Abstract
Autophagy is an evolutionarily conserved, intracellular self-defense mechanism where organelles and proteins are sequestered into autophagic vesicles (AVs) that are subsequently degraded through fusion with lysosomes. Cells thereby prevent the toxic accumulation of damaged or unnecessary components, but also recycle these components to sustain metabolic homoeostasis. Heightened autophagy is a mechanism of resistance for cancer cells faced with metabolic and therapeutic stress, revealing opportunities for exploitation as a therapeutic target in cancer. We summarize recent developments in the field of autophagy and cancer, and build upon the results presented at the Cancer Therapeutics and Evaluation Program (CTEP) Early Drug Development meeting in March, 2010. Herein, we describe our current understanding of the core components of the autophagy machinery, the functional relevance of autophagy within the tumor microenvironment and outline how this knowledge has informed preclinical investigations combining the autophagy inhibitor hydroxychloroquine (HCQ) with chemotherapy, targeted therapy and immunotherapy. Finally, we describe ongoing clinical trials involving HCQ as a first generation autophagy inhibitor, as well as strategies for the development of novel, more potent and specific inhibitors of autophagy.
Introduction
The anatomy of autophagy
Autophagy is a lysosomal degradative pathway characterized by the formation of double-membrane autophagic vesicles (AVs), also known as autophagosomes, which engulf portions of the cytosol, damaged organelles, protein aggregates and bacteria. AVs are typically transported along microtubule tracks to a perinuclear location. The outer membrane of the AV subsequently fuses with the lysosome, resulting in degradation of the AV contents and inner membrane (Figure 1) (1, 2). Autophagy occurs at basal levels in virtually all cells, performing homeostatic functions such as protein and organelle turnover. Autophagy is upregulated when cells require intracellular nutrients and energy such as during starvation and growth factor withdrawal or in the context of high bioenergetic demand. Additionally, autophagy is upregulated under other stress conditions, such as when there is a need to clear aggregated proteins, damaged organelles, or intracellular pathogens. A number of signaling pathways intersect with the autophagy system. This intersection allows a tightly regulated and dynamic autophagic response to environmental perturbations.
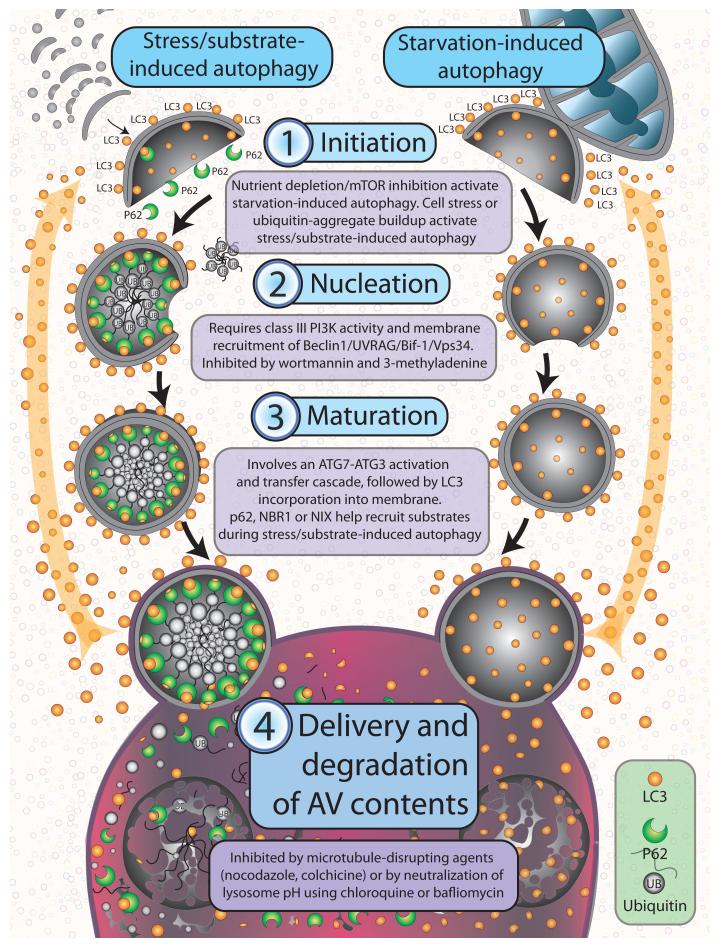
Figure 1. Anatomy of autophagy.
Autophagy occurs through a multi-step process including 4 control points: 1) initiation, 2) nucleation, 3) maturation, and 4) delivery and degradation of AV contents. These steps occur irrespective of whether autophagy has been induced through stress/ubiquitinated substrate accumulation, or through starvation. During initiation, nascent AV membranes derived from multiple potential sources (including isolated membranes, ER or mitochondria outer membranes) form a cup-like structure onto which autophagosomal machinery, including LC3, dynamically associates. As the cup-like structure enlarges, it sequesters substrate, which includes ubiquitinated proteins or organelles in the case of stress/substrate induced autophagy, and soluble cytoplasm in the case of starvation induced autophagy. The double membrane comprising the nascent AV then closes to form the mature autophagic vesicle, which then targets and fuses with the lysosome. In the lysosome, hydrolytic enzymes digest the contents and inner membrane of the AV, with autophagic machinery (i.e., LC3) recycled through the cytoplasm for recruitment to other nascent autophagosomes.
AV production and turnover
The anatomy, physiology, and molecular machinery of autophagy is highly conserved among eukaryotic cells. It includes distinct steps for AV production and turnover, including 1) initiation, 2) nucleation of 3) maturation of AVs, and 4) fusion and degradation of AV contents in lysosomes (3, 4) (Figure 1). The ULK1 (ATG1) kinase complex consisting of ULK1 (and/or possibly ULK2), Atg13, and Atg17 integrates stress signals from mammalian target of rapamycin complex 1 (mTORC1) and controls the initiation of autophagy (5, 6). Once mTORC1 kinase activity is inhibited, the cytoplasmic autophagy machinery described below is recruited onto phospholipid membranes derived from the endoplasmic reticulum (7) and trans-golgi network (8). More recently the mitochondrial outer membrane (9) and plasma membrane (10) were identified as additional important sources of phosphatidylethenolamine (PE)-rich membranes, which are characteristic of AVs.
