Abstract
SHORT-ROOT (SHR) is a well-characterized regulator of radial patterning and indeterminacy of the Arabidopsis (Arabidopsis thaliana) primary root. However, its role during the elaboration of root system architecture remains unclear. We report that the indeterminate wild-type Arabidopsis root system was transformed into a determinate root system in the shr mutant when growing in soil or agar. The root growth behavior of the shr mutant results from its primary root apical meristem failing to initiate cell division following germination. The inability of shr to reactivate mitotic activity in the root apical meristem is associated with the progressive reduction in the abundance of auxin efflux carriers, PIN-FORMED1 (PIN1), PIN2, PIN3, PIN4, and PIN7. The loss of primary root growth in shr is compensated by the activation of anchor root primordia, whose tissues are radially patterned like the wild type. However, SHR function is not restricted to the primary root but is also required for the initiation and patterning of lateral root primordia. In addition, SHR is necessary to maintain the indeterminate growth of lateral and anchor roots. We conclude that SHR regulates a wide array of Arabidopsis root-related developmental processes.
Higher plants exhibit an amazing diversity of root architectures at both the systems and anatomical levels (Waisel et al., 2002). Many dicotyledonous plants like Arabidopsis (Arabidopsis thaliana) have a primary root that repeatedly branches to generate several orders of lateral roots, whereas the root systems of cereal crops such as rice (Oryza sativa) and maize (Zea mays) are predominantly composed of adventitious roots (Hochholdinger et al., 2004; Osmont et al., 2007). Despite these anatomical differences, with the exception of the highest orders, roots from all plant species exhibit indeterminate growth behavior (i.e. are able to maintain growth indefinitely; Waisel et al., 2002).
The cellular basis of indeterminate root growth has been best studied in the Arabidopsis primary root. In this experimental system, stem cells (also termed initials) abut specialized organizing cells (often denoted as the quiescent center [QC]) within the root apical meristem (RAM; Dolan et al., 1993). When initial cells divide, they generate two daughter cells; the one directly abutting the QC remains a stem cell, while the remaining daughter cell (also termed a transit amplifying cell) undergoes a finite number of cell divisions before exiting the meristem and elongating (Scheres, 2007). Mutants that disrupt initial cell/QC function often exhibit a determinate root growth phenotype, resulting in the termination of organ growth (Benfey et al., 1993; Di Laurenzio et al., 1996).
Genetic studies have identified several genes that regulate indeterminate root growth in Arabidopsis. One of these genes encodes the SHORT-ROOT (SHR) protein, which has been shown to be essential for RAM function (Benfey et al., 1993; Scheres et al., 1995). In mutant plants lacking SHR, primary roots are dramatically shortened (Benfey et al., 1993; Scheres et al., 1995). Another related protein, SCARECROW (SCR), has a similar role alongside SHR (Benfey et al., 1993; Di Laurenzio et al., 1996). SHR and SCR genes encode closely related transcription factors belonging to the GRAS gene family (Di Laurenzio et al., 1996; Helariutta et al., 2000). SHR has been demonstrated to directly regulate the expression of genes including SCR (Levesque et al., 2006) and a number of cell cycle components including the D-type cyclin, CYCD6;1 (Sozzani et al., 2010).
The spatiotemporal roles of SHR, SCR, and various others genes such as PLETHORA1 (PLT1) and PLT2 in establishing and maintaining the RAM have been extensively studied. In the model proposed by Blilou et al. (2005), redistribution of polar localized auxin efflux carriers of the PIN-FORMED (PIN) family results in the accumulation of auxin at the prospective meristem stem cell niche; this maximum is maintained throughout postembryonic elaboration of the root (Sabatini et al., 1999; Aida et al., 2002; Friml et al., 2003, Blilou et al., 2005). The distal auxin maximum then determines the expression pattern of the transcription factors PLT1 and PLT2, which are required for QC identity and stem cell specification from embryogenesis onward (Aida et al., 2004). The distal auxin maximum maintained by PIN proteins restricts PLT expression to the basal pole of the proembryo; this facilitates the activation of the root primordium. In turn, PLT positively regulates PIN expression (PIN3, PIN4, and PIN7; Blilou et al., 2005), which reinforces the flux of auxin into the RAM, thereby maintaining the postembryonic position of the distal stem cell niche. The radial expression domains of the transcription factors SHR and SCR participate in the positioning of the stem cell niche. The SHR gene is transcribed in the stele, but the mobile SHR protein migrates outward one cell layer to the adjacent QC, cortex-endodermis initial (CEI) cell, and endodermis layer (Helariutta et al., 2000; Nakajima et al., 2001), where it activates the expression of SCR (Nakajima et al., 2001). The endodermal/QC domain of SHR/SCR protein expression overlaps with the distal PLT expression domain, with the highest level of both components defining the stem cell niche.
Despite of the importance of SHR for regulating RAM function and indeterminate primary root growth, no studies have addressed its impact on root system architecture (RSA) to date. The progression of a plant root system from a single meristem-containing primary root, formed during embryogenesis, to the extensively elaborated RSA of a mature plant involves numerous exogenous abiotic and biotic factors plus endogenous genetic factors. One of the most important factors determining total RSA is the postembryonic appearance of lateral root primordia (LRP) and ultimately lateral roots that branch off from the primary root (Nibau et al., 2008). Given the fact that lateral root development is considered to be a broad recapitulation of equivalent processes in the primary root, we investigated whether SHR plays a role in LRP development and, consequently, on global RSA. We report that SHR is indeed a key regulator, controlling primary, lateral, and anchor root development at the macroscopic scale and the patterning of lateral (but not anchor) roots at the microscopic scale.
RESULTS
The shr Mutation Causes RSA to Become Determinate
The shr mutants were initially identified based on their severely reduced primary root growth and radial patterning defects (Benfey et al., 1993; Scheres et al., 1995). Nevertheless, shr mutant seedlings are able to grow and complete their life cycle, somehow compensating for the dramatic reduction of their primary root length.
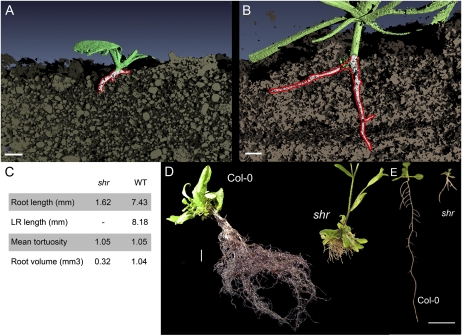
We initially investigated the morphological effect the shr mutation had on the development of the root system in soil. X-ray computed tomography (CT) was employed to investigate the three-dimensional (3D) root architecture of 3-week-old wild-type and shr seedlings grown in a sandy loam soil (Fig. 1, A–C; Supplemental Movies S1 and S2). This technique relies on discriminating differences in attenuation density between the soil matrix and live roots. Difficulties can arise when discriminating differences in attenuation density between a water-filled pore space and live roots. Nevertheless, the CT scan and subsequent 3D reconstruction allowed us to retrieve more than 50% of the root tissues of the scanned plants. This amounted to a total root volume of 0.3 mm3 for shr and 1 mm3 for the wild type (root volume was automatically estimated by the 3D reconstruction software; for additional details, see “Materials and Methods”). Roots were relatively straight, with a mean tortuosity of 1.05 both in shr and the wild type (tortuosity being the ratio of the root length over the shortest distance between the two extremities of the measured root). However, shr did not exhibit a strong 3D branching phenotype when grown in the sandy loam soil compared with the wild type.
Figure 1.
RSA becomes determinate in the shr mutant. A and B, 3D x-ray microtomography analysis of RSA of 3-week-old soil-grown shr (A) and wild-type (B) seedlings grown in a sandy loam soil. Bars = 1 mm. C, Quantitative description of root architecture extracted from A and B. WT, Wild type. D, RSA of 4-week-old compost-grown wild-type and shr seedlings. Bar = 1 cm. E, RSA of 12-d-old agar-grown wild-type and shr seedlings. Bar = 1 cm.
