Abstract
Potato (Solanum tuberosum) is relatively vulnerable to abiotic stress conditions such as drought, but the tolerance mechanisms for such stresses in potato are largely unknown. To identify stress-related factors in potato, we previously carried out a genetic screen of potato plants exposed to abiotic environmental stress conditions using reverse northern-blot analysis. A cDNA encoding a putative R1-type MYB-like transcription factor (StMYB1R-1) was identified as a putative stress-response gene. Here, the transcript levels of StMYB1R-1 were enhanced in response to several environmental stresses in addition to drought but were unaffected by biotic stresses. The results of intracellular targeting and quadruple 9-mer protein-binding microarray analysis indicated that StMYB1R-1 localizes to the nucleus and binds to the DNA sequence G/AGATAA. Overexpression of a StMYB1R-1 transgene in potato plants improved plant tolerance to drought stress while having no significant effects on other agricultural traits. Transgenic plants exhibited reduced rates of water loss and more rapid stomatal closing than wild-type plants under drought stress conditions. In addition, overexpression of StMYB1R-1 enhanced the expression of drought-regulated genes such as AtHB-7, RD28, ALDH22a1, and ERD1-like. Thus, the expression of StMYB1R-1 in potato enhanced drought tolerance via regulation of water loss. These results indicated that StMYB1R-1 functions as a transcription factor involved in the activation of drought-related genes.
As plants are sessile organisms, environmental stresses such as drought and high salinity conditions can compromise economic output and the overall human food supply (Barnabás et al., 2008). Drought stress gives rise to biochemical, molecular, physiological, and morphological changes that adversely affect plant growth, development, and productivity (Reynolds and Tuberosa, 2008; Moore et al., 2009). Under drought-stress conditions, the phytohormone abscisic acid (ABA) is synthesized. ABA modulates the expression of stress-related genes and activates signal transduction pathways that lead to a variety of physiological responses, including changes in stomatal aperture (McCourt and Creelman, 2008). ABA accumulation in guard cells triggers an increase in cytosolic Ca2+, resulting in the activation of calcium-dependent protein kinases (CDPKs; Pei et al., 1996). These CDPKs, especially CPK23, in turn, regulate the activity of anion channels such as slow anion channel 1 (SLAC1; Mori et al., 2006; Geiger et al., 2010). The consequent efflux of anions and depolarization of the membrane induce stomatal closure to prevent transpirational water loss (Israelsson et al., 2006). In Arabidopsis (Arabidopsis thaliana), slac1 mutants exhibit reduced stomatal closure in response to ABA, Ca2+, and hydrogen peroxide (Vahisalu et al., 2008), and SLAC1 anion channel activity is directly controlled by the ABA-activated protein kinase OPEN STOMATA1 (OST1/SRK2E/SnRK2.6) and ABI1/PP2C phosphatase complexes (Geiger et al., 2009; Lee et al., 2009; Vahisalu et al., 2010). Recently, it was reported that RCAR/PYR1/PYL family of START proteins can bind ABA through a gate-latch-lock mechanism and interacts with ABI1/PP2C as coreceptors in Arabidopsis (Melcher et al., 2009; Nishimura et al., 2009; Santiago et al., 2009). ABA perception by the RCAR/PYR1/PYL proteins suppresses PP2C-mediated dephosphorylation of the SnRKs and allows their activation (Umezawa et al., 2009). The pyr1/pyl1/pyl2/pyl4 quadruple-mutant plants show insensitivity in ABA-induced stomatal closure and ABA inhibition of stomatal opening (Nishimura et al., 2010).
The expression of various stress-response genes by ABA is mediated by a number of transcription factors (TFs), such as the MYB family of TFs and ABA response element-binding factors (Fujii et al., 2009). MYB TFs are composed of one, two, or three imperfect helix-turn-helix repeats that recognize the major groove of DNA (Yanhui et al., 2006). MYB TFs are grouped into three subfamilies according to their MYB domain arrangement: R1R2R3, R2R3, and MYB related (containing a single MYB-like domain; Du et al., 2009). In animals, MYB TFs are typically of the R1R2R3 type, which contain three repeats of the MYB domain (Ramsay and Gonda, 2008). In contrast, most plant MYB TFs are of the R2R3 type, which contain two repeats of the MYB domain (Du et al., 2009). Over the past decade, R2R3-type MYB TFs have been implicated in a variety of plant-specific processes, including cell morphogenesis, secondary metabolism, cell differentiation, and stress responses (Nesi et al., 2001; Baumann et al., 2007; Ishida et al., 2007; Zhao et al., 2008; Lippold et al., 2009; Ma et al., 2009). In Arabidopsis, MYB60 and MYB61 are specifically expressed in guard cells. The atmyb60 mutant exhibits reduced light-induced stomatal opening; whereas, the atmyb61 mutant exhibits reduced ABA-induced stomatal closing (Liang et al., 2005).
In Arabidopsis and rice (Oryza sativa), 49 and 84 R1-type MYB genes, respectively, have been described (Yanhui et al., 2006). Only one contains the typical two- or three-Trp repeat in the MYB domain (Yanhui et al., 2006). To date, 66 MYB-like TFs have been described in potato (Solanum tuberosum) plants (http://planttfdb.cbi.pku.edu.cn/), but the potato genome project is ongoing and will likely yield a more precise number upon completion. Compared with R2R3-type MYB TFs, there are few reports of functional studies of single MYB-like domain TFs in plants. Potato MybSt1, the first reported single MYB-like domain TF in plants, recognizes a target sequence located between nucleotides −73 and −48 of the cauliflower mosaic virus (CaMV) 35S promoter and functions as a transcriptional activator (Baranowskij et al., 1994). In Arabidopsis, the single MYB-like domain TF CIRCADIAN CLOCK ASSOCIATED1 (CCA1) has been shown to bind to two imperfect repeats in the light-harvesting chlorophyll a/b promoter and acts as a specific activator of phytochrome signal transduction (Wang et al., 1997). Constitutive expression of CCA1 results in longer hypocotyls and substantially delayed flowering (Wang and Tobin, 1998). In rice, ANTHER INDEHISCENCE1 (AID1) is closely related to other single MYB-like domain TFs in plants. A Ds-tagged recessive aid1 mutant, in which the transposon is inserted in the coding region of AID1, was identified in a genetic screen as playing a role in partial to complete spikelet sterility (Zhu et al., 2004). Three rice MYB proteins have been identified that interact with the promoter of α-amylase, an important component of the GA and sugar responses (Lu et al., 2002). Interestingly, OsMYBS1 and 2 positively regulate the α-amylase promoter when sugar is present, whereas OsMYBS3 represses transcription from the same promoter in response to sugar starvation (Lu et al., 2002). Recently, it was reported that expression of an OsMYBS3 transgene in rice confers cold tolerance without a significant penalty in yield under normal field conditions (Su et al., 2010).