AV formation begins with the generation of phosphoinositide signals on the surface of source membranes by multi-protein complexes that include the class III phosphoinositide 3-kinase (PI3K) Vps34 and Beclin1 (11). The cytoplasmic ubiqituin-like protein Atg8 (LC3) is conjugated to PE on these membranes, which identifies them as incipient AVs. Lipidation of LC3 occurs by a ubiquitin-like protein (UBL) conjugation cascade involving an E1-like enzyme (ATG7) and E2-like enzyme (ATG3), following cleavage by a cysteine protease (ATG4) (12). Once LC3 is integrated into the bilayer, it recruits cargo adaptor proteins (also known as autophagy receptors) such as p62, Nbr1 or NIX. These proteins in turn recruit cargo from the cytoplasm (i.e., ubiquinated protein aggregates in the case of p62 (13) and damaged organelles in the case of NIX (14)) to promote AV closure (15). AVs are then delivered to lysosomes where their luminal and inner membrane constituents are broken down by lysosomal hydrolases. Lysosomal permeases then release the degradation products into the cytosol for re-utilization (16). AV components not exposed to lysosomal hydrolases are recycled via a system involving multiple components of the outer membrane ATG9, ATG2, ATG18 and ATG21 (17). Alternatively, autophagosomes may also fuse with the plasma membrane and release their contents (18).
Pharmacological targeting of control points in the autophagy system
Autophagy initiation is associated with down-regulation of mTORC1 activity. Activated mTORC1 inhibits autophagy by causing hyperphosphorylation of ATG13, reducing its interaction with ATG1/ULK1, and by controlling phosphorylation of autophagy effectors such as the Vps34-Beclin1 complex. Proteomic studies investigating how inhibition of the mTORC1 pathway affects the global features of autophagy control showed no large-scale changes in core conjugation, lipid kinase and recycling complexes. This implies that post-translational modifications may be involved in AV accumulation when the autophagy pathway is activated (4) and may be a potential means to control autophagy.
AV nucleation represents a second major autophagy control point, involving Vps34, and interacting partners, Beclin1 and p150 (19). Drugs that interfere with recruitment of Vps34 to membranes, including wortmannin and 3-methyladenine, are powerful (although non- specific) proximal inhibitors of autophagy. Direct inhibitors of Vps34 and drugs that sequester or free-up Beclin1 may also be deployed for autophagy inhibition (20). Multiple PI3K/Beclin1 complexes may be involved in mammalian autophagy (11). For example, PI3K/Beclin1 complexed with UVRAG and Bif-1(21) can activate autophagy on membranes, whereas PI3K/Beclin1 complexed to Rubicon (22) plays an inhibitory role in membrane trafficking of AV to lysosomes. Therefore, care must be taken in interpreting results when Vps34 or Beclin1 are pharmacologically or genetically suppressed in autophagy studies.
UBL-containing ATG8 family proteins are central coordinators of AV maturation (4), and represent a third autophagy control point. LC3 the most widely studied ATG8 family member is cleaved by ATG4 and conjugated to PE by an ATG7- and ATG3-dependent activation and transfer cascade. In this manner, LC3 is incorporated into the membrane where it orchestrates AV growth and cargo recruitment. Cargo recruitment involves a conserved surface on LC3 (23) interacting with motifs in cargo binding proteins. Mutations in these motifs reduce the binding of cargo adaptor proteins such as p62, Nbr1 and Nix to ATG8 proteins, and disrupt transfer of AV cargo to lysosomes (13, 24, 25). Nbr1 and p62 contain ubiquitin-binding domains in addition to the motif that interacts with LC3. This allows these adaptor proteins to bind both ubiquinated cargo and LC3, enabling tight sequestration of ubiquinated cargo by surrounding LC3-containing membranes, with little cytosolic content included (26). The cargo adaptor protein NIX similarly recruits mitochondria to LC3-containing membranes (25). ATG8 family members such as LC3 dictate cargo binding through cargo adaptor interaction thereby determining the type of cargo sequestered during autophagy.
Delivery and degradation of AV contents represents a fourth autophagy control point. Because AVs and lysosomes move along microtubules, drugs that disrupt microtubules, such as nocodazole, colchicines, taxanes, and vinca alkaloids, inhibit AV fusion with lysosomes, resulting in AV accumulation. Rab GTPases likely play a role in vesicle maturation and fusion with lysosomes (27). Lysosomes are acidic organelles, with their digesting hydrolases dependent on low pH. Consequently agents such as bafilomycin or chloroquine derivatives, which disrupt the vacuolar H+ ATPase responsible for acidifying lysosomes, block autophagy in its final step, resulting in the accumulation of AVs (28-30).
Autophagy subtypes
Types of autophagy vary depending on the stimulus and requirement for substrate degradation (4, 31, 32). Inhibition of mTOR for example, decreases association of p62 with LC3-containing membranes (4). This type of autophagy, occurring when food supply is limiting, is likely different from autophagy activated when cells are stressed from buildup of damaged organelles and protein aggregates (which utilizes p62). Differences in starvation- versus stress-induced autophagy are also manifested by the site of AVs origination and by the type of cargo sequestered. For instance, starvation-induced autophagy is characterized by AV membrane budding off of the mitochondrial outer membrane and once formed, starvation-induced AVs are more likely to contain free, soluble cytosol (9).
The breadth of autophagy’s crucial roles in survival, adaptability and overall physiology suggests multiple subtypes of autophagy that are location- and cargo-specific within the cell, and tissue-specific within the organism. Thus, therapeutic strategies for inhibiting or inducing autophagy need to be tailored towards stress- versus starvation-induced autophagy. Further analysis of the physiological conditions under which different subtypes of autophagy are utilized, and further clarification of which autophagy pathway is targeted by specific inducers or inhibitors will guide development of autophagy modulators in cancer therapeutics.