To investigate further the capability of shr to develop a branched root architecture, we then compared 4-week-old compost-grown wild-type and shr seedlings (Fig. 1D). Seedlings grown in compost exhibited a much bigger root system compared with soil-grown plants. The dimorphism between shr and the wild type was also amplified compared with soil-grown plants. Whereas wild-type plants had an indeterminate tap root system, the shr mutant exhibited a fibrous determinate root system. The shr seedling had numerous short roots of equivalent lengths, each one thicker than individual wild-type roots. Microscopic observation of the shr roots revealed a low degree of branching toward the apex of roots, with no discernible primordia or young laterals along the bottom half of roots (data not shown). Instead, new roots appeared to originate and branch near the seedling root-shoot junction. Similar alterations of RSA can be similarly observed in agar-grown shr seedlings, which exhibit reduced growth and a characteristic “tripod” architecture after 1 week of growth (Fig. 1E).
The shr Mutant Fails to Reinitiate Cell Division in the Primary Root following Germination
We next investigated how the shr mutation could lead to this dramatic change in overall RSA. To understand the basis for the determinacy of the RAM in shr, we studied the mitotic activity of the mutant before and after germination using the cyclinB1:1:GUS marker (Fig. 2).
Figure 2.
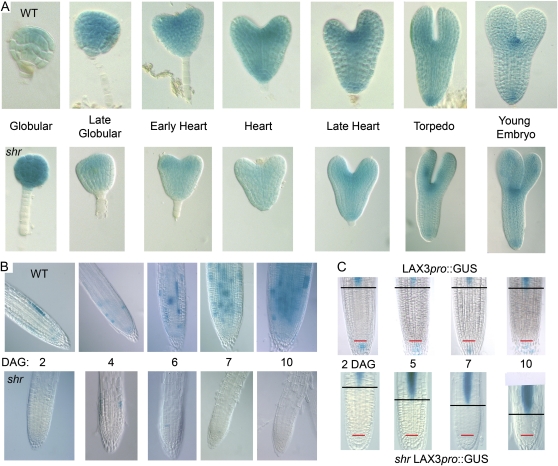
The shr mutation blocks mitotic activity in the RAM following germination. A, CyclinB1:1:GUS expression during wild-type (WT) and shr embryo development. B, CyclinB1:1:GUS postgermination expression in the RAM of the wild type and shr at different DAG. C, LAX3pro:GUS expression in the RAM of shr at different DAG. The red line indicates the position of the QC, and the black bar indicates the start of the elongation zone coinciding with the LAX3pro:GUS expression.
As the Arabidopsis embryo develops, almost every cell is continuously dividing, as revealed by the mitotic marker cyclinB1:1:GUS (Fig. 2A). No major differences in mitotic marker expression were observed between shr and the wild type during embryogenesis. After seedling germination, cell division must be reinitiated in the RAM (Masubelele et al., 2005). SHR and SCR gene products are required to maintain the activity of this population of root stem cells (Sabatini et al., 2003). However, we observed that in the shr mutant background, expression of the mitotic marker was largely absent in cells close to the root apex postgermination (Fig. 2B). Hence, SHR appears to be required for the reactivation of the Arabidopsis RAM following germination. Despite this root division defect, shr seedlings germinated normally (Supplemental Fig. S1). However, root length was severely reduced compared with the wild type or the scr mutant (Supplemental Fig. S2).
As a result of the failure to reactivate mitotic activity in the primary root, the RAM became progressively smaller (Fig. 2C). This reduction in RAM size was monitored using several markers, including the LAX3pro:GUS reporter (Swarup et al., 2008), which is expressed in stele cells entering the elongation zone. We observed that the LAX3pro:GUS marker moves progressively closer to the root tip in older shr seedlings (Fig. 2C). We conclude that as the shr root grows, cells exit the arrested meristem for the elongation zone until the meristem is largely depleted of cells. SHR is required to maintain indeterminate growth (and therefore appears to function as an indeterminacy factor) in the postembryonic primary root meristem.
The shr Mutant Is Perturbed in Auxin Abundance and Biosynthesis
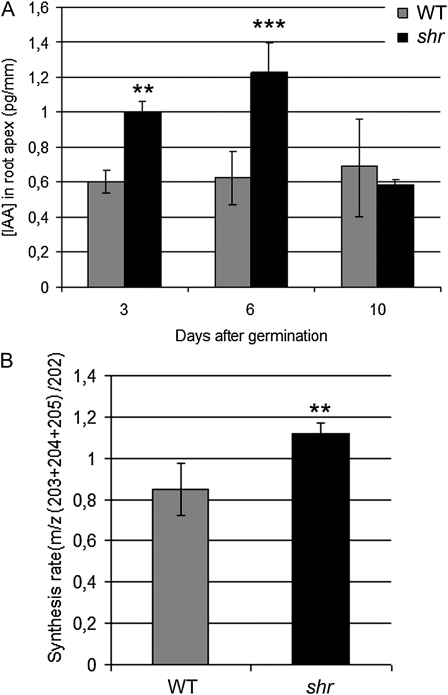
The shr mutation disrupts the maintenance of RAM activity, a process that is closely associated with the mitotic signal auxin. To investigate whether there was a reduction in the accumulation and/or synthesis of auxin in mutant root tissues, we analyzed indole-3-acetic acid (IAA) concentration and biosynthesis rate in the root apex of wild-type and shr seedlings during the first 10 d of germination using mass spectrometry (as described by Edlund et al. [1995] and Ljung et al. [2005]). Surprisingly, IAA concentration was significantly elevated in shr root tips compared with the wild type in seedlings at 3 and 6 d after germination (DAG; pooled 1-mm root tips; Fig. 3A). However, no significant difference in IAA concentration between the wild type and shr was observed in root tips from 10-DAG seedlings.
Figure 3.
Auxin abundance and biosynthesis are perturbed in shr root apical tissues. A, IAA concentration in the root apex of the wild type (WT) and shr at 3, 6, and 10 DAG. B, IAA biosynthesis in the root apex of the wild type and shr at 6 DAG. IAA quantifications and biosynthesis measurements were performed by gas chromatography-selected reaction monitoring-mass spectrometry after 24 h of incubation with deuterated water and compared using Student’s t test (two-sample assuming equal variances, two-tailed distribution; ** 0.001 < P < 0.01, *** P ≤ 0.001). Values represent means ± sd.
In parallel, the IAA synthesis rate in root tips from wild-type and shr seedlings was also analyzed. Five-day-old excised seedling roots were incubated in liquid medium containing 30% deuterated water. Excised roots were incubated instead of whole seedlings in order to separate IAA synthesized de novo in the root system from IAA synthesized in the aerial part of the seedling and then transported to the root. After 24 h of incubation, 1-mm root tips were pooled and analyzed (Fig. 3B). We observed a significantly higher IAA synthesis rate in shr root tips compared with the wild type, suggesting that the higher IAA concentration that we observed in the root apex is in part due to higher root-specific IAA synthesis and is not necessarily caused by the accumulation of IAA coming from the aerial part of the seedling.
We conclude that shr primary root determinancy is not due to either a reduced level or synthesis of auxin. Instead, mutant RAM tissues exhibit a higher rate of biosynthesis of auxin and transient accumulation of this hormone during the first week after germination.
Root Determinacy Is Associated with a Loss of PIN Auxin Carrier Accumulation
In order to understand why auxin accumulates in the root apex during the first 10 DAG, we monitored the expression patterns of AUX1 and PIN classes of auxin influx and efflux carriers in wild-type versus shr root apical tissues (Fig. 4).
Figure 4.
