Abstract
C4 photosynthesis involves alterations to the biochemistry, cell biology, and development of leaves. Together, these modifications increase the efficiency of photosynthesis, and despite the apparent complexity of the pathway, it has evolved at least 45 times independently within the angiosperms. To provide insight into the extent to which gene expression is altered between C3 and C4 leaves, and to identify candidates associated with the C4 pathway, we used massively parallel mRNA sequencing of closely related C3 (Cleome spinosa) and C4 (Cleome gynandra) species. Gene annotation was facilitated by the phylogenetic proximity of Cleome and Arabidopsis (Arabidopsis thaliana). Up to 603 transcripts differ in abundance between these C3 and C4 leaves. These include 17 transcription factors, putative transport proteins, as well as genes that in Arabidopsis are implicated in chloroplast movement and expansion, plasmodesmatal connectivity, and cell wall modification. These are all characteristics known to alter in a C4 leaf but that previously had remained undefined at the molecular level. We also document large shifts in overall transcription profiles for selected functional classes. Our approach defines the extent to which transcript abundance in these C3 and C4 leaves differs, provides a blueprint for the NAD-malic enzyme C4 pathway operating in a dicotyledon, and furthermore identifies potential regulators. We anticipate that comparative transcriptomics of closely related species will provide deep insight into the evolution of other complex traits.
C4 photosynthesis is a complex biological trait that enables plants to either accumulate biomass at a much faster rate or live in adverse environments compared with “ordinary” plants (Hatch, 1987; Osborne and Freckleton, 2009). These C4 plants have added a CO2 concentration mechanism on top of their regular photosynthetic carbon fixation that makes them not only more efficient at assimilating inorganic carbon; they frequently also have higher water and nitrogen use efficiencies (Black, 1973; Oaks, 1994; Osborne and Freckleton, 2009). Beyond the basic biochemistry, our understanding of C4 photosynthesis is limited.
The principle of C4 photosynthesis is deceivingly simple: instead of using Rubisco as the primary carbon-fixing enzyme, C4 plants use phosphoenolpyruvate carboxylase (PEPC). Unlike Rubisco, PEPC is more specific for inorganic carbon (Hatch, 1987). Since the C4 cycle is an add-on rather than a replacement for Rubisco and the Calvin-Benson cycle, the prefixed CO2 is transported in a bound form, a C4 acid (hence the name), to the site of Rubisco. The C4 cycle generates high concentrations of CO2 around Rubisco (Hatch, 1987), and this increases the rate of photosynthesis because competition between CO2 and oxygen at the active site of Rubisco is reduced (Jordan and Ogren, 1984). In most C4 plants, concentrating CO2 around Rubisco involves the reactions of photosynthesis being partitioned between bundle sheath (BS) and mesophyll (M) cells as well as changes to cell biology and leaf development (Hatch, 1987; Sage, 2004), although in some lineages, C4 photosynthesis operates within individual cells (Reiskind et al., 1989; Keeley, 1998; Voznesenskaya et al., 2001, 2002, 2003).
In all known C4 plants, CO2 enters M cells and is converted into bicarbonate by carbonic anhydrase. PEPC then combines HCO3− with PEP to generate the C4 oxaloacetic acid, which is rapidly converted into either Asp or malate. These C4 acids then diffuse to the site of Rubisco through abundant plasmodesmata, where C4 acid decarboxylases release CO2 (Hatch, 1987). Three distinct C4 acid decarboxylases, known as NADP-dependent malic enzyme (NADP-ME), NAD-dependent malic enzyme (NAD-ME), and PEP carboxykinase, have been coopted into the C4 pathway, and this has been used to define three biochemical subtypes of C4 photosynthesis. The three-carbon compound released after decarboxylation diffuses back to the M cells and is converted to PEP catalyzed by pyruvate,orthophosphate dikinase (PPDK; Hatch and Slack, 1968). Because the enzymes involved in the C4 cycle are found in the cytosol, chloroplasts, and mitochondria, a significant amount of transport across organellar membranes is required for the C4 cycle to operate. However, few genes encoding transporters that allow the increased intracellular flux of metabolites required for C4 photosynthesis have been identified (Bräutigam et al., 2008a; Majeran and van Wijk, 2009). In addition, we have a very limited understanding of the mechanisms controlling the altered cell biology and morphology associated with C4 leaves. The C4 cycle likely affects not only the relatively small number of enzymes and transport proteins needed to perform the core reactions but, given the consequences on the ecological performance of the plants, also a range of other processes.
The gaps in our understanding of the mechanisms underlying C4 photosynthesis limit insight into a metabolic pathway that has evolved repeatedly at least 45 times in plants (Sage, 2004) and so is of interest in terms of understanding a remarkable example of convergent evolution. In addition, because C4 plants are among the most productive on the planet and the pathway is associated with increased water and nitrogen use efficiencies (Brown, 1999), it has been suggested that characteristics of C4 photosynthesis should be placed into C3 crops (Matsuoka et al., 2001; Mitchell and Sheehy, 2006; Hibberd et al., 2008). A more complete understanding of genes involved in C4 photosynthesis is fundamental to attempts at placing components of the C4 pathway into C3 crops to increase yield.
Recently, a new set of tools has become available to analyze species without sequenced genomes on a genomic scale: next generation sequencing (NGS) technology (summarized in Metzker, 2010). With NGS, the transcriptome of a tissue can be sequenced and quantified at the same time (RNA-Seq; Wang et al., 2009). The 454 FLX genome sequencer provides a quarter million sequence reads of 230 bases in each run from a cDNA template generated from mRNA (http://www.454.com/; Metzker, 2010). The resulting reads can be mapped onto a closely related reference to quantify the number of reads matching a gene locus, thus providing a measure of transcript abundance (Flicek and Birney, 2009; Bräutigam and Gowik, 2010). We chose to compare the C4 plant Cleome gynandra with the C3 plant Cleome spinosa, since they are members of the same genus and are closely related to Arabidopsis (Arabidopsis thaliana; Brown et al., 2005; Marshall et al., 2007). Given the close phylogenetic relationship, we can take advantage of the well-annotated Arabidopsis genome (Swarbreck et al., 2008) and its known genome history (Bowers et al., 2003; Haberer et al., 2004; Thomas et al., 2006) to identify and quantify the biological functions regulated at the level of transcript abundance in the C4 species compared with the C3 species. Although the experiment will also capture variation in the abundance of transcripts associated with differences between the species that do not relate to C4 photosynthesis, the close proximity of the Cleome species should reduce this effect. We chose to use mature fully differentiated leaves for the analysis, since we wanted to minimize the influence of species-specific effects during leaf differentiation but rather focus on transcript profiles when C4 photosynthesis is fully operational. Once this profile is defined, analysis of developmental stages may reveal how the profile is achieved during differentiation.
By comparing the transcriptomes of closely related C3 and C4 species, we will test (1) whether cross-species transcriptomic comparisons are feasible, (2) the degree to which the core C4 cycle enzymes and transport proteins are regulated at the level of transcript abundance, and (3) whether the changes in metabolism associated with C4 photosynthesis are associated with additional unexpected shifts in transcript profiles in leaves of C4 compared with C3 plants, and (4) define candidates for additional functions critical to C4 photosynthesis based on unbiased observation of the data. By analyzing the complete transcriptome, we define the maximal extent to which the C4 pathway alters leaf transcript profiles.
RESULTS
Physiological Analysis of C3 and C4 Leaves Confirms C4 Metabolism in C. gynandra
To confirm that the C. spinosa and C. gynandra leaves we used for transcriptomic analysis were using C3 and C4 photosynthesis, respectively, we analyzed the steady-state levels of metabolites associated with the C4 cycle. For example, large quantities of Asp, Ala, and pyruvate are produced in M and BS cells of NAD-ME C4 leaves, and they were 19, 3.9, and 3.6 times more abundant, respectively, in C. gynandra compared with C. spinosa (Supplemental Table S1). In contrast, and in agreement with the lower demand for the photorespiration in C4 leaves, glycerate and glycolate, intermediates of the photorespiratory cycle, were 4.5 and 1.9 times more abundant in C. spinosa (Supplemental Table S1). We also determined the extractable activities of PEPC, aspartate aminotransferase (AspAT), NAD-dependent malate dehydrogenase (NAD-MDH), NAD-ME, and alanine aminotransferase (AlaAT). Except for NAD-MDH, significantly higher activities of the enzymes required for the C4 cycle were measured in C. gynandra leaf extracts (Supplemental Fig. S1). The metabolite profiling of leaf extracts using gas chromatography-electron impact-time of flight (GC-EI-TOF) and the enzyme activity assays showed that the plants we used for digital gene expression analysis had clear differences in their metabolite profiles and enzyme activities, and these were consistent with functional C3 and C4 photosynthesis operating in leaves of C. spinosa and C. gynandra, respectively.