Autophagy suppresses tumor development while supporting survival of established tumors
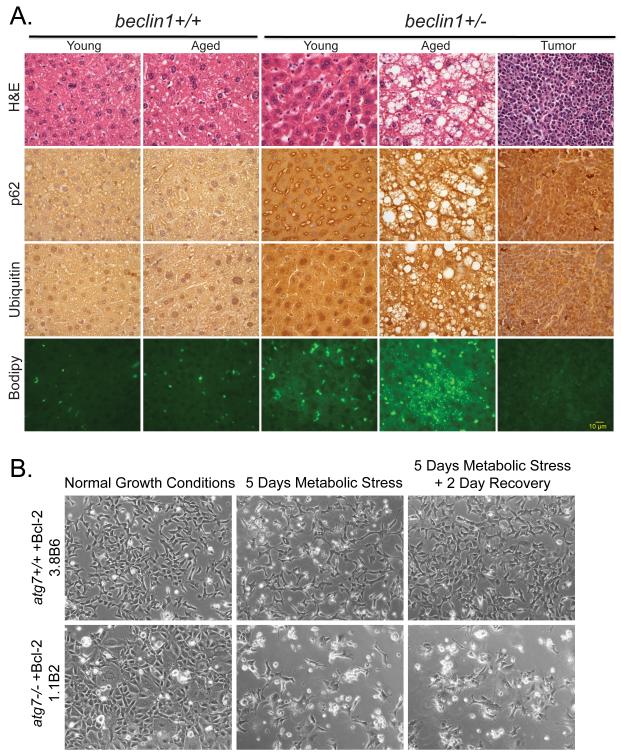
Comparison of normal and autophagy-defective mice and cells has illuminated the role of autophagy in suppression of tumor development. Mice with autophagy defects accumulate ubiqutinated keratins, the autophagy cargo adaptor p62, and abnormal mitochondria (33-35). High levels of p62 in many tissues and tumors and phospho-keratin 8 in mammary tissues and tumors are potential biomarkers for autophagy defects (34, 36). These damaged cellular components accumulate, often in large aggregates or inclusions, and are linked to reactive oxygen species (ROS) production, activation of the DNA damage response, cell damage, and death that can lead to a chronic inflammatory state (35, 37). Progressive cell and tissue damage due to failure of autophagy-mediated cellular garbage disposal provokes degenerative and inflammatory diseases (38, 39) and may contribute to cancer. Chronic tissue damage and inflammation is associated with DNA-damaging ROS production, contributing to mutations that can initiate cancer and promote tumor progression (40). Mice with allelic loss of the essential autophagy gene beclin1 display defective autophagy, altered protein homeostasis (accumulation of ubiqutinated proteins and p62) and gross morphologic tissue damage that is particularly striking in liver where there is also an accelerated incidence of hepatocellular carcinoma (33-35) (Figure 2A). These findings suggest that autophagy stimulators may prevent both degenerative diseases and cancers arising from chronic tissue damage and inflammation, such hepatocellular carcinomas (41, 42).
Figure 2. Role of autophagy in suppressing liver damage and cell death.
(A) Elevated p62, ubiquitin and accumulation of lipids in aged beclin1+/− mouse liver. Sections of liver from young (16 months old) and aged (> 24 months old) beclin1 +/+ and +/− mice and a representative spontaneous liver tumor from a beclin1+/− mouse were stained with H&E, and by immunohistochemistry for p62 and ubiquitin, and with BODIPY to indicate lipid droplet accumulation by fluorescence. (B) Autophagy promotes cell viability in metabolic stress. Representative images of immortal baby mouse kidney epithelial (iBMK) cells derived from atg7+/+ and −/− mice that were untreated, treated with metabolic stress (ischemia: no glucose and 1% oxygen or hypoxic conditions) and allowed to recover (35). Images generously provided by Dr. C. Karp and H.-Y. Chen from the White laboratory.
While autophagy can suppress tumor development, it clearly plays a role in promoting the survival of tumor cells within the tumor microenvironment. Although autophagy induction can be associated with cancer cell death, this may be due to a futile attempt of the cancer cells to survive through autophagy, also known as cell death with autophagic features (43). This underscores the importance of interrogating the functional role of autophagy when autophagosomes are present. In some cases, knockdown of essential autophagy genes by RNAi enhances survival. Whether this is due the absence of autophagic cell death by and prevention of over-activation of autophagy and cell death by fatal self-consumption, or another unknown mechanism, is not yet known. In other settings autophagic cell death was limited to an in vitro conditions and not manifested in vivo. The most prevailing and convincing evidence, however, is that in vivo, autophagy is induced by cellular stress, including nutrient, growth factor and oxygen deprivation, and functions to maintain survival of normal cells, mice and also tumor cells (37, 44, 45). When in vitro models incorporate stresses commonly encountered in vivo, autophagy’s contribution to cell survival becomes more clear. For example autophagy-defective tumor cells undergoing metabolic stress (ischemia) showed impaired survival in comparison with autophagy-proficient cells (Figure 2B). Furthermore, autophagy localizes to hypoxic regions within tumors, and genetic ablation of autophagy promotes the selective death of those metabolically stressed cells (41). The mechanism by which autophagy enables survival of normal or tumor cells in stress is not known. In oxidative stress, the clearance of damaged proteins and organelles, particularly mitochondria, may limit cellular damage and death through ROS production. When nutrients are limiting, autophagy may promote viability by maintaining cellular metabolism through intracellular recycling (46).
Regardless of how autophagy increases survival in stress, concurrent inhibition of autophagy may improve outcomes in cancer therapy. Cytotoxic cancer therapeutics induce autophagy, most likely by causing damage to DNA, cellular proteins and organelles. Inhibition of autophagy in preclinical models improves the response of tumors to alkylating agents, suggesting that autophagy promotes survival (47). Targeted cancer therapies also stimulate autophagy, often by mimicking signaling of starvation or factor deprivation. Inhibitors of mTOR, in particular, are potent activators of autophagy, yet the functional consequences of this activation in cancer therapy is not fully understood (48). An important future direction is to establish the functional consequence of autophagy stimulation by cancer therapeutics. Three additional areas of intense focus critical to understanding the role of autophagy in cancer are: 1) The role of commonly activated oncogenes and inactivated tumor suppressor genes in determining autophagy levels and function within the tumor cell; 2) The role of role of autophagy activation by targeted therapies; 3) Network interactions among the proteasome, the endoplasmic reticulum stress response and autophagy; 4) Extracellular control of autophagy by the immune system, tumor stroma and vasculature.
As outlined in the sections below, knowledge gained from these studies will guide more sophisticated approaches to the therapeutic manipulation of autophagy, with the common aim of limiting the development of cancer, and reducing mortality in patients presenting with overt malignancies. Furthermore, identifying the “autophagic switch” that mediates the transition from suppressed autophagy, important early in the neoplasia, to enhanced autophagy, contributing to malignant progression, is critical to understand this complicated process and to develop rational therapeutic strategies.