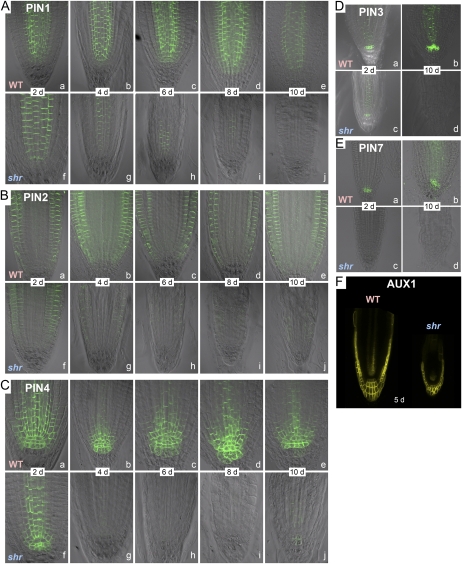
The shr mutation causes a progressive reduction of PIN auxin transporter abundance. A to E, Dynamic changes of PIN1 (A), PIN2 (B), PIN4 (C), PIN3 (D), and PIN7 (E) expression as shown by immunolocalization in the wild type (WT) and shr. F, Expression of AUX1 in the wild type and shr at 5 DAG. PIN expression in A to E was assessed by immunolocalization, while AUX1 expression in F was investigated using the fusion protein AUX1-YFP.
In wild-type root apical tissues, PIN1 was detected predominantly in stele and endodermal cells, and its pattern of expression did not change significantly during root development between 2 and 10 DAG (Fig. 4A). In shr root apical tissues, the PIN1 signal was observed only in stele cells, since this mutant lacks an endodermis (Fig. 4A). Most strikingly, PIN1 abundance progressively decreased in shr mutant seedlings from 4 DAG, and the signal had almost completely disappeared by 10 d (Fig. 4A).
In the case of PIN2, its signal was significantly reduced from 4 DAG in shr versus the wild type (Fig. 4B). However, the polarity of the PIN2 protein in epidermal cells was the same in shr as in the wild type (i.e. shootward/basipetal [pointing away from the root apex] in the membrane of epidermal cells and rootward/acropetal [pointing toward the root apex] in the membrane of cortical cells) until the transition to the elongation zone, where PIN2 switches to a basipetal orientation in the cortex (Supplemental Fig. S3). Nevertheless, the switch in PIN2 orientation in the cortex was shifted toward the RAM in shr (data not shown). This shift is consistent with the reduction of root meristem size in the shr mutant (Fig. 2).
In wild-type root apical tissues from 2 to 10 DAG, PIN4 was expressed in the QC, the surrounding initials, and their abutting daughters with a polar localization toward the QC (Fig. 4C), consistent with previous reports (Friml et al., 2002; Blilou et al., 2005). In contrast, PIN4 was absent in the QC of shr seedling roots at 2 DAG but was detected in initials and their daughter cells plus the stele (Fig. 4C). However, PIN4 was no longer detectable in these shr root tissues from 4 DAG (Fig. 4C). Immunolocalization (Fig. 4D) revealed that PIN7 was one of the first PIN proteins to be down-regulated in the shr background, being almost undetectable at 2 DAG. By day 10, neither PIN7 (nor PIN3) proteins were detectable (Fig. 4, D and E).
In addition to PINs, we monitored the expression of an AUX1pro::AUX1-YFP (for yellow fluorescent protein) translational fusion in the shr background. Contrary to PINs, AUX1-YFP expression was maintained in the shr root apex (Fig. 4F). The only significant difference detected was the absence of AUX1-YFP signal in the vascular strand and a smaller domain of expression in the lateral root cap (reflecting the loss of protophloem cells and the smaller size of the meristem, respectively).
In summary, shr root apical tissues exhibit a progressive loss of PIN (but not AUX1) auxin transporter expression, which is likely to negatively impact meristem function (Blilou et al., 2005) and may contribute to the loss of primary root indeterminancy.
The scr mutant must clearly reinitiate RAM activity, based on its ability to grow to 50% of the wild-type root length (Supplemental Fig. S2). Intriguingly, unlike shr, the scr mutant does not lose PIN protein expression (Supplemental Fig. S7), consistent with a link between cell proliferation and PIN protein abundance.
SHR Is Not Required for the Initiation and Radial Patterning of Anchor Root Primordia
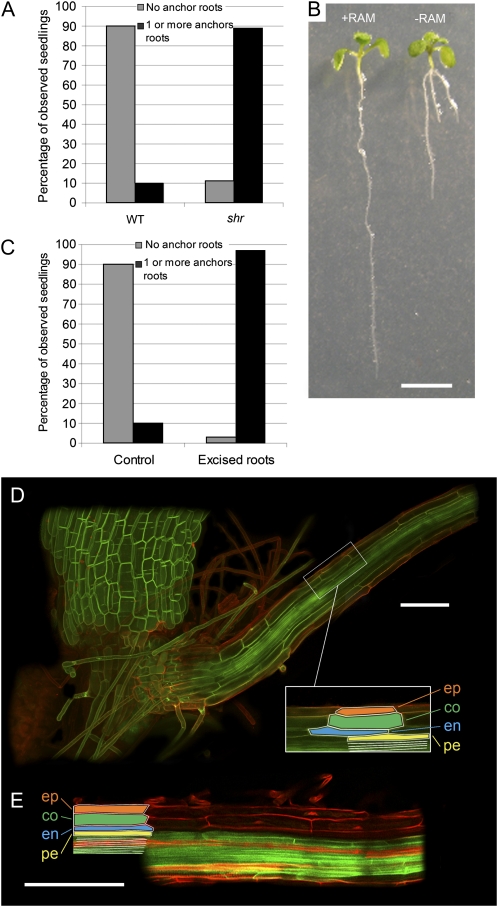
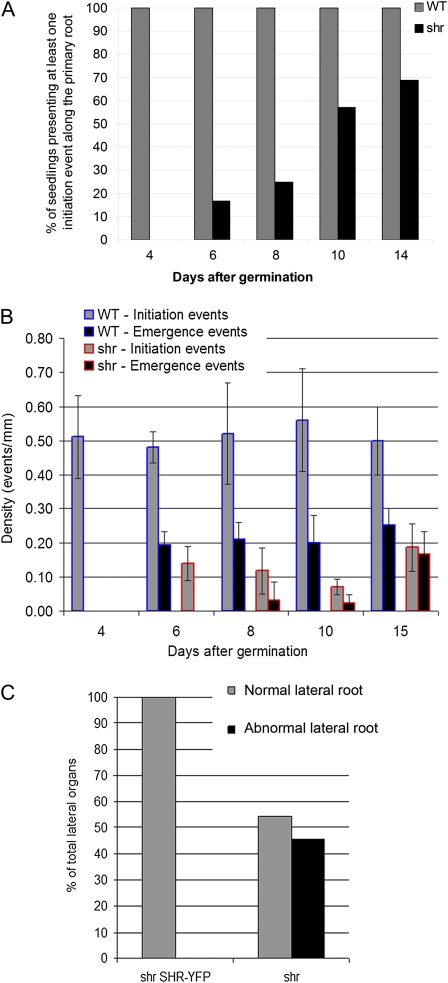
We next investigated the impact of the loss of SHR and primary root meristem activity on root architecture. The majority of shr seedlings exhibit a distinctive tripod-like root phenotype, with nearly all (89% at 15 DAG; n = 394) forming at least one anchor root originating at the root-hypocotyl junction (Fig. 1E). In contrast, wild-type Arabidopsis seedlings rarely activate anchor root primordia at the root-hypocotyl junction (Fig. 5B). Indeed, only 10% (n = 40) of wild-type seedlings had formed one or more anchor roots by 8 DAG (Fig. 5A). Anchor root activation in the shr seedlings may represent a generic mechanism to compensate for the loss of RAM activity. To test this, we examined the impact of loss of the RAM in wild-type seedlings by excising the bottom 2 to 3 mm of the wild-type root at 3 DAG, then scoring numbers of anchor roots at 8 DAG (Fig. 5C). The percentage of seedlings that formed one or more anchor roots by 8 DAG increased dramatically from 10% (n = 40) in nonexcised wild-type seedlings to 97% (n = 39) in RAM-excised wild-type seedlings (Fig. 5C). A representative image is shown in Figure 5B. We conclude that this represents a generic mechanism present in wild-type Arabidopsis to compensate for the loss of growth potential in the primary root. In the case of shr, the progressive loss of RAM activity most likely results in the activation of anchor root primordia. Hence, unlike the primary RAM, SHR is clearly not required for the postembryonic activation of anchor root primordia, based on its mutant phenotype (Fig. 1E).
