proteinaceous enzymes, in contrast to our ribozyme, develop concerted oxocarbenium-like transition states with appreciable bond orders (5, 6). The observed primary isotope effect for uncatalyzed hydrolysis of PRPP is consistent with a dissociative mechanism but was measured at a pH of 2 (5). It is reasonable to expect that hydrolysis becomes more concerted at higher pH; however, studies of PRPP hydrolysis in basic solutions are precluded by the propensity of the molecule to form 1,2-cyclic phosphates in alkali (23). The hydrolysis of purine nucleotides is weakly concerted, even when measured at low pH (24), whereas hydrolysis of pyrimidine nucleotides is difficult to study because of the kinetic stability of the pyrimidine glycosidic linkage (25).
Taken as a whole, a comparison with the available theoretical and experimental data indicates that our ribozyme transiently stabilizes an oxocarbenium ion during nucleotide synthesis, which has very little, if any, residual bond order with the leaving or attacking moieties. Our KIE measurements are most consistent with a dissociative reaction mechanism more resembling that of protein-catalyzed nucleotide hydrolysis than that of protein-catalyzed nucleotide synthesis. This raises the question as to how effectively the ribozyme prevents hydrolysis during nucleotide synthesis. The original ribozyme isolate hydrolyzes PRPP 50–60 times faster than the uncatalyzed hydrolysis rate (1). Additional in vitro evolution, which generated the improved ribozyme used in the KIE experiments reported here, led to a 12-fold increase in catalysis with an accompanying 2- to 3-fold increase in hydrolysis. Thus, mutations able to increase catalytic efficiency were not obtained without also increasing undesired hydrolytic activity, even though there was strong selective pressure against hydrolysis, because molecules that hydrolyze their tethered PRPP are no longer reactive. Nonetheless, when taking into account the much lower concentration of 4SUra compared with water, the improved ribozyme shows a discrimination of ≈5 × 105-fold for 4SUra over water and has an apparent first-order rate enhancement for the desired reaction of at least 108-fold, compared with <200-fold for hydrolysis (2).
Why does RNA use a dissociative mechanism most similar to hydrolytic enzymes when protein-based nucleotide-synthesis enzymes use a more concerted reaction? A concerted strategy might appear more appropriate, because such a catalytic strategy would naturally tend to limit harmful side reactions such as hydrolysis. Indeed, the need to avoid hydrolysis has been used to explain why the protein enzymes whose function requires the attack of a specific nucleophile might use reactions that are more concerted than those of the hydrolytic enzymes (9). The mechanism of uracil-DNA glycosylase provides a possible explanation for why the dissociative pathway might be more accessible to nascent ribozymes. Uracil-DNA glycosylase decreases the activation energy for deoxyuridine hydrolysis by ≈110 kJ/mol; ≈90 kJ/mol of this total is contributed by four phosphate groups found within the backbone of the DNA substrate. These anionic charges are able to stabilize the positively charged oxocarbenium ion formed during hydrolysis (11) and assist with the departure of the negatively charged uracil leaving group. A very similar catalytic strategy can be imagined for an RNA catalyst, with several of its many phosphate groups positioned to stabilize the oxocarbenium ion and assist with departure of the negatively charged pyrophosphate leaving group. The notion of phosphates pointing toward the ribozyme catalytic site would also explain why an alignment of active ribozyme variants has only two invariant residues outside of Watson–Crick helices, despite the large size of the ribozyme catalytic motif (2). However, in contrast to the uracil-DNA glycosylase active site, the ribozyme active site would need to both restrict access to water and position 4SUra to promote detectable nucleotide synthesis.
The transition-state structure of our nucleotide synthase ribozyme, inferred by KIE measurement, demonstrates that a highly dissociative reaction can be mediated by a ribozyme and indicates that RNA is likely to be able to promote other small-molecule metabolic reactions, particularly those involving reactive cation-like transition states, such as glycosyl-transfer reactions, likely to have been required in an RNA world (26, 27).
Acknowledgments
We thank V. Schramm and P. Berti for helpful discussions, M. Lawrence for comments on the manuscript, and P. Berti and J. Williamson for enzymes and constructs. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (to P.J.U.) and the National Aeronautics and Space Administration Exobiology Program (to D.P.B.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PRPP, 5-phosphoribosyl 1-pyrophosphate; AppRpp, 5-adenylyl-PRPP; KIE, kinetic isotope effect; APM, [(N-acryloylamino)phenyl]mercuric chloride; 4SUra, 4-thiouracil.