Autophagy inhibition can overcome therapeutic resistance to PI3K/mTOR signaling inhibitors
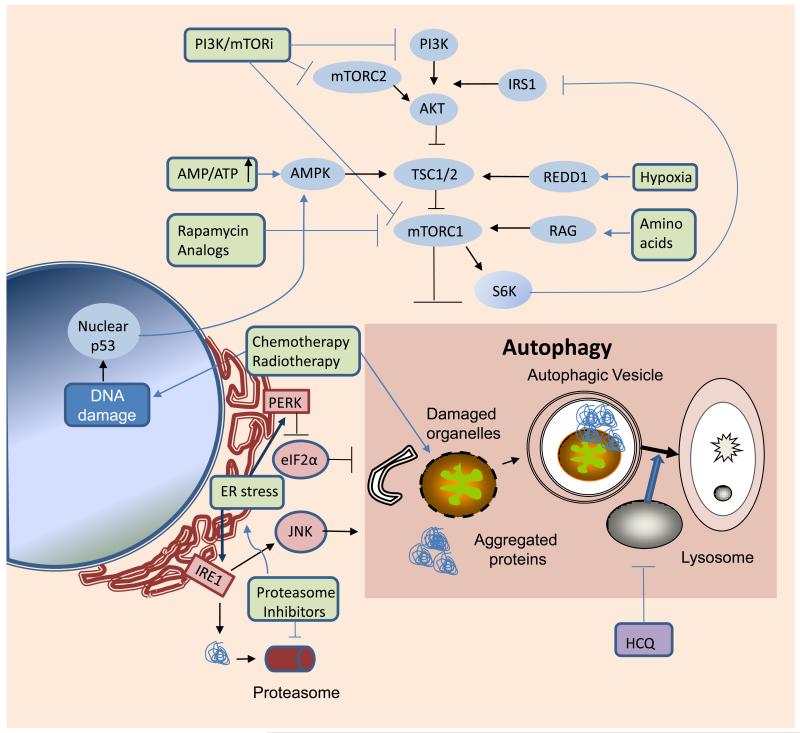
In a recently fed organism, growth factors bind to their cognate receptors (typically a receptor tyrosine kinase-RTK) and signal through class I PI3Ks, leading to phosphorylation of the pleckstrin homology domain kinase Akt, subsequently activating mTOR. The serine-threonine kinase mTOR plays a prominent role in regulating growth and proliferation in both normal and tumor cells (Figure 3). It integrates signals from key environmental sensors such as the AMPK (cellular energy status), Rag GTPases (amino acid availability), REDD1 (oxygen availability), and p53 (DNA damage) (49); and modulates the rate of translation of proteins required for growth, proliferation and metastases. Activated mTOR thus engages anabolic pathways, while in parallel down-regulating catabolic pathways including autophagy. The axis linking growth factor receptors, PI3K, Akt and mTOR is tightly regulated in cells.
Figure 3. Network interactions between kinase signaling, protein metabolism and autophagy.
Metabolic and therapeutic stresses (green) converge on mTORC1 and mediators of ER stress to induce autophagy. Autophagy serves a cytoprotective role in response to these stresses, by clearing damaged organelles, aggregated proteins, and recycling macromolecules to sustain survival. Arrows:activation; flat lines: inhibition. PI3K: phosphotidyl inositol-3 kinase IRS1: insulin receptor substrate 1; mTORC2: mammalian target of rapamycin complex 2; TSC: tuberous sclerosis complex; AMPK: AMP-activated protein kinase; REDD1: regulated in development and DNA damage 1; mTORC1: mammalian target of rapamycin complex 1; PERK: protein kinase-like endoplasmic reticulum kinase; eIF2α: eukaryotic initiation factor 2α: ER: endoplasmic reticulum; IRE-1: inositol requiring enzyme-1; JNK: Jun N-terminal kinase; HCQ: hydroxychloroquine; PI3K/mTORi: dual PI3K/mTOR inhibitors
As central integrators of nutrient and growth factor signaling, PI3K, Akt, and mTOR represent critical nodes regulating cell growth, proliferation and survival. It is therefore not surprising that inappropriate activation of this signaling cascade is commonly found in cancer. Mutational activation of PI3K occurs commonly in cancers, whereas mutational activation of Akt is relatively infrequent. Amplification or epigenetic activation of RTK’s such as EGFR, and mutational or epigenetic inactivation of negative regulators of this pathway, including the phosphatase and tensin homolog (PTEN), are other common aberrations across malignancies Thus, the vast majority of cancers have some degree of aberrant activation of PI3K-Akt-mTOR signaling axis (50).
Activation of mTOR represents the most down-stream target in this signaling pathway, suggesting mTOR inhibition as a critical strategy for cancer therapy. When these findings were translated into clinical trials however, mTOR inhibitors have failed to produce clinical benefit in many cancers. Activation of growth factor receptors ultimately leads to activation of mTOR and increased anabolic functions. This increase is short-lived however, as the target S6K in-turn feeds back to phosphorylate and drive degradation of insulin receptor substrate 1, repressing upstream signaling through PI3K (49). Thus inhibition of mTOR leads to activation of PI3K and Akt. Since Akt has over 150 downstream targets, the net effect is to inhibit a single target, at the expense of activating a multitude of additional targets.
One way to circumvent this problem is to use a dual inhibitor of PI3K and mTOR, a single molecule that effects mTOR inhibition, while simultaneously blocking feedback activation of Akt. In preclinical models of glioblastoma, treatment with dual PI3K/mTOR inhibitors is associated with a cytostatic rather than cytotoxic response (51). How can a drug that blocks signaling through three key survival kinases, PI3K, Akt, and mTOR, fail to affect survival in cancer?
Recent studies have demonstrated that dual inhibitors of PI3K/mTOR activate autophagy, that this activation is regulated by both mTOR complexes (mTORC1 and mTORC2), and that blockade of autophagy in early or late stages can cooperate with dual inhibitors of PI3K/mTOR to promote cell death (52). This cell death is prominent even in glioma cells mutant for PTEN. In contrast, in the glioma models, inhibitors of mTORC1 did not cooperate with inhibitors of autophagy to induce cell death, possibly due to the S6K-IRS1-PI3K feedback loop described above. Whereas dual inhibitors of PI3K/mTOR (including mTORC1 and mTORC2) induce autophagy as a central survival signal, selective inhibitors of mTORC1 activate both autophagy and Akt as separate survival signals. Effecting cell death in preclinical models of PTEN mutant glioma (in vitro and in vivo) thus requires blockade of three targets: Akt, mTOR and autophagy. Further studies are underway to determine if this finding is specific to glioma or can be generalized to all cancers with activated PI3K/AKt/mTOR signaling.
Future directions include elucidating mechanisms through which autophagy blockade cooperates with dual inhibitors of PI3K/mTOR to induce cell death, identifying key Akt targets that block this effect when Akt is activated, and identifying new, and more selective autophagy inhibitors, that circumvent toxicities associated with chloroquine derivatives. While PTEN mutation is generally associated with therapeutic resistance in glioma and other cancers (53), dual inhibitors of PI3K/mTOR when combined with chloroquine, readily induce apoptosis of PTEN mutant glioma in-vivo . This combination of agents could be tested in the near future in patients with this generally lethal tumor.
Crosstalk of the proteasome and the autophagy networks in cancer therapy
The autophagy-lysosome system and the ubiquitin-proteasome system (UPS) constitute the two major intracellular degradation systems. While UPS mainly targets short-lived proteins and soluble misfolded proteins, autophagy is particularly important for the turnover of long-lived proteins, aggregated and misfolded proteins, and organelles (39, 54-56).