Potato is an important food crop worldwide, with annual production approaching 300 million tons (Camire et al., 2009). Potato is relatively vulnerable to abiotic stresses such as drought and high salinity and is classified as an environmentally sensitive crop along with paddy rice and sugarcane (Saccharum officinarum; Levy, 1985; Bray et al., 2000; Vasquez-Robinet et al., 2008). Potato tuber initiation, bulking, and tuber growth stage especially are sensitive to drought stress (Evers et al., 2010). Therefore, water availability is an important factor in increasing tuber formation and potato crop yields. Several studies in potato were done to overcome crop yield loss under environmental stress conditions through genetic and molecular biological approaches (Jeong et al., 2001; Schafleitner et al., 2007; Stiller et al., 2008; Evers et al., 2010). Recently, drought-related elements in potato are profiled through a combined transcriptomic and targeted metabolite approach (Vasquez-Robinet et al., 2008; Evers et al., 2010). However, little is known about stress-tolerance mechanisms in potato plants. In this work, we identified an environmental stress-responsive R1-type MYB TF in potato and characterized its biological role in detail by examining its expression and physiological phenotype in potato plants.
RESULTS
Isolation of a Stress-Responsive Single MYB-Like Domain TF from Potato
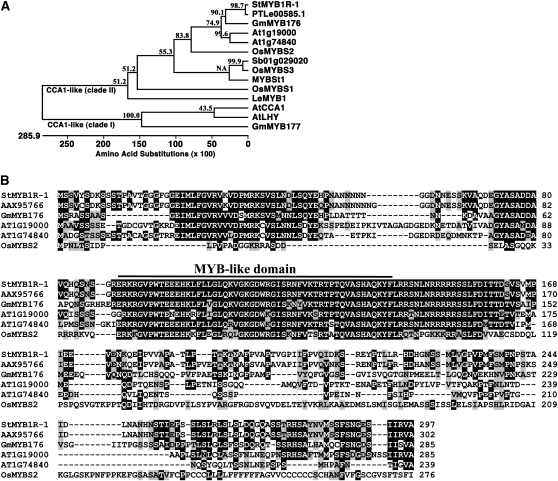
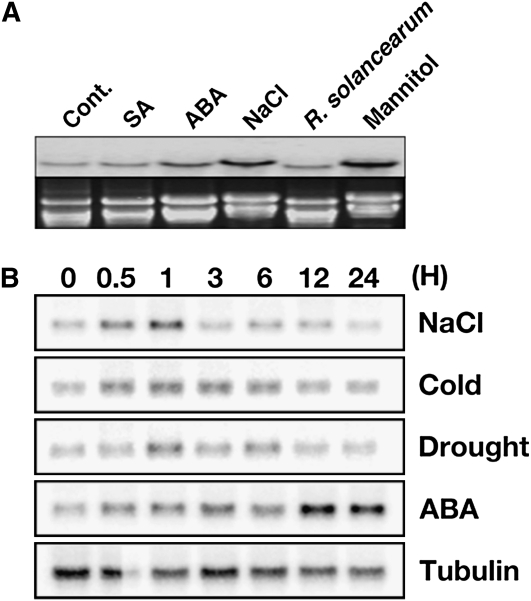
To explore the molecular mechanisms of tolerance in potato (cv Superior), we previously identified 17 genes by reverse northern-blot analysis that were up-regulated by cold, drought, and salt stress (Lee et al., 2007). Here, we carried out a series of molecular experiments to characterize one of these abiotic-responsive genes, clone SOT171183 (accession no. AU279205). Clone SOT171183 was 434 bp in length and contained no ATG start codon. Using the sequence of SOT171183 as a probe in a colony hybridization screening assay, we identified and sequenced six putative clones of different lengths that were all derived from the same gene. The longest clone was 1,166 bp in length, with an open reading frame that encoded a putative protein of 297 amino acids and a calculated molecular mass of 32.5 kD (Supplemental Fig. S1). An in silico database search revealed that the sequence of this clone was similar to single MYB-like domain TFs from Arabidopsis, rice, pepper (Capsicum annuum), and soybean (Glycine max) belonging to the CCA1-like clade II subgroup (Yanhui et al., 2006). Outside of the MYB domain, the sequence similarity between our clone and other members of the CCA1-like clade II family was lower than the MYB domain itself, with the exception of GmMYB176 and AT1G74840, which showed a high degree of conservation within and outside of the MYB domain with our clone (Fig. 1, A and B). The gene mapped to chromosome 6 and contained three exons and two introns (http://solanaceae.plantbiology.msu.edu/). To begin to characterize the biological role of this putative stress-responsive gene in potato, we analyzed RNA transcript levels under various stress conditions. Expression was increased under environmental stress conditions such as drought, but not biotic stress conditions such as salicylic acid (SA) treatment (Fig. 2). These results indicated that this novel gene, which we designated StMYB1R-1, encodes a putative stress-responsive R1-type MYB TF in potato.
Figure 1.
Sequence comparison of single MYB-like domain TFs and StMYB1R-1. A, Phylogenetic analysis of StMYB1R-1. The phylogenic tree was constructed with the DNASTAR program (DNASTAR, Inc.) using the deduced amino acid sequence of StMYB1R-1 and other single MYB-like domain protein sequences. B, Amino acid sequence alignment of StMYB1R-1 and other single MYB-like domain proteins. Accession numbers are as follows: StMYB1R-1, ABB86258; At1g19000, BAH19529; At1g74840, BAH56970; AtCCA1, AAB40525; AtLHY, NP_00103092; GmMYB176, ABH02865; GmMYB177, ABH02866; LeMYB1, CAB65169; MYBSt1, AAB32591; OsMYBS1, AAN63152; OsMYBS2, AAN63153; OsMYBS3, AAN63154; PTLe00585.1, AAX95766; and Sb01g029020, XP_002464930.
Figure 2.
Transcript levels of StMYB1R-1 are up-regulated in response to environmental stress, but not biotic stress conditions. A, Transcript levels of StMYB1R-1 are enhanced in response to ABA, NaCl, and mannitol stress, but not SA or R. solanacearum treatment. Total RNA was extracted from 3-week-old potatoes treated with ABA (3 μm), NaCl (100 mm), mannitol (200 mm), SA (10 μm), and R. solanacearum for 1 h. A gel prestained with ethidium bromide (bottom) was used to confirm equal loading in all wells. B, Transcript levels of StMYB1R-1 increase within 1 h in response to environmental stress conditions. Total RNA was extracted from 3-week-old potato progenies treated for various lengths of time with ABA (3 μm), NaCl (100 mm), cold (4°C), and drought (on 3MM paper). Total RNA (20 μg) was analyzed by northern-blot analysis using full-length StMYB1R-1 as the probe. Potato β-tubulin was used as a quantitative control.
