Abstract
Xyloglucan endo-transglycosylases (XETs) encoded by xyloglucan endo-transglycosylases/hydrolase (XTH) genes modify the xyloglucan-cellulose framework of plant cell walls, thereby regulating their expansion and strength. To evaluate the importance of XET in wood development, we studied xyloglucan dynamics and XTH gene expression in developing wood and modified XET activity in hybrid aspen (Populus tremula × tremuloides) by overexpressing PtxtXET16-34. We show that developmental modifications during xylem differentiation include changes from loosely to tightly bound forms of xyloglucan and increases in the abundance of fucosylated xyloglucan epitope recognized by the CCRC-M1 antibody. We found that at least 16 Populus XTH genes, all likely encoding XETs, are expressed in developing wood. Five genes were highly and ubiquitously expressed, whereas PtxtXET16-34 was expressed more weakly but specifically in developing wood. Transgenic up-regulation of XET activity induced changes in cell wall xyloglucan, but its effects were dependent on developmental stage. For instance, XET overexpression increased abundance of the CCRC-M1 epitope in cambial cells and xylem cells in early stages of differentiation but not in mature xylem. Correspondingly, an increase in tightly bound xyloglucan content was observed in primary-walled xylem but a decrease was seen in secondary-walled xylem. Thus, in young xylem cells, XET activity limits xyloglucan incorporation into the tightly bound wall network but removes it from cell walls in older cells. XET overexpression promoted vessel element growth but not fiber expansion. We suggest that the amount of nascent xyloglucan relative to XET is an important determinant of whether XET strengthens or loosens the cell wall.
Xylem cell morphology is an important determinant of wood properties. The size and shape of all wood cells, including (in angiosperms) vessel elements, fibers, and radial and axial parenchyma, are determined during their development from initials in the vascular cambium and the primary-walled stage of differentiation (Larson, 1994; Mellerowicz et al., 2001). Xylem fibers are long, slender cells that become significantly longer than the fusiform cambium cells from which they originate. In contrast, the diameter (but not length) of vessel elements markedly increases, and they develop perforations between neighboring elements to form vessels. Developing wood cells expand via both symplastic and intrusive growth, which involves modification of pectins, hemicelluloses, and cellulose (Mellerowicz, 2006; Gray-Mitsumune et al., 2008; Siedlecka et al., 2008).
Xyloglucan is an important hemicellulose of the primary cell wall in developing wood of both softwoods and hardwoods that binds noncovalently to cellulose, coating and cross-linking adjacent cellulose microfibrils, thereby forming the load-bearing network of the wall (Hayashi, 1989; McCann et al., 1990). Remodeling of this network by expansins, cellulases, xyloglucanases, and xyloglucan endo-transglycosylases (XETs) is thought to play a major role in the regulation of wall stress relaxation, which drives cell growth (Cosgrove, 2005). XET breaks a β-(1→4) glycosidic bond in the xyloglucan backbone and transfers the xyloglucanyl segment to O-4 of the nonreducing terminal Glc residue of an acceptor, which may be either xyloglucan or a xyloglucan oligosaccharide (XGO; Fry et al., 1992; Nishitani and Tominaga, 1992). XET enzymes of plants belong to glycoside hydrolase family 16 (GH16). Most members of this family in plants that have been biochemically characterized exhibit strict XET activity. However, a notable exception is the nasturtium (Tropaeolum majus) xyloglucan-specific endo-(1,4)-β-D-glucanase (NGX1) enzyme, a xyloglucan endo-hydrolase (XEH) for which water rather than xyloglucan serves as an acceptor (for review, see Baumann et al., 2007). Therefore, enzyme family members are called xyloglucan endo-transglycosylases/hydrolases (XTHs) until their substrate preferences are known (Rose et al., 2002; Eklöf and Brumer, 2010). Plant genome sequencing has revealed that XTH families in plant species are large; 33 members have been identified in Arabidopsis (Arabidopsis thaliana; Yokoyama and Nishitani, 2001), 29 in rice (Oryza sativa; Yokoyama et al., 2004), and 41 in poplar (Populus spp.; Geisler-Lee et al., 2006). XEH activity is restricted to a small subfamily, IIIA, members of which have a short conserved sequence in the catalytic domain, while the other XTH members are thought to encode enzymes with XET activity (Baumann et al., 2007; Eklöf and Brumer, 2010). The XTH family members are subject to tissue-specific, environmental, and hormonal regulation (Yokoyama and Nishitani, 2001; Yokoyama et al., 2004; Becnel et al., 2006). However, despite extensive data on the expression of XET genes, their roles in plant growth and development are not clear. Heterologously expressed tomato (Solanum lycopersicum) pericarp XET has been shown to increase the creep of cellulose-xyloglucan composites (Chanliaud et al., 2004), and various XET isoforms reportedly induce either strengthening or loosening of heat-inactivated cell walls (Van Sandt et al., 2007; Maris et al., 2009). It has been suggested that individual XET isoforms may act as either wall-loosening or wall-strengthening agents, depending on the isoform, but no isoform-specific properties have been identified that could explain the differences in their effects (Herbers et al., 2001; Cho et al., 2006; Osato et al., 2006; Shin et al., 2006; Liu et al., 2007; for review, see Cosgrove, 2005). Moreover, conflicting effects of XTH gene expression on xyloglucan chain length have been observed in transgenic plants. For example, Herbers et al. (2001) found that down-regulation of NtXET1 in tobacco (Nicotiana tabacum) resulted in increases in xyloglucan size compared with the wild type, whereas Liu et al. (2007) found a positive correlation between the presence of AtXTH21 transcript and xyloglucan size in Arabidopsis. The reasons for the apparently varying effects of XET activities on xyloglucan size have not yet been elucidated.
The importance of XET activity for vascular tissue differentiation and/or function is indicated by the high expression of certain XTH genes (Antosiewicz et al., 1997; Herbers et al., 2001; Dimmer et al., 2004; Romo et al., 2005; Yun et al., 2005; Jiménez et al., 2006) and XET activity in these tissues (Bourquin et al., 2002). Interestingly, XET activity has been detected in developing xylem cells during secondary wall deposition in aspen (Populus spp.), implying that it plays a role in carbohydrate transglycosylation within and between different cell wall layers of xylem cells (Bourquin et al., 2002; Nishikubo et al., 2007; Baba et al., 2009). In addition, Matsui et al. (2005) showed that Arabidopsis XTH27 is required for the differentiation of xylem conduits in tertiary veins in the leaves and speculated that XTH degradation in differentiating tracheary elements might be important for vein differentiation.
To elucidate the roles of different XTH genes during wood formation, we identified XTH genes expressed in wood-forming tissues of hybrid aspen (Populus tremula × tremuloides) and determined their expression patterns during primary and secondary wall biosynthesis. To assess the role of XET activity in developing wood cells, we also ectopically overexpressed PtxtXET16-34, which has been previously shown to encode a XET (Kallas et al., 2005). We show that increased XET activity has differing effects on the xyloglucan content of expanding and maturing wood cells. We also show that XET effects on cell expansion differ between cell types, according to their xyloglucan epitope content, and propose a model that links XET effects on growth to the relative level of xyloglucan secreted into cell walls.
RESULTS
Identification of Poplar XTH Gene Family Members Expressed in Wood-Forming Tissues
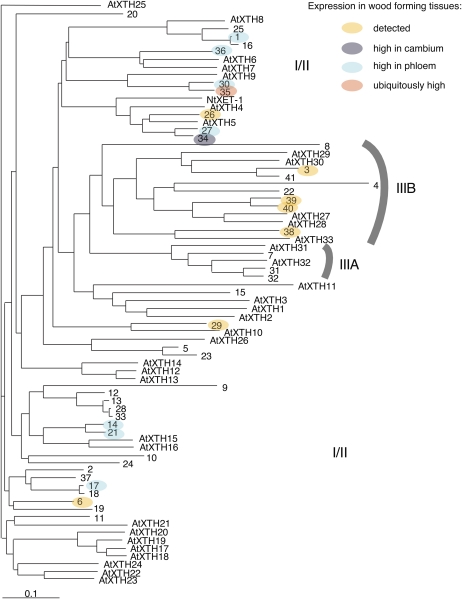
Geisler-Lee et al. (2006) identified 41 gene models of the GH16 XTH family in version 1.0 of the Populus trichocarpa genome. The updated list of these models is included in Supplemental Table S1. Among 22 Populus XTH genes in the Swedish Populus EST database, 10 (XTH3, -26, -27, -30, -34, -35, -36, -38, -39, and -40) were represented in libraries from developing wood/bark tissues, and transcripts of one (XTH6) were abundant in the dormant cambium library. We found five additional genes (XTH1, -14, -17, -21, and -29) to be expressed in the wood-forming tissues by cDNA cloning using degenerate primers corresponding to the highly conserved catalytic domain DEIDFEFLG of XETs. Thus, at least 16 XTH genes were found to be expressed in the wood-forming tissues (Fig. 1; Supplemental Table S1).
Figure 1.
Distribution of XTH genes expressed in the wood-forming tissues of Populus plants within the XTH gene family. The phylogenetic tree presents predicted protein sequences for the XTH family of P. trichocarpa, numbered according to Geisler-Lee et al. (2006), and the gene models used are listed in Supplemental Table S1. Arabidopsis XTH proteins, numbered according to Yokoyama and Nishitani (2001), and NtXET1 of tobacco (Herbers et al., 2001) are included for comparison. The subfamily IIIA represents putative XEH proteins (Baumann et al., 2007).