A number of studies indicate functional connections between these two degradation systems (57, 58). Inhibition of UPS compensatively activates autophagy. Notably, endoplasmic reticulum (ER) stress plays a critical role in the cross-talk between the two systems. The ER is the major site for processing protein conformation. Misfolded proteins are normally exported out of the ER lumen and degraded by the proteasome, via the ER-associated degradation pathway (ERAD) (59). Autophagy is another important mitigating mechanism that clears misfolded proteins in response to ER stress (57, 60). Drug-induced ER stress can also induce autophagy (61-65). Interestingly, ER-associated autophagy (ERAA), like ERAD, can also be regulated by the unfolded protein response, particularly PERK-eIF2α, IRE-1-JNK signaling pathways (57, 58, 60-63, 65, 66) (Figure 3). ERAA is particularly important if ER stress is caused by proteasome inhibition, resulting in loss of ERAD’s critical degradation machinery.
Since uncompensated ER stress can lead to cell death (59, 67), the compensatory activation of autophagy provides a pro-survival mechanism (64). This notion is particularly relevant for cancer therapy with proteasome inhibitors, such as bortezomib. Indeed, genetic ablation of autophagy sensitizes tumor cells to proteasome inhibitors (35); with combined use of bortezomib and chloroquine increasing tumor cell death in vitro and in vivo (68). Tumor cells are more sensitive to this combination than normal cells (68), highlighting autophagy’s role in the survival of cancer cells, perhaps reflecting a higher metabolic rate and stress status in cancer relative to normal cells. Bortezomib is approved by the FDA for the treatment of multiple myeloma, a tumor type likely prone to elevated ER stress due to abundant synthesis of immunoglobulin, and autophagy inhibitors may similarly be effective in this setting. Thus understanding of the relationship of the proteasome and autophagy via ER stress is not only important for the development of novel cancer therapies, but to understand the nature and causes of increased cellular stress in cancer cells and resulting adaptive responses.
Autophagy, Immunity and Cancer
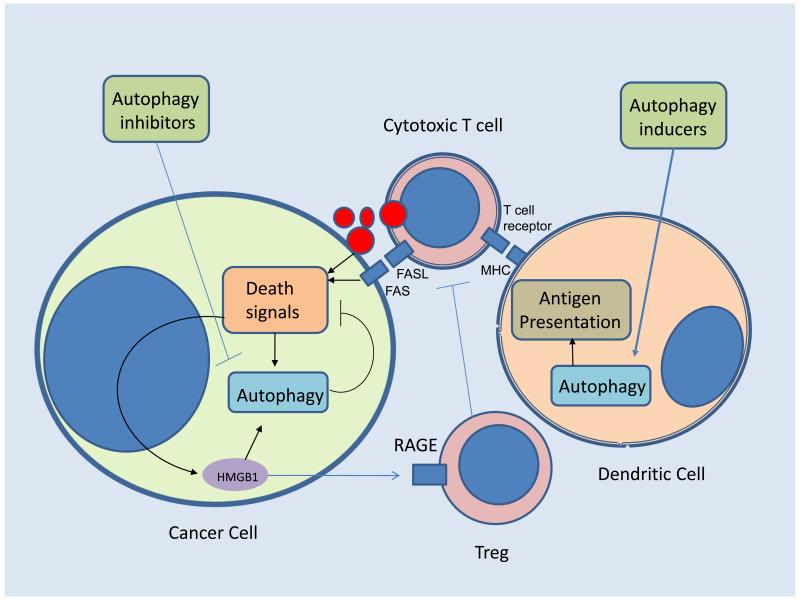
Cancer in adults (69), but not in children (70), arises in the setting of chronic inflammation. The tumor microenvironment is characterized by a disordered state associated with hypoxia, glycolysis, perpetual autophagy and resultant necrosis under conditions of heightened stress. A good example of how intimately linked tumor cell metabolism, autophagy and immune tolerance can be is highlighted by recent studies focused on the pleiomorphic functions of the highly conserved nuclear protein high mobility group B1 (HMGB1). Autophagic stimuli promote cytosolic and mitochondrial translocation and extracellular release of HMGB1 (71) (Figure 4). As a cytosolic factor, HMGB1 itself promotes autophagy, enhances ATP production, and limits apoptosis (72). Extracellular HMGB1 serves as a damage-associated molecular pattern molecule [DAMP], which interacts with the receptor for advanced glycation end products (RAGE) (73) and toll-like receptors (74) to recruit inflammatory cells to the site of damage. Thus HMGB1 represents one of likely many molecules that critically link cellular metabolism, cell death decisions and immunity.
Figure 4. Cancer Immunity and autophagy.
Imbalance of autophagy leads to immune tolerance in cancer patients. Heightened autophagy in tumor cells prevents immune effector cell mediated cytotoxicity. In addition, Stress-induced release of the damage-associated molecular pattern molecule HMGB1 induces cytoprotective autophagy and once extruded into the extracellular matrix recruits regulatory T cells (Treg) resulting in anergy. Suppressed autophagy in dendrtic cells limits effective priming of antigen presentation that trains cytotoxic T cells. Simultaneous or sequential pharmacological induction of autophagy in dendritic cells, and autophagy inhibition in the tumor cell would ideally reverse this imbalance and enhance antitumor immunity. FAS: FS7-associated cell surface antigen; FASL: Fas ligand; MHC: major histocompatability complex; RAGE: receptor for advanced glycation endproducts
Recently, three randomized studies have demonstrated survival benefits for immunotherapy in refractory cancers, (75-77). These limited successes come on the heels of decades of failures. Studies of autophagy in tumor tissue and immune cells suggests that cancer patients are suffering from a systemic autophagic syndrome in which autophagy is pathologically increased within the cancer cell and suppressed in the immune cells. Adoptive transfer of T-cells, dendritic cell (DC) vaccines, administration of antibodies or administration of human recombinant cytokines such as IL-2, only hold promise for immunotherapy, if the imbalance between host and tumor autophagic response can be ameliorated. DC vaccines involve isolation of the patient’s antigen presenting cells (APCs), followed by a procedure of ex-vivo gene therapy or incubation with targeted tumor associated and specific antigen (TAA, TSA), and subsequent re-introduction of the matured DCs so that they may mediate a highly specific anti-tumor immune response facilitated by DC-activated CD8+ T cells (78-81). Within professional antigen presenting cells (Figure 3) antigen processing and delivery to MHC Class I and Class II molecules is directed by the proteasome and autophagy. Autophagic cargo which can be extruded into the extracellular matrix from tumor cells should be superior sources from which DCs can derive antigen for T cell priming (78, 82). Thus systemic induction of autophagy early in the course of adaptive immunity may prevent the emergence of immune tolerance, and ex vivo induction of autophagy in the presence of antigen may improve the efficacy of cellular immunotherapies.
Autophagy inhibition may augment the cytotoxicity of effector T cells and NK cells once they have been activated to lyse the tumor, similar to the notion that autophagy limits the effectiveness of chemo- and radiation therapy. Currently autophagy inhibition with HCQ in combination with IL-2 is being tested in a murine tumor model and is poised to be rapidly translated into a multi-institution clinical trial. Future approaches may include combining ex-vivo induction of autophagy in DCs and systemic autophagy inhibition, delivered at the time of adjunctive treatment and designed to stimulate cytotoxic effectors. This approach may facilitate improved anti-tumor immunity with DC delivery, and enhanced antitumor efficacy of the activated immune system (Figure 4).