Figure 5.
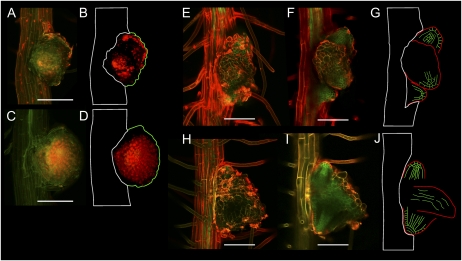
The shr mutation causes the activation of anchor root primordia. A, Percentage of wild-type (WT; n = 40) and shr (n = 394) seedlings having zero (gray bars) or one or more anchor roots (black bars) at 8 DAG. B, Excision of the RAM of wild-type seedlings. Seedlings were grown vertically, and half the seedlings had 2 to 3 mm of the root (from the apex upward) excised at 8 DAG. Representative wild-type (+RAM) and wild-type minus RAM (−RAM) seedlings are shown at 5 d after excision. Bar = 5 mm. C, Percentage of wild-type control (n = 40) and excised (n = 39) seedlings having zero (gray bars) or one or more anchor roots (black bars) 5 d after excision of the RAM. D, Root structure of wild-type anchor root. Epidermis (ep), cortex (co), endodermis (en), pericycle (pe), and stele (white) tissue structures (inset) are revealed by the LTi6a membrane marker (green) and propidium iodide (red). Bar = 100 μm. E, Root structure of shr anchor root. Epidermis, cortex, endodermis, and stele tissue structures are revealed by propidium iodide (red). Pericycle is revealed by the AUX1-YFP membrane marker. Bar = 100 μm.
We went on to investigate whether SHR was necessary for radial patterning of anchor roots. Confocal imaging of anchor roots in the wild type revealed a radial patterning identical to that observed in primary and lateral roots (Fig. 5D). Surprisingly, shr anchor roots also contained a wild-type number of layers, as revealed by the presence of two cell layers between the AUX1-YFP-marked epidermis and pericycle layers in shr AUX1pro::AUX1-YFP seedlings (Fig. 5E). This observation is in contrast to the radial organization of mutant primary root tissues, where loss of SHR results in the loss of a cell layer (Benfey et al., 1993; Scheres et al., 1995). Hence, activation and radial patterning of anchor root primordia appears not to be dependent on SHR function. Nevertheless, shr anchor root growth behavior is determinate, where the new organ ceases growing at approximately the same length as the primary root (Fig. 1E). This implies that, as in the case of primary root meristem, SHR also functions as an indeterminacy factor for anchor root development.
SHR Is Required for Lateral Root Initiation and Patterning of New Primordia
We next investigated whether SHR plays a role during lateral root development. We initially determined whether SHR was expressed during lateral root formation employing a GUS reporter regulated by 2.5 kb of the native SHR promoter (SHRpro:GUS). SHRpro:GUS was expressed exclusively in the central zone of the LRP throughout the initiation and emergence processes (Supplemental Fig. S4, A–E). Upon emergence, reporter expression extended into the root cap, in addition to the stele and the base of the lateral root (Supplemental Fig. S4, E–J). SHRpro:GUS expression in the lateral root then resumed a pattern similar to that observed in the primary root (i.e. stele exclusive; Supplemental Fig. S4, K and L). Thus, SHR was expressed during all stages of lateral root development.
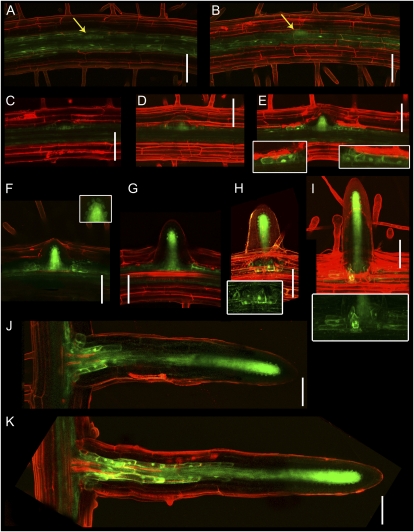
As the SHR protein regulates primary root radial patterning in a non-cell-autonomous manner by migrating from the stele to the QC and future endodermis tissues (Nakajima et al., 2001), we examined whether SHR also moved to cells outside its expression domain during lateral root development. To do this, we created a SHR-YFP translational fusion driven by 2.5 kb of the native SHR promoter (SHRpro::SHR-YFP) for confocal microscopy observation. The SHR-YFP fusion was functional, based on its ability to fully rescue the shr mutant primary and lateral root phenotypes (Supplemental Fig. S5). The lateral root expression of SHR-YFP in shr SHRpro::SHR-YFP was then analyzed by confocal microscopy (Fig. 6). SHR-YFP localization was first observed in the lower half of young LRP (Fig. 6, A and B). As primordia developed, the SHR-YFP protein accumulated in the inner cell layers of the primordia, corresponding to the prospective stele and pericycle (Fig. 6, C–E). In addition, the SHR-YFP protein was also targeted to the nuclei and cytoplasm of the flanking cells in the LRP (Fig. 6E, inset). During lateral root emergence, the SHR-YFP protein was also targeted to the nuclei of the QC and putative endodermis (Fig. 6F, inset). After emergence, the SHR-YFP protein was located in the mature lateral root as described for the primary root (Nakajima et al., 2001). However, expression was prolonged at the base of the lateral root following organ outgrowth and elongation (Fig. 6, J and K).
Figure 6.
SHR is expressed throughout lateral root development. Confocal images of LRP and lateral roots of 8-d-old shr SHRpro::SHR-YFP Arabidopsis are shown. The SHR-YFP fusion protein is functional and rescues the shr mutant when expressed under the control of the SHR promoter (Supplemental Fig. S5). A and B, The SHR protein (green) is expressed in the lower half of the primordium during initial developmental stages. C to E, As the primordium acquires its dome shape, the SHR protein accumulates in the future stele of the primordium and nuclei of the flanking cells of the primordia (insets). F and G, Upon emergence, the SHR protein is targeted to the nuclei and cytoplasm of the QC and ground tissues (inset). H and I, Once the lateral root is fully emerged, the SHR protein is targeted to an annulus of cells around the base of the new vascular bundle neighboring the flanking cells of the original primordium (insets). J and K, The SHR protein is located in the mature lateral root as it is located in the primary root. Localization in the flanking cells of the young primordium is maintained along the ground tissue following lateral root outgrowth. Root tissue structure is revealed by propidium iodide counterstaining (red). Bars = 50 μm.
LRP originate from pericycle cells that are considered to represent part of an extended meristem that (unlike other root tissues) retains its mitotic potential once exiting the primary root meristem (Péret et al., 2009). Given the impact of the shr mutation on RAM mitotic activity, we investigated whether SHR was also required for lateral root development. The density of lateral root initiation in wild-type and shr seedlings was scored from 4 to 14 DAG. Less than 40% (n = 49) of 14-d-old mutant seedlings exhibited any lateral root initiation events (Fig. 7A). In addition, initiation events in shr seedlings that resulted in emerged lateral roots were greatly reduced compared with the wild type (over a 3-fold reduction; Fig. 7B). Although the density of emergence events scored after 1 week of growth was also reduced in shr, the proportion (density of emergence event/density of initiation event) appeared higher in shr (70%) than in the wild type (40%) after 2 weeks of growth (Fig. 7B), consistent with the existence of equilibrium between initiation and emergence (Lucas et al., 2008). However, over 40% (n = 46) of emerging lateral roots did not develop any further following emergence (Fig. 7C).