The Leaf Transcriptomes for Closely Related C3 and C4 Species Are Qualitatively Similar
To obtain sequence tags for digital gene expression (DGE) analysis from C. spinosa (C3) and C. gynandra (C4), RNA was isolated from mature leaves of each species and prepared for 454 sequencing. One sequencing run on a Genome Sequencer FLX (GS FLX; Roche) sequencing system was conducted on leaf cDNA isolated from either C. gynandra or C. spinosa. From C. spinosa, we obtained 70,564,592 nucleotides, and from C. gynandra, 91,851,136 nucleotides of raw sequence were obtained; after quality control, these corresponded to 65,525,139 and 85,681,233 nucleotides, respectively (Table I). The mean read length of the cleaned sequence reads was 232 nucleotides for C. gynandra and 230 nucleotides for C. spinosa (Table I).
Table I. Massively parallel signature sequencing allows large-scale assembly of transcripts in both C. spinosa and C. gynandra after comparison with the TAIR 8 Arabidopsis database.
One GS FLX sequencing run allowed significant generation of sequence for both species, and the vast majority of these could be used to assemble contigs and then matched to Arabidopsis genes.
| Data | C. spinosa | C. gynandra |
| Raw reads | 313,807 | 402,674 |
| Raw nucleotides | 70,564,592 | 91,851,136 |
| Raw mean length | 225 | 228 |
| Clean reads | 284,318 | 368,333 |
| Clean nucleotides | 65,525,139 | 85,681,233 |
| Clean mean length | 230 | 232 |
| Contigs | 17,655 | 18,992 |
| Total length (nucleotides) | 7,746,894 | 9,062,043 |
| Total reads | 245,324 | 319,732 |
| Percent assembled | 86.3 | 86.8 |
To exclude program-specific mapping artifacts and to test whether the C. gynandra and C. spinosa libraries behave robustly during mapping, two different programs, BLAST and BLAT (BLAST-Like Alignment Tool), were used to align the reads to Arabidopsis as the reference genome. To define the most suitable mapping parameters, an array of parameters for mappings in both the DNA and protein space were tested (Table II). Neither the C. gynandra nor the C. spinosa library mapped well to Arabidopsis cDNAs in the DNA space using BLAT or BLAST, although the differences are more dramatic for BLAT (Table II). In the protein space, however, the proportion of mapped reads increased dramatically. When 75% amino acid sequence identity was required, three-quarters of the reads could be mapped with BLAT, resulting in 1.48 and 1.57 average mappings per read, respectively. Even with the most lenient mapping parameters, the proportion of mapped reads did not exceed 83% with BLAT and 78.8% with BLAST (Table II). In all mapping attempts, the C. gynandra and C. spinosa read libraries yielded qualitatively similar mapping results, irrespective of mapping program or parameters.
Table II. Mapping the sequence reads with different BLAT and BLAST parameters to empirically determine suitable mapping conditions.
The percentage of AGI codes with at least one mapped read and the average mappings per read were determined prior to parsing the tables to retain only the best match. Suitable mapping conditions are printed in bold; for BLAT, the cutoff value is the minimal number of matching bases; for BLAST, it is the minimal accepted e-value.
| Mapping Program | Library | Search Space | Cutoff Value | Percentage Reads with at Least One Hit in the Reference | Percentage AGI Codes with at Least One Mapped Read | Average Mappings per Read |
| BLAT | C. gynandra | DNA | 60 | 40.9 | 42.0 | 1.19 |
| 75 | 40.7 | 41.7 | 1.19 | |||
| 85 | 30.2 | 35.8 | 1.15 | |||
| 90 | 7.7 | 19.5 | 1.09 | |||
| Protein | 25 | 82.6 | 70.4 | 2.35 | ||
| 50 | 82.6 | 70.4 | 2.35 | |||
| 75 | 75.4 | 62.6 | 1.48 | |||
| 80 | 56.4 | 52.2 | 1.27 | |||
| C. spinosa | DNA | 60 | 40.8 | 38.9 | 1.29 | |
| 75 | 40.6 | 38.5 | 1.28 | |||
| 85 | 29.7 | 32.3 | 1.21 | |||
| 90 | 8.5 | 17.1 | 1.15 | |||
| Protein | 25 | 83.0 | 67.7 | 2.49 | ||
| 50 | 83.0 | 67.7 | 2.46 | |||
| 75 | 76.0 | 58.9 | 1.57 | |||
| 80 | 57.9 | 48.4 | 1.32 | |||
| BLAST | C. gynandra | DNA | 1e-05: | 68.9 | 56.5 | 30.7 |
| 1e-10: | 58.8 | 49.1 | 27.7 | |||
| 1e-30: | 29.6 | 30.5 | 18.9 | |||
| 1e-50: | 9.9 | 15.9 | 11.5 | |||
| Protein | 1e-05: | 78.0 | 76.9 | 106.6 | ||
| 1e-10: | 67.8 | 71.0 | 64.6 | |||
| 1e-30: | 29.0 | 39.5 | 22.9 | |||
| 1e-50: | 0.1 | 0.3 | 7.5 | |||
| C. spinosa | DNA | 1e-05: | 69.7 | 53.0 | 28.2 | |
| 1e-10: | 59.6 | 46.3 | 25.1 | |||
| 1e-30: | 29.4 | 28.3 | 16.2 | |||
| 1e-50: | 9.8 | 14.4 | 9.8 | |||
| Protein | 1e-05: | 78.8 | 75.3 | 93.7 | ||
| 1e-10: | 68.3 | 68.7 | 56.4 | |||
| 1e-30: | 29.3 | 36.0 | 21.2 | |||
| 1e-50: | 0.1 | 0.3 | 4.6 |
To obtain a stringent yet inclusive mapping, the mapping conducted in protein space at 75% or greater identity with BLAT was chosen, and this mapping file was parsed by in-house scripts to keep only the read match with the highest number of matching bases. For a more lenient mapping, a BLAST mapping at a cutoff of 1e−5 was chosen and parsed to keep only the best BLAST hit for each read. For each Arabidopsis Genome Initiative (AGI) code, the number of matching reads was counted and the hit count was then transformed to reads per million (RPM) to normalize for the number of reads available for each species. After parsing, the sequenced libraries matched between 50.5% and 55.3% of the genes in the Arabidopsis reference (Supplemental Table S2).
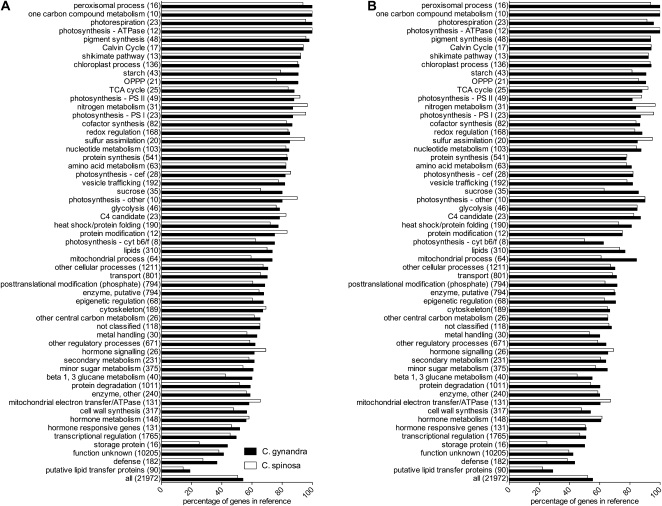
To assess whether the data sets for the two different species and the two different mappings were qualitatively similar, we tested the coverage of the functional classes. Overall, about 50% of all genes were represented in both species with the BLAT (Fig. 1A) and the BLAST mapping (Fig. 1B). Although the majority of gene classes were represented by more than 50% of genes in each class for both mappings, the classes function unknown, putative lipid transfer protein, storage protein, and defense were underrepresented compared with all genes (Fig. 1). Genes present in the organellar genomes were not well represented (Supplemental Table S3). Genes classified into primary metabolism including photosynthesis, central carbon, nitrogen metabolism, amino acid, and nucleotide metabolism as well as many cellular processes were well-represented categories, and about four-fifths of genes predicted to be involved in the C4 pathway were detected in both species. Overall, the pattern of detection in the different gene classes was similar for both species and independent of the program used for the mapping (Fig. 1).
Figure 1.
The qualitative patterns of transcript abundance between C. gynandra and C. spinosa are very similar, with the same classes underrepresented and overrepresented in both libraries. A, Analysis based on BLAT mapping. B, Analysis based on BLAST mapping. Black bars refer to the C4 plant C. gynandra, and white bars refer to the C3 plant C. spinosa.