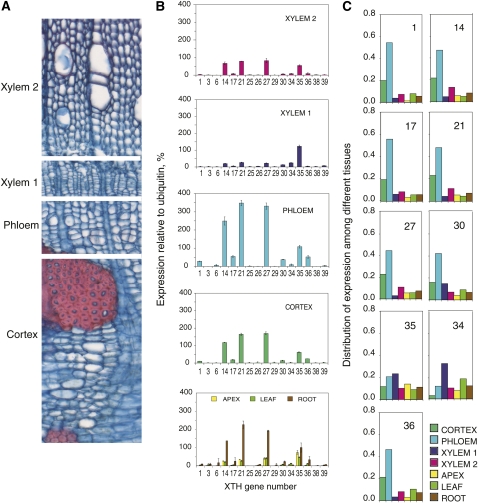
The expression profile of these genes in the stem was analyzed using the gene-specific macroarray for wood-expressed XTH genes (Nishikubo et al., 2007). Fractions enriched in cortex, phloem, cambial zone cells, and expanding primary-walled xylem elements (denoted xylem 1) and secondary wall-developing xylem (denoted xylem 2) were obtained by peeling the bark and carefully scraping the exposed surfaces of the bark and the wood. Phloem tissues showed the highest expression of XTH genes, with XTH14, -21, -27, and -35 showing the strongest signals relative to ubiquitin (Fig. 2). The same genes were also expressed in the cortex and secondary-walled xylem as well as other vegetative tissues. Thus, these genes tended to be ubiquitously expressed in vegetative tissues. The primary-walled xylem fraction showed a somewhat different expression pattern compared with other stem tissues. Here, XTH35 was the most abundant and XTH34 was significantly expressed. High expression of XTH14, -17, -21, -27, -34, and -35 in the wood-forming tissues was also evident from the frequency of cDNA clones detected in these tissues (Supplemental Table S1).
Figure 2.
Expression profiles of wood-expressed XTH genes in the Populus stem, determined with a gene-specific macroarray. A, Cross-section through a P. tremula stem showing the sampled tissue fractions. B, XTH gene signal intensity in relation to the ubiquitin signal in the indicated stem tissue fractions and other tissues for comparison. Values shown are means ± se; n = 3 technical replicates. The same expression patterns were observed in samples from two independent trees, and representative data are shown. C, Distribution of the expression of selected XTH genes among different vegetative tissues. For each gene, the sum of expression in all tissues is set to 1. Genes with very low expression levels (greater than 20% of ubiquitin expression) are not included. Xylem 1 indicates primary-walled developing xylem comprising the vascular cambium and radial expansion zone, and xylem 2 indicates secondary-walled developing xylem.
Comparison of the expression profile across different tissues (Fig. 2C) showed that the majority of XTH genes (XTH14, -17, -21, -27, -30, and -36) were most strongly expressed in the phloem, but transcripts of XTH35 were broadly distributed among all the examined tissues and XET16-34 was most strongly expressed in primary-walled xylem. No gene from the analyzed pool showed preferential expression in secondary-walled xylem, and no member of the IIIA subfamily of putative XEHs (Baumann et al., 2007; Eklöf and Brumer, 2010) was expressed in the wood-forming tissues (Fig. 1).
Overexpression of PtxtXET16-34 Affects Wood Cell Expansion
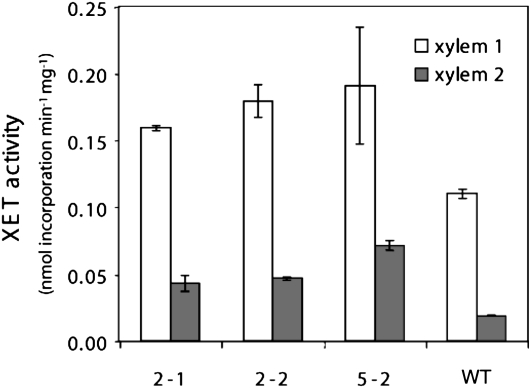
To investigate the role of XET activity during wood development, we overexpressed PtxtXET16-34 in aspen. Transgenic lines with PtxtXET16-34 cDNA controlled by the 35S promoter were generated, and three lines with 2- to 3-fold increased expression over wild-type levels, as determined by competitive reverse transcription (RT)-PCR (Supplemental Fig. S1), were selected for phenotyping. The presence of PtxtXET16-34 protein in the transgenic lines was confirmed by probing with anti-XET16A antibody (Bourquin et al., 2002), but no clear differences in signal intensities in transgenic lines compared with the wild type were observed by this technique (Supplemental Fig. S2). Therefore, to investigate if XET activity was effectively increased in these lines, the soluble proteins were extracted from developing xylem tissues and the incorporation of radiolabeled XGO into insoluble xyloglucan (Fry et al., 1992) was measured in the extracts. Increases in XET activity of 1.5- and 3-fold compared with wild-type levels were recorded in the primary- and secondary-walled developing xylem, respectively (Fig. 3), accompanied by positive correlations between XET activities and PtxtXET16-34 expression levels, with r2 values of 0.89 and 0.68, respectively. In addition, we measured XEH activity in PtxtXET16-34-overexpressing (OE) lines, since some members of the higher plant XTH family have both XET and XEH activities (Eklöf and Brumer, 2010). XEH activity was detected only in the secondary-walled xylem, where it appeared to be negatively correlated with PtxtXET16-34 expression (Supplemental Fig. S3), possibly because negative subsites (−1 to −4) of the PtxtXET16-34 active center have a high propensity to bind the reducing end of xyloglucan donors (Eklöf and Brumer, 2010), thus effectively reducing the number of reducing ends created by the background activity of unrelated xylogucanases. Hence, the PtxtXET16-34 gene product clearly exhibits XET rather than XEH activity, confirming conclusions from a previous study in Pichia pastoris (Kallas et al., 2005).
Figure 3.
XET activity in developing xylem of PtxtXET16-34-OE lines and wild type (WT) plants. XET activity was measured as incorporation of [1-3H]XLLGol into xyloglucan. Means ± ranges from duplicate analyses of pooled samples from 10 trees are shown. Xylem 1 indicates primary-walled developing xylem comprising vascular cambium and the radial expansion zone, and xylem 2 indicates secondary-walled developing xylem.
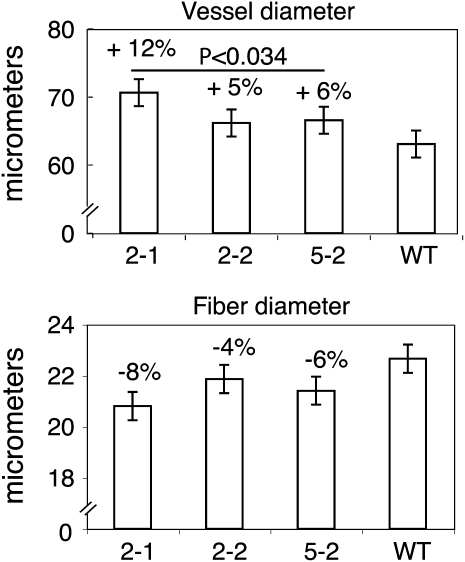
The overexpression of XET activity did not alter the morphology of the transgenic plants, except for a slight reduction of stem elongation (Supplemental Fig. S4). To evaluate the impact of higher XET activity on xylem cell expansion, wood macerates were prepared from the transgenic and wild-type lines, and the lengths and diameters of fibers and vessel elements were measured. Fiber diameter was not significantly affected by overexpression of PtxtXET16-34, although fibers tended to be narrower in all the transgenic lines than in wild-type counterparts. In contrast, vessel elements were significantly wider in the transgenic lines than in the wild-type controls (Fig. 4). Thus, the two cell types of the axial wood system, which have contrasting rates and types of growth (Mellerowicz et al., 2001; Mellerowicz, 2006; Siedlecka et al., 2008), showed different reactions to the increased XET activity. Fiber and vessel element lengths were not significantly affected (data not shown).
Figure 4.
PtxtXET16-34 overexpression stimulates vessel element, but not fiber, radial expansion. Line designation is as in Figure 3. Error bars represent least square mean se values calculated for the ANOVA model. The effect of genotype was significant only for dimensions of the vessel elements, for which the P values of the contrasts between the transgenic lines and the wild type (WT) are given above the bars.
Overexpression of PtxtXET16-34 Affects Xyloglucan Content
To explore the effects of XET on xyloglucans in cell walls, we examined xyloglucan contents in primary- and secondary-walled xylem. For this purpose, cell wall polysaccharides were isolated as starch-free ethanol-insoluble carbohydrates, fractionated into pectins (EDTA extracts), loosely bound hemicelluloses (4% KOH extracts), and tightly bound hemicelluloses (24% KOH extracts), and then the xyloglucan content in each of these fractions was determined by iodine staining, which detects polymeric xyloglucan in destarched cell wall preparations (Kooiman, 1960; Sulová et al., 1995). Xyloglucan was detected only in 4% and 24% KOH extracts. In primary-walled xylem, PtxtXET16-34 overexpression increased the tightly bound xyloglucan fraction (up to 3-fold) while slightly decreasing the loosely bound fraction (Table I). Thus, the proportion of tightly bound xyloglucan in the total xyloglucan increased from 68% in the wild type to approximately 90% in the OE lines, and their overall xyloglucan content increased. In contrast, in secondary-walled xylem, PtxtXET16-34 overexpression decreased the tightly bound xyloglucan content, which was the only form of xyloglucan found.