Autophagy inhibition with HCQ in combination anticancer regimens for patients with refractory malignancies
Autophagy inhibition augments the efficacy of anticancer agents in a variety of tumor histologies in multiple preclinical models (37, 47, 68, 83-85). Based on reports that effective autophagy inhibition can be achieved in vivo with the antimalarial drug chloroquine (CQ) (47), clinical trials for patients with refractory malignancies were undertaken. For the past 60 years, CQ derivatives have been prescribed for malaria (86), rheumatoid arthritis (87), and HIV (88). They are inexpensive oral drugs that cross the blood-brain barrier. Case reports of infant deaths associated with single tablet ingestions suggest high peak concentrations of CQ may result in significant toxicity. In contrast, suicide attempts involving hydroxychloroquine (HCQ) did not result in fatalities (89), suggesting HCQ can be safely dose-escalated in cancer patients (90). In vitro studies indicate these two drugs are equipotent at autophagy inhibition.
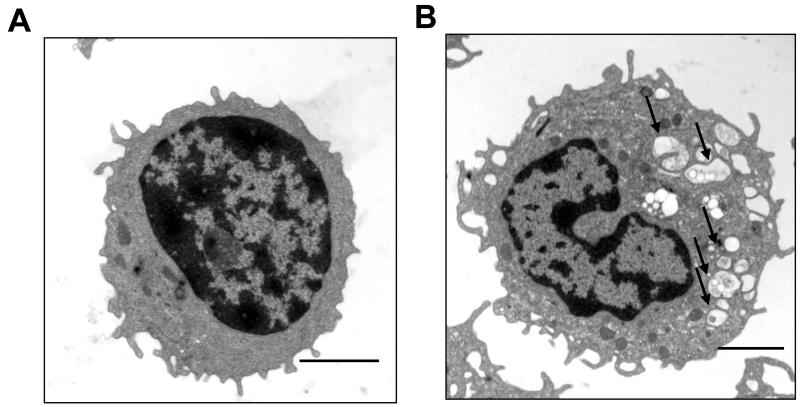
A phase III trial in glioblastoma patients treated with radiation and carmustine with or without daily CQ found a median overall survival of 24 and 11 months in CQ- and placebo-treated patients, respectively (91). This single institution study was not adequately powered to detect a significant difference in survival, but established the safety of adding low dose CQ to DNA damaging therapy. Key issues remain that the pharmacology of HCQ (characterized by a long half-life resulting in weeks to achieve peak concentration) and the low potency of the drug (micromolar concentrations are required to inhibit autophagy) may limit its efficacy as an autophagy inhibitor in patients (92). To address these concerns, a phase I/II trial of HCQ with temozolomide and radiation for glioblastoma patients was launched through the American Brain Tumor Consortium, and included pharmacodynamic (PD) and pharmacokinetic (PK) analyses. PD evidence of HCQ dose –dependent autophagy inhibition was observed using a novel electron microscopy assay on serial blood mononuclear cells (Figure 5) (93). Overall survival is the primary endpoint for this phase I/II trial so information about the antitumor activity of this combination should be forthcoming.
Figure 5. Pharmacodynamic assay for autophagy inhibition.
Electron micrographs of peripheral blood mononuclear cells from a glioma patient enrolled on the phase I trial of temozolomide, radiation and hydroxychloroquine; (A) Pretreatment and (B) 3 weeks of combined therapy. Arrows: autophagic vesicles, scale bar 200 μm.
Currently there are more than 20 trials involving HCQ accruing cancer patients nationwide and many of them have evidence of preliminary antitumor activity (Table 1). The knowledge gained from the PD, PK and predictive biomarkers in these studies will guide the development of more potent and specific autophagy inhibitors that are being developed by academic and industry discovery programs.
Table 1.
Examples of clinical trials combining the autophagy inhibitor HCQ with anticancer therapies
| Condition | Intervention | Phase | Sponsors | Identifier | Title |
|---|---|---|---|---|---|
| Multiple myeloma |
HCQ + Bortezomib |
I/II | UPenn, Millenium |
NCT00568880 | A Phase I/II Trial of HCQAdded to Bortezomib for Relapsed/Refractory Myeloma |
| Brain, central nervous system tumors |
HCQ+ Temozolomide/ RT |
I/II | UPenn, CTEP, NCI |
NCT00486603 | A Phase I/II Trial of HCQ in Conjunction with Radiation Therapy and Concurrent and Adjuvant Temozolomide in Patients With Newly Diagnosed Glioblastoma Multiforme |
| Adult solid tumors |
HCQ+ temozolomide |
I | UPenn, Merck |
NCT00714181 | A Phase I Study of HCQ in Combination with temozolomide in Patients with Advanced Solid Tumors |
| Adult solid tumors |
HCQ+ temsirolimus |
I | UPenn, Pfizer |
NCT00909831 | A Phase I Study of HCQ in Combination with temsirolimus in Patients with Advanced Solid Tumors |
| Adult Solid Tumors |
HCQ + Vorinostat |
I | San Antonio, NCI, Merck |
NCT01023737 | A phase I pharmacokinetic and Pharmacodynamic Study of Hydroxychloroquine in combination with vorinostat for the treatment of patients with advanced solid tumors |
| Prostate cancer |
HCQ + Docetaxel | II | CINJ, NCI | NCT00786682 | A Phase II Study of Docetaxel and Modulation of Autophagy with HCQ for Metastatic Hormone Refractory Prostate Cancer |
| Prostate cancer |
HCQ | II | CINJ, NCI | NCT00726596 | Autophagic Cell Death in Patients with Hormone- Dependent Prostate-Specific Antigen Progression after Local Therapy for Prostate Cancer |
| Breast cancer |
HCQ+ ixabepilone |
I/II | CINJ, NCI | NCT00765765 | Phase I/II Study of Ixabepilone in Combination with the Autophagy Inhibitor HCQ for the Treatment of Patients with Metastatic Breast Cancer |
| Lung cancer | HCQ+ Bevacizumab carboplatin paclitaxel |
I/II | CINJ, NCI | NCT00728845 | Modulation of Autophagy with HCQ in Combination with Carboplatin, Paclitaxel and Bevacizumab in Patients with Advanced/Recurrent Non-Small Cell Lung Cancer -A Phase I/II Study |
| Advanced cancer |
HCQ + sunitinib |
I | CINJ, CTEP, NCI |
NCT00813423 | Anti-Angiogenic Therapy in Patients with Advanced Malignancies: A Phase I Trial of Sunitinib and HCQ |
| Pancreas Cancer |
HCQ+ Gemcitabine |
I | U. Pittsburgh |