Transcripts of Known C4 Genes Are More Abundant with One Exception
Detailed analysis of known C4 genes showed that all but one gene necessary for the core C4 cycle of NAD-ME-type plants were massively up-regulated in C. gynandra compared with C. spinosa. Transcripts encoding PEPC were up-regulated 78-fold, those encoding AspAT were up-regulated 343-fold, the transcripts for the two isoforms of NAD-ME were up-regulated 27- and 21-fold, respectively, and AlaAT were up-regulated 29-fold (Table III). The results for the BLAT and the BLAST mappings were similar with one exception. In the BLAST mapping, the reads mapping to PEPC were split onto two genes in the Arabidopsis reference genome, whereas they mapped to only one gene in the BLAT mapping (Table III). Transcripts encoding mitochondrial malate dehydrogenases were increased only 1.3-fold (Supplemental Table S3). Not only were genes associated with the C4 pathway up-regulated compared with C3, but they also had high absolute read counts between 1,800 and 4,806 RPM.
Table III. Transcript abundance of C4 cycle genes that have significantly higher transcript abundance in C4 leaf tissue.
Asterisks denote changes significant only in BLAST mapping.
| Enzyme | Locus | BLAT Mapping |
BLAST Mapping |
||||
| C. gynandra RPM | C. spinosa RPM | Fold Change | C. gynandra RPM | C. spinosa RPM | Fold Change | ||
| AspAT | AT2G30970 | 4,806 | 14 | 343.3 | 4,601 | 18 | 257.9 |
| PPDK | AT4G15530 | 3,262 | 14 | 233.0 | 3,216 | 13 | 240.3 |
| PEPC | AT2G42600 | 9,702 | 124 | 78.2 | 8,321 | 169 | 49.1 |
| AlaAT | AT1G17290 | 7,610 | 267 | 28.5 | 7,242 | 259 | 28.0 |
| NAD-ME1 | AT4G00570 | 1,357 | 51 | 26.6 | 1,326 | 49 | 27.0 |
| NAD-ME2 | AT2G13560 | 1,800 | 87 | 20.7 | 1,723 | 85 | 20.3 |
| PEPC kinase | AT1G08650 | 230 | 37 | 6.2 | 226 | 36 | 6.3 |
| NADP-ME* | AT1G79750 | 227 | 60 | 3.8 | 216 | 45 | 4.8 |
| PEPC* | AT1G53310 | 94 | 248 | 0.4 | 950 | 192 | 5.0 |
| PPDK regulatory protein* | AT4g21210 | 148 | 32 | 4.6 | 198 | 27 | 7.3 |
The Leaf Transcriptomes for Closely Related C3 and C4 Species Are Quantitatively Different
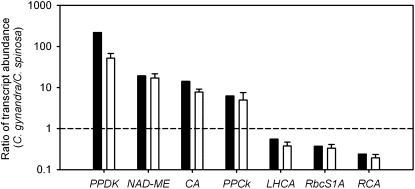
Before undertaking detailed analysis of differences in transcript abundance between C. gynandra and C. spinosa, we used quantitative (q)PCR to confirm estimates of transcript abundance identified by RNA-Seq. We chose genes whose transcript abundance differed over 4 orders of magnitude and used qPCR to assess their abundance. qPCR was performed on both the cDNA used for RNA-Seq and cDNA generated from RNA isolated from leaves in a separate experiment. This approach provided strong support for the differences in abundance of transcripts between the two species that we determined from RNA-Seq (Fig. 2). Overall, this showed that the ratios of transcript abundance obtained by RNA-Seq-based DGE are suitable for calling differentially expressed genes between two related species.
Figure 2.
Massively parallel sequencing of mRNAs (RNA-Seq) and qPCR generate similar profiles of transcript abundance in C. gynandra and C. spinosa. Ratios of transcript abundance in C. gynandra and C. spinosa were calculated, and transcripts selected for this analysis spanned 4 orders of magnitude. CA, Carbonic anhydrase; PPCk, PEPC kinase; LHCA, light-harvesting complex subunit A; RbcS1a, ribulose bisphosphate carboxylase oxygenase 1a; RCA, Rubisco activase. Black bars represent data from RNA-Seq, and white bars represent data from qPCR. The horizontal dashed line represents a ratio of 1 and indicates no difference in transcript abundance between the two species.
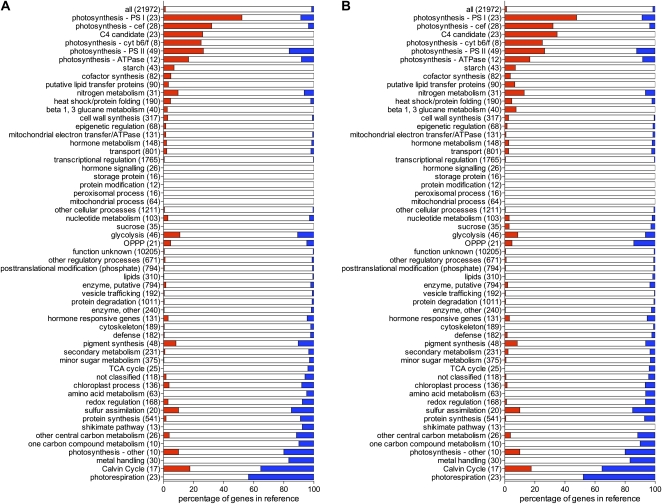
Of the 13,662 transcripts for which we captured quantitative data (Supplemental Table S3), we identified 583 (BLAT) or 603 (BLAST) transcripts whose abundance differed significantly (P ≤ 0.01) between C. spinosa and C. gynandra, with 256/258 (1.2%/1.2%) transcripts being more abundant in C. gynandra and 327/345 (1.5%/1.6%) transcripts being more abundant in C. spinosa (Fig. 3, “all”). We tested whether significantly changed transcripts are enriched in functional categories and whether they were more highly expressed in the C4 or the C3 species. While the qualitative classification of detected genes showed a very similar pattern between C. spinosa and C. gynandra (Fig. 1), the quantitative analysis revealed massive differences in representation between gene classes in the C3 and the C4 species (Fig. 3). The transcript profile generated by the BLAT mapping (Fig. 3A) is similar to the one generated by the BLAST mapping (Fig. 3B), although not all genes called as significantly regulated were identical (Supplemental Table S3). The classes containing the highest percentage of changed genes are the photosynthetic classes as well as the C4 cycle, Calvin-Benson cycle, and photorespiration (Fig. 3). The latter two have lower steady-state mRNA levels in C4 leaf tissue (Fig. 3, bottom), while the photosynthetic classes of PSI, cyclic electron flow, and cytochrome b6/f complex as well as the C4 cycle have higher levels in C4 leaf tissue (Fig. 3, top). A number of classes involved in primary metabolism also have lower steady-state transcript levels in C4 tissues: one-carbon compound metabolism, other central carbon metabolism, shikimate pathway, and amino acid metabolism. Protein synthesis also has lower steady-state transcript levels, which are limited to cytosolic and plastidic protein synthesis genes (Supplemental Fig. S3). Among the classes with higher steady-state transcript levels are starch metabolism, cofactor synthesis, putative lipid transfer proteins, nitrogen metabolism, and β-1,3 glucan metabolism. The quantitative pattern (Fig. 3) is similar to the qualitative pattern (Fig. 1) with regard to the influence of the mapping program; the BLAT and BLAST mappings look remarkably similar with the exception of shikimate metabolism.
Figure 3.
The quantitative patterns of transcript accumulation in C. gynandra and C. spinosa are distinct. A, Analysis based on BLAT mapping. B, Analysis based on BLAST mapping. Shown are the percentages of genes with significantly higher abundance of transcripts in C4 (red bars), unchanged (white bars, including genes not detected), and significantly lower abundance of transcripts in C4 (blue bars) based on the total number of genes in each annotation class (in parentheses on the y axis).
Transcripts with Similar Patterns of Abundance Compared with Bona Fide C4 Genes and Rubisco
The list of 13,662 transcripts detected in either C. spinosa or C. gynandra tissues and the list of 603 transcripts that are differentially regulated between both species (Supplemental Table S3, BLAST mapping) prompted us to determine which transcripts showed changes in abundance similar to the core C4 genes or Rubisco subunit-encoding genes. Such transcripts display both a large fold change between the C3 and the C4 plants and large absolute read numbers. For example, among the transcripts encoding putative transport proteins, three plastidic transport proteins, the PEP phosphate translocator PPT, a putative bile acid:sodium symporter, and a putative proton:sodium antiporter, two mitochondrial dicarboxylate carriers, and one plasma membrane intrinsic protein were massively up-regulated in C4 C. gynandra (Table IV). No transcripts encoding transport proteins were found to be down-regulated to a comparable degree. Among metabolic genes, two cytosolic carbonic anhydrases, one of which (CA4; Table IV) is likely tethered to the plasma membrane, an adenylate kinase, and a pyrophosphatase were up-regulated at levels comparable to those of C4 cycle genes. Many proteins of unknown function showed differential expression, the most striking case being a putative lipid transfer protein, also annotated as an extensin-like protein. Based on annotation and differential expression pattern, several transcripts predicted to encode known C4 functions that have not yet been assigned to genes, such as CHLOROPLAST UNUSUAL POSITIONING1 (CHUP1) and actin for chloroplast positioning or callose-degrading enzymes for regulating plasmodesmatal opening, were identified (Table IV).