Table I. Xyloglucan content and molecular mass in transgenic PtxtXET16-34-OE lines and wild-type plants.
Xyloglucan was extracted from primary and secondary wall-developing xylem (xylem 1 and xylem 2, respectively) with both 4% KOH and 24% KOH. Sequential extractions were performed using tissues pooled from five to six individuals for each line with two (xylem 1) or three (xylem 2) technical replicates. The average xyloglucan content of each transgenic line was significantly different from that of the wild type based on ANOVA followed by t tests (P ≤ 0.05), as indicated by asterisks. Weight-averaged molecular mass [M(w)] was calculated for the main peak illustrated in Figure 5. ND, Not detected.
| Line | Xylem 1 |
Xylem 2 |
||||
| 4% KOH | 24% KOH | 24% KOH M(w) | 24% KOH in Total Xyloglucan | 4% KOH | 24% KOH | |
| mg g−1 dry wt | kD | % | mg g−1 dry wt | |||
| 2-1 | 1.13* | 12.63* | 126 | 91.8 | ND | 0.95* |
| 2-2 | 1.74* | 18.43* | 110 | 91.4 | ND | 1.09* |
| 5-2 | 1.50* | 8.58* | 152 | 85.1 | ND | 0.56* |
| Wild type | 2.91 | 6.18 | 148 | 68.0 | ND | 1.40 |
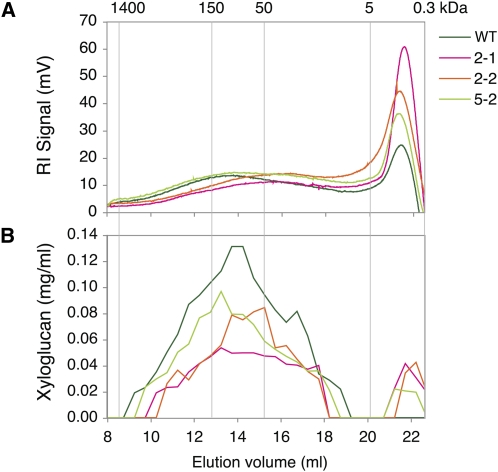
The 24% KOH hemicellulose fraction from primary-walled xylem was further analyzed by size exclusion chromatography. The elution profile of wild-type samples showed a main polydispersive peak of polysaccharides, with the most abundant size around 150 kD and a sharp peak of compounds of low molecular mass (Fig. 5A). The size distribution profile of samples from the OE lines showed a shift toward lower molecular mass within the main peak and higher contents of the low molecular mass compounds. The iodine staining of collected fractions indicated the presence of xyloglucan in the main peak, with estimated sizes between 150 and 50 kD in extracts from all the lines, and in the low molecular mass peak in extracts from the OE lines (Fig. 5B). The weight-averaged molecular mass of the main xyloglucan peak tended to be lower in the OE lines (Table I) and negatively correlated with the tightly bound xyloglucan content (r2 = 0.91). The low molecular mass iodine-stainable compound of OE lines that was absent in the wild-type extracts had an average molecular mass of approximately 2.7 kD, according to calibration with dextran standards and maltose. The nature of this compound is not clear, but it constituted between 6% and 15% of the total amount of iodine-stainable material extracted from the OE lines.
Figure 5.
Molecular size distribution of the hemicellulose fraction of polysaccharides from xylem 1 tissues of OE and wild-type (WT) lines (pooled samples from five to six trees per line) extracted with 24% KOH and analyzed by size exclusion chromatography. Polysaccharides were detected by monitoring the refractive index (RI) of the eluate (A), and xyloglucan concentrations in different fractions were determined by iodine staining (B). Line designation is as in Figure 3. The molecular masses of dextran standards in kD are shown above the graph.
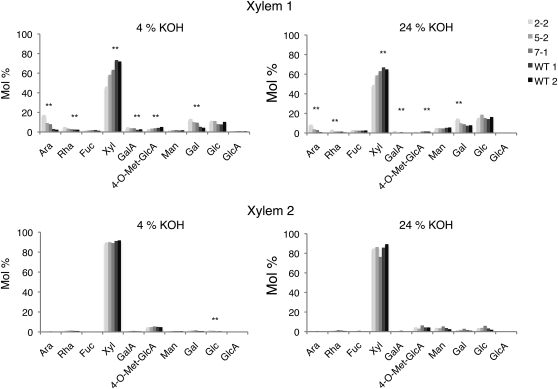
The monosaccharide composition of the loosely and tightly bound fractions was determined by methanolysis followed by trimethylsilyl derivatization and gas-liquid chromatography to elucidate the polysaccharides in these fractions and to assess the possibility that another compound could be present in the hemicellulose fractions, such as glucomannan, that could potentially affect the iodine staining. The dominant sugar was found to be Xyl, indicating the presence of glucuronoxylan in all samples, while other typical xyloglucan monosaccharides were extracted primarily from the primary-walled xylem by 24% KOH (Fig. 6). These findings are consistent with xyloglucan being responsible for the iodine staining (Table I). Assuming ratios of Glc:Xyl:Gal:Fuc of 50:29:12:7 for poplar xyloglucan (Hayashi and Takeda, 1994), at most approximately 10 mol % of Xyl in the 24% KOH hemicellulose fraction extracted from the primary-walled xylem could be attributable to xyloglucan. Ara, Rha, and GalUA were present as minor compounds that were more abundant in the primary-walled than in secondary-walled xylem, where they were detected in both hemicellulose fractions, suggesting that some pectin coextracted with the hemicelluloses. Man, indicative of glucomannan, was a minor component in the 24% and 4% KOH fractions of both primary- and secondary-walled xylem.
Figure 6.
Molecular composition of hemicellulose fractions extracted from primary- and secondary-walled xylem tissues (denoted xylem 1 and 2, respectively) of OE and wild-type lines. The analyzed lines include two OE lines (2-2 and 5-2), a transgenic line with the same construct but having a wild-type level of PtxtXE16-34 expression (7-1; compare with Supplemental Fig. S1), and two independent samples of the wild type (WT 1 and WT 2). Data represent averages obtained from two technical replicates of pooled samples from three to six trees per line, sequentially extracted with EDTA, followed by 4% KOH and 24% KOH, and analyzed by methanolysis followed by trimethylsilyl derivatization and gas-liquid chromatography. Line designation is as in Supplemental Figure S1. Asterisks indicate significant differences between the transgenic lines and the wild type according to the ANOVA test with a contrast (P ≤ 0.05).
The molecular composition of hemicelluloses extracted from the primary-walled xylem was affected by PtxtXET16-34 overexpression, whereas it was not much altered by the transgene in the secondary-walled xylem (Fig. 6). For example, PtxtXET16-34 overexpression in the primary-walled xylem significantly reduced Xyl and 4-O-methyl-GlcUA contents in both hemicellulose fractions. Considering the Glc and Gal contents in these samples, these reductions indicate a decrease in glucuronoxylan rather than xyloglucan. By contrast, Ara, Rha, GalUA, and Gal contents increased in the loosely bound fraction and (to a lesser degree) in the tightly bound fraction. These changes most likely reflect increases in pectins coextracted with hemicelluloses from the transgenic lines, and the increase in Gal in the tightly bound fraction could also reflect an increase in xyloglucan, as indicated by the iodine staining (Table I). No significant change in Man content was found in any fraction, expressed as either mol % (Fig. 6) or per unit cell wall weight (data not shown), excluding the possibility that glucomannan significantly contributed to the variability in iodine staining seen in the OE lines (Table I).