Figure 7.
The shr mutation disrupts lateral root initiation, patterning, and emergence. A, Time course of the percentage of wild-type (WT) and shr seedlings having initiated at least one lateral root (n = 49). B, Time course of densities of lateral root initiation and lateral root emergence events in wild-type and shr seedlings with at least one initiation event (in events per mm; n = 20). Values represent means ± sd. C, Percentage of abnormal lateral roots after emergence in shr SHRpro::SHR-YFP and shr seedlings (n = 46). [See online article for color version of this figure.]
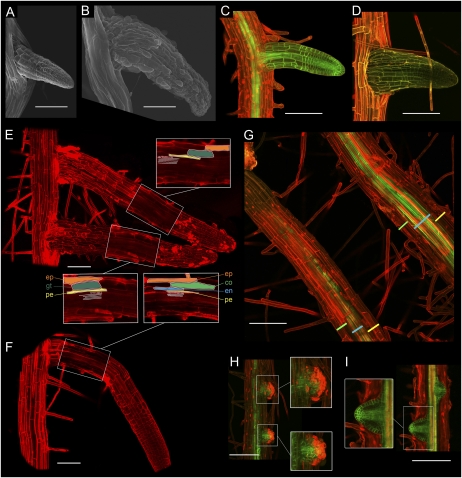
To understand why such a large proportion of lateral roots did not develop in shr, we investigated lateral root development in the mutant employing scanning electron and confocal microscopy techniques (Fig. 8). Electron microscopic analysis of developing lateral roots revealed that lateral roots emerging from shr primary roots were approximately double the thickness of wild-type lateral roots (Fig. 8, compare A and B). We employed the AUX1-YFP marker (AUX1pro::AUX1-YFP) in conjunction with confocal microscopy to study the cellular organization of LRP in the wild type versus the shr mutant (Fig. 8, C and D). In wild-type plants, primary and lateral roots are patterned in the following order of cell layers from the outside to the inside: epidermis, cortex, endodermis, pericycle, stele cylinder. In the shr mutant primary root, the cortex/endodermis initials fail to divide, resulting in the cortex and endodermis being replaced by a single ground tissue layer (Scheres et al., 1995). We observed that this is also the case in shr LRP (Fig. 8, E and F). Confocal imaging revealed that lateral root cells in the outer tissue layers of the wild type and shr were of equal dimension (Fig. 8, E and F). However, the increased thickness in developing shr lateral root appeared to be due to increased radial vascular development (Fig. 8G). A similar ectopic lignification phenotype was observed in the shr hypocotyl (Supplemental Fig. S6).
Figure 8.
The shr mutant exhibits radial and vascular patterning defects. A and B, Electron microscopy images of Col-0 (A) and shr (B) emerging lateral roots. Emerging lateral roots in shr are approximately two times wider than wild-type lateral roots. C and D, Confocal images of Col-0 AUX1pro::AUX1-YFP (C) and shr AUX1pro::AUX1-YFP (D). E, Root structure of lateral root in shr. Lateral roots in shr exhibit the same radial patterning defect as the primary root, with a single ground tissue layer (gt) replacing cortex and endodermis between pericycle and epidermis. Stele tissues are marked in white. Root structure is revealed by confocal imaging with propidium iodide staining (red). F, Root structure of lateral root in Col-0. From outer to inner layer: epidermis (ep), cortex (co), endodermis (en), pericycle (pe), stele tissues (white). Root structure is revealed by confocal imaging with propidium iodide staining (red). G, Confocal images of shr AUX1pro::AUX1-YFP primary (bottom left) and lateral (top right) roots. The thickness gain in the developing lateral root of shr is linked with excessive radial development of vascular tissues (blue bar), with no visible changes in the thickness of outer tissue layers (green and yellow bars). H and I, Confocal images of AUX1pro::AUX1-YFP in shr and the wild type. Propidium iodide (red) stains cells in the outer tissue layers of young LRP, while the AUX1-YFP marker (green) is expressed in lateral root stele cells. Overlaying the two markers reveals that outer cells in shr LRP are nonviable (H, insets) compared with the wild type (I, inset). Bars = 100 mm.
In addition to an extravascular development patterning defect, we observed that around half of developing shr LRP exhibited severe patterning defects compared with the wild type (Fig. 8, H and I). Prior to emergence, such LRP lacked the cellular organization of wild-type LRP (Fig. 8I) and exhibited disorganized expression of the AUX1-YFP marker (Fig. 8H). Postemergence, we distinguished two types of aborted lateral roots. In the first category, termed “globular LRP” due to their symmetrical shape, the AUX1-YFP marker was not significantly expressed and propidium iodide rapidly stained the nuclei of lateral root cells (Fig. 9, A–D), indicating that cells were no longer viable. In the second category, AUX1-YFP expression was maintained and revealed that despite their altered outer structure (Fig. 9, E and H), these aborted organs retained some internal organization (Fig. 9, F and I). In contrast to globular LR, the second class of lateral root formed clusters of fused LRP (Fig. 9, F and I). These “cluster LRP” expressed the AUX1-YFP marker in a pattern similar to normal LRP (Fig. 9, G and J).
Figure 9.
SHR is required for lateral root patterning and viability. Upon emergence, approximately 50% of shr lateral roots fail to elongate. Two types of patterning defects can be distinguished within these abnormal lateral roots. A to D, Globular lateral roots lack inner structure (B and D, schematic representations of the inner organization of the lateral root), and their cells appear to be nonviable based on their permeability to propidium iodide (red, strong nuclei staining). Images of shr AUX1pro::AUX1-YFP (green) were taken by confocal microscopy with propidium iodide counterstaining. E to J, Fused lateral roots appeared outwardly amorphous (E and H) yet contained internal structures (F and I). Optical sectioning by confocal microscopy of shr AUX1pro::AUX1-YFP with propidium iodide counterstaining reveals multiple fused LRP (G and J, schematic representation of the fused LRP). The LRP comprising the fused lateral roots exhibit standard patterns of AUX1pro::AUX1-YFP expression (epidermis, lateral root cap, columella, and stele tissues). Bars = 100 μm.
We conclude that SHR is required for the initiation, radial patterning, and organization of new LRP. In the absence of SHR, lateral roots exhibit radial and vascular patterning defects. This may reflect, as observed in the primary root, that shr mutant cells differentiate into a state where they are no longer competent to fully form lateral roots.
DISCUSSION
The shr mutant was originally identified based on its radial patterning and determinate RAM phenotype (Benfey et al., 1993). However, the impact of the shr mutation on other root developmental programs is unclear. In this paper, we describe the important roles played by SHR during primary, lateral, and adventitious root development.
SHR Is Essential for Mitotic Reactivation of the RAM after Germination
SHR has been described to be essential for maintaining RAM activity (Benfey et al., 1993; Di Laurenzio et al., 1996; Fig. 1). In mutant plants lacking SHR, primary root growth is severely reduced (Benfey et al., 1993; Scheres et al., 1995; Fig. 1; Supplemental Fig. S2). Mutant roots exhibit QC defects and loss of activity of surrounding stem (initial) cells, resulting in the disruption of formative cell division events (Sozzani et al., 2010) and the depletion of proliferating cells in the RAM (Sabatini et al., 2003).