NCT01128296 | Phase I/II study of Preoperative Gemcitabine in Combination with Oral HCQ in Subjects With Resectable Stage IIb Or III Pancreatic Adenocarcinoma |
| Renal Cancer |
HCQ | I | U. Pittsburgh |
NCT01144169 | Neoadjuvant Study of Preoperative HCQ in Patients with Resectable Renal Cell Carcinoma |
Future drug development of autophagy modulators
In the era of targeted drug development, efforts to understand, modulate and develop biomarkers of autophagy, as a survival mechanism used by tumor cells to tolerate stress, are critically important. As an addition to the hallmarks of cancer originally proposed by Weinberg et al.(94), new basic hallmarks of cancer cells were recently highlighted and included the ability to tolerate metabolic, oxidative, DNA damage, mitotic and proteotoxic stresses (95). Given that autophagy can allow tumor cells to tolerate these multiple stresses, and many novel agents under development in clinical trials have been found to modulate autophagy, the assessment of autophagy and its relevance to a particular agent will likely help improve effectiveness. In fact, multiple agents under development within pharmaceutical companies or CTEP (http://ctep.cancer.gov/branches/idb/default.htm) have been demonstrated to modulate autophagy including histone deacetylase inhibitors, anti-angiogenic agents, mTOR inhibitors, BH3 domain mimetics and glycolytic inhibitors (83, 96-98). In a phase I clinical trial, 2-deoxyglucose, a prototypical agent that inhibits glycolysis, was well tolerated and reduced p62 in peripheral blood mononuclear cells consistent with induction of autophagy (99). Preclinical studies with 2-deoxyglucose demonstrated induction of autophagy, and modulaton of autophagy increased cytotoxicity, supporting the hypothesis that further studies of agents such as 2-deoxyglucose that induce autophagy should be tested in combination with autophagy inhibition (100-102). To date there has not been a comprehensive study that compared multiple classes of inhibitors for their ability to induce autophagy in the same model system.
In addition to focusing research efforts on identifying which anticancer therapeutics are most limited by therapy-induced autophagy, there is a growing interest in developing more potent and specific autophagy inhibitors. Academic and industry efforts are underway to develop tools that will enable high-throughput screening of chemical libraries to identify novel candidate compounds that inhibit autophagy at various points of control described above. Compounds have been found that unexpectedly inhibit autophagy such as 2-phenylethynesulfonamide (PES), a small molecule HSP70 inhibitor that results in misfolding of a number of lysosomal proteins (103). A critical component to drug development is the development of assays that can be translated into pharmacodynamic and predictive biomarkers of response to autophagy induction and inhibition. Although studies of biomarkers of autophagy are early in development with additional markers emerging, preliminary data supports the ability to measure Beclin1 by immunohistochemistry as a measure of autophagy competence, and the measurement of AV number directly by electron microscopy, LC3 and p62 levels as markers of autophagy modulation (35) (104). Given the basic biological importance of autophagy as a cellular mechanism of survival during multiple forms of cancer and therapeutic induced stress, an ongoing dialogue between emerging laboratory and clinical research will be imperative to address autophagy as a targetable resistance mechanism in advanced disease and the induction of autophagy as chemoprevention strategy in early phase disease.
Statement of Translational Relevance.
Intrinsic mechanisms of resistance to cancer therapy are a key limitation to improving cure rates across malignancies. This review highlights recent advances in our knowledge of autophagy as a resistance mechanism to metabolic stress and multiple anticancer agents, and current strategies to block autophagy as an approach to enhancing the efficacy of anticancer therapy. A detailed understanding of the over 100 components of this complex, multistep process can identify new targets for drug development. Moreover, new biomarkers of the functional status of autophagy in tumors can be developed to guide clinical development of autophagy inhibitors. The emerging appreciation for autophagy’s role as a common resistance mechanism to metabolic stress, kinase inhibitors, disrupted protein metabolism, and chemotherapy will necessitate new approaches to drug combinations. Fundamental insights into the role of autophagy in the immune system may provide clues for how to improve immunotherapies that have started to demonstrate incremental survival benefits across diseases. Highlights of ongoing phase I and phase II clinical trials involving the first generation autophagy inhibitor hydroxychloroquine illuminate the potential for clinical translation in this field. The unmet needs for additional research in autophagy and cancer are detailed.
Acknowledgements
Grants supporting this work include: RKA: 1K23CA120862-01A2, WAW: NIH P50CA097257, Burroughs Wellcome Fund, American Brain Tumor Association, The Brain Tumor Society, Accelerate Brain Cancer Cure; Pediatric Brain Tumor, and Samuel G. Waxman Foundations; RD: W81XWH-09-1-0394, W81XWH-09-1-0145, and U01 CA132194; XMY: CA83817, CA111456; and EW: R37CA53370, RO1CA130893, and RC1CA147961.