Overexpression of PtxtXET16-34 Increases the CCRC-M1 Labeling of Xylem Cells in Early Developmental Stages
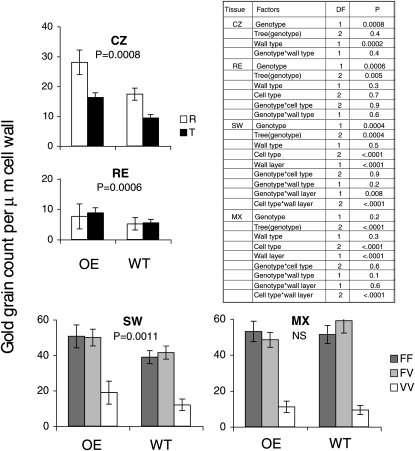
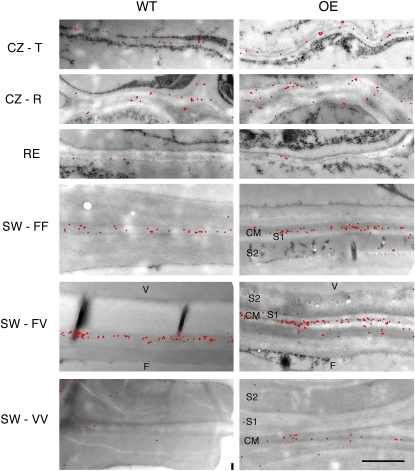
To study the effects of PtxtXET16-34 overexpression on xyloglucan distribution in developing fibers and vessel elements, ultrathin sections of cambial tissues of two OE lines and wild-type plants were analyzed by xyloglucan immunolabeling using the monoclonal antibody CCRC-M1. This antibody detects terminal Fuc-linked α-(1→2) to a galactosyl residue, which is primarily found in xyloglucan (Puhlmann et al., 1994). Bourquin et al. (2002) observed that its label density and distribution were strongly influenced by wood cell developmental stage. Therefore, the effects of PtxtXET16-34 overexpression were analyzed separately for each stage, and factors potentially affecting the labeling were considered. In the cambial zone (meristematic stage), the label was approximately two times higher in sections of OE lines than in wild-type sections, and in both cases the label was higher in radial walls than in tangential walls (Fig. 7), because the radial walls were much thicker (Fig. 8). During radial expansion, the label remained significantly higher in OE sections compared with wild-type counterparts, but the difference was smaller than in the previous stage (Fig. 7). In sections of both OE and wild-type lines, the label was substantially weaker in the radial expansion zone compared with the cambial zone, especially in radial walls, reflecting the radial wall thinning during cell expansion (Fig. 8). During the early stage of secondary wall formation, labeling dramatically increased in the walls between adjacent fibers (fiber-fiber walls) and between fiber and vessel elements (fiber-vessel walls) compared with the earlier stages (Fig. 7) and was concentrated in the primary wall layer (Fig. 8). In the walls between vessel elements (vessel-vessel walls), the label was much less abundant and distributed more evenly between the primary and secondary cell wall layers. Significantly higher labeling was still observed in sections of transgenic lines compared with the wild type, but the difference was even less pronounced than in the radial expansion stage. At a more advanced developmental stage, when secondary wall deposition was almost complete in fibers, no apparent effects of XET overexpression were observed, but fiber-fiber and fiber-vessel walls contained five to six times more label than vessel-vessel walls in both genotypes. Thus, the effects of PtxtXET16-34 overexpression on the abundance of accessible CCRC-M1 epitopes were developmental stage dependent; they were strongest during the primary wall stage of xylem cell development (meristematic and radial cell expansion stages), weaker during early secondary wall development, and undetectable in more mature xylem. However, no significant differences in trends were detected between fibers and vessel elements, radial and tangential walls, or primary and secondary wall layers, since no significant “genotype × cell type,” “genotype × wall type,” and “genotype × wall layer” interactions were detected (Fig. 7).
Figure 7.
Semiquantitative transmission electron microscopy analysis of xyloglucan labeling in situ with the monoclonal antibody CCRC-M1. Ultrathin sections of the wood-forming tissues of two trees representing PtxtXET16-34-OE lines (2-1 and 5-2) and two wild-type (WT) trees were probed with CCRC-M1 antibody, and the gold particles were counted along 3 μm of the common cell wall between pairs of adjacent cells in the cambial zone (CZ), radial expansion zone (RE), early secondary wall formation zone (SW), and nearly mature xylem (MX). For each cell type, both radial (R) and tangential (T) walls were examined. In RE, SW, and MX zones, labeling in primary and secondary wall layers of common fiber-fiber (FF), fiber-vessel (FV), and vessel-vessel (VV) walls was separately scored. P values indicate the significance of the genotype (wild-type versus transgenic) on the measured variables according to the ANOVA; NS indicates nonsignificant. Significances of other factors analyzed are given in the ANOVA table. DF, Degrees of freedom.
Figure 8.
Distribution of CCRC-M1-labeled xyloglucan (red dots) in cell walls along the wood development gradient in the wild type (WT) and transgenic PtxtXET16-34-OE lines for which quantitative labeling data are presented in Figure 7. CM, Compound middle lamella; F, fiber; S1 and S2, successive secondary wall layers; V, vessel element. Other labels are as in Figure 7. Except for the cambial zone, all presented walls are radial. Bar = 1 μm.
DISCUSSION
Several XTH Genes Are Involved in the Development of Secondary Vascular Tissues
In this paper, we show that the high XET activity detected in the wood-forming tissues of aspen (Bourquin et al., 2002) is due to the expression of at least 16 genes identified by ESTs and cloning (Supplemental Table S1; Geisler-Lee et al., 2006). All these genes probably encode proteins with XET rather than XEH activity, according to the sequence analysis (Baumann et al., 2007; Eklöf and Brumer, 2010). The survey revealed that, surprisingly, no set of XTH genes appears to be specific to the developing woody tissues, since the same XTH genes (PtxtXTH14, -21, -27, and -35) were highly expressed in most nongrowing tissues (Fig. 2). Similarly, in Arabidopsis, few XTH genes are both highly and ubiquitously expressed (Yokoyama and Nishitani, 2001), and several XTH genes are coexpressed in the same tissue or even the same cell (Becnel et al., 2006; http://atted.jp/). Some of them also have very similar sequence and expression patterns and may have overlapping functions (Osato et al., 2006). However, the abundant transcripts in developing wood tissues of aspen clearly represent different clades (Fig. 1) and therefore may have specialized functions.
Remarkably, we found that most XTH genes expressed in the cambial region are also strongly expressed in the developing phloem (Fig. 2). Phloem sieve cells develop specialized nacreous walls that have been found to strongly react with CCRC-M1 and XET16A antibodies in aspen, but (surprisingly) they do not incorporate fluorescently labeled XGOs (Bourquin et al., 2002). This suggests that xyloglucan metabolism is fundamentally different in the sieve tube cells than in other cell types, an implication that warrants closer investigation.
Developing primary-walled xylem, comprising meristematic and expanding cells, was found to contain a distinct set of XTH transcripts compared with those found in the adjacent nongrowing tissues. In particular, PtxtXTH35 was highly expressed, mimicking the pattern seen in the shoot apical meristem (Fig. 2). Since PtxtXTH35 is orthologous to AtXTH9, a highly expressed gene in Arabidopsis root and shoot meristems (Yokoyama and Nishitani, 2001; Becnel et al., 2006), we speculate that it has a specific function that is required in all meristematic and expanding tissues. By contrast, PtxtXET16-34 seems to be specific to the cambial meristem and developing phloem and xylem and to be expressed in both primary- and secondary-walled xylem cells (Fig. 2C; Bourquin et al., 2002). Close homologs of this gene in other plant species, including VaXTH1 and VaXTH2 in adzuki bean (Vigna angularis; Nakamura et al., 2003), CaXTH1 in chickpea (Cicer arietinum; Jiménez et al., 2006), NtXET1 in tobacco (Herbers et al., 2001), and BvXTH1 and BvXTH2 in sugar beet (Beta vulgaris; Dimmer et al., 2004), also show expression in nongrowing vascular tissues.
Role of XET Activity in Xyloglucan Rearrangements in Primary- and Secondary-Walled Xylem
Xyloglucan is reportedly extensively modified after deposition in the cell wall. Observed changes include increases in size within 30 min of deposition (Talbott and Ray, 1992; Thompson and Fry, 1997; Kerr and Fry, 2003; Popper and Fry, 2008), depolymerization and increases in relative amounts during cell expansion (Thompson and Fry, 1997; Pauly et al., 2001; Tokumoto et al., 2002), and increases in size accompanied by reductions in relative amounts during cell maturation (Herbers et al., 2001). We detected several xyloglucan rearrangements that occur during the development of wood cells. During the transition from the primary- to the secondary-walled stage of wood cells, all xyloglucan becomes tightly bound to cellulose microfibrils as a result of either removal of loosely bound xyloglucan or its incorporation into the tightly bound network (Table I). Another developmental change is that the density of the fucosylated epitope in the primary cell wall layers dramatically increases during secondary wall deposition (Figs. 7 and 8; Bourquin et al., 2002). This could be caused by unmasking of the epitope, for example by removal of the masking pectins (Marcus et al., 2008), the deposition of new xyloglucan in the primary wall layers during this stage, or both.
XET is thought to be a major player in these developmental xyloglucan rearrangements, but previous studies of plants with modified XET activity have provided limited (Herbers et al., 2001; Genovesi et al., 2008; Miedes et al., 2010) or conflicting (Liu et al., 2007) indications of the effects of XET on xyloglucan. Here, we have analyzed the effects of XET overexpression on xyloglucan content and size in developing wood. Using two complementary approaches (iodine staining combined with sugar analysis of sequentially extracted hemicelluloses, and the reaction of tissue sections with CCRC-M1 antibodies), we found that in primary-walled xylem, XET activity promotes xyloglucan incorporation into the cell wall and the tightly bound hemicellulose fraction, with a concomitant reduction in xyloglucan size (Figs. 5–8; Table I). Moreover, the monosaccharide composition of hemicellulose fractions suggests that XET activity also increases the content of rhamnogalacturonan I-type pectin in these fractions. These observations provide experimental support for the hypothesis that in cells synthesizing xyloglucan, XET activity incorporates nascent xyloglucan linked to rhamnogalacturonan I-type pectin in the xyloglucan network of cell walls by polymer-oligomer transglycosylation, leading to a net decrease in cell wall xyloglucan chain length (Talbott and Ray, 1992; Thompson and Fry, 1997; Thompson et al., 1997; Herbers et al., 2001; Kerr and Fry, 2003; Popper and Fry, 2008).
In cells that do not synthesize xyloglucan, XET activity may only lead to xyloglucan rearrangement. In a closed system, this would increase the polydispersivity of xyloglucan without affecting its content or average Mr. However, a plant cell wall constitutes an open system, since matrix components are lost from cells to the surrounding medium (Kerr and Fry, 2003; Popper and Fry, 2008) or internalized by endocytosis (Baluška et al., 2005). Therefore, in xylem cells forming secondary walls, which probably have relatively low xyloglucan biosynthetic activity (Mellerowicz and Sundberg, 2008), the overexpression of XET activity would be expected to remove some xyloglucan from the tightly bound fraction, as observed (Table I), and to increase its average Mr, as recently reported in ripening fruits (Miedes et al., 2010).