StMYB1R-1 Functions as a TF
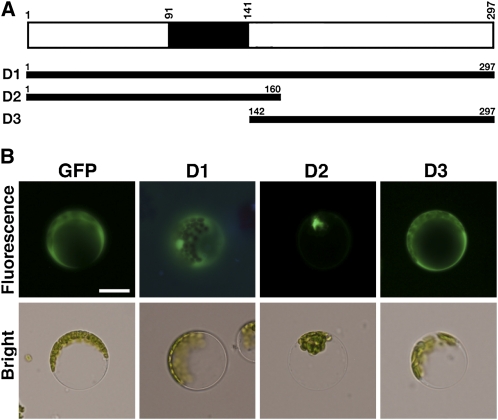
Most TFs, including MYB, localize to the nucleus and bind specific DNA sequences to regulate gene expression. Sequence analysis indicated that StMYB1R-1 belongs to the MYB family of TFs. To determine whether StMYB1R-1 functioned as a TF, we first examined the subcellular localization of StMYB1R-1 in Arabidopsis protoplasts and onion (Allium cepa) epidermal cells. Recombinant full-length StMYB1R-1, expressed as a transgene encoding full-length StMYB1R coupled to GFP (StMYB1R-1:GFP), localized predominantly to the nucleus, and to a lesser extent the cytosol (Fig. 3B; Supplemental Fig. S2). An in silico database search yielded no evidence of a nuclear localization signal or nuclear export signal peptide in the StMYB1R-1 protein. We generated variants of StMYB1R-1 that contained only the N- or C-terminal region and then examined the subcellular localization of each domain. Only the N-terminal domain of StMYB1R-1, which contained the MYB-like domain, localized to the nucleus, whereas the C-terminal segment localized to the cytosol (Fig. 3B). These results indicated that StMYB1R-1 localizes to the nucleus, where it could function as a TF.
Figure 3.
StMYB1R-1 localizes to the nucleus. A, Physical map of the StMYB1R-1 segment cloned into p326GFP. Black box indicates putative single MYB-like domain. B, Protoplasts prepared from Arabidopsis leaves were transfected with StMYB1R-1:GFP and then observed by fluorescence microscopy 12 and 24 h after transformation. Protoplasts expressing GFP were used as a control. Scale bar indicates 20 μm. [See online article for color version of this figure.]
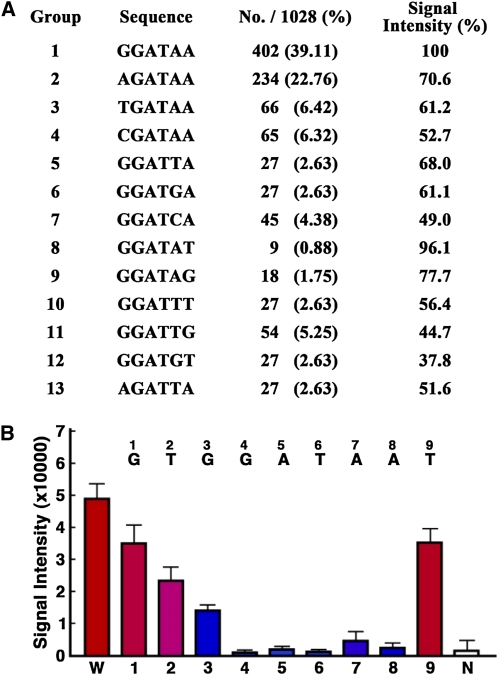
We took advantage of a recently developed protein-binding microarray to determine the putative DNA-binding sequence of StMYB1R-1. The quadruple 9-mer (Q9) protein-binding array was developed as a screening tool for rapid identification of DNA-binding sequences of TFs and contains more than 100,000 unique double-stranded DNA oligomers synthesized as quadruples of all possible 9-mer combinations (Kim et al., 2009). Using a DsRed fusion protein of StMYB1R-1 (StMYB1R-1:DsRed) to monitor binding, Q9 protein-binding microarray analysis yielded 1,028 putative DNA-binding sequences (>3,500 relative signal intensity). The DNA sequences were segregated into 13 groups, and two sequences, GGATAA and AGATAA, emerged as the predominant DNA-binding sequences (Fig. 4A; Supplemental Tables S1 and S2). The signal intensity of GGATAA was 21,860, while the signal intensities of the other groups, with the exception of group 8, were below 16,000. To test whether the GGATAA sequence was essential for DNA binding by StMYB1R-1, we analyzed the relative signal intensities of single nucleotide substitution variants of the putative binding sequence (with the exception of AGATAA). Individual substitutions at all positions of the GGATAA sequence significantly reduced the signal intensity of binding, whereas substitutions outside this core sequence had no effect (Fig. 4B). To confirm these results, we performed an electrophoresis mobility shift assay (EMSA) using StMYB1R-1:DsRed as the probe. Recombinant StMYB1R-1 was able to bind to and cause a shift in the mobility of an oligonucleotide containing the GGATAA sequence, but not any of the single nucleotide substitution variant sequences (Supplemental Fig. S4). These results indicated that StMYB1R-1 selectively binds to DNA at G/AGATAA sequences and may function as a TF.
Figure 4.
Identification of DNA-binding sequences of StMYB1R-1. A, Summary of putative StMYB1R-1 DNA-binding sequences. Q9 protein-binding microarray analysis was performed using a StMYB1R-1-DsRed fusion protein. A total of 1,028 probe sequences that produced high signal intensities were selected for further analysis. B, Signal intensities of the GGATAA sequence and single nucleotide substitution derivatives using the Q9 protein-binding microarray. W, Wild-type sequence; 1 to 9, single nucleotide substitution variants (number corresponds to the position of the nucleotide substitution). One of the probe sets that is irrelevant with the GGAAA sequences was used as negative control (N). [See online article for color version of this figure.]
Expression of StMYB1R-1 Enhances Drought Tolerance
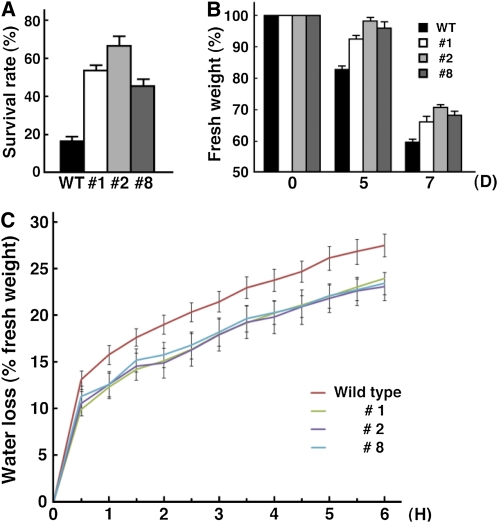
Expression analysis of StMYB1R-1 indicated that it is positively regulated under drought-stress conditions. We generated transgenic potato plant lines that overexpressed StMYB1R-1 to investigate whether overexpression of StMYB1R-1 increases drought tolerance. We first examined the effect of StMYB1R-1 expression on potato agricultural traits by morphological analysis. The expression of StMYB1R-1 did not significantly alter the growth characteristics of transgenic potato plants as compared with wild-type plants under greenhouse conditions (Supplemental Fig. S5; Supplemental Results S2). To assess the effect of StMYB1R-1 overexpression on drought tolerance, 8-week-old potato plants were deprived of water for 15 d, and then watering was resumed in the greenhouse. After 12 d without water, foliar tissue drooping was evident in wild-type plants, but not transgenic plants. After 15 d without water, most of the leaves of wild-type potato plants had wilted, while in transgenic plants, drooping of foliar tissue was just beginning to become apparent. After watering was resumed, some of the leaves of transgenic potatoes turned yellow brown in color as a result of drought stress, but the plants survived and eventually developed green leaves. By comparison, wild-type potato plants did not go on to develop green leaves (Fig. 5). Seven days after the resumption of watering and growth under normal conditions, survival rates were determined for wild-type and transgenic potato plants. More than 50% of the transgenic potato plants survived after the period of drought stress compared with 19% of wild-type potato plants (Fig. 6A). We also measured the weight of fresh leaves before and after drought stress. Before drought stress treatment, the leaf weights of wild-type and transgenic plants were similar. After 5 d of water deprivation, the water content of wild-type leaves decreased by 20%, whereas that of transgenic leaves decreased by only 5%. After 7 d without water, leaf water content in wild-type and transgenic leaves decreased by 40% and 30%, respectively (Fig. 6B).