Table IV. Transcript abundance of selected genes with an expression similar to that of C4 cycle genes and Rubisco.
All changes are significant at P ≤ 0.01. n/a, Not available.
| Function | Locus | Annotation (TAIR 9) | C. gynandra RPM | C. spinosa RPM | Ratio |
| Transport proteins | |||||
| AT2G26900 | Bile acid:sodium symporter family protein | 4,774 | 55 | 86.8 | |
| AT2G22500 | Mitochondrial dicarboxylate carrier | 324 | 0 | n/a | |
| AT4G24570 | Mitochondrial dicarboxylate carrier | 148 | 0 | n/a | |
| AT2G45960 | Plasma membrane intrinsic protein subfamily protein | 2,686 | 133 | 20.2 | |
| AT5G33320 | Phosphoenolpyruvate/phosphate translocator | 1,955 | 97 | 20.2 | |
| AT1G49810 | Member of Na+/H+ antiporter family | 1,321 | 83 | 15.9 | |
| Metabolism | |||||
| AT3G52720 | α-Carbonic anhydrase 1 | 227 | 152 | 1.5 | |
| AT1G23730 | β-Carbonic anhydrase 4 | 497 | 87 | 5.7 | |
| AT5G35170 | Adenylate kinase family protein | 1,994 | 235 | 8.5 | |
| AT5G09650 | Inorganic pyrophosphatase | 2,664 | 833 | 3.2 | |
| Proteins of unknown function | |||||
| AT1G12090 | Extensin-like protein (ELP) | 6,278 | 147 | 42.7 | |
| Callose-degrading enzymes | |||||
| AT3G57240 | Member of glycosyl hydrolase family 17, likely β-1,3 glucanase | 436 | 0 | n/a | |
| AT1G32860 | Member of glycosyl hydrolase family 17, likely β-1,3 glucanase | 50 | 0 | n/a | |
| AT5G42100 | Plasmodesmal-associated β-1,3-glucanase | 173 | 32 | 5.4 | |
| Cell biology | |||||
| AT3G25690 | CHUP1 | 22 | 170 | 0.13 | |
| AT3G12110 | ACTIN | 122 | 727 | 0.2 | |
Regulatory Genes That Are Significantly Changed
The transcript profiles of these C3 and C4 species identify a number of regulatory proteins that are candidates for maintaining C4 status. Among transcripts encoding proteins with regulatory functions, 43 were significantly up-regulated in either C. gynandra or C. spinosa (Fig. 3). These include bona fide transcription factors, protein phosphatases and kinases, and the regulatory proteins of the pyruvate dehydrogenase complex (up-regulated in C4), of PPDK (up-regulated in C4), and of Rubisco (down-regulated in C4). Only 17 transcription factors are significantly changed; seven of those have higher steady-state mRNA levels compared with the C3 leaf tissue, while 10 have lower steady-state mRNA levels (Table V).
Table V. Transcription factors that are significantly changed between the leaf tissue samples.
Asterisks denote changes significant only in BLAST mapping. n/a, Not available.
| Locus | Transcription Factor Type | BLAT Mapping |
BLAST Mapping |
Segmentally Duplicated? | ||||
| C. gynandra RPM | C. spinosa RPM | Ratio | C. gynandra RPM | C. spinosa RPM | Ratio | |||
| AT1G25560 | AP2-EREBP | 176 | 9 | 19.6 | 219 | 9 | 24.3 | Yes |
| AT5G07580 | AP2-EREBP | 223 | 51 | 4.4 | 292 | 36 | 8.1 | Yes |
| AT1G53910 | AP2-EREBP* | 32 | 138 | 0.2 | 84 | 268 | 0.3 | Yes |
| AT5G10570 | bHLH | 0 | 83 | n/a | 0 | 112 | n/a | Yes |
| AT3G21330 | bHLH* | 0 | 74 | n/a | 0 | 107 | n/a | |
| AT3G62420 | bZIP | 11 | 138 | 0.1 | 10 | 138 | 0.1 | |
| AT2G20570 | G2-like | 220 | 0 | n/a | 292 | 0 | n/a | |
| AT1G72030 | GNAT | 11 | 179 | 0.1 | 10 | 330 | 0.0 | |
| AT2G22430 | HB | 515 | 106 | 4.9 | 505 | 116 | 4.4 | Yes |
| AT1G10200 | LIM | 22 | 230 | 0.1 | 21 | 205 | 0.1 | |
| AT4G30410 | Not specified* | 0 | 32 | n/a | 0 | 76 | n/a | |
| AT1G32700 | PLATZ | 176 | 9 | 19.6 | 115 | 4 | 28.8 | |
| AT5G02810 | Pseudo ARR-B | 0 | 106 | n/a | 10 | 112 | 0.1 | |
| AT2G36990 | Sigma70-like | 130 | 0 | n/a | 143 | 0 | n/a | |
| AT1G48500 | Tify | 11 | 147 | 0.1 | 10 | 174 | 0.1 | Yes |
| AT1G17380 | Tify* | 18 | 110 | 0.2 | 24 | 161 | 0.1 | Yes |
| AT3G02790 | Zinc finger | 374 | 87 | 4.3 | 407 | 112 | 3.6 | Yes |
In addition to the detailed quantitative and qualitative analysis of read mappings to generate ESTs for both species, contigs were assembled from cleaned reads for each species as described previously (Weber et al., 2007; Bräutigam et al., 2008b) and then annotated by BLASTX versus The Arabidopsis Information Resource (TAIR) 9 protein models. A total of 18,992 and 17,655 contigs representing total sequence lengths of 9,062,043 and 7,746,894 nucleotides were obtained for C. gynandra and C. spinosa, respectively (Table I).
DISCUSSION
Transcriptomic Comparisons of Different Species with NGS Technology Are Feasible
Read mapping by alignment is a well-established tool to quantify transcript abundance and thus determine mRNA steady-state levels (Wall et al., 2009; Metzker, 2010). The concept of mapping to a cross-species reference has also been established theoretically (Palmieri and Schlotterer, 2009), although the potential has not been experimentally explored to date (Bräutigam and Gowik, 2010).
To explore cross-species mapping, the transcriptome sequencing was carried out using 454 FLX, a long-read technology, since theoretical work had established that at least BLAT is capable of mapping reads that contain alterations in comparison with the reference if the reads are at least 100 bases long (Palmieri and Schlotterer, 2009). We also established a reference database, which removes the genome history of Arabidopsis as far as it is known (Bowers et al., 2003; Haberer et al., 2004; Thomas et al., 2006). Tandem duplicated genes and segmentally duplicated genes (remnants of the last whole genome duplications) were removed to prevent genome history from interfering with comparative quantitative mapping (Bräutigam and Gowik, 2010).
Both BLAT and BLAST mappings indicate that using a minimal reference does not diminish read mappings (Supplemental Table S4) while avoiding mapping problems based on genome history (Bräutigam and Gowik, 2010). The mappings in protein space allowed more successful read mappings, because protein sequences diverge more slowly than nucleotide sequences. Although the proportion of reads mapped varied with changing mapping parameters (Table II; Supplemental Table S4), the C. spinosa and C. gynandra libraries yielded similar results, indicating that, evolutionarily, both species are approximately equally distant from Arabidopsis, with mapping incurring similar penalties depending on parameters.
Since no read alignment program has emerged as the consensus program for NGS data analysis, two different programs were used for mapping and the output was compared in all cases. The output proved robust against changing the mapping program both qualitatively and quantitatively. When we mapped the quarter million reads obtained from each species of Cleome to a minimized TAIR 9 release of the Arabidopsis genome, they corresponded to approximately 11,000 loci. As the minimized TAIR 9 data set contains 21,972 gene loci, the reads we collected in C. gynandra and C. spinosa represent approximately 50% of the transcriptome. In Arabidopsis seedlings, approximately 60% of the loci represented in the TAIR 8 release were detectable (Weber et al., 2007); hence, we have likely captured a large proportion of the transcripts associated with leaves of C. spinosa and C. gynandra.
The qualitative representation of gene classes detected reflects that leaf tissues were analyzed. While photosynthetic genes as well as primary metabolism are well represented in all data sets, genes implicated in cell walls, secondary metabolism, and defense responses are underrepresented (Fig. 1). These classes contain genes that are likely specific to certain tissues, developmental stages, or environmental challenges. For example, cell wall genes may be better represented if our sampling had included expanding leaf or stem material (Schmid et al., 2005), and stress-response genes may be better represented if plants were sampled after exposure to extreme conditions (Kilian et al., 2007).