XET Activity Stimulates Cell Expansion in Vessel Elements But Not in Fibers
In the presence of XGOs, XET is thought to be a major player in the depolymerization of xyloglucan (Chanliaud et al., 2004), which is traditionally believed to be a key process in cell expansion (Nishitani and Masuda, 1981; Hayashi, 1989; Talbott and Ray, 1992). Indications that XET stimulates cell expansion have been found in studies of Atxth21 mutants (Liu et al., 2007), rice OsXTH8 RNA interference lines (Jan et al., 2004), and Arabidopsis lines expressing BcXTH1 (Shin et al., 2006). However, Maris et al. (2009) recently reported that exogenous applications of AtXTH14 and AtXTH26 rapidly inhibit root and root hair elongation, accompanied by abnormal broadening of root hair bases. Similar broadening of root hair bases can also be caused by xyloglucan deficiency (Cavalier et al., 2008) and a loss of functionality of AtXTH21 (Liu et al., 2007). Thus, the functions of XET appear to be more complex than simple catalysis of depolymerization leading to cell expansion. Accordingly, the only marked effect of increasing XET activity we observed on xylem cell expansion in our OE lines was enhanced radial expansion of vessel elements (Fig. 4).
It has been argued that effects of XET on cell expansion may depend on the length of secreted xyloglucan chains, because the incorporation of labeled XGOs in cell walls accelerates growth while incorporation of labeled long-chain xyloglucan inhibits cell expansion (Takeda et al., 2002). Indeed, the size and composition of xyloglucan residing in cell walls can vary substantially as a result of transglycosylation, endohydrolysis, and trimming by wall-residing XETs, α-xylosidases and β-glucosidases, and endoglucanases, respectively (Kaida et al., 2010; Sampedro et al., 2010). However, there have been no demonstrations that different plant tissues synthesize and secrete xyloglucan chains of differing length. An alternative possibility is that the effects of XET on cell expansion could depend on the relative amounts of XET and xyloglucan secreted into the cell wall. The nascent xyloglucan forms a pool of binding/cutting sites that compete with xyloglucan tethers in the cell wall for XET. Thus, when nascent xyloglucan is abundant, most nascent XET may cleave it and enter the cell wall in covalent xyloglucan-XET complexes. Such covalently bound XET may only transglycosylate available nonreducing ends of wall-attached xyloglucan, thus augmenting and strengthening the wall xyloglucan network. However, when more XET is synthesized than xyloglucan, the surplus XET may enter the cell wall in a free form that is capable of cleaving the load-bearing xyloglucan tethers. This, in turn, may weaken the cell wall, favoring expansion. Hence, the effects of XET on cell expansion may depend on rates of xyloglucan biosynthesis and secretion into the cell wall and the relative amounts of xyloglucan and XET, which may differ between cell types, being low in vessel elements and high in fibers, as suggested by the CCRC-M1 labeling (Figs. 7 and 8).
Putative Roles of XTH Genes in Nongrowing Xylem Cells
The abundance of CCRC-M1 antibody epitopes is very high in the fibers and substantially increases (in the primary cell wall layer) during cell differentiation (Figs. 7 and 8; Bourquin et al., 2002). An important role of fibers, particularly in tension wood, is the generation of maturation (growth) stresses (Plomion et al., 2001). There are emerging indications that XET may play important roles in the development of these stresses (Nishikubo et al., 2007; Mellerowicz et al., 2008) and that the xyloglucan network is essential for bringing inclined woody stems to the upright position (Baba et al., 2009). Maturation stress develops gradually in the xylem of both upright and inclined trees and probably involves polymer rearrangements that are poorly understood in secondary walls. We explored some of the rearrangements of the xyloglucan network and found that during xylem cell development, loosely bound xyloglucan disappears and all xyloglucan becomes tightly bound (Table I). Moreover, we have demonstrated that XET activity decreases the loosely bound xyloglucan fraction while increasing the tightly bound fraction. These findings are consistent with the observation that XET generates xyloglucan fragments that rapidly become inaccessible to enzymes (Vissenberg et al., 2005). In the gelatinous layers of tension wood fibers, the newly deposited xyloglucan also rapidly becomes inaccessible to antibodies and XET (Nishikubo et al., 2007; Baba et al., 2009). Hence, we hypothesize that in maturing xylem, XET trims xyloglucan to a size that can be accommodated between coalescing cellulose microfibrils, thereby contributing to the generation of tensile stress in cellulose macrofibrils (Mellerowicz et al., 2008).
In addition, it has been proposed that xyloglucan tethers different cell wall layers in wood fibers and that XET constantly repairs the tethers (Bourquin et al., 2002; Mellerowicz et al., 2008; Baba et al., 2009). XET activity has been shown to be maintained for years in dead gelatinous fibers (Nishikubo et al., 2007) and may actively repair cross-links when they (presumably) break due to uneven shrinking of different layers in mature xylem (Mellerowicz et al., 2008). Such prolonged activity would also lead to the release of xyloglucan oligosaccharins that could be distributed via apoplast routes to growing parts and signal mechanical stress in the plant. These proposed functions in secondary-walled xylem cells might be similar to XET functions in nongrowing primary-walled cells in other tissues. Thus, in nongrowing cells, XET might participate in the generation of tensile stress in cellulose macrofibrils, by trimming xyloglucan; provide a means to withstand the generated tensile stress by xyloglucan transglycosylation between cell wall layers; and generate signals of mechanical stress in the plant. These functions may be important for the wall strengthening required to accommodate mechanical challenges during growth and in thigmomorphogenesis.
CONCLUSION AND PERSPECTIVE
We have shown that wood development in aspen involves extensive xyloglucan rearrangements that appear to be mediated by several generally expressed XTH genes, most likely encoding XET enzymes, and at least one (PtxtXET16-34) that is up-regulated during this process. We have investigated the effects of overexpression of this gene, but effects of XET isoforms encoded by other genes might be slightly different due to differences in their catalytic parameters and binding specificities. However, our study clearly shows that the same isoform can have varying effects on xyloglucan contents and cell expansion, depending on the developmental context, indicating that further factors modulate their effects. We propose that one such factor may be the abundance of newly deposited xyloglucan, which may effectively compete with xyloglucan load-bearing tethers for the nascent XET, leading to contrasting effects on the mechanical properties of the growing wall and supramolecular cell wall assembly. These hypotheses should be further tested using simpler models, for example, cellulose-xyloglucan composites (Chanliaud et al., 2004). Intriguingly, XET activity appeared to affect not only cell wall xyloglucan but also pectins, supporting the hypothesis that pectin-xyloglucan complexes are formed in the Golgi apparatus (Popper and Fry, 2008). Finally, a role for XET activity in mature nongrowing wood cells is emerging (Nishikubo et al., 2007; Mellerowicz et al., 2008), and we provide some insights regarding the nature of xyloglucan rearrangements mediated by XET in these cells. Further exploration of the roles of XET and other cell wall enzymes during the development and adjustment of the fine structure and micromechanics of maturing walls is required for thorough elucidation of the processes involved in growth stress development, which is highly important for the mechanical design of plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Hybrid aspen (Populus tremula × tremuloides clone T89) plants were propagated in vitro on a complete Murashige and Skoog medium in 0.9% agar (Sigma) under an 18-h photoperiod with 80 μmol m−2 s−1 flux density provided by Philips Master TLD 58W lamps under a 22°C/18°C day/night temperature regime. Rooted cuttings were transferred to soil and grown initially in a climate chamber providing 20°C, 16-h/16°C, 8-h day/night cycles, illuminated by Osram HQI-TS 400-W lamps giving a minimum quantum flux density of 150 μmol m−2 s−1 during the light periods. After approximately 2 weeks of adjustment to soil, cuttings were transferred to the greenhouse, with 22°C/18°C day/night temperatures, under natural daylight supplemented with light from high-pressure sodium lamps when the outdoor photoperiod was shorter than 18 h. Plants were watered daily and fertilized once a week with a SuperbraS nutrient solution (Supra Hydro).
For macroarray analysis, stem segments were collected from two field-grown aspen trees (P. tremula) growing in a natural stand near Umea, Sweden (63°50′N, 20°20′E) in June 2002, immediately frozen in liquid nitrogen, and stored at −70°C. Cambial region tissues were collected by peeling the bark and scraping the exposed surfaces with a scalpel. The fraction scraped from the xylem side (denoted xylem 2) consisted mostly of xylem elements that were depositing secondary wall, lignifying, and undergoing programmed cell death (Gray-Mitsumune et al., 2004), while the fraction gently scraped from the bark side (denoted xylem 1) consisted of expanding primary-walled xylem elements and cambial zone cells. The tissue obtained by the second scraping of the bark (denoted phloem) consisted of differentiating and mature phloem and phloem fibers. The remaining tissue (denoted cortex) consisted of primary phloem, primary phloem fibers, cortex, and periderm. The other tissues were obtained from greenhouse-grown hybrid aspen plants as described by Takahashi et al. (2009).