Figure 5.
Transgenic potato plants overexpressing StMYB1R-1 are tolerant to drought-stress conditions. Eight-week-old T0 plantlets were deprived of water for 15 d and then watering was resumed for 7 d. n = 3 independent experiments (20 plants per experiment). D, Day; RW, rewatering.
Figure 6.
Physiological traits of transgenic potato plants overexpressing StMYB1R-1 under drought-stress conditions. A, Survival rates of transgenic potato plants overexpressing StMYB1R-1 under drought-stress conditions. Eight-week-old T0 plantlets were deprived of water for 15 d, watering was resumed for 7 d, and then the plants were scored for viability. Plants were considered dead if all the leaves were brown and there was no growth after 7 d of watering. n = 3 independent experiments (20 plants per experiment). B, Fresh weight of leaves from transgenic potato plants overexpressing StMYB1R-1 under drought-stress conditions. Detached leaves from transgenic and wild-type potato plants were measured. n = 3 independent experiments (20 detached leaves per data point). C, The kinetics of water loss in detached leaves from wild-type and transgenic potato plants overexpressing StMYB1R-1. Water loss is presented as the percentage of weight loss versus initial fresh weight. Values represent the means ± se of three independent experiments. [See online article for color version of this figure.]
Expression of StMYB1R-1 Prevents Water Loss and Enhances Hypersensitivity to ABA-Induced Stomatal Closure
To determine whether StMYB1R-1 was involved in regulating water loss under conditions of drought stress, we performed a water loss assay of wild-type and transgenic plants. The rate of water loss was determined by measuring the fresh weight of detached leaves at 30-min intervals. The fresh weight of transgenic and wild-type leaves was decreased by approximately 22% and 27%, respectively, after drought stress (Fig. 6C). When we examined transpiration rate and stomatal conductance of detached potato leaves, the time to reach a 50% reduction in transpiration rate was approximately 9 min for wild-type plants, and less than 7 min for transgenic plants. Transgenic plants also exhibited a more rapid loss of stomatal conductance than wild-type plants (Supplemental Fig. S6). Transgenic line 2 in particular had a more rapid physiological response under drought-stress conditions. These results indicated that StMYB1R-1 overexpression results in a significant decrease in the rate of water loss in potato plants.
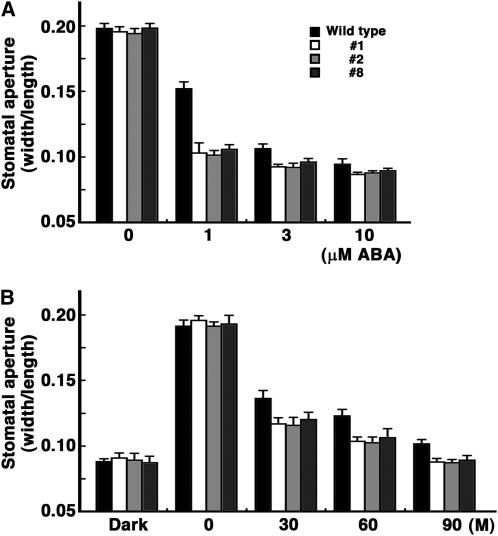
Water loss in plants is regulated by guard cell density, guard cell size, and/or stomatal opening and closing. To determine whether StMYBR-1 was involved in regulating these parameters in potato plants, we first compared guard cell density and guard cell size in wild-type and transgenic plants. Guard cell density and size were similar between wild-type and transgenic plants under normal growth conditions (data not shown). To determine the effect of StMYB1R-1 overexpression on stomatal opening, leaves from wild-type and transgenic potato plants were incubated in the light for 3 h to allow stomata to open fully, and then the widths of the stomatal apertures were determined. In wild-type leaves, stomata measured 12.10 ± 0.017 μm in width as compared with 12.13 ± 0.024 μm (L1, 12.47 ± 0.027 μm; L2, 12.07 ± 0.021 μm; L8, 12.21 ± 0.025 μm) in transgenic plants. Thus, overexpression of StMYB1R-1 did not appear to significantly affect stomatal opening. However, after treatment with ABA for 1 h, there was a marked dose-dependent decrease in the size of stomatal pores (ratio of width to length) in leaves from transgenic plants as compared with wild type (Fig. 7A).
Figure 7.
Stomatal closure of transgenic potato plants overexpressing StMYB1R-1 is more sensitive to ABA. A, Stomatal apertures were measured following exposure to the indicated concentrations of ABA for 1 h. ABA-induced stomatal closing was analyzed by measuring stomatal apertures. Detached leaves from 4-week-old T0 plantlets were floated on opening solution for 3 h before the addition of ABA. B, After the addition of 1 μm ABA, stomatal apertures were measured over time. n = 3 independent experiments (>100 stomatal apertures per data point). M, Minutes.
As indicated above, in response to treatment with 1 μm ABA, there was a marked difference in stomatal aperture between wild-type and transgenic plants. We performed a kinetic analysis of stomatal aperture closing in wild-type and transgenic plants in the presence of 1 μm ABA. After 30 min of ABA treatment, stomatal aperture size in wild-type and transgenic plants decreased to 0.132 ± 0.005 and 0.113 ± 0.02 (L1, 0.115 ± 0.004; L2, 0.112 ± 0.005; L8, 0.118 ± 0.004), respectively. After an additional 60-min incubation period in the presence of ABA, the stomatal aperture size of transgenic plants was close to that observed under dark conditions, while the stomatal aperture of wild-type leaves remained slightly open (Fig. 7B). These results suggested that the expression of StMYB1R-1 potentiates the ABA response of guard cells, leading to reduced water loss under drought conditions. This drought tolerance phenotype of transgenic plants was consistent with the slow rate of water loss from leaves.