Likewise, certain pathways of secondary metabolism are likely restricted to defined tissues or developmental stages, making it unlikely that we would pick up many of these genes when profiling leaf libraries. Based on the gene detection pattern, the two plant species did not encounter different biotic or abiotic stresses or were not in different stages of growth, as very similar genes were detected in both species (Figs. 1 and 3).
Finally, only a very small proportion of transcripts showed significant differences in abundance between the two different species (Supplemental Tables S2 and S3), and these changes were enriched in a limited number of functional classes (Fig. 3). We conclude that cross-species mapping in protein space is a feasible strategy to compare different species as long as an equidistant reference is available.
Transcripts Derived from Core C4 Cycle Genes Are More Abundant in the C4 Species
C4 photosynthesis has evolved convergently in many different lineages of plants (Sage, 2004), and in many cases the alterations to expression of specific genes has been related to transcriptional regulation (summarized in Sheen, 1999). Our genome-scale analysis allowed us to compare the steady-state transcript levels for all candidate C4 genes at the same time. For all of the enzymes where a change in total extractable activity could be shown (Supplemental Fig. S1), a higher mRNA level of at least one isoform as judged from the read count was also present (Table III). The only enzyme showing no changes in transcript level is the mitochondrial NAD-MDH. Possibly, the activity of the mitochondrial NAD-MDH is high enough already in C3 plants to support a C4-type metabolic flux. The only transport protein known to date that is involved in the C4 cycle, the PEP phosphate translocator (Fischer et al., 1997; Bräutigam et al., 2008a), is also up-regulated 20-fold, indicating that this transport protein is regulated at the level of mRNA abundance. Based on similarities in transcript abundance to known C4 genes, our comparative RNA-Seq also identified likely additional components needed for C4 photosynthesis. When PPDK was characterized, it was proposed that adenylate kinase as well as inorganic pyrophosphatase need to be abundant in C4 chloroplasts (Hatch and Slack, 1968). RNA-Seq confirmed this prediction and showed that the up-regulation also occurs at the level of transcript abundance. Taken together, we found that almost all transcripts encoding the proteins required for the core C4 cycle have higher steady-state mRNA levels, and we propose that, at least in C. gynandra, the activity of C4 cycle enzymes and transport proteins is controlled at least partially at the level of transcript abundance.
Alterations to the Abundance of Transcripts Associated with Other Metabolic Processes
Changes in the abundance of transcripts that are not associated with the core C4 cycle are also detectable in leaves of C. gynandra and C. spinosa. The high-flux C4 cycle poses additional demands in terms of ATP and reduction equivalents on the light reaction (Hatch, 1987). Specifically, the recycling of the initial CO2 acceptor PEP requires additional ATP molecules (Hatch, 1987). In C4 leaf tissue, one-third to one-half of the genes in the photosynthetic gene classes that contribute to ATP production by cyclic electron flow are up-regulated compared with C3 leaf tissue: PSI, the cytochrome b6/f complex, and the genes mediating cyclic electron flow themselves (Fig. 3). It remains an open question whether these higher steady-state levels are caused by higher ATP demand or whether C4 photosynthesis requires up-regulation of these genes to meet the ATP demand prior to establishing C4 photosynthesis.
On the other hand, the classes of Calvin-Benson cycle genes and photorespiratory genes are those with the highest number of genes with significantly lower steady-state mRNA levels. It is a well-established fact that most C4 plants have less Rubisco protein compared with C3 plants (Ku et al., 1979) and that flux through the photorespiratory pathway is reduced compared with C3 species (Chollet and Ogren, 1975; Leegood, 2002). Transcripts encoding the large and small subunits of Rubisco were reduced from 22,968 and 15,442 RPM to 6,984 and 4,900 RPM in C. spinosa and C. gynandra, respectively. Overall, the trend for Calvin-Benson cycle genes was for them to be down-regulated in C. gynandra compared with C. spinosa (Fig. 3). Likewise, a large number of genes encoding photorespiratory proteins, proteins involved in one-carbon compound metabolism, and the genes involved in ammonia reassimilation, Gln synthetase, and Glu synthase have lower steady-state transcriptional levels (Fig. 3; Supplemental Table S3). The reduced flow through the photorespiratory pathway obviously decreases the demand on the expression system to maintain high steady-state levels of mRNA for many Calvin-Benson cycle and photorespiratory genes. The photosynthetic genes, the Calvin-Benson cycle and photorespiratory genes (in C3), and the C4 cycle genes (in C4) are those with the highest read counts of the genes with known function (Supplemental Table S3). Although it is currently not possible to quantify absolute transcript levels, since the genome of neither Cleome species has been sequenced, the high read counts obtained for the genes of central carbon metabolism and photosynthesis indicate that the steady-state levels of transcripts are high. Since the most altered gene classes are also those that contain the genes with the highest absolute read counts, it is not clear whether C4 photosynthesis lowers or raises the demand on protein synthesis and accessory pathways such as amino acid synthesis. However, both the protein synthesis and the amino acid metabolism classes contain more genes that have lower steady-state levels in C4 leaf tissue (Fig. 3). Within the protein synthesis gene class, many transcripts encoding structural components of plastidic and cytosolic ribosomes were reduced (Supplemental Fig. S3). This was not the case for components of mitochondrial ribosomes (Supplemental Fig. S3), indicating that there is not a general effect on translation but that the effect is likely specific to ribosomes involved in translation for the Calvin-Benson cycle and photorespiration. The protein-to-fresh weight ratio is also lower in C4 leaf tissue compared with C3 leaf tissue (Supplemental Fig. S2). We propose that plastidic ribosomes are relieved of the high translation load associated with the large subunit of Rubisco and that the cytosolic ribosomes need to translate fewer transcripts associated with central carbon metabolism as well as the small subunit of Rubisco. The reduced production of proteins in the leaves of C4 plants is considered important in increasing nitrogen use efficiency, because the rate of photosynthesis per unit of nitrogen in the leaf is increased (Oaks, 1994). Our data indicate that there is also likely a significant saving in the nitrogen provision in the leaf, because fewer ribosomes as well as fewer proteins for central carbon metabolism are required.
The data set contains two additional gene classes, β-1,3 glucan metabolism and putative lipid transfer proteins, that showed differences in transcript abundance between C. gynandra and C. spinosa that could be explained within the current framework of knowledge of C4 photosynthesis. The C4 pathway requires efficient exchange of metabolites between M and BS cells via large numbers of plasmodesmata connecting both cell types, while the BS cell wall of many C4 plants is suberized to reduce diffusion of CO2 away from Rubisco (Hatch, 1987). Transcripts encoding three distinct glucan 1,3-β-glucosidases (Table IV) involved in governing plasmodesmatal conductivity by regulating the turnover of the β-1,3-glucan callose (Levy et al., 2007) were up-regulated in leaves of C. gynandra compared with C. spinosa. Therefore, it is possible that these genes are involved in increasing the open probability of plasmodesmata (Roberts and Oparka, 2003), which allows the efficient flux of organic acids between M and BS cells required during C4 photosynthesis (Evert et al., 1977; Botha, 1992; Roberts and Oparka, 2003). A transcript annotated as a putative lipid transfer protein is among those that are most highly up-regulated in C. gynandra compared with C. spinosa. Lipid transfer proteins are required for the export of lipids to the cell wall during cutin biosynthesis (DeBono et al., 2009). Interestingly, in Arabidopsis, some lipid transfer proteins are exclusively and abundantly expressed in the root endodermis, where suberin biosynthesis is required to establish the Casparian strip.
There are additional changes in the transcript profile that are less easily explained. Among the gene classes containing more genes with significantly higher transcript levels in C4 leaf tissue are starch metabolism, cofactor synthesis and nitrogen metabolism, and heat shock/protein folding (in order of decreasing number of significantly different genes). On the other hand, it is difficult to conceive why genes involved in metal handling are frequently lower in transcript level in C4 leaf tissues (Fig. 3). These changes may be connected to currently unknown phenomena relating to the C4 pathway or may be part of differences not relating to C4 photosynthesis between the two species. Overall, the global analysis of transcription on the level of functional classes reveals unexpected shifts in transcript profiles that can be explained based on the current knowledge about the C4 pathway, while a range of smaller changes remain enigmatic.
Finally, our global transcriptional analysis of C4 and C3 leaf tissues not only allows testing hypotheses about the C4 pathway on a global scale but also allows genes with expression patterns similar to those of known C4 genes to be identified. The phylogenetic proximity of the Cleomaceae to Arabidopsis allows the identification of the orthologs in Arabidopsis, which will facilitate translational research into the model species (Brown et al., 2005).