Cloning of PtxtXTH cDNAs
Thirteen wood-related XTH cDNAs were identified among entries in the Swedish Populus EST database (http://www.populus.db.umu.se/). Sequence tags were also obtained by cloning and sequencing expressed XTH genes, using whole poplar stem RNA as a template with the degenerate primer XET-F1 (5′-GAYGARATHGAYTTYGAGTTYYTDGGDAA-3′) and dt18XKB (5′-CTGATCTAGAGGTACCGGATCCTTTTTTTTTTTTTTTTTT-3′) for conserved regions in the XTH gene family. Populus trichocarpa sequence information was retrieved by searching public databases with the BLAST algorithm at the Joint Genome Institute (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). Multiple sequence alignments were constructed using ClustalW (http://www.clustal.org/), and a dendrogram was generated using ClustalW and TreeViewPPC software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html), based on the predicted poplar and Arabidopsis (Arabidopsis thaliana) XTH amino acid sequences.
Macroarray Analysis of XTH Gene Expression
For this analysis, the gene-specific macroarray for wood-expressed XTH genes constructed by Nishikubo et al. (2007) was used. In addition to wood-related XTH genes, this array contains three loading controls that are constitutively expressed in poplar, according to results of microarray studies (http://www.upscbase.db.umu.se/).
mRNA was extracted directly from powdered frozen tissue using paramagnetic oligo(dT)25 beads (Dynal) according to the manufacturer’s instructions, and 32P-labeled cDNAs were generated by RT-PCR using a mixture of primers specific for each target XTH gene and control gene. Samples from two aspen trees were used in this analysis, and the hybridizations were repeated three times.
Generation of PtxtXET16-34-OE Lines
The coding region of PtxtXET16-34 (AF515607) was cloned in the sense orientation in the pPCV702kana vector between the cauliflower mosaic virus 35S promoter and the nopaline synthase terminator, and hybrid aspen was transformed by Agrobacterium tumefaciens infiltration, as described previously (Gray-Mitsumune et al., 2008). Ten independent lines carrying the sense PtxtXET16-34 construct were recovered, multiplied in vitro, and rooted. Wild-type plants (clone T89) were multiplied and rooted in the same way. Three lines with reproducible high expression were randomly selected for detailed characterization.
Maceration of Xylem
Debarked stem segments from internode 45 of five to six plants representing each of the transgenic lines and wild-type controls, approximately 2 to 3 mm in diameter and 10 mm long, were macerated in 3% hydrogen peroxide and 50% glacial acetate acid at 95°C for 8 h (Gray-Mitsumune et al., 2008). The resulting solutions were decanted, and the samples were washed three times in water, resuspended in 3 mL of water, and sodium bicarbonate was slowly added until the pH was neutralized. The samples were washed three times in water and resuspended in 2 mL of 70% ethanol. The samples were vigorously shaken to release individual cells, which were examined under differential interference contrast using a Zeiss Axioplan microscope (Carl Zeiss), and their lengths and widths were measured from images captured by an AxioVision camera using the image-analysis program supplied by AxioVision. One hundred fibers and 50 vessel elements from each tree were measured, and the effects of genotype (OE lines versus the wild type) on the dimensions of the measured cells were assessed by ANOVA using the following mixed model, with tree (within-genotype) as a random effect:
Least square mean se values were calculated for the ANOVA model to represent the variability in the data. For variables significantly affected by the factor genotype (P < 0.10), the contrasts between the transgenic lines and the wild type were tested, and their significance is presented in Figure 4. All statistics were calculated using JMP software, version 8.0.1 (SAS).
XET Activity Assay
Xylem 1 and xylem 2 fractions from internodes 40 to 50 from five or six trees representing each line were scraped into liquid nitrogen and ground. To 100 mg of each tissue powder, 0.2 mL of ice-cold extraction buffer (350 mm sodium citrate, 10 mm CaCl2, pH 5.5) was added. The samples were kept on ice for 15 min and then centrifuged for 15 min at 4°C in a table centrifuge at maximum speed. The supernatants were collected and recentrifuged as above and then kept at 4°C prior to analysis. The total protein content of each sample was between 1.1 and 2.3 mg mL−1, as determined by the Bradford assay. The XET activity in the samples was assayed by measuring their ability to catalyze the incorporation of radioactive xylooligosaccharides into xyloglucan (Fry et al., 1992), as described by Henriksson et al. (2003). Briefly, 10 mL of each extracted enzyme solution was added to a mixture of 10 mL of a stock solution of 2.2 mm [1-3H]XLLGol (reduced xyloglucan-derived nonasaccharide) with a specific radioactivity of 3.7 TBq mol−1 in 100 mm sodium citrate (pH 5.5) and 10 mL of xyloglucan solution (3.0 mg mL−1 in sodium citrate buffer, 100 mm, pH 5.5). Following incubation at 25°C for 1 h, the reaction was stopped by adding 50% (v/v) formic acid in water (20 mL). The incorporated radioactivity was measured in a Packard Tri-Carb 1500 scintillation counter, and the obtained values (cpm) were converted into molar activity values. The activity in duplicate samples of each line was measured, and their average specific activity was calculated and expressed in nmol XLLGol incorporated min−1 mg−1 total protein.
Fractionation of Cell Wall Polysaccharides
Xylem 1 and xylem 2 tissues were collected from five to six trees of each genotype, including wild-type and three OE lines (2-1, 2-2, 5-2), ground in liquid nitrogen, and freeze dried. Equal weights of dry powders from all trees of the same genotype were pooled, then each pool was split into two (xylem 1) or three (xylem 2) technical replicates for fractionation by the method of Tokumoto et al. (2002). Briefly, the pooled samples were boiled in methanol at 70°C for 5 min to extract the soluble sugars, followed by rehydration in ice-cold water and homogenization using an ice-cold mortar and pestle. The homogenates were then extracted with water, acetone, and a methanol:chloroform mixture (1:1, v/v) and treated with porcine pancreas α-amylase (29 units g−1; A-6255; Sigma) in 100 mm Tris-HCl buffer (pH 7.0) at 37°C for 3 h. Following the amylase treatment, the reaction mixtures were centrifuged, as above, and bovine pancreas chymotrypsin (418 μL g−1; sequencing grade; Roche Diagnostics) in 10 mm CaCl2 and 100 mm Tris-HCl buffer (pH 7.8) was added to the pellets after thorough washing with water. After centrifugation, removal of the supernatant, and a further wash with water, the destarched, deproteinized residues were ready for fractionation.
Polysaccharides were fractionated into four fractions, consisting of pectin and two hemicellulose fractions, as follows. Pectins were extracted by incubating the destarched, deproteinized alcohol-insoluble residues in three portions of 50 mm EDTA (pH 6.8) at 100°C for 15 min and combining the extracts. A hemicellulose fraction denoted the 4% KOH extract was then extracted with three portions of 4% (w/v) KOH at room temperature on a rocker platform (first for 2 h, then overnight, and finally for 2 min) and then pooling these three extracts. Hemicellulosic material in the remaining solid residue was extracted with three portions of a solution containing 24% (w/v) KOH and 0.02% (w/v) NaBH4 in a similar manner, and the resulting (pooled) extract was designated the 24% KOH extract. The 4% KOH and 24% KOH extracts were neutralized with concentrated acetic acid on ice and dialyzed in dialysis tubing (Spectra/Por6 1000 MWCO; Spectrum) against water until the conductivity was less than 1 mS. The dialyzed material was freeze dried and either dissolved again in water, with vigorous stirring, at 100°C for approximately 10 min before xyloglucan assay and size exclusion chromatography or used for monosaccharide composition analysis.
Determination of Xyloglucan Content
Xyloglucan was assayed in all polysaccharide fractions by iodine staining (Kooiman, 1960) as follows. A 250-μL portion of each sample was mixed with 125 μL of a 20 mm iodine:60 mm potassium iodide solution and 1,250 μL of a sodium sulfate solution (1.4 m), then after incubation for 1 h at room temperature, the absorbance was determined at 640 nm. Xyloglucan contents of the samples were then calculated and expressed in mg g−1 freeze-dried tissue using tamarind (Tamarindus indica) xyloglucan (Megazyme) as a concentration standard. The detection limit of these assays was 0.02 mg xyloglucan mL−1.
The effects of genotype on xyloglucan contents in each hemicellulose fraction were analyzed with ANOVA by applying the model Y = μ + genotype + error, followed by a t test for the preplanned comparisons between the transgenic and wild-type lines, setting the probability threshold for significance at P ≤ 0.05. JMP software version 8.0.1 (SAS) was used to calculate the statistics.
Size Exclusion Chromatography
Freeze-dried 4% KOH and 24% KOH fractions were dissolved in 0.5 m NaOH to a concentration of 2.5 mg mL−1 with vigorous stirring at 100°C for 10 min. Samples (0.5 mL) were filtered (using 0.2-μm GHP Acrodiscs) and injected onto a Superose 6 10/300 column (GE Healthcare) connected to a FPLC system (GE Healthcare) with a refractive index detection system (Erma). The analytes were then eluted using 0.5 m NaOH as a mobile phase at a flow rate of 0.5 mL min−1, 0.5-mL fractions were collected, and 0.25-mL portions were assayed for xyloglucan. Dextran molecular mass standards (Fluka Chemie) and maltose (molecular mass 342 kD; Sigma) were also applied and eluted as calibration standards. The weight-averaged molecular mass of xyloglucan was calculated according to Lundqvist et al. (2003), and the reproducibility of the resulting patterns was assessed by analyzing several technical replicates.