Expression of StMYB1R-1 Enhances the Expression of Drought Stress-Related Genes
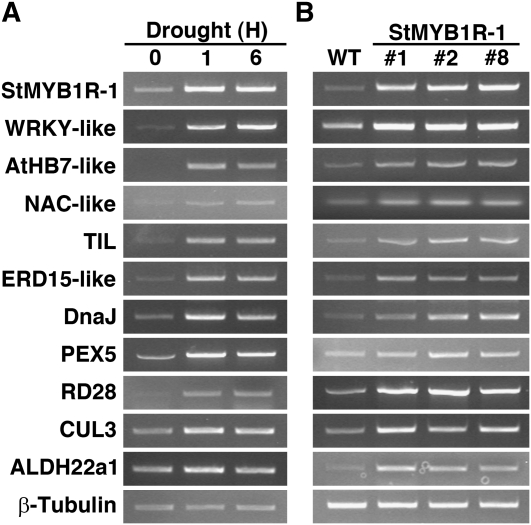
Given that overexpression of StMYB1R-1 led to drought tolerance in potato, we explored whether the phenotypic changes in the transgenic plants correlated with changes in the expression patterns of stress-responsive genes. Microarray analysis using The Institute for Genomic Research (TIGR) 10 K potato chip (Lee et al., 2007) revealed that 295 genes were up-regulated 2-fold or more in transgenic plants as compared with wild-type plants. Most of the annotations were hypothetical or unknown proteins (Supplemental Table S3), but 16 of the genes were abiotic stress-responsive genes. We analyzed these 16 genes by semiquantitative reverse transcription (RT)-PCR to determine whether they were regulated by drought stress and to validate the results of the microarray analysis. Expression of the Na+-dependent bile acid transporter (AtBAT5-like) was similar in the presence and absence of drought-stress treatment, and trehalose-6-P synthase (TPS1-like) and fumarylacetoacetate hydrolase (FAH1-like) were undetectable by RT-PCR under these experimental conditions. Notably, the transcript levels of several genes with high similarity to AtHB-7, RD28, ALDH22a1, and ERD15-like, which are involved in plant stress tolerance, were dramatically up-regulated after drought-stress treatment and were enhanced in transgenic plants (Fig. 8).
Figure 8.
Up-regulation of drought stress-responsive genes in transgenic potato plants overexpressing StMYB1R-1. A, Transcript levels of putative stress-responsive genes selected based on cDNA microarray analysis. Three-week-old wild-type potatoes were incubated on 3MM paper for 1 or 6 h to simulate drought stress, and then total RNA was extracted. Transcript levels for each gene were analyzed using gene-specific primers and semiquantitative RT-PCR. B, Transcript levels of drought stress-responsive genes are enhanced in transgenic potato plants overexpressing StMYB1R-1. Potato β-tubulin was used as a quantitative control (Nicot et al., 2005).
DISCUSSION
Potato is one of the world’s major food crops (Gopal and Iwama, 2007). Despite this, there are a few reports in the literature of molecular engineering to improve or manipulate stress tolerance in potato plants (Stiller et al., 2008; Tang et al., 2008). A limited number of single MYB-like domain TFs have been identified in plants (Penfield and Hall, 2009). Here, we report the identification and characterization of a putative single MYB-like domain TF, StMYB1R-1, in potato.
Various TFs are rapidly induced during the early phases of environmental stress conditions such as drought and high salinity. AtMYB2, GmMYB92, and CpMYB10, which encode R2R3-type MYB TFs, are induced by ABA as well as drought (Urao et al., 1993; Villalobos et al., 2004; Liao et al., 2008), and AtMYB44 and AtMYB102 are induced after ABA and drought treatment. These genes are also up-regulated by jasmonic acid and wounding stress (Denekamp and Smeekens, 2003; Jung et al., 2008). Although functional roles for single MYB-like domain TFs such as CCA1 have been identified in light-related and other developmental processes (Wang et al., 1997; Zhu et al., 2004), other MYB family TFs, such as GmMYB117, an R1-type MYB TF found in soybean, and OsMYBS3 in rice are enhanced by ABA and abiotic stresses (Liao et al., 2008) and cold stress conditions (Su et al., 2010), respectively. Here, we showed that the expression of StMYB1R-1 is induced after ABA treatment and by various abiotic stresses, but is unaffected by SA and Ralstonia solanacearum treatment (Fig. 2). These results suggest that single MYB-like domain TFs may function specifically in abiotic stress responses, and that StMYB1R-1, a novel R1-type member of the MYB family in potato, is involved in regulating these types of stress responses (Figs. 2 and 3; Supplemental Fig. S3; Supplemental Results S1).
TFs interact with specific DNA sequences (cis-acting elements) in target genes to modulate the transcription process (Priest et al., 2009). Protein DNA-binding properties are traditionally investigated by methods such as EMSAs and filter-binding assays (Garner and Revzin, 1981; Söderman and Reichard, 1986). The development of the Q9 protein-binding microarray along with the availability of whole-genome sequence information and advances in microarray technology have greatly facilitated our ability to characterize protein DNA-binding specificities in vitro (Kim et al., 2009). The results of Q9 protein-binding microarray analysis and EMSA demonstrated that StMYB1R-1 binds DNA mainly at G/AGATAA sequences (Fig. 4; Supplemental Fig. S4). Previously, it was shown StMYB1 binds specifically to the sequence GGATA within a region (nucleotides −73 to −48) of the CaMV 35S promoter to activate transcription in tobacco (Nicotiana tabacum) protoplasts (Baranowskij et al., 1994). Three single MYB-like domain proteins in rice (OsMYBSs) interact with a TATCCA element in the promoter of αAmy3, a rice α-amylase gene that is strongly induced in response to sugar starvation (Lu et al., 2002). In addition, OsMYBS3 in rice regulates the expression of cold-related genes such as WRKY77 and Glu decarboxylase, both of which contain a TATCCA element in their promoters (Su et al., 2010). The fact that the DNA-binding sequences of StMYB1 and the OsMYBSs are similar to the target-binding sequence identified here for StMYB1R-1 supports the use of the Q9 protein-binding microarray as a reliable technique for studying genome-wide protein-DNA interactions. These results also indicate that the DNA-binding sequences of plant single MYB-like domain TFs may be similar, and that binding specificity may be regulated by interactions with other regulatory proteins.
Overexpression of StMYB1R-1 in potato plants induced the expression of several genes, including AtHB-7, RD28, ALDH, and ERD15, and follow-up RT-PCR analysis confirmed that these genes are up-regulated under drought stress conditions. AtHB-7 is up-regulated in response to ABA treatment and water deficit, and expression of AtHB-7 has been shown to increase ABA sensitivity (Olsson et al., 2004). RD28 is a member of the major intrinsic protein 2 family of water channels (Chaumont et al., 1997). The expression of water channels in Arabidopsis is up- or down-regulated under conditions of abiotic stress such as drought (Jang et al., 2004). In particular, PIP2;2, which encodes a water channel protein, is activated predominantly in roots, and PIP2;2 functions in root water uptake under conditions of reduced transpiration (Javot et al., 2003). Gly betaine is an important osmolyte source in plants, and ALDH genes, which encode betaine aldehyde dehydrogenases, are activated under saline or water-deficit conditions (Kirch et al., 2005). ERD15 is rapidly expressed in response to abiotic and biotic stresses, particularly dehydration (Kariola et al., 2006). Thus, the enhanced drought tolerance observed in StMYB1R-1 transgenic plants might depend in part on changes in the expression levels of these genes. That some of these up-regulated genes such as RD28 contain an A/GGATAA box in their promoters (data not shown) supports a mechanism in which binding of StMYB1R-1 to cis-elements in the promoters of target genes results in transcriptional regulation of environmental stress-inducible genes.