Candidates for Additional C4-Related Genes
The identification of transport proteins involved in the C4 cycle lags behind that of enzymes, considering that the C4 cycle requires the intracellular transport of pyruvate, PEP, Asp, and Ala across different organellar membranes (Bräutigam and Weber, 2011). A wide range of C4 plants take up pyruvate into chloroplasts from the M in cotransport with sodium (Aoki et al., 1994; Aoki and Kanai, 1997), which might explain the requirement for sodium as a micronutrient in many C4 species (Brownell and Crossland, 1972). Since the rate of pyruvate transport into C4 M cell chloroplasts occurs at or exceeds the apparent rate of CO2 assimilation, sodium-coupled pyruvate import implies a large influx of sodium into these chloroplasts, but the transporter has not yet been identified at the molecular level (Aoki and Kanai, 1997). Our finding that a putative plastidic proton:sodium symporter (NHD1) is 16-fold up-regulated in C. gynandra prompts us to hypothesize that it functions in exporting sodium from the chloroplast in order to maintain the sodium gradient required for import of pyruvate. In addition, we found strong up-regulation of a putative bile acid:sodium cotransporter in C. gynandra. Interestingly, up-regulation of the putative bile acid:sodium cotransporter or of NHD1 was not observed in maize (Zea mays; Bräutigam et al., 2008a), which belongs to a group of C4 plants that show proton-dependent, not sodium-dependent, transport of pyruvate into M cell chloroplasts (Aoki et al., 1994; Aoki and Kanai, 1997). PEP generated from pyruvate in M cell chloroplasts is exported from these chloroplasts by PPT, thereby providing the substrate for the cytosolic PEPC reaction. Accordingly, transcripts encoding PPT are 20-fold up-regulated in C. gynandra, likely reflecting the increased requirement for transport of PEP (Table III). In contrast to what has been observed for the NADP-ME-type C4 plant maize by quantitative proteomic analysis (Bräutigam et al., 2008a), we did not detect increased transcript abundance of the putative M chloroplast oxaloacetate/malate exchanger DiT1 (Taniguchi et al., 2002, 2004; Renne et al., 2003; Supplemental Table S3). This is consistent with the fact that oxaloacetic acid/malate shuttling across the M cell chloroplast envelope membrane is not required for NAD-ME-type C4 photosynthesis (Weber and von Caemmerer, 2010; Bräutigam and Weber, 2011). The mitochondrial dicarboxylate carriers are prime suspects for the C4 acid importer into the mitochondria, where decarboxylation takes place (Table IV). The initial uptake of inorganic carbon and its conversion to bicarbonate may be facilitated by the concerted action of a membrane intrinsic protein channeling the gas and a carbonic anhydrase that is predicted to be membrane bound (Table IV).
Chloroplasts in the BS of C. gynandra are larger than those in the BS of C3 species and, as in many other C4 plants, are positioned in a strictly centripetal pattern (Marshall et al., 2007; Voznesenskaya et al., 2007). Transcripts derived from the GIANT CHLOROPLAST1 (GC1) gene were more abundant in C. gynandra than in C. spinosa (Table IV). Although overexpression of GC1 in Arabidopsis is reported not to effect chloroplast division (Maple et al., 2004), it is possible that it does so in C. gynandra. In addition, we also detected reduced accumulation of transcripts derived from the CHUP1 and ACTIN11 genes. In Arabidopsis, the outer chloroplast envelope membrane protein CHUP1 contains an actin-binding motif and is required for preventing chloroplast aggregation (Oikawa et al., 2003). Differential positioning of chloroplasts in BS and M cells of the C4 plants finger millet (Eleusine coracana) and maize requires the actomyosin system (Kobayashi et al., 2009). Since AtCHUP1 is involved in positioning chloroplasts at the periclinal plasma membrane during the weak-light acclimation response via a coiled-coil domain and interaction with the cytoskeleton (Oikawa et al., 2003), it is possible that the centripetal positioning of chloroplasts in BS cells is linked to lower expression of the CgCHUP1 and ACTIN11 genes.
Controlling and Maintaining a C4 State in Leaf Tissue
Our estimate that around 603 transcripts accumulate differentially in leaves of C3 and C4 species provides insight into the extent to which gene expression profiles change in C4 leaves. For example, the fact that 258 transcripts were more abundant in the leaves of C4 compared with C3 species indicates that about 2.8% of the leaf transcriptome differentially accumulates in C4 leaves (Supplemental Tables S2 and S6). To compare the complexity of the C4 pathway with other multigenic traits, we assessed the number of transcripts that are known to be regulated by sugars, cold, diurnal and circadian rhythms, as well as attack by pests and pathogens (Table VI). Interestingly, the alterations in transcript abundance of leaves of C. gynandra compared with those of C. spinosa were greater than those observed in response to cold treatment and lower than those induced by Glc feeding, those occurring during pathogen attack, and the response to both diurnal and circadian rhythms. As significant progress has been made in understanding sugar signaling (Rolland et al., 2006), pathogen attack (Wise et al., 2007), and the control of gene expression in response to the diurnal cycle and circadian rhythms (Imaizumi et al., 2007), it should be possible to identify the regulators responsible for these alterations in transcript abundance in a C4 leaf compared with a C3 leaf. The changes in transcript abundance that we document in a C4 leaf compared with a C3 leaf likely overrepresent the changes in transcript abundance actually associated with C4 photosynthesis on a whole leaf basis, as some differences in gene expression are likely due to the phylogenetic distance between C. gynandra and C. spinosa. A more confident estimate of the extent to which the leaf transcriptome is altered in association with C4 photosynthesis will be generated when additional congeneric pairs of C3 and C4 species are subjected to deep transcriptome analysis and shared transcripts are identified. Between M and BS cells, the alterations in gene expression may be greater than those that we have defined for whole leaves. For example, up to 18% of genes are estimated to be differentially expressed between M and BS cells of maize (Sawers et al., 2007). However, it is not clear how different the transcript profiles of M and BS cells are in a dicot C3 leaf, and until this is defined, it is not possible to infer the extent to which transcript abundance alters in these cell types in association with C4 photosynthesis.
Table VI. Comparison of alterations in transcript abundance in C4 and C3 leaves with those induced by cold, sugar feeding, attack by pests or pathogens, diurnal changes to light, or circadian rhythms.
| Cause | Estimated Change in Transcriptome | Change | Reference |
| % | |||
| Cold treatment | 514 (24,000) ATH1 | 2.1 | Vogel et al. (2005) |
| C4 leaves and C3 leaves | 583/603 (13,443/13,662) | 2.7/2.8 | This study |
| Glc feeding | 978 (22,500) ATH1 | 4.4 | Price et al. (2004) |
| Pseudomonas syringae | 2,034 (23,750) ATH1 | 8.6 | De Vos et al. (2005) |
| Myzus persicae | 2,181(23,750) ATH1 | 9.1 | De Vos et al. (2005) |
| Diurnal regulation | 1,115 (11,521) cDNA array | 11 | Schaffer et al. (2001) |
| Circadian regulation | 2,282 (18,890) Galbraith | 12 | Dodd et al. (2007) |
As we sampled from mature leaves to capture the differences between C3 and C4 leaves at the point of fully differentiated pathways, we likely also captured regulatory genes needed to maintain C4 architecture and metabolism in mature leaves. Of the 17 transcription factors significantly altered (Table V), GOLDEN2-LIKE1 (GLK1) has previously been implicated in regulating genes important in C4 photosynthesis. In maize, GOLDEN2 controls functional differentiation of chloroplasts in BS cells (Langdale and Kidner, 1994), and GLK1 has been implicated in the expression of photosynthesis genes in M cells (Rossini et al., 2001). The fact that GLK1 transcripts are significantly more abundant in leaves of C. gynandra would not necessarily be predicted, as previous work indicates that it becomes specialized in BS cells of C4 leaves but not that its abundance is altered significantly. This implies that the increase in abundance of GLK1 transcripts may not simply be due to its involvement in C4 photosynthesis. When overexpression of GLK1 was induced in Arabidopsis, the abundance of 114 transcripts was altered (Waters et al., 2009). We assessed the extent to which the genes that are controlled by GLK1 change in abundance in leaves of C. gynandra compared with C. spinosa and found that only 19 genes were shared between the two data sets. This may be due to a number of factors that could include the following: that there are differences in the targets of GLK1 in Arabidopsis and C. gynandra; that a number of other transcriptional regulators are more important than GLK1 in maintaining patterns of photosynthesis gene expression in C. gynandra; and that a rapid induction of GLK1 gene expression has more impact than increasing the steady-state level of GLK1. This analysis is also subject to the caveat that in neither case was the amount of GLK1 protein measured.