Monosaccharide Composition of Hemicellulose Fractions
The neutral and acidic sugars of the 4% KOH and 24% KOH hemicellulose fractions were quantified by methanolysis followed by trimethylsilyl derivatization and gas-liquid chromatography analysis (Sweeley et al., 1963). Approximately 500 μg of the freeze-dried hemicelluloses was methanolyzed with 2 m HCl in dry methanol at 85°C for 24 h. The released sugars were washed with methanol and derivatized with Tri-sil reagent (hexamethyldisilazane + trimethylchlorosilane + pyridine 3:1:9 [Sylon HTP; Supelco]) at 80°C for 20 min. The derivatized sugars were analyzed by gas-liquid chromatography using an Agilent Technologies 7890A gas chromatography system equipped with a J&W (Agilent Technologies) DB-5 capillary column (30 m × 0.25 mm i.d., 0.25-μm film thickness) with the following temperature program: 80°C for 2 min, followed by a linear 20°C min−1 gradient to 140°C, held for 2 min, then further gradients of 2°C to 200°C and 30°C to 250°C, which was held for 5 min. The sugars were detected using a flame ionization detector. The monosaccharide concentration in each sample were calculated from standard curves for each of the analyzed sugars except 4-O-Me-GlcUA (for which the GlcUA standard curve was used). Pooled samples, with technical replicates from three to six trees, were used in these analyses, and two independent wild-type samples were analyzed to evaluate the biological variation.
Data were analyzed by ANOVA using the model Y = μ + genotype + error, followed by the contrast between the OE transgenic lines and the lines with wild-type PtxtXET16-34 level, calculated for consistently different variables, setting the probability threshold for significance at P ≤ 0.05. JMP software version 8.0.1 (SAS) was used to calculate the statistics.
Xyloglucan Immunolocalization
The location of xyloglucan in the aspen stem was visualized by transmission electron microscopy using the monoclonal antibody CCRC-M1 (Puhlmann et al., 1994), which recognizes terminal Fuc-linked α-(1→2) to a galactosyl residue of xyloglucan. For this purpose, small blocks of tissue (height: 5 mm; tangential width: 2 mm; radial width: 3 mm), including cambium and developing xylem and phloem, were taken from the 21st internode of two wild-type trees and two PtxtXET16-34-OE trees, one from line 2-1 and the other from line 5-2, and fixed in 4% paraformaldehyde and 0.05% glutaraldehyde (Analytical Standards) in 25 mm phosphate buffer, pH 7.2. The blocks were dehydrated in a graded ethanol series (30%, 50%, 70%, 80%, 90%, 95%, and 3× 100%), each for 10 min, and embedded in LR White resin (TAAB). Transverse sections (80–100 nm thick) were cut with an ultramicrotome (MT-7000; RMC) and transferred to 200-mesh nickel grids (TAAB) covered with formvar. Sections were blocked with blocking solution (0.5% skim milk in phosphate-buffered saline buffer) for 20 min and then incubated with CCRC-M1 antibody obtained from Dr. M. Hahn (CCRC) diluted 10-fold for 2 h at room temperature. Sections were washed with the blocking solution for 15 min with gentle shaking and incubated with 10-nm colloidal gold-conjugated anti-mouse antibody (BB International) at a dilution of 1:20 for 1 h at room temperature. Sections were then washed with distilled water for 20 min, dried on filter paper, stained with 3% uranyl acetate to increase the contrast of the image, washed with distilled water, and dried on filter paper once again. Finally, they were observed using a JEM-1230 transmission electron microscope (JEOL). Two independent labeling experiments were performed, in each case using a different tissue block and one or two labeled sections per block. Labeling density was scored in tissues representing four developmental stages that could be observed in each section: meristematic-cambial zone (CZ), radial expansion (RE), early secondary wall deposition (SW), and mature xylem (MX). In tissues representing meristematic and radial expansion stages, labeling in primary cell walls between fusiform cells was scored, while in secondary-walled stages, labeling was separately scored in walls between adjacent fiber cells (FF), between fibers and vessel elements (FV), and between adjacent vessel elements (VV). For each cell type, separate counts were obtained in both tangential (T) and radial (R) directions for both primary and secondary wall layers. In each case, gold grains were counted along a 3-μm line in walls of five randomly selected cells in each block. The effects of developmental stage, cell type (FF, FV, or VV), cell wall type (R or T), and cell wall layer (primary or secondary), in addition to the genotype, on the labeling were then evaluated by ANOVA according to the following models:
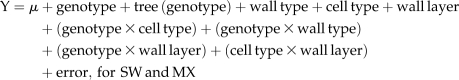 |
The data were analyzed by a general linear model procedure by SAS version 6.12 (SAS) using type 1 statistics.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF515607 for PtxtXET16-34.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PtxtXET16-34 transcript levels in the wood-forming tissues of PtxtXET16-34 sense-oriented transgenic lines relative to wild-type (T89) levels as determined by RT-PCR.
Supplemental Figure S2. Western blotting of proteins extracted from developing xylem tissues of PtxtXET16-34-OE and wild-type lines.
Supplemental Figure S3. Relationship between XEH (EC 3.2.1.151) activity and PtxtXET16-34 expression level in the transgenic lines.
Supplemental Figure S4. Effects of PtxtXET16-34 overexpression on plant growth.
Supplemental Table S1. Populus XTH gene models identified by Geisler-Lee et al. (2006), updated to the current genome version (1.1) and corresponding EST clones found in the indicated tissue-specific libraries in the Swedish EST database (http://www.populus.db.umu.se/) or obtained by RT-PCR amplification from the cambial region mRNA using XET-specific degenerate primers.
Supplementary Material
Acknowledgments
We thank Kjell Olofsson, Nancy Suorsa, and Lenore Johansson for technical assistance. Dr. M. Hahn is acknowledged for providing the CCRC-M1 antibody and Dr. P. Immerzeel for advice on sugar analysis.
References
- Antosiewicz DM, Purugganan MM, Polisensky DH, Braam J. (1997) Cellular localization of Arabidopsis xyloglucan endotransglycosylase-related proteins during development and after wind stimulation. Plant Physiol 115: 1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Park YW, Kaku T, Kaida R, Takeuchi M, Yoshida M, Hosoo Y, Ojio Y, Okuyama T, Taniguchi T, et al. (2009) Xyloglucan for generating tensile stress to bend tree stem. Mol Plant 2: 893–903 [DOI] [PubMed] [Google Scholar]
- Baluška F, Liners F, Hlavacka A, Schlicht M, Van Cutsem P, McCurdy DW, Menzel D. (2005) Cell wall pectins and xyloglucans are internalized into dividing root cells and accumulate within cell plates during cytokinesis. Protoplasma 225: 141–155 [DOI] [PubMed] [Google Scholar]
- Baumann MJ, Eklöf JM, Michel G, Kallas ÅM, Teeri TT, Czjzek M, Brumer H., III (2007) Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell 19: 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J. (2006) Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol 61: 451–467 [DOI] [PubMed] [Google Scholar]
- Bourquin V, Nishikubo N, Abe H, Brumer H, Denman S, Eklund M, Christiernin M, Teeri TT, Sundberg B, Mellerowicz EJ. (2002) Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 14: 3073–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanliaud E, De Silva J, Strongitharm B, Jeronimidis G, Gidley MJ. (2004) Mechanical effects of plant cell wall enzymes on cellulose/xyloglucan composites. Plant J 38: 27–37 [DOI] [PubMed] [Google Scholar]
- Cho SK, Kim JE, Park JA, Eom TJ, Kim WT. (2006) Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580: 3136–3144 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Dimmer E, Roden L, Cai DG, Kingsnorth C, Mutasa-Göttgens E. (2004) Transgenic analysis of sugar beet xyloglucan endo-transglucosylase/hydrolase Bv-XTH1 and Bv-XTH2 promoters reveals overlapping tissue-specific and wound-inducible expression profiles. Plant Biotechnol J 2: 127–139 [DOI] [PubMed] [Google Scholar]
- Eklöf JM, Brumer H. (2010) The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 153: 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunnerås S, et al. (2006) Poplar carbohydrate-active enzymes: gene identification and expression analyses. Plant Physiol 140: 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovesi V, Fornalé S, Fry SC, Ruel K, Ferrer P, Encina A, Sonbol FM, Bosch J, Puigdomènech P, Rigau J, et al. (2008) ZmXTH1, a new xyloglucan endotransglucosylase/hydrolase in maize, affects cell wall structure and composition in Arabidopsis thaliana. J Exp Bot 59: 875–889 [DOI] [PubMed] [Google Scholar]
- Gray-Mitsumune M, Blomquist K, McQueen-Mason S, Teeri TT, Sundberg B, Mellerowicz EJ. (2008) Ectopic expression of a wood-abundant expansin PttEXPA1 promotes cell expansion in primary and secondary tissues in aspen. Plant Biotechnol J 6: 62–72 [DOI] [PubMed] [Google Scholar]
- Gray-Mitsumune M, Mellerowicz EJ, Abe H, Schrader J, Winzéll A, Sterky F, Blomqvist K, McQueen-Mason S, Teeri TT, Sundberg B. (2004) Expansins abundant in secondary xylem belong to subgroup A of the α-expansin gene family. Plant Physiol 135: 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. (1989) Xyloglucan in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol 40: 139–168 [Google Scholar]
- Hayashi T, Takeda T. (1994) Compositional analysis of the oligosaccharide units of xyloglucan from suspension-cultured poplar cells. Japan Society for Bioscience Biotechnology and Agrochemistry 58: 1707–1708 [DOI] [PubMed] [Google Scholar]