The expression of stress-inducible genes, including MYB TFs, affects stress tolerance in plants. AtMYB2 is induced by dehydration and ABA treatments, and overexpression of AtMYB2 results in increased sensitivity to ABA (Abe et al., 2003). AtMYB44 functions in leaf epidermal guard cells, and AtMYB44 transgenic plants exhibit a more rapid ABA-induced stomatal closure response, reduced rate of water loss, and markedly enhanced drought tolerance as compared with wild-type plants (Jung et al., 2008). Ectopic expression of GmMYB177 confers salt and freezing tolerance in Arabidopsis (Liao et al., 2008). OsMYBS3 transgenic rice plants acquire cold tolerance with no penalty in terms of yield under normal field conditions (Su et al., 2010). AtMYB41 is expressed in response to drought and salt treatment in an ABA-dependent manner, but transgenic plants that overexpress AtMYB41 lose water more rapidly than wild-type plants and are hypersensitive to desiccation (Cominelli et al., 2008). Furthermore, AtMYB41 has been shown to negatively regulate salt-induced genes such as AtDREB2a and AtNCED3 (Lippold et al., 2009). Here, overexpression of StMYB1R-1 greatly improved drought tolerance in potato plants, as demonstrated by increased survival rates, decreased water loss, and enhanced stomatal closure in transgenic plants as compared with wild type (Figs. 6–8). StMYB1R-1 transgenic plants also exhibited salt tolerance, as observed by comparing root elongation in Murashige and Skoog (MS) media under high saline conditions between wild-type and transgenic plants (Supplemental Fig. S7). These results indicate that StMYB1R-1 functions in drought and salt tolerance to regulate water content via transcriptional regulation of stress-responsive genes in potato plants.
Trehalose-6-P synthase (TPS1) is involved in the synthesis of trehalose. Expression of Saccaharomyces cerevisiae trehalose-6-P synthase (TPS1) from the constitutive CaMV35S promoter resulted in drought tolerance in tobacco and potato plants (Goddijn et al., 1997; Romero et al., 1997). Transgenic potato plants expressing TPS1 have 30% to 40% fewer guard cells than the wild-type plants, and this correlates with their ability to retain significantly more water than wild-type plants (Stiller et al., 2008). Ectopic expression of Arabidopsis AtCBF genes under the control of the constitutive CaMV 35S promoter in potato enhances freezing tolerance; however, plant growth is inhibited, as is development, particularly tuber formation and flowering time (Pino et al., 2007). Expression of AtCBF genes under the control of the rd29A promoter, on the other hand, ameliorated these agricultural traits without affecting freezing tolerance (Pino et al., 2007). Transgenic potato plants expressing AtNDPK2 under the control of the stress-inducible SWPA2 promoter or CaMV 35S promoter exhibit enhanced tolerance to oxidative stress and salt stress. However, expression of AtNDPK2 under the control of the SWPA2 promoter resulted in much less leaf damage than nontransgenic plants, whereas under the control of the CaMV 35S promoter, AtNDPK2 expression resulted in an intermediate tolerance phenotype between AtNDPK2-SWPA2 transgenic and nontransgenic plants (Tang et al., 2008). Here, the overexpression of StMYB1R-1 did not affect plant growth and development. In StMYB1R-1 transgenic plants, foliar tissue size and biomass were similar to wild type (Fig. 5). Importantly, tuber shape and size in StMYB1R-1 transgenic lines were largely unaffected; tuber yield, on the other hand, was slightly reduced. These results suggest that stress-inducible promoters such as the rd29A and SWPA2 promoters are more efficient than the CaMV 35S promoter in terms of developing stress-tolerant transgenic plants without negatively affecting other agricultural traits (Pino et al., 2007; Tang et al., 2008).
In summary, StMYB1R-1 functions as a TF to improve drought tolerance in potato. Although the detailed mechanism of StMYB1R-1 function in response to abiotic stresses is not yet clear, this report provides valuable information for molecular breeding that could lead to improved stress tolerance in agricultural crops. Loss-of-function analysis of StMYB1R-1 transgenic lines using RNA interference, currently under way in our laboratory, will further improve our understanding of the biological and molecular functions of StMYB1R-1.
MATERIALS AND METHODS
Plant Growth Conditions and Stress Treatments
Potato (Solanum tuberosum ‘Superior’) was used throughout this study. Plants were grown in soil in a greenhouse or in hormone-free MS agar medium in a growth chamber maintained at 21°C to 23°C and 60% relative humidity under long-day conditions (16-h light/8-h dark cycle). Three micrometers of (±) ABA or 10 μm of SA were applied to the surface of solid MS agar medium containing 3-week-old potato progenies. Abiotic stresses were applied to the 3-week-old progenies by either treating them with 100 mm NaCl, incubating them at 4°C under continuous light, or drying them on Whatman 3MM paper (Lee et al., 2007). Ralstonia solanacearum KACC10699 bacterial cells were grown on nutrient broth agar medium with phosphate for 24 h at 28°C, suspended in sterile distilled water, and adjusted to 108 colony forming units/mL. The roots of the potato plants were immersed in 50 mL of bacterial suspension for 1 h.
Northern-Blot Analyses, Microarray, and Semiquantitative RT-PCR
Total RNA from 200 mg of tissue was extracted using the RNeasy plant mini kit (Qiagen) following the manufacturer’s instructions. Samples (10 μg lane−1) were separated by 1.2% formaldehyde agarose gel electrophoresis, transferred to nylon membranes (Hybond N+; Amersham) by capillary blotting, and UV cross-linked. Hybridization was performed in Church buffer using a full-length StMYB1R-1 probe, after which the membranes were washed with 2×, 1×, and 0.5× SSC for 10 min each, and then rewashed with 0.1× SSC/0.1% SDS at 65°C for 10 min. The blots were then exposed to x-ray film (Eastman Kodak).
For microarray experiments, total RNAs were extracted from StMYB1R-1 overexpression transgenic plants and wild-type plants using TRIzol (Invitrogen) as described by the manufacturer and were used for preparation of Cy5- and Cy3-labeled cDNA probes. Microarray was conducted with the TIGR cDNA potato microarray chip. Probe labeling and chip hybridization were carried out through the Korea TAKARA custom service (http://takara.co.kr/index.asp) by following the TIGR standard protocol (http://jcvi.org/potato/sol_ma_protocols.shtml). Normalization was performed according to the standard TIGR protocol to allow the comparison of the samples for each set of experiments. The Excel for significance analysis of microarrays was used to identify differentially expressed genes between StMYB1R-1 overexpression transgenic plants and wild-type plants.
For semiquantitative RT-PCR, 2 μg of total RNA extracted from potato progenies were reverse transcribed with SuperScript III reverse transcriptase (Invitrogen). The PCR program consisted of 25 to 30 cycles of amplification (20 s, 95°C; 30 s, 56°C; and 30 s, 72°C) using ExTaq polymerase (TAKARA) with gene-specific primers. RT-PCR reactions were repeated three times. All of the gene-specific primers are listed in Supplemental Table S4.