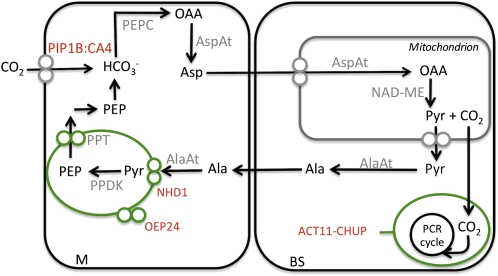
In all of our analyses, differences in transcript abundance between the leaves of C. gynandra and C. spinosa may reflect the operation of the C4 and C3 photosynthetic pathways; alternatively, they may be due to differences in metabolism and cell biology associated with the phylogenetic distance between the two species. However, in many cases, it is striking that our analysis has identified differences in the abundance of transcripts derived from genes that have been documented to be involved in processes known to alter in a C4 leaf. Taken together, the analysis allows us to significantly extend the number of C4-related genes controlled at the level of transcript abundance and to extend the current model for C4-related processes in NAD-ME C4 plants (Fig. 4). Analysis of additional pairs of C3 and C4 species will likely facilitate the identification of genes specifically involved in the C4 pathway and exclude genes that are modified for other reasons.
Figure 4.
Schematic of components associated with the C4 cycle in the NAD-ME subtype based on interpretation of RNA-Seq. Proteins that have been described previously are in gray, and novel proteins are marked in red. Metabolites are in black. PIP1B:CA4, PIP1B plasma membrane aquaporin:membrane-tethered carbonic anhydrase; OAA, oxaloacetic acid; ACT11-CHUP11, ACTIN11-CHUP1 complex; Pyr, pyruvate; OEP24, chloroplast outer envelope protein 24.
MATERIALS AND METHODS
Plant Material and 454 Sequencing
Cleome spinosa and Cleome gynandra plants for transcript profiling by RNA-Seq were grown in standard potting mix in a glasshouse in August and September 2007. To obtain sequence tags for DGE analysis from C. spinosa and C. gynandra, total RNAs were isolated from fully expanded leaves sampled from 56-d-old plants of each species. mRNA was reverse transcribed to cDNA after two consecutive rounds of oligo(dT) purification and prepared for 454 sequencing as described previously (Weber et al., 2007).
Mapping and Quantification of the Sequence Reads
Evolution did not stop in the lineage to the reference genome of Arabidopsis (Arabidopsis thaliana) after the Cleomaceae branch diverged. Hence, there may be genes that were tandem duplicated or retained after the whole genome duplication event of the Brassicaceae that are absent in either of the Cleomaceae species (Bräutigam and Gowik, 2010). To avoid mapping problems such as splitting of reads or mapping errors due to differential retention of genes in either Cleomaceae or Arabidopsis, we created a minimal genome for mapping. The remnants of the last whole genome duplication in the lineage of the Brassicaceae (Bowers et al., 2003; Thomas et al., 2006) and the tandem duplicated genes (Haberer et al., 2004) were reduced to one representative for each based on the TAIR 9 coding sequence set. In each case, the gene with the lowest AGI code was retained for mapping. For each gene, the Supplemental Data store whether there are duplicates and which duplicates match the gene (Supplemental Tables S3 and S5). We recommend recovery of the associated duplicated genes followed by a detailed analysis with phylogenetic trees to define the true ortholog when translating the results of Cleomaceae analyses to Arabidopsis research.
The 454 sequence reads were mapped onto coding sequences of the minimalized TAIR 9 genome by BLAT (Kent, 2002) and BLAST (Altschul et al., 1997) with varying parameters, and the output was parsed with in-house PERL scripts to retain only the best matching AGI codes for each sequence read and the best BLAST hit, respectively. Differentially expressed transcripts were identified using the Poisson statistics developed by Audic and Claverie (1997) followed by a Bonferroni correction to account for the accumulation of α-type errors when conducting multiple pair-wise comparisons (Audic and Claverie, 1997).
Plant Material and qPCR Analysis
Both species were grown in a growth chamber in long-day conditions (16 h of light/8 h of dark) under 350 μmol photons m−2 s−1, at 22°C, and 65% relative humidity prior to samples being taken for qPCR. qPCR was conducted on the same samples used for RNA-Seq and also on mature leaves collected at noon grown in the growth cabinet. For qPCR, RNA was isolated using TriPure reagent (Roche Applied Science). RNA was treated with DNase I (Promega) and purified with the RNeasy Mini Kit (Qiagen). First-strand cDNA was then synthesized with SuperScriptII reverse transcriptase (Invitrogen) using 4 μg of RNA and oligo(dT) primers (Roche Applied Science). Quantitative reverse transcription-PCR was carried out with 96-well plates using a DNA Engine thermal cycler, Chromo4 real-time detector (Bio-Rad), SYBR Green JumpStart Taq Ready Mix (Sigma), and 15-fold dilution of the cDNA as a template. Initial denaturation was carried out at 94°C for 2 min, followed by 40 cycles of 94°C for 20 s, 60°C for 30 s, 72°C for 30 s, and 75°C for 5 s. Primers were designed to have melting temperatures of 60°C ± 0.5°C and to produce amplicons of 91 to 189 bp. The specificity of the primers and lack of primer dimers in the PCR were verified using agarose gel electrophoresis and melting curve analysis. For each product, the threshold cycle CT, where the amplification reaction enters the exponential phase, was determined for three technical replicates and four independent biological replicates per species. The comparative 2−ΔΔCT method was used to quantify relative abundance of transcripts (Livak and Schmittgen, 2001). ACTIN7 was chosen as a reference because the 454 sequencing data showed equal, intermediate levels of ACTIN7 transcripts in both species. For the qPCR, se values were calculated from 2−ΔΔCT values of each combination of biological replicates.
Polar Metabolite, Chlorophyll, Protein, and Enzyme Activity Analyses
For metabolite analysis, mature leaves from 56-d-old plants were collected in the middle of the light period and immediately frozen in liquid nitrogen. Three independent biological replicates were used. The tissues were ground in a mortar, and a 50-mg fresh weight aliquot was extracted using the procedure described by Lee and Fiehn (2008). Ribitol was used as an internal standard for data normalization. For GC-EI-TOF analysis, samples were processed and analyzed according to Lee and Fiehn (2008). Enzyme activities, chlorophyll, and protein content were determined according to Hausler et al. (2001).
The Cleome read data have been submitted to the National Center for Biotechnology Information Short Read Archive: C. spinosa = SRS002743.1 and C. gynandra = SRS002744.2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantitation of marker enzyme activities in leaf extracts of C. spinosa and C. gynandra.
Supplemental Figure S2. Protein-to-fresh weight and protein-to-chlorophyll ratios in leaves of C. gynandra and C. spinosa.
Supplemental Figure S3. Changes in transcript abundance for ribosomal proteins.
Supplemental Table S1. Relative abundance of predominant metabolites detected by GC-EI-TOF in C. gynandra and in C. spinosa.
Supplemental Table S2. Number of gene loci and number of differentially expressed genes detected with BLAT and BLAST.
Supplemental Table S3. Quantitative information for all reads mapped onto the reference genome from Arabidopsis.
Supplemental Table S4. Comparison of mapping parameters.
Supplemental Table S5. Segmental and tandem duplicates in the Arabidopsis genome.