References
- 1.Unrau, P. J. & Bartel, D. P. (1998) Nature 395, 260–263. [DOI] [PubMed] [Google Scholar]
- 2.Chapple, K., Bartel, D. P. & Unrau, P. J. (2003) RNA 9, 1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joyce, G. F. (2002) Nature 418, 214–221. [DOI] [PubMed] [Google Scholar]
- 4.Orgel, L. E. (1998) Trends Biochem. Sci. 23, 491–495. [DOI] [PubMed] [Google Scholar]
- 5.Goitein, R. K., Chelsky, D. & Parsons, S. M. (1978) J. Biol. Chem. 253, 2963–2971. [PubMed] [Google Scholar]
- 6.Tao, W., Grubmeyer, C. & Blanchard, J. S. (1996) Biochemistry 35, 14–21. [DOI] [PubMed] [Google Scholar]
- 7.Kyte, J. (1995) Mechanism in Protein Chemistry (Garland, New York).
- 8.Chen, X. Y., Berti, P. J. & Schramm, V. L. (2000) J. Am. Chem. Soc. 122, 1609–1617. [Google Scholar]
- 9.Berti, P. J. & Tanaka, K. S. E. (2002) Adv. Phys. Org. Chem. 37, 239–314. [Google Scholar]
- 10.Mazzella, L. J., Parkin, D. W., Tyler, P. C., Furneaux, R. H. & Schramm, V. L. (1996) J. Am. Chem. Soc. 118, 2111–2112. [Google Scholar]
- 11.Dinner, A. R., Blackburn, G. M. & Karplus, M. (2001) Nature 413, 752–755. [DOI] [PubMed] [Google Scholar]
- 12.Chen, X. Y., Berti, P. J. & Schramm, V. L. (2000) J. Am. Chem. Soc. 122, 6527–6534. [Google Scholar]
- 13.Rupert, P. B., Massey, A. P., Sigurdsson, S. T. & Ferre-D'Amare, A. R. (2002) Science 298, 1421–1424. [DOI] [PubMed] [Google Scholar]
- 14.Shan, S. O., Yoshida, A., Sun, S. G., Piccirilli, J. A. & Herschlag, D. (1999) Proc. Natl. Acad. Sci. USA 96, 12299–12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berti, P. J. (1999) Methods Enzymol. 308, 355–397. [DOI] [PubMed] [Google Scholar]
- 16.Schramm, V. L. (1999) Methods Enzymol. 308, 301–355. [DOI] [PubMed] [Google Scholar]
- 17.Schramm, V. L. (1998) Annu. Rev. Biochem. 67, 693–720. [DOI] [PubMed] [Google Scholar]
- 18.Tolbert, T. J. & Williamson, J. R. (1996) J. Am. Chem. Soc. 118, 7929–7940. [Google Scholar]
- 19.Lohrmann, R. & Orgel, L. E. (1978) Tetrahedron 34, 853–855. [Google Scholar]
- 20.Werner, R. M. & Stivers, J. T. (2000) Biochemistry 39, 14054–14064. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, Y. L., Drohat, A. C., Ichikawa, Y. & Stivers, J. T. (2002) J. Biol. Chem. 277, 15385–15392. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Y., Bommuswamy, J. & Sinnott, M. L. (1994) J. Am. Chem. Soc. 116, 7557–7563. [Google Scholar]
- 23.Khorana, H. G., Fernandes, J. F. & Kornberg, A. (1958) J. Biol. Chem. 230, 941–948. [PubMed] [Google Scholar]
- 24.Parkin, D. W., Leung, H. B. & Schramm, V. L. (1984) J. Biol. Chem. 259, 9411–9417. [PubMed] [Google Scholar]
- 25.Prior, J. J. & Santi, D. V. (1984) J. Biol. Chem. 259, 2429–2434. [PubMed] [Google Scholar]
- 26.Gesteland, R. F., Cech, T. R. & Atkins, J. F. (1999) The RNA World (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 27.Bartel, D. P. & Unrau, P. J. (1999) Trends Cell Biol. 9, M9–M13. [PubMed] [Google Scholar]
- 28.Swain, C. G., Stivers, J. F., Reuwer, J. & Schaad, L. J. (1958) J. Am. Chem. Soc. 80, 5885. [Google Scholar]