References
- 1.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 21:683–90. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 22:132–9. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 8.Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 9.Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–67. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 20:355–62. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satoo K, Noda NN, Kumeta H, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–50. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 14.Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young AR, Chan EY, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 18.Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol. 188:537–46. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–6. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller S, Tavshanjian B, Oleksy A, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 327:1638–42. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 23.Noda NN, Kumeta H, Nakatogawa H, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–8. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 24.Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–16. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–74. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jager S, Bucci C, Tanida I, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 28.Marceau F, Bawolak MT, Bouthillier J, Morissette G. Vacuolar ATPase-mediated cellular concentration and retention of quinacrine: a model for the distribution of lipophilic cationic drugs to autophagic vacuoles. Drug Metab Dispos. 2009;37:2271–4. doi: 10.1124/dmd.109.028480. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Polo RA, Boya P, Pauleau AL, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci. 2005;118:3091–102. doi: 10.1242/jcs.02447. [DOI] [PubMed] [Google Scholar]
- 30.Poole B, Ohkuma S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J Cell Biol. 1981;90:665–9. doi: 10.1083/jcb.90.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 22:157–68. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 22:140–9. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 34.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kongara S, Kravchuk O, Teplova I, et al. Autophagy regulates keratin 8 homeostasis in mammary epithelial cells and in breast tumors. Mol Cancer Res. 8:873–84. doi: 10.1158/1541-7786.MCR-09-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 40.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–7. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 22:212–7. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 45.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 46.Rabinowitz JD, White E. Autophagy and metabolism. Science. doi: 10.1126/science.1193497. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13:7271–9. doi: 10.1158/1078-0432.CCR-07-1595. [DOI] [PubMed] [Google Scholar]
- 49.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 50.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan QW, Cheng C, Hackett C, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3:ra81. doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degtyarev M, De Maziere A, Orr C, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101–16. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–50. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 55.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–50. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 56.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding WX, Ni HM, Gao W, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–24. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu K, Dunner K, Jr., McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2009 doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–99. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 60.Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–9. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 61.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–31. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ding WX, Ni HM, Gao W, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–10. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 65.Hoyer-Hansen M, Bastholm L, Szyniarowski P, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 66.Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6 doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- 67.Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–80. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- 68.Ding WX, Ni HM, Gao W, et al. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol Cancer Ther. 2009;8:2036–45. doi: 10.1158/1535-7163.MCT-08-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nat Rev Immunol. 2004;4:641–8. doi: 10.1038/nri1415. [DOI] [PubMed] [Google Scholar]
- 70.Vakkila J, Jaffe R, Michelow M, Lotze MT. Pediatric cancers are infiltrated predominantly by macrophages and contain a paucity of dendritic cells: a major nosologic difference with adult tumors. Clin Cancer Res. 2006;12:2049–54. doi: 10.1158/1078-0432.CCR-05-1824. [DOI] [PubMed] [Google Scholar]
- 71.Tang D, Kang R, Livesey KM, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 190:881–92. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang D, Kang R, Cheh CW, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang R, Tang D, Schapiro NE, et al. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ. 17:666–76. doi: 10.1038/cdd.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Apetoh L, Ghiringhelli F, Tesniere A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–9. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 75.Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 363:411–22. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 76.Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuster SJNS, Gause BL, Muggia FM, Gockerman JP, Sotomayor EM, Winter JN, Flowers CR, Stergiou AM, Kwak LW. Idiotype vaccine therapy (BiovaxID) in follicular lymphoma in first complete remission: Phase III clinical trial results. J Clin Oncol. 2009;27(suppl):18s. abstr 2. 2009. [Google Scholar]
- 78.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 2009;16:991–1005. doi: 10.1038/cdd.2009.8. [DOI] [PubMed] [Google Scholar]
- 80.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr., Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–76. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 81.Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis, and necrotic signals promotes T-cell homeostasis. Immunol Rev. 236:95–109. doi: 10.1111/j.1600-065X.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y, Wang LX, Pang P, et al. Cross-presentation of tumor associated antigens through tumor-derived autophagosomes. Autophagy. 2009;5:576–7. doi: 10.4161/auto.5.4.8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–58. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 85.Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–22. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Neill PM, Bray PG, Hawley SR, Ward SA, Park BK. 4-Aminoquinolines--past, present, and future: a chemical perspective. Pharmacol Ther. 1998;77:29–58. doi: 10.1016/s0163-7258(97)00084-3. [DOI] [PubMed] [Google Scholar]
- 87.Kremer JM. Rational use of new and existing disease-modifying agents in rheumatoid arthritis. Ann Intern Med. 2001;134:695–706. doi: 10.7326/0003-4819-134-8-200104170-00013. [DOI] [PubMed] [Google Scholar]
- 88.Romanelli F, Smith KM, Hoven AD. Chloroquine and hydroxychloroquine as inhibitors of human immunodeficiency virus (HIV-1) activity. Curr Pharm Des. 2004;10:2643–8. doi: 10.2174/1381612043383791. [DOI] [PubMed] [Google Scholar]
- 89.Smith ER, Klein-Schwartz W. Are 1-2 dangerous? Chloroquine and hydroxychloroquine exposure in toddlers. J Emerg Med. 2005;28:437–43. doi: 10.1016/j.jemermed.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 90.Gunja N, Roberts D, McCoubrie D, et al. Survival after massive hydroxychloroquine overdose. Anaesth Intensive Care. 2009;37:130–3. doi: 10.1177/0310057X0903700112. [DOI] [PubMed] [Google Scholar]
- 91.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337–43. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 92.Carmichael SJ, Charles B, Tett SE. Population pharmacokinetics of hydroxychloroquine in patients with rheumatoid arthritis. Ther Drug Monit. 2003;25:671–81. doi: 10.1097/00007691-200312000-00005. [DOI] [PubMed] [Google Scholar]
- 93.Rosenfeld MRGS, Brem S, Mikkelson T, Wang D, Piao S, Davis L, O’Dwyer PJ, Amaravadi RK. Pharmacokinetic analysis and pharmacodynamic evidence of autophagy inhibition in patients with newly diagnosed glioblastoma treated on a phase I trial of hydroxychloroquine in combination with adjuvant temozolomide and radiation (ABTC 0603) J Clin Oncol. 2010;28 Abstract # 3086. [Google Scholar]
- 94.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 95.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Watanabe M, Adachi S, Matsubara H, et al. Induction of autophagy in malignant rhabdoid tumor cells by the histone deacetylase inhibitor FK228 through AIF translocation. Int J Cancer. 2009;124:55–67. doi: 10.1002/ijc.23897. [DOI] [PubMed] [Google Scholar]
- 97.Munoz-Gamez JA, Rodriguez-Vargas JM, Quiles-Perez R, et al. PARP-1 is involved in autophagy induced by DNA damage. Autophagy. 2009;5:61–74. doi: 10.4161/auto.5.1.7272. [DOI] [PubMed] [Google Scholar]
- 98.Gao P, Bauvy C, Souquere S, et al. The Bcl-2 homology domain 3 mimetic gossypol induces both Beclin 1-dependent and Beclin 1-independent cytoprotective autophagy in cancer cells. J Biol Chem. 2010;285:25570–81. doi: 10.1074/jbc.M110.118125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stein M, Lin H, Jeyamohan C, et al. Targeting tumor metabolism with 2-deoxyglucose in patients with castrate-resistant prostate cancer and advanced malignancies. Prostate. 70:1388–94. doi: 10.1002/pros.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.DiPaola RS, Dvorzhinski D, Thalasila A, et al. Therapeutic starvation and autophagy in prostate cancer: a new paradigm for targeting metabolism in cancer therapy. Prostate. 2008;68:1743–52. doi: 10.1002/pros.20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matsuda F, Fujii J, Yoshida S. Autophagy induced by 2-deoxy-D-glucose suppresses intracellular multiplication of Legionella pneumophila in A/J mouse macrophages. Autophagy. 2009;5:484–93. doi: 10.4161/auto.5.4.7760. [DOI] [PubMed] [Google Scholar]
- 102.Sahra IB, Tanti JF, Bost F. The combination of metformin and 2-deoxyglucose inhibits autophagy and induces AMPK dependent apoptosis in prostate cancer cells. Autophagy. 2010;6 doi: 10.4161/auto.6.5.12434. [DOI] [PubMed] [Google Scholar]
- 103.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Amaravadi R. Autophagy can contribute to cell death when combining targeted therapy. Cancer Biol Ther. 2009;8:130–3. doi: 10.4161/cbt.8.21.10416. [DOI] [PubMed] [Google Scholar]