Supplementary Material
Acknowledgments
We thank Tom Hardcastle for help with R.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Kanai R. (1997) Reappraisal of the role of sodium in the light-dependent active transport of pyruvate into mesophyll chloroplasts of C4 plants. Plant Cell Physiol 38: 1217–1225 [Google Scholar]
- Aoki N, Ohnishi JI, Kanai R. (1994) Proton/pyruvate cotransport into mesophyll chloroplasts of C-4 plants. Plant Cell Physiol 35: 801–806 [Google Scholar]
- Audic S, Claverie JM. (1997) The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Black CC. (1973) Photosynthetic carbon fixation in relation to net CO2 uptake. Annu Rev Plant Physiol Plant Mol Biol 24: 253–286 [Google Scholar]
- Botha CEJ. (1992) Plasmodesmatal distribution, structure and frequency in relation to assimilation in C3 and C4 grasses in southern Africa. Planta 187: 348–358 [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong JK, Paterson AH. (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Gowik U. (2010) What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol 12: 831–841 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Hofmann-Benning S, Weber APM. (2008a) Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol 148: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Shrestha RP, Whitten D, Wilkerson CG, Carr KM, Froehlich JE, Weber APM. (2008b) Comparison of the use of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. J Biotechnol 136: 44–53 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Weber APM. (2011) Transport processes: connecting the reactions of C4 photosynthesis. Raghavendra AS, Sage RF, , C4 Photosynthesis and Related CO2 Concentrating Mechanisms. Advances in Photosynthesis and Respiration, Vol 32. Springer, Berlin, pp 199–219 [Google Scholar]
- Brown NJ, Parsley K, Hibberd JM. (2005) The future of C4 research: maize, Flaveria or Cleome? Trends Plant Sci 10: 215–221 [DOI] [PubMed] [Google Scholar]
- Brown RH. (1999) Agronomic implications of C4 photosynthesis. Sage RF, Monson RK, , C4 Plant Biology. Academic Press, San Diego, pp 473–507 [Google Scholar]
- Brownell PF, Crossland CJ. (1972) The requirement for sodium as a micronutrient by species having the C4 dicarboxylic photosynthetic pathway. Plant Physiol 49: 794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R, Ogren WL. (1975) Regulation of photorespiration in C3 and C4 species. Bot Rev 41: 137–179 [Google Scholar]
- DeBono A, Yeats TH, Rose JKC, Bird D, Jetter R, Kunst L, Samuelsa L. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux JP, Van Loon LC, Dicke M, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, Assie JM, Robertson FC, Jakobsen MK, Goncalves J, et al. (2007) The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 318: 1789–1792 [DOI] [PubMed] [Google Scholar]
- Evert RF, Eschrich W, Heyser W. (1977) Distribution and structure of plasmodesmata in mesophyll and bundle-sheath cells of Zea mays L. Planta 136: 77–89 [DOI] [PubMed] [Google Scholar]
- Fischer K, Kammerer B, Gutensohn M, Arbinger B, Weber A, Hausler RE, Flugge UI. (1997) A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Birney E. (2009) Sense from sequence reads: methods for alignment and assembly. Nat Methods 6: S6–S12 [DOI] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KFX. (2004) Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136: 3009–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD. (1987) C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta 895: 81–106 [Google Scholar]
- Hatch MD, Slack CR. (1968) A new enzyme for interconversion of pyruvate and phosphopyruvate and its role in C4 dicarboxylic acid pathway of photosynthesis. Biochem J 106: 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausler RE, Rademacher T, Li J, Lipka V, Fischer KL, Schubert S, Kreuzaler F, Hirsch HJ. (2001) Single and double overexpression of C4-cycle genes had differential effects on the pattern of endogenous enzymes, attenuation of photorespiration and on contents of UV protectants in transgenic potato and tobacco plants. J Exp Bot 52: 1785–1803 [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Sheehy JE, Langdale JA. (2008) Using C4 photosynthesis to increase the yield of rice: rationale and feasibility. Curr Opin Plant Biol 11: 228–231 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA, Schroeder JI. (2007) Circadian rhythms: daily watch on metabolism. Science 318: 1730–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL. (1984) The CO2/O2 specificity of ribulose 1,5-bisphosphate carboxylase oxygenase: dependence on ribulosebisphosphate concentration, pH and temperature. Planta 161: 308–313 [DOI] [PubMed] [Google Scholar]
- Keeley JE. (1998) C4 photosynthetic modifications in the evolutionary transition from land to water in aquatic grasses. Oecologia 116: 85–97 [DOI] [PubMed] [Google Scholar]
- Kent WJ. (2002) BLAT: the BLAST-Like Alignment Tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Yamada M, Taniguchi M, Kawasaki M, Sugiyama T, Miyake H. (2009) Differential positioning of C4 mesophyll and bundle sheath chloroplasts: recovery of chloroplast positioning requires the actomyosin system. Plant Cell Physiol 50: 129–140 [DOI] [PubMed] [Google Scholar]
- Ku MSB, Schmitt MR, Edwards GE. (1979) Quantitative determination of ribulose bisphosphate carboxylase oxygenase protein in leaves of several C3 and C4 plants. J Exp Bot 114: 89–98 [Google Scholar]
- Langdale JA, Kidner CA. (1994) Bundle-sheath defective, a mutation that disrupts cellular differentiation in maize leaves. Development 120: 673–681 [Google Scholar]
- Lee DY, Fiehn O. (2008) High quality metabolomic data for Chlamydomonas reinhardtii. Plant Methods 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC. (2002) C4 photosynthesis: principles of CO2 concentration and prospects for its introduction into C3 plants. J Exp Bot 53: 581–590 [DOI] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL. (2007) A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Majeran W, van Wijk KJ. (2009) Cell-type-specific differentiation of chloroplasts in C4 plants. Trends Plant Sci 14: 100–109 [DOI] [PubMed] [Google Scholar]
- Maple J, Fujiwara MT, Kitahata N, Lawson T, Baker NR, Yoshida S, Moller SG. (2004) GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Curr Biol 14: 776–781 [DOI] [PubMed] [Google Scholar]
- Marshall DM, Muhaidat R, Brown NJ, Liu Z, Stanley S, Griffiths H, Sage RF, Hibberd JM. (2007) Cleome, a genus closely related to Arabidopsis, contains species spanning a developmental progression from C3 to C4 photosynthesis. Plant J 51: 886–896 [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Furbank RT, Fukuyama H, Miyao M. (2001) Molecular engineering of C4 photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 52: 297–314 [DOI] [PubMed] [Google Scholar]
- Metzker ML. (2010) Applications of next-generation sequencing technologies: the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Mitchell PL, Sheehy JE. (2006) Supercharging rice photosynthesis to increase yield. New Phytol 171: 688–693 [DOI] [PubMed] [Google Scholar]
- Oaks A. (1994) Efficiency of nitrogen utilization in C3 and C4 cereals. Plant Physiol 106: 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K, Kasahara M, Kiyosue T, Kagawa T, Suetsugu N, Takahashi F, Kanegae T, Niwa Y, Kadota A, Wada M. (2003) CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CP, Freckleton RP. (2009) Ecological selection pressures for C4 photosynthesis in the grasses. Proc R Soc Lond B Biol Sci 276: 1753–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri N, Schlotterer C. (2009) Mapping accuracy of short reads from massively parallel sequencing and the implications for quantitative expression profiling. PLoS ONE 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC. (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiskind JB, Berg RH, Salvucci ME, Bowes G. (1989) Immunogold localization of primary carboxylases in leaves of aquatic and a C3-C4 intermediate species. Plant Sci 61: 43–52 [Google Scholar]
- Renne P, Dressen U, Hebbeker U, Hille D, Flugge UI, Westhoff P, Weber APM. (2003) The Arabidopsis mutant dct is deficient in the plastidic glutamate/malate translocator DiT2. Plant J 35: 316–331 [DOI] [PubMed] [Google Scholar]
- Roberts AG, Oparka KJ. (2003) Plasmodesmata and the control of symplastic transport. Plant Cell Environ 26: 103–124 [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rossini L, Cribb L, Martin DJ, Langdale JA. (2001) The maize Golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 13: 1231–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. (2004) The evolution of C4 photosynthesis. New Phytol 161: 341–370 [DOI] [PubMed] [Google Scholar]
- Sawers RJH, Liu P, Anufrikova K, Hwang JTG, Brutnell TP. (2007) A multi-treatment experimental system to examine photosynthetic differentiation in the maize leaf. BMC Genomics 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1999) C4 gene expression. Annu Rev Plant Physiol Plant Mol Biol 50: 187–217 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Taniguchi Y, Kawasaki M, Takeda S, Kato T, Sato S, Tahata S, Miyake H, Sugiyama T. (2002) Identifying and characterizing plastidic 2-oxoglutarate/malate and dicarboxylate transporters in Arabidopsis thaliana. Plant Cell Physiol 43: 706–717 [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Nagasaki J, Kawasaki M, Miyake H, Sugiyama T, Taniguchi M. (2004) Differentiation of dicarboxylate transporters in mesophyll and bundle sheath chloroplasts of maize. Plant Cell Physiol 45: 187–200 [DOI] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M. (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Edwards GE, Kiirats O, Artyusheva EG, Franceschi VR. (2003) Development of biochemical specialization and organelle partitioning in the single-cell C4 system in leaves of Borszczowia aralocaspica (Chenopodiaceae). Am J Bot 90: 1669–1680 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Franceschi VR, Kiirats O, Artyusheva EG, Freitag H, Edwards GE. (2002) Proof of C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae). Plant J 31: 649–662 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Franceschi VR, Kiirats O, Freitag H, Edwards GE. (2001) Kranz anatomy is not essential for terrestrial C4 plant photosynthesis. Nature 414: 543–546 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Koteyeva NK, Chuong SDX, Ivanova AN, Barroca J, Craven LA, Edwards GE. (2007) Physiological, anatomical and biochemical characterisation of photosynthetic types in genus Cleome (Cleomaceae). Funct Plant Biol 34: 247–267 [DOI] [PubMed] [Google Scholar]
- Wall PK, Leebens-Mack J, Chanderbali AS, Barakat A, Wolcott E, Liang HY, Landherr L, Tomsho LP, Hu Y, Carlson JE, et al. (2009) Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genomics 10: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM, von Caemmerer S. (2010) Plastid transport and metabolism of C3 and C4 plants: comparative analysis and possible biotechnological exploitation. Curr Opin Plant Biol 13: 256–264 [DOI] [PubMed] [Google Scholar]
- Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. (2007) Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol 144: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RP, Moscou MJ, Bogdanove AJ, Whitham SA. (2007) Transcript profiling in host-pathogen interactions. Annu Rev Phytopathol 45: 329–369 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.