SHR is also required for reinitiating mitotic activity in the RAM following germination. We report that in the absence of SHR, mutant RAM cells fail to express the mitotic marker, cyclinB1:1:GUS (Fig. 2B; Ferreira et al., 1994). Consistent with a role as a key root mitotic regulator, Sozzani et al. (2010) recently reported that SHR controls the expression of a number of genes encoding components of the cell cycle machinery. Direct targets of SHR include the D-type cyclin gene CYCD6;1 and the cyclin-dependent kinase genes CDKB2;1 and CDK2;2 (Sozzani et al., 2010). While ectopic expression of two of these genes in shr was able to partially restore formative divisions, it was not able to rescue mutant root growth (Sozzani et al., 2010). Hence, further genes are likely to be required. Indeed, 266 genes were identified by Sozzani et al. (2010) to be direct targets of SHR, of which 65 were differentially expressed in a SHR-inducible system. In total, the expression of approximately 2,500 genes was observed to change over a 12-h time course following the induction of SHR expression in a shr background. Hence, SHR appears to control the expression of a very large number of genes, consistent with its role as a key regulator linking patterning and growth (Sozzani et al., 2010).
We report that the shr mutant exhibits a progressive reduction and eventual loss of detectable PIN auxin efflux carrier protein following germination (Fig. 4). PIN protein localization was performed up to 10 DAG on seedling roots from multiple independent shr null alleles isolated from different accessions (for details, see “Materials and Methods”). Every null allele tested was observed to exhibit a progressive loss of PIN1, PIN2, PIN3, PIN4, and PIN7 proteins (Fig. 4). Hence, PIN protein abundance appeared to be dependent on SHR activity. None of these PIN genes were identified among the direct targets of SHR regulation (Levesque et al., 2006; Sozzani et al., 2010); hence, protein abundance was likely to be regulated indirectly by SHR. Surprisingly, quantitative reverse transcription (qRT)-PCR profiling revealed that SHR does not regulate PIN abundance at the transcriptional level, since their mRNAs are still detectable (Supplemental Fig. S8). Our qRT-PCR results are consistent with recently published array data for shr and scr root apical tissues, which detected PIN expression in both mutant backgrounds (Sozzani et al., 2010). Hence, SHR must regulate PIN abundance at the posttranscriptional level. As in the wild type, PIN abundance is maintained in the scr mutant (Supplemental Fig. S7), suggesting that the abundance of these proteins is regulated via a gene product(s) controlled by SHR (but not SCR). It is intriguing that PIN abundance in shr root apical cells decreases (Fig. 4), despite elevated auxin abundance and biosynthesis in the mutant (Fig. 3) and contrary to the observed auxin inducibility of PIN expression (Vieten et al., 2005). One simple explanation is that the loss of PIN abundance in shr (but not scr) may represent an indirect consequence of mutant RAM cells differentiating as a result of their failure to reinitiate cell division following germination.
SHR Is Required for Lateral Root Formation in a Manner Distinct from the Primary Root
LRP in Arabidopsis originate from the pericycle (Malamy and Benfey, 1997). LRP initiation involves the asymmetric division of xylem pole pericycle founder cells (Dubrovsky et al., 2001). Based on data showing that the pericycle remains in G1 phase of the cell cycle (Beeckman et al., 2001) and that the xylem pole pericycle continues to cycle after leaving the RAM (Dubrovsky et al., 2000), it has been postulated that the pericycle acts as an extended monolayered meristem (Casimiro et al., 2003). While pericycle division is first visible in the differentiation zone well above the dividing RAM, xylem pole pericycle founder cells first need to be primed in the basal meristem (close to the elongation zone) in order to trigger LRP initiation (De Smet et al., 2007). Despite the loss of mitotic activity in the shr RAM, pericycle cells retain their ability to initiate new LRP (Figs. 1 and 7). Nevertheless, the number of newly initiated LRP in shr was significantly reduced, suggesting that SHR facilitates (rather than is essential for) lateral root initiation.
LRP development and patterning are generally considered to be a postembryonic recapitulations of the equivalent processes in the primary root (Malamy and Benfey, 1997; Scheres et al., 2002). We investigated the expression of SHR employing transcriptional and translational reporter fusions during LRP development. We first observed that SHR-YFP moved to adjacent QC, CEI, and endodermal cell layers when the dome-like structure of the LRP is formed (stages IV/V onward; Fig. 6). This expression event coincides with the restricted expression domain of SCR in emerging primordia and prior to LRP tissues adopting a radial organization equivalent to the primary root (Malamy and Benfey, 1997). After LRP emergence, the expression domain of the SHR protein appears identical to that described for SHR in the RAM (Fig. 6; Nakajima et al., 2001).
Our study also revealed that, in addition to its radial patterning function, SHR is required to coordinate the overall morphology of new LRP. In the absence of SHR, over half of the initiated LRP exhibited severe patterning defects (Fig. 9). These LRP were either classified as cluster LRP or globular LRP. Cluster LRP were composed of multiple fused LRP, while globular LRP failed to develop any properly organized inner architecture or boundaries. Consistent with such functions, in addition to being expressed in inner LRP tissues, the SHR-YFP marker was also detected in the nuclei and cytoplasm of the outermost cells flanking the LRP as early as stage III/IV (Fig. 6). This novel domain of SHR expression in cells flanking the LRP could play a role in the specification of primordia boundaries, akin to its regulation of radial patterning in QC, CEI, and endodermal cell layers. Hence, SHR appears to regulate lateral root patterning in an overlapping yet distinct manner to the primary root.
SHR Is Not Required for Correct Patterning of Anchor Root Tissues
SHR is best known as a regulator of radial patterning in Arabidopsis primary root apical tissues (Benfey et al., 1993). The SHR gene is first transcribed in the stele, and then the SHR protein moves outward one cell layer to adjacent QC, CEI, and endodermal cells (Nakajima et al., 2001). SHR triggers SCR expression, causing CEI cells to divide longitudinally and generate endodermal and cortical layers. In the absence of SHR, mutant roots form a single layer of cells with a cortical identity (Benfey et al., 1993; Scheres et al., 1995). Hence, SHR is necessary for radial patterning of root tissues plus specifying endodermal cell fate. In contrast to defective radial patterning in the shr RAM, mutant anchor roots retain a wild-type-like tissue organization (Fig. 5). Nevertheless, SHR is required for radial patterning of LRP tissues (Fig. 8). These differences are unlikely to reflect that SHR is essential for radial patterning only in root-derived organs, since this gene product is also required for the formation of the starch-sheath layer in shoot tissues (Fukaki et al., 1998). Instead, it is most likely that another GRAS family member (Pysh et al., 1999) performs a SHR-like patterning function in this tissue.
Loss of SHR Activity Impacts RSA
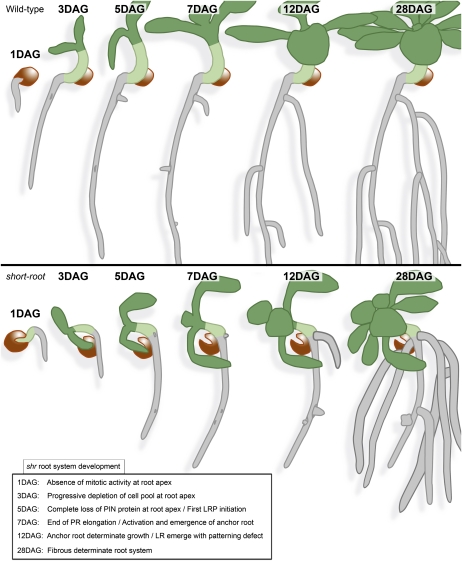
We report that the loss of SHR activity leads to major changes in RSA (summarized in Fig. 10). Micro-CT imaging of RSA in soil revealed that the wild-type tap root architecture is converted to a dwarfed fibrous-like root system in shr mutants (Fig. 1). This dramatic change in shr RSA is caused by changes in the relative contribution of RAM, lateral root, and anchor roots (Fig. 10). We have reported that the shr RAM fails to reinitiate mitotic activity postembryonically (Fig. 2). Consequently, the mutant primary root elongates until the pool of cells at its RAM is depleted and no longer sufficient to maintain growth. Loss of RAM activity in shr promotes the initiation of anchor root primordia (Fig. 7). We propose that this activation represents a mechanism to compensate for RAM death, as it could be phenocopied in the wild type following RAM excision. The underlying factors responsible for anchor root activation upon loss of the RAM remain unclear. It is possible that an as yet unidentified inhibitory signal(s) is produced in the RAM that normally acts at the root-hypocotyl junction to prevent anchor root formation in the presence of an active RAM. Alternatively, anchor root activation may simply be a response to the supply of nutrients and/or proliferative signals from the aerial portions of the plant that are in excess of that able to be consumed by the determinate mutant primary root. The activation of determinate anchor roots leads to repeated ramification near the base of the root-hypocotyl junction, as observed in soil-grown shr (Fig. 1). Moreover, shr exhibits lower density of lateral root initiation, with a significant fraction of initiated (globular and fused) lateral roots aborting postemergence. The activation of anchor roots and the lower level of lateral root initiation and development give rise to the fibrous architecture of the shr mature root system, with high branching density near the base of the root system and low branching density along the roots (Fig. 10).