Subcellular Localization of StMYB1R-1
For subcellular localization, the cDNA fragments without the stop codon were amplified by PCR with specific primers containing appropriate restriction sites and subcloned directly upstream from the N terminus of the GFP coding region in the p326GFP vector (Niwa et al., 1999). The plasmids were introduced into Arabidopsis (Arabidopsis thaliana) protoplasts that had been prepared from leaf tissue by polyethylene glycol-mediated transformation (Jin et al., 2001). Expression of the fusion constructs was monitored at various time points after transformation and images were captured with a cooled CCD camera through a Zeiss Axioplan fluorescence microscope (Carl Zeiss). Data were then processed using Adobe Photoshop software (Adobe Systems) and presented in pseudo-color format.
Determining of Putative DNA-Binding Sequence of StMYB1R-1
To determine the DNA-binding sequence of StMYB1R-1, a full-length cDNA of StMYB1R-1 was inserted into the pET32(a) vector (Novagen) with an N-terminal fusion to a polyhistidine tag and DsRed-monomer fluorescent protein (Novagen). The StMYB1R-1 fusion protein was purified from Escherichia coli strain BL21-CodonPlus (Stratagene). A total of 200 nm of the StMYB1R-1 protein was incubated with Q9 protein-binding microarray, included 101,073 features from each 9-mers, in phosphate-buffered saline-2% bovine serum albumin, 50 ng/μL salmon testes DNA (Sigma), and 50 μm zinc acetate at 25°C for 1 h (Kim et al., 2009). Fluorescence images were obtained with a 4000B microarray scanner (Molecular Devices). The signal intensities were ranged from 0 to 70,000. The significant probes were selected by statistical analysis modified (Supplemental Materials and Methods S1; Kim et al., 2009). Probe sequence showing a >3,500 signal intensity from microarray fluorescence image were extracted and analyzed.
Stomatal Aperture Bioassays
To measure stomatal closure, detached leaves from 3-week-old potato progenies cultured in MS medium were floated in stomatal opening solution (15 mm KCl, 10 μm CaCl2, and 10 mm MES-KOH, pH 6.15) in the light (200 μmol m−2 s−1, 22°C). After 3 h, the buffer was replaced with fresh stomatal opening solution containing various concentrations of ABA. After an additional 1 h incubation, the leaves were cut into 3-mm pieces, placed on a microscope slide, and covered with a coverslip. The abaxial epidermal layers of the leaves were observed using bright-field microscopy (Axioskop 2, Carl Zeiss) and images were captured using a CCD camera (Axio Cam, Carl Zeiss). Aperture size was measured from the photographs using the Interactive Measurement software package AxioVision 3.0.6 (Carl Zeiss).
Water Loss
To determine water loss, nine leaves from 3-week-old potato progenies cultivated in the growth chamber were detached and placed in a weighing dish. The dishes were maintained in the growth chamber, and the loss of fresh weight was determined at 30-min intervals during 6 h (Bouchabke et al., 2008).
Generation of Transgenic Plants and Phenotype Observation
To generate StMYB1R-1 overexpression plants, the full-length cDNA of StMYB1R-1 was cloned into the pBIN121 vector (CLONTECH) under the control of the CaMV 35S promoter. The recombinant plasmid was introduced into Agrobacterium tumefaciens LBA4404, and transgenic potato plants were obtained by internode transformation (Lee et al., 2007). Twenty-four independent transgenic potato lines were generated and confirmed by hygromycin selection and northern-blot analysis. Among these transgenic potato lines, three independent transgenic lines were selected and grown in growth chamber before being used in later experiments.
To observe the phenotype under drought-stress conditions, 4-week-old potato plants cultured on MS medium and hydroponic chamber were transferred to soil (each 900 g of soil). To adapt the potato plants to the soil, potato progenies were grown for 4 more weeks in a greenhouse. Water was withheld from 8-week-old progenies for 15 d before resuming watering and everyday pot position was changed to minimize side effects. Photographs were taken, and the survival rate was determined on the 7th d after watering resumed. All stress assays were performed in three independent experiments, and mean values and ses are presented.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AU279205 (StMYB1R-1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide and predicted amino acid sequence of StMYB1R-1 cDNA.
Supplemental Figure S2. StMYB1R-1 localizes to the nucleus.
Supplemental Figure S3. Transcriptional activation test for StMYB1R-1 in yeast.
Supplemental Figure S4. StMYB1R-1 binds specifically to the GGATAA sequences.
Supplemental Figure S5. Growth characteristics of transgenic potato plants overexpressing StMYB1R-1.
Supplemental Figure S6. Physiological characteristics of transgenic potato plants overexpressing StMYB1R-1.
Supplemental Figure S7. Transgenic potato plants overexpressing StMYB1R-1 are tolerant to salt stress conditions.
Supplemental Table S1. Nine probes presenting the same DNA sequences were grouped as one probe set.
Supplemental Table S2. Total DNA-binding sequences of StMYB1R-1.
Supplemental Table S3. Up-regulated genes in transgenic plant overexpressing StMYB1R-1.
Supplemental Table S4. Primer sequences used for semiquantitative RT-PCR.
Supplemental Results S1. C-terminal domain of StMYB1R-1 functions as transcriptional activator.
Supplemental Results S2. Effect of StMYB1R-1 expression in potato.
Supplemental Materials and Methods S1. Data analysis of the Q9 protein-binding microarray.