- Henriksson H, Denman SE, Campuzano IDG, Ademark P, Master ER, Teeri TT, Brumer H., III (2003) N-Linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16A. Biochem J 375: 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Lorences EP, Barrachina C, Sonnewald U. (2001) Functional characterisation of Nicotiana tabacum xyloglucan endotransglycosylase (NtXET-1): generation of transgenic tobacco plants and changes in cell wall xyloglucan. Planta 212: 279–287 [DOI] [PubMed] [Google Scholar]
- Jan A, Yang G, Nakamura H, Ichikawa H, Kitano H, Matsuoka M, Matsumoto H, Komatsu S. (2004) Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiol 136: 3670–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez T, Martín I, Labrador E, Dopico B. (2006) The immunolocation of a xyloglucan endotransglucosylase/hydrolase specific to elongating tissues in Cicer arietinum suggests a role in the elongation of vascular cells. J Exp Bot 57: 3979–3988 [DOI] [PubMed] [Google Scholar]
- Kaida R, Serada S, Norioka N, Norioka S, Neumetzler L, Pauly M, Sampedro J, Zarra I, Hayashi T, Kaneko TS. (2010) Potential role for purple acid phosphatase in the dephosphorylation of wall proteins in tobacco cells. Plant Physiol 153: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallas ÅM, Piens K, Denman SE, Henriksson H, Fäldt J, Johansson P, Brumer H, Teeri TT. (2005) Enzymatic properties of native and deglycosylated hybrid aspen (Populus tremula × tremuloides) xyloglucan endotransglycosylase 16A expressed in Pichia pastoris. Biochem J 390: 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr EM, Fry SC. (2003) Pre-formed xyloglucans and xylans increase in molecular weight in three distinct compartments of a maize cell-suspension culture. Planta 217: 327–339 [DOI] [PubMed] [Google Scholar]
- Kooiman P. (1960) A method for the determination of amyloid in plant seeds. Recl Trav Chim Pays Bas 79: 675–678 [Google Scholar]
- Larson PR. (1994) The Vascular Cambium. Springer Verlag, Berlin [Google Scholar]
- Liu YB, Lu SM, Zhang JF, Liu S, Lu YT. (2007) A xyloglucan endotransglucosylase/hydrolase involves in growth of primary root and alters the deposition of cellulose in Arabidopsis. Planta 226: 1547–1560 [DOI] [PubMed] [Google Scholar]
- Lundqvist J, Jacobs A, Palm M, Zacchi G, Dahlman O, Stålbrand H. (2003) Characterizations of galactoglucomannan extracted from spruce (Picea abies) by heat fractionation at different conditions. Carbohydr Polym 51: 203–211 [Google Scholar]
- Marcus SE, Verhertbruggen Y, Hervé C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WGT, Knox JP. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris A, Suslov D, Fry SC, Verbelen JP, Vissenberg K. (2009) Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J Exp Bot 60: 3959–3972 [DOI] [PubMed] [Google Scholar]
- Matsui A, Yokoyama R, Seki M, Ito T, Shinozaki K, Takahashi T, Komeda Y, Nishitani K. (2005) AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J 42: 525–534 [DOI] [PubMed] [Google Scholar]
- McCann MC, Wells B, Roberts K. (1990) Direct visualization of cross-links in the primary plant cell wall. J Cell Sci 96: 323–334 [Google Scholar]
- Mellerowicz EJ. (2006) Xylem cell expansion: lessons from poplar. Hayashi T, , The Science and Lore of the Plant Cell Wall. Universal Publishers Brown Walker Press, Boca Raton, FL, pp 267–275 [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W. (2001) Unravelling cell wall formation in the woody dicot stem. Plant Mol Biol 47: 239–274 [PubMed] [Google Scholar]
- Mellerowicz EJ, Immerzeel P, Hayashi T. (2008) Xyloglucan: the molecular muscle of trees. Ann Bot (Lond) 102: 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Sundberg B. (2008) Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties. Curr Opin Plant Biol 11: 293–300 [DOI] [PubMed] [Google Scholar]
- Miedes E, Herbers K, Sonnewald U, Lorences EP. (2010) Overexpression of a cell wall enzyme reduces xyloglucan depolymerization and softening of transgenic tomato fruits. J Agric Food Chem 58: 5708–5713 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yokoyama R, Tomita E, Nishitani K. (2003) Two adzuki bean XTH genes, VaXTH1 and VaXTH2, with similar tissue-specific expression profiles, are differently regulated by auxin. Plant Cell Physiol 44: 16–24 [DOI] [PubMed] [Google Scholar]
- Nishikubo N, Awano T, Banasiak A, Bourquin V, Ibatullin F, Funada R, Brumer H, Teeri TT, Hayashi T, Sundberg B, et al. (2007) Xyloglucan endo-transglycosylase (XET) functions in gelatinous layers of tension wood fibers in poplar: a glimpse into the mechanism of the balancing act of trees. Plant Cell Physiol 48: 843–855 [DOI] [PubMed] [Google Scholar]
- Nishitani K, Masuda Y. (1981) Auxin-induced changes in the cell-wall structure: changes in the sugar compositions, intrinsic viscosity and molecular weight distributions of matrix polysaccharides of the epicotyl cell wall of Vigna angularis. Physiol Plant 52: 482–494 [Google Scholar]
- Nishitani K, Tominaga R. (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J Biol Chem 267: 21058–21064 [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K. (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res 119: 153–162 [DOI] [PubMed] [Google Scholar]
- Pauly M, Qin Q, Greene H, Albersheim P, Darvill A, York WS. (2001) Changes in the structure of xyloglucan during cell elongation. Planta 212: 842–850 [DOI] [PubMed] [Google Scholar]
- Plomion C, Leprovost G, Stokes A. (2001) Wood formation in trees. Plant Physiol 127: 1513–1523 [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. (2008) Xyloglucan-pectin linkages are formed intra-protoplasmically, contribute to wall assembly, and remain stable in the cell wall. Planta 227: 781–794 [DOI] [PubMed] [Google Scholar]
- Puhlmann J, Bucheli E, Swain MJ, Dunning N, Albersheim P, Darvill AG, Hahn MG. (1994) Generation of monoclonal antibodies against plant cell-wall polysaccharides: I. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiol 104: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romo S, Jiménez T, Labrador E, Dopico B. (2005) The gene for a xyloglucan endotransglucosylase/hydrolase from Cicer arietinum is strongly expressed in elongating tissues. Plant Physiol Biochem 43: 169–176 [DOI] [PubMed] [Google Scholar]
- Rose JK, Braam J, Fry SC, Nishitani K. (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Pardo B, Gianzo C, Guitián E, Revilla G, Zarra I. (2010) Lack of α-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol 154: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YK, Yum H, Kim ES, Cho H, Gothandam KM, Hyun J, Chung YY. (2006) BcXTH1, a Brassica campestris homologue of Arabidopsis XTH9, is associated with cell expansion. Planta 224: 32–41 [DOI] [PubMed] [Google Scholar]
- Siedlecka A, Wiklund S, Péronne MA, Micheli F, Lesniewska J, Sethson I, Edlund U, Richard L, Sundberg B, Mellerowicz EJ. (2008) Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of Populus. Plant Physiol 146: 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulová Z, Lednická M, Farkas V. (1995) A colorimetric assay for xyloglucan-endotransglycosylase from germinating seeds. Anal Biochem 229: 80–85 [DOI] [PubMed] [Google Scholar]
- Sweeley CC, Bentley R, Makita M, Wells WW. (1963) Gas-liquid chromatography of trimethylsilyl derivatives of sugars and related substances. J Am Chem Soc 85: 2497–2507 [Google Scholar]
- Takahashi J, Rudsander UJ, Hedenström M, Banasiak A, Harholt J, Amelot N, Immerzeel P, Ryden P, Endo S, Ibatullin FM, et al. (2009) KORRIGAN1 and its aspen homolog PttCel9A1 decrease cellulose crystallinity in Arabidopsis stems. Plant Cell Physiol 50: 1099–1115 [DOI] [PubMed] [Google Scholar]
- Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T. (2002) Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci USA 99: 9055–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbott LD, Ray PM. (1992) Changes in molecular size of previously deposited and newly synthesized pea cell wall matrix polysaccharides: effects of auxin and turgor. Plant Physiol 98: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Fry SC. (1997) Trimming and solubilization of xyloglucan after deposition in the walls of cultured rose cells. J Exp Bot 48: 297–305 [Google Scholar]
- Thompson JE, Smith RC, Fry SC. (1997) Xyloglucan undergoes interpolymeric transglycosylation during binding to the plant cell wall in vivo: evidence from 13C/3H dual labelling and isopycnic centrifugation in caesium trifluoroacetate. Biochem J 327: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumoto H, Wakabayashi K, Kamisaka S, Hoson T. (2002) Changes in the sugar composition and molecular mass distribution of matrix polysaccharides during cotton fiber development. Plant Cell Physiol 43: 411–418 [DOI] [PubMed] [Google Scholar]
- Van Sandt VS, Suslov D, Verbelen JP, Vissenberg K. (2007) Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot (Lond) 100: 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg K, Fry SC, Pauly M, Höfte H, Verbelen JP. (2005) XTH acts at the microfibril-matrix interface during cell elongation. J Exp Bot 56: 673–683 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JK, Nishitani K. (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice: classification and expression analysis. Plant Physiol 134: 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun HS, Kwon C, Kang BG, Lee JS, Han TJ, Chang SC, Kim SK. (2005) A xyloglucan endotransglycosylase/hydrolase1, VrXTH1, is associated with cell elongation in mungbean hypocotyls. Physiol Plant 125: 106–113 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.