Figure 10.
Comparative summary of root system development of the wild type and shr. The inset lists the main events of root system development impacted by the shr mutation.
In summary, our results reveal that SHR function is not restricted to the primary root. Instead, SHR is required for the initiation and patterning of LRP and for maintaining the indeterminate growth of primary, lateral, and adventitious roots.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized for 8 min in 50% (v/v) bleach and then washed twice in 0.1% (v/v) Triton X-100. The seeds were then plated on square plates containing 0.5× Murashige and Skoog salt mixture in 1% (w/v) bacto agar. The plates were cold treated for 2 d at 4°C in the dark to synchronize germination. Plates were then incubated in a nearly vertical position at 23°C in 150 μmol m−2 s−1 constant light with a cycle of 18 h of light/6 h of dark. For the analysis of the loss of the RAM on ecotype Columbia (Col-0) seedlings, 2 to 3 mm of the root tip was excised with a sterile razor blade at 3 DAG, and the seedlings were returned to growth conditions.
To generate the SHRp::SHR-YFP construct, 2-kb upstream sequence of the SHR gene (AT4G37650) was PCR amplified and cloned into BamHI and XhoI sites of pBluescript SK+ (Stratagene) to create pBS-SHRpro. Subsequently, full-length SHR gene (including the 3′ untranslated region) was PCR amplified with an in-frame XhoI restriction enzyme site at the N terminus and cloned between XhoI and KpnI sites of pBS-SHRpro to create pBS-SHR-X. Full-length EYFP sequence (excluding the stop codon) was then PCR amplified using primers with in-frame XhoI restriction enzyme sites at the ends and subsequently cloned in frame at the engineered XhoI site of the SHR gene to create pBS-SHR-YFP. The SHR-YFP sequence was then cloned into a binary vector (pMOG; Mogen International) between BamHI and KpnI to create pMOG-SHR-YFP. Transformation of Agrobacterium tumefaciens (C58) and Arabidopsis was done as described by Swarup et al. (2005).
X-Ray CT Scanning
Plants (shr and wild-type Col-0) were grown in a sandy loam soil (Dunnington Heath series; Stagno-gleyic Luvisol) from the University of Nottingham experimental farm at Sutton Bonington (52.5°N 1.3°W), sieved to 2 mm, and packed loosely (bulk density approximately 1.1 g cm−3) into 30-mm-diameter, 55-mm-high columns. The base of the column was covered with “micropore” tape and placed in a tray to allow watering from below to prevent gas entrapment. Seeds were sown directly on the soil surface after surface watering, and columns were subsequently watered from below. The plants were provided with supplemental nutrients by adding 1 mL of commercial Miracle-Gro (diluted as per instructions) to the top of the columns on two occasions, 11 and 25 d after sowing. Plants were grown in a controlled-environment room with 16-h day/8-h night and temperature of 23°C day and 18°C night. The columns with live plants were scanned in a Nanotom X-Ray Micro-Computed Tomography scanner (Phoenix X-Ray). Samples were scanned at 80 mV and 220 mA, with 1,440 images collected over a 50-min period. The spatial resolution was set at 18 μm pixel−1. All samples were scanned at an approximated field capacity soil moisture status. Image slices were reconstructed into 3D volumes using DatosX Rec version 1.5 (Phoenix X-Ray) with beam-hardening and ring artifact reduction algorithms applied and then manipulated and analyzed in VGStudioMax 2.0 (Volume Graphics). The 3D volume was median filtered, and the roots were manually segmented through a region growing tool and visual identification. Tortuosity is defined here as the ratio of the root length over the shortest distance between the two extremities of the measured root.
Phenotypic Analysis
Primary root length was determined from digital images of the plates by measuring from root tip to hypocotyl base using ImageJ 1.40 software (http://rsb.info.nih.gov/ij/). Emerged lateral and anchor roots were counted using a binocular. Two-sample unpaired t test was used for comparison of primary root length and lateral root density, using a confidence interval of 95%.
GUS Assay and Microscopy
GUS activity of LAX3pro:GUS Col-0 and shr seedlings was assayed by incubating whole seedlings in a staining solution comprising 1 mm 5-bromo-4-chloro-3-indolyl β-d-glucuronide in 0.5% (v/v) dimethylformamide, 0.5% (v/v) Triton X-100, 1 mm EDTA (pH 8), and 500 mm phosphate buffer (Na2PO4, pH 7) for 2 to 3 h at 37°C. To limit the diffusion of the blue stain and therefore maintain high staining specificity, 0.5 mm K3Fe(CN)6 and K4Fe(CN)6 were added. Both stained and unstained roots were cleared by immersion in 20% (v/v) methanol/4% (v/v) hydrochloric acid at 57°C for 20 min, followed by immersion in 7% (w/v) NaOH/60% (v/v) ethanol at room temperature for 15 min. Roots were then rehydrated for 5 min each in 40%, 20%, and 10% (v/v) ethanol and infiltrated in 5% (v/v) ethanol/25% (v/v) glycerol for 15 min. Roots were mounted in 50% (v/v) glycerol on glass microscope slides and were imaged using a Nikon differential interference contrast optics microscope.
For analysis of AUX1pro::AUX1-YFP and SHRpro::SHR-YFP seedlings, whole mounts were prepared on microscope slides in water, and the roots were imaged either on a Leica SP2 confocal microscope using the 514-nm line of the argon laser or a Nikon eC1/TE2000U inverted confocal microscope using the 488-nm line of the argon laser. For general morphological analysis, seedlings were stained by immersion in 5 μL mL−1 propidium iodide for 5 min, whole mounted in water, and imaged using a Nikon eC1/TE2000U inverted confocal microscope.
IAA Quantification and Biosynthesis Measurements
Seedlings were grown on vertical agar plates under long-day conditions as described by Ljung et al. (2005). IAA quantification was performed on shr and wild-type seedlings at 3, 6, or 10 DAG. For IAA synthesis measurements, excised roots (cut at the hypocotyl-root junction) coming from 5-d-old shr and wild-type seedlings were transferred to liquid medium containing 30% deuterated water and incubated in long days for 24 h. The most apical 1 mm of wild-type and shr seedling roots was pooled for IAA quantification (50 sections per sample, five to eight replicates) and biosynthesis measurements (100 sections per sample, four replicates). IAA quantifications and biosynthesis measurements were done by gas chromatography-selected reaction monitoring-mass spectrometry (Edlund et al., 1995; Ljung et al., 2005) and compared using Student’s t test (two-sample assuming equal variances, two-tailed distribution; ** 0.001 < P < 0.01, *** P ≤ 0.001).