Supplementary Material
Acknowledgments
We are grateful to Prof. Dae-Jin Yun (Gyeongsang National University) and Dong-Hern Kim (National Academy of Agricultural Science) for their critical reading, and Soo In Lee (National Academy of Agricultural Science) for assisting with microscope techniques.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowskij N, Frohberg C, Prat S, Willmitzer L. (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J 13: 5383–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabás B, Jäger K, Fehér A. (2008) The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ 31: 11–38 [DOI] [PubMed] [Google Scholar]
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701 [DOI] [PubMed] [Google Scholar]
- Bouchabke O, Chang F, Simon M, Voisin R, Pelletier G, Durand-Tardif M. (2008) Natural variation in Arabidopsis thaliana as a tool for highlighting differential drought responses. PLoS One 3: e1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Wertilnyk E. (2000) Responses to abiotic stresses. Buchanan BB, Gruissem W, Jones RL, , Biochemistry & Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 1162–1170 [Google Scholar]
- Camire ME, Kubow S, Donnelly DJ. (2009) Potatoes and human health. Crit Rev Food Sci Nutr 49: 823–840 [DOI] [PubMed] [Google Scholar]
- Chaumont F, Loomis WF, Chrispeels MJ. (1997) Expression of an Arabidopsis plasma membrane aquaporin in Dictyostelium results in hypoosmotic sensitivity and developmental abnormalities. Proc Natl Acad Sci USA 94: 6202–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C. (2008) Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J 53: 53–64 [DOI] [PubMed] [Google Scholar]
- Denekamp M, Smeekens SC. (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132: 1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX. (2009) Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry (Mosc) 74: 1–11 [DOI] [PubMed] [Google Scholar]
- Evers D, Lefèvre I, Legay S, Lamoureux D, Hausman JF, Rosales RO, Marca LR, Hoffmann L, Bonierbale M, Schafleitner R. (2010) Identification of drought-responsive compounds in potato through a combined transcriptomic and targeted metabolite approach. J Exp Bot 61: 2327–2343 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner MM, Revzin A. (1981) A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res 9: 3047–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, et al. (2010) Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddijn OJ, Verwoerd TC, Voogd E, Krutwagen RW, de Graaf PT, van Dun K, Poels J, Ponstein AS, Damm B, Pen J. (1997) Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol 113: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal J, Iwama K. (2007) In vitro screening of potato against water-stress mediated through sorbitol and polyethylene glycol. Plant Cell Rep 26: 693–700 [DOI] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R, Inoue K, Shirano Y, Hayashi H, Shibata D, Sato S, Kato T, Tabata S, et al. (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19: 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsson M, Siegel RS, Young J, Hashimoto M, Iba K, Schroeder JI. (2006) Guard cell ABA and CO2 signaling network updates and Ca2+ sensor priming hypothesis. Curr Opin Plant Biol 9: 654–663 [DOI] [PubMed] [Google Scholar]
- Jang JY, Kim DG, Kim YO, Kim JS, Kang H. (2004) An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol Biol 54: 713–725 [DOI] [PubMed] [Google Scholar]
- Javot H, Lauvergeat V, Santoni V, Martin-Laurent F, Güçlü J, Vinh J, Heyes J, Franck KI, Schäffner AR, Bouchez D, et al. (2003) Role of a single aquaporin isoform in root water uptake. Plant Cell 15: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong MJ, Park SC, Byun MO. (2001) Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3 phosphate dehydrogenase gene transfer. Mol Cells 12: 185–189 [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ. (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariola T, Brader G, Helenius E, Li J, Heino P, Palva ET. (2006) EARLY RESPONSIVE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol 142: 1559–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Lee TH, Pahk YM, Kim YH, Park HM, Choi YD, Nahm BH, Kim YK. (2009) Quadruple 9-mer-based protein binding microarray with DsRed fusion protein. BMC Mol Biol 10: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch HH, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D. (2005) Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol Biol 57: 315–332 [DOI] [PubMed] [Google Scholar]
- Lee HE, Shin D, Park SR, Han SE, Jeong MJ, Kwon TR, Lee SK, Park SC, Yi BY, Kwon HB, et al. (2007) Ethylene responsive element binding protein 1 (StEREBP1) from Solanum tuberosum increases tolerance to abiotic stress in transgenic potato plants. Biochem Biophys Res Commun 353: 863–868 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. (1985) The response of potatoes to a single transient heat or drought stress imposed at different stages of tuber growth. Potato Res 28: 415–424 [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. (2005) AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol 15: 1201–1206 [DOI] [PubMed] [Google Scholar]
- Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS, Chen SY. (2008) Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res 18: 1047–1060 [DOI] [PubMed] [Google Scholar]
- Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK. (2009) AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol 149: 1761–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Ho TH, Ho SL, Yu SM. (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, et al. (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt P, Creelman R. (2008) The ABA receptors—we report you decide. Curr Opin Plant Biol 11: 474–478 [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Le NT, Brandt WF, Driouich A, Farrant JM. (2009) Towards a systems-based understanding of plant desiccation tolerance. Trends Plant Sci 14: 110–117 [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H. (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18: 455–463 [DOI] [PubMed] [Google Scholar]
- Olsson AS, Engström P, Söderman E. (2004) The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol Biol 55: 663–677 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Ward JM, Harper JF, Schroeder JI. (1996) A novel chloride channel in Vicia faba guard cell vacuoles activated by the serine/threonine kinase, CDPK. EMBO J 15: 6564–6574 [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Hall A. (2009) A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21: 1722–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino MT, Skinner JS, Park EJ, Jeknić Z, Hayes PM, Thomashow MF, Chen TH. (2007) Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol J 5: 591–604 [DOI] [PubMed] [Google Scholar]
- Priest HD, Filichkin SA, Mockler TC. (2009) Cis-regulatory elements in plant cell signaling. Curr Opin Plant Biol 12: 643–649 [DOI] [PubMed] [Google Scholar]
- Ramsay RG, Gonda TJ. (2008) MYB function in normal and cancer cells. Nat Rev Cancer 8: 523–534 [DOI] [PubMed] [Google Scholar]
- Reynolds M, Tuberosa R. (2008) Translational research impacting on crop productivity in drought-prone environments. Curr Opin Plant Biol 11: 171–179 [DOI] [PubMed] [Google Scholar]
- Romero C, Bellés JM, Vayá JL, Serrano R, Culiáñez-Macià FA. (1997) Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta 201: 293–297 [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Schafleitner R, Gutierrez Rosales RO, Gaudin A, Alvarado Aliaga CA, Martinez GN, Tincopa Marca LR, Bolivar LA, Delgado FM, Simon R, Bonierbale M. (2007) Capturing candidate drought tolerance traits in two native Andean potato clones by transcription profiling of field grown plants under water stress. Plant Physiol Biochem 45: 673–690 [DOI] [PubMed] [Google Scholar]
- Söderman K, Reichard P. (1986) A nitrocellulose filter binding assay for ribonucleotide reductase. Anal Biochem 152: 89–93 [DOI] [PubMed] [Google Scholar]
- Stiller I, Dulai S, Kondrák M, Tarnai R, Szabó L, Toldi O, Bánfalvi Z. (2008) Effects of drought on water content and photosynthetic parameters in potato plants expressing the trehalose-6-phosphate synthase gene of Saccharomyces cerevisiae. Planta 227: 299–308 [DOI] [PubMed] [Google Scholar]
- Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM. (2010) A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol 153: 145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Kim MD, Yang KS, Kwon SY, Kim SH, Kim JS, Yun DJ, Kwak SS, Lee HS. (2008) Enhanced tolerance of transgenic potato plants overexpressing nucleoside diphosphate kinase 2 against multiple environmental stresses. Transgenic Res 17: 705–715 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K. (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, et al. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Puzõrjova I, Brosché M, Valk E, Lepiku M, Moldau H, Pechter P, Wang YS, Lindgren O, Salojärvi J, et al. (2010) Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J 62: 442–453 [DOI] [PubMed] [Google Scholar]
- Vasquez-Robinet C, Mane SP, Ulanov AV, Watkinson JI, Stromberg VK, De Koeyer D, Schafleitner R, Willmot DB, Bonierbale M, Bohnert HJ, et al. (2008) Physiological and molecular adaptations to drought in Andean potato genotypes. J Exp Bot 59: 2109–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos MA, Bartels D, Iturriaga G. (2004) Stress tolerance and glucose insensitive phenotypes in Arabidopsis overexpressing the CpMYB10 transcription factor gene. Plant Physiol 135: 309–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM. (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60: 107–124 [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999 [DOI] [PubMed] [Google Scholar]
- Zhu QH, Ramm K, Shivakkumar R, Dennis ES, Upadhyaya NM. (2004) The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol 135: 1514–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.