PIN Localization
Seedlings of the wild type, scr, and shr were sampled at different stages of development (see “Results”). PIN localization was investigated in three different alleles of shr mutants (shr1 [ecotype Wassilewskija], shr3, and tpd [Col-0]). All alleles demonstrated similar dynamics of changes in PIN localization. Immunolocalization in roots was performed as described (Friml et al., 2002). Rabbit anti-PIN1 (Gälweiler et al., 1998), anti-PIN2 (Müller et al., 1998), affinity-purified anti-PIN3 (Friml et al., 2002), anti-PIN4 (Friml et al., 2002), and mouse anti-PIN7 (Paponov et al., 2005) antibodies were diluted 1:500, 1:400, 1:100, 1:400, and 1:50, respectively. The secondary antibody, Alexa 488-conjugated anti-rabbit or anti-mouse (for PIN7) antibody, was diluted 1:400. Solutions during the immunolocalization procedures were changed using a pipetting robot (Insitu Pro; Intavis).
Quantitative PCR
Plants were grown on 0.5× Murashige and Skoog and 1% bacto agar at 23°C and 150 μmol m−2 s−1 in long days (16 h of light). Total RNA was extracted from 6-DAG roots using the Qiagen RNeasy Plant Mini Kit with on-column DNase treatment (RNase free DNase set; Qiagen). Poly(dT) cDNA was prepared from 2 μg of total RNA using the Transcriptor first-strand cDNA synthesis kit (Roche). Quantitative PCR was performed using SYBR Green Sensimix (Quantace) on a Stratagene Mx3005P apparatus. PCR was carried out on 96-well optical reaction plates heated for 5 min to 95°C, followed by 40 cycles of denaturation for 10 s at 95°C and annealing-extension for 30 s at 60°C. Target quantifications were performed with the following specific primer pairs: for TIR1 (AT3G62980), TIR1forward (5′-CCTAAACTGCAGCGCCTCT-3′) and TIR1reverse (5′-GGTTGAAGCAAGCACCTCA-3′); for AFB1 (AT4G03190), AFB1forward, (5′-ACTGATGGTATCGCTGCTATTG-3′) and AFB1reverse (5′-AGTTGAACTCTCTGGAAAATAGCTAAG-3′); for AFB2 (AT3G26810), AFB2forward (5′-CGTGCCTCGAAGGAGAAAC-3′) and AFB2reverse (5′-TTTGGAGACCTAGCAACAAGC-3′); for AFB3 (AT1G12820), AFB3forward (5′-TGATAAACTTTACCTCTACCGAACAG-3′) and AFB3reverse (5′-CCTAACATATGGTGGTGCATCTT-3′); for PIN3 (AT1G70940), PIN3forward (5′-CCCAGATCAATCTCACAACG-3′) and PIN3reverse (5′-CCGGCGAAACTAAATTGTTG-3′); for PIN7 (AT1G23080), PIN7forward (5′-TGGGCTCTTGTTGCTTTCA-3′) and PIN7reverse (5′-TCACCCAAACTGAACATTGC-3′); for PIN1 (AT1G73590), PIN1forward (5′-CCTCAGGGGAATAGTAACGACA-3′) and PIN1reverse (5′-TCATCGTCTTTGTTACCGAAACT-3′); for PIN2 (AT5G57090), PIN2forward (5′-GGCGAAGAAAGCAGGAAGA-3′) and PIN2reverse (5′-GGTGGGTACGACGGAACA-3′); for PIN4 (AT2G01420), PIN4forward (5′-TTGTCTCTGATCAACCTCGAAA-3′) and PIN4reverse (5′-ATCAAGACCGCCGATATCAT-3′); and for IAA2 (AT3G23030), IAA2forward (5′-GAAGAATCTACACCTCCTACCAAAA-3′) and IAA2reverse (5′-CACGTAGCTCACACTGTTGTTG-3′).
Expression levels were normalized to UBA (AT1G04850) using the following primers: UBAforward (5′-AGTGGAGAGGCTGCAGAAGA-3′) and UBAreverse (5′-CTCGGGTAGCACGAGCTTTA-3′). All qRT-PCR experiments were performed in triplicate, and the values presented represent means ± sd.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At4g37650 (SHR), At3g62980 (TIR1), At4g03190 (AFB1), At3g26810 (AFB2), At1g12820 (AFB3), At1g70940 (PIN3), At1g23080 (PIN7), At1g73590 (PIN1), At5g57090 (PIN2), At2g01420 (PIN4), At3g23030 (IAA2), and At1g04850 (UBA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Germination of the shr mutant is comparable with the wild type.
Supplemental Figure S2. Root length in shr is severely reduced compared with the wild type and scr.
Supplemental Figure S3. Inversion of polarity of PIN2 in the cortical layer.
Supplemental Figure S4. SHRpro:GUS expression pattern during lateral root development.
Supplemental Figure S5. The SHRpro::SHR-YFP transgene rescues the shr mutation.
Supplemental Figure S6. The shr mutation causes ectopic lignification in the hypocotyl.
Supplemental Figure S7. The loss of auxin carrier expression in shr is independent of SCR.
Supplemental Figure S8. Expression of auxin-related genes in wild-type, shr, and scr 6-d-old seedlings.
Supplemental Movie S1. 3D movie of a 3-week-old shr seedling grown in sandy loam.
Supplemental Movie S2. 3D movie of a 3-week-old wild-type seedling grown in sandy loam.
Supplementary Material
Acknowledgments
We thank Philip Benfey (Duke University) for providing seeds for shr1 (Wassilewskija) and shr3 alleles and Laurent Laplaze (IRD Montpellier) and members of our team for their helpful comments on the manuscript.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Aida M, Vernoux T, Furutani M, Traas J, Tasaka M. (2002) Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129: 3965–3974 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inzé D. (2001) The peri-cell-cycle in Arabidopsis. J Exp Bot 52: 403–411 [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang HM, Casero P, Sandberg G, Bennett MJ. (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8: 165–171 [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NFD, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Doerner PW, Colón-Carmona A, Rost TL. (2000) Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol 124: 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colón-Carmona A, Doerner P. (2001) Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214: 30–36 [DOI] [PubMed] [Google Scholar]
- Edlund A, Eklof S, Sundberg B, Moritz T, Sandberg G. (1995) A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol 108: 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PCG, Hemerly AS, Engler JdA, Van Montagu M, Engler G, Inzé D. (1994) Developmental expression of the Arabidopsis cyclin gene cyc1At. Plant Cell 6: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M. (1998) Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J 14: 425–430 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Park WJ, Sauer M, Woll K. (2004) From weeds to crops: genetic analysis of root development in cereals. Trends Plant Sci 9: 42–48 [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui HC, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. (2005) Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Guédon Y, Jay-Allemand C, Godin C, Laplaze L. (2008) An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS ONE 3: e3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Masubelele NH, Dewitte W, Menges M, Maughan S, Collins C, Huntley R, Nieuwland J, Scofield S, Murray JA. (2005) D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc Natl Acad Sci USA 102: 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Guan CH, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Coates JC. (2008) Branching out in new directions: the control of root architecture by lateral root formation. New Phytol 179: 595–614 [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou I, Palme K. (2005) The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci 10: 170–177 [DOI] [PubMed] [Google Scholar]
- Péret B, Larrieu A, Bennett MJ. (2009) Lateral root emergence: a difficult birth. J Exp Bot 60: 3637–3643 [DOI] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. (2007) Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol 8: 345–354 [DOI] [PubMed] [Google Scholar]
- Scheres B, Benfey P, Dolan L. (2002) Root development. Somerville CR, Meyerowitz EM, , The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0101, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Di Laurenzio L, Willemsen V, Hauser MT, Janmaat K, Weisbeek P, Benfey PN. (1995) Mutations affecting the radial organization of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62 [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JAH, Benfey PN. (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ. (2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Waisel Y, Eshel A, Kafkafi U. (2002) Plant Roots: The Hidden Half. Marcel Dekker, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.