Abstract
The cricket paralysis virus internal ribosome entry site (IRES) can, in the absence of canonical initiation factors and initiator tRNA (Met-tRNAi), occupy the ribosomal P-site and assemble 80S ribosomes. Here we show that the IRES assembles mRNA-80S ribosome complexes by recruitment of 60S subunits to preformed IRES-40S complexes. Addition of eukaryotic elongation factors eEF1A and eEF2 and aminoacylated elongator tRNAs resulted in the synthesis of peptides, implying that the IRES RNA itself mimics the function of Met-tRNAi in the P-site to trigger the first translocation step without peptide bond formation. IRES-80S complexes that contained a stop codon in the A-site recruited eukaryotic release factor eRF1, resulting in ribosome rearrangements in a surprisingly eEF2-dependent manner. Thus, this P-site-occupying IRES directs the assembly of 80S ribosomes, sets the translational reading frame, and mimics the functions of both Met-tRNAi and peptidyl tRNA to support elongation and termination.
Unsolved questions in eukaryotic protein biosynthesis include the positioning of the mRNA start codon in the ribosomal P-site, the mechanism of dynamics of peptide bond formation, and ribosomal translocation during elongation of the growing chain. Many of these questions have remained unanswered because the eukaryotic translation apparatus is a complex machinery that functions through a concerted action of many key and auxiliary factors.
For most mRNAs, translation is initiated by a cap-dependent mechanism in which 40S ribosomal subunits, carrying eukaryotic initiation factors eIF3 and eIF2-GTP and initiator tRNA (Met-tRNAi), are recruited to the capped 5′ end of the mRNA through an interaction between eIF3 and the cap binding complex eIF4F (1). 40S subunits then scan the mRNA until an appropriate start codon (i.e., AUG, GUG, or CUG) is encountered. Hydrolysis of eIF2-GTP by factor eIF5 releases eIF2 from the 40S subunit, and subsequently, in the presence of eIF5B-GTP, the 60S joins the 40S subunit to assemble the 80S ribosome with the start codon Met-tRNAi positioned in the P-site of the ribosome. eIF5B-GTP is hydrolyzed, releasing eIF5B from the ribosome, and elongation commences (1).
Some aspects of translation initiation have been uncovered by studying internal ribosome entry sites (IRESs), in which highly structured RNA elements directly recruit 40S subunits without the requirement for a full set of canonical translation factors (2). However, all IRESs examined so far require most of the canonical initiation factors and Met-tRNAi for translation initiation (2). In contrast, the cricket paralysis virus (CrPV) IRES and related insect viruses can mediate translation initiation in the absence of canonical eIFs such as eIF4F, eIF2, eIF3, and even Met-tRNAi (3–5). The CrPV IRES, located in an intergenic region (IGR) of the CrPV RNA genome, is ≈200 nt long and contains three overlapping pseudoknot structures (3, 6, 7), which direct the efficient synthesis of viral capsid proteins when eIF2-Met-tRNAi levels are low (8, 9). Toeprint analyses have shown that the IGR IRES itself occupies the P-site of the ribosome in place of Met-tRNAi and mediates translation initiation from an Ala GCU codon located in the ribosomal A-site (3). This model implies that the IGR IRES must mediate the first translocation step, whereby the GCU codon is translocated into the P-site without peptide bond formation. Here, we present direct evidence that this divergent IRES fulfills the functional properties of translational initiation factors, recruiting 40S and 60S subunits to assemble elongation-competent 80S ribosomes. Furthermore, we show that the P-site-occupying IRES mimics the roles of both Met-tRNAi and peptidyl tRNA during protein synthesis.
Materials and Methods
DNA Constructs. Dicistronic and monocistronic luciferase plasmids containing the IGR IRES have been described (3, 9). Mutated IGR IRESs and stop codon mutations were generated by using the QuikChange kit (Stratagene). Mutations were confirmed by sequencing. Human elongator Met-tRNA and Ala-tRNA were generated as described (10).
In Vitro Transcription and Translation. To generate full-length dicistronic RNAs, dicistronic luciferase plasmids were linearized with BamHI. For in vitro transcription of monocistronic RNAs, plasmids were linearized with NarI, which cleaves 33 nt downstream of the ATG start codon of the firefly luciferase gene. For in vitro transcription of WT IGR IRES small ORF (ORFS), mutated IGR IRES-ORFS, and encephalomyocarditis virus (EMCV) IRES-ORFS RNAs, dicistronic luciferase plasmids containing the respective IRESs were linearized with NarI. These RNAs are predicted to produce a 17-aa peptide. For in vitro transcription of WT IGR IRES large ORF (ORFL) RNAs, dicistronic luciferase construct containing the WT IGR IRES was linearized with ScaI, which has a predicted result of an ORF of 58 aa. The mutated IGR IRES-ORFS RNA contains mutations that disrupt pseudoknot I (6). The mutated IGR IRES RNA used in the gel-mobility shift assays contains mutations that disrupt pseudoknots I and III (6). For in vitro transcription of tRNAs, tRNA constructs were linearized with BbsI. The integrity and purity of the RNAs were confirmed by gel analysis.
Aminoacylation of tRNAs. Bulk liver tRNAs and in vitro synthesized tRNAs were aminoacylated by using a tRNA synthetase extract from HeLa cells (10). The extent of aminoacylation was monitored by [35S]Met and [3H]Ala incorporation and by gel filtration analysis. Aminoacylation of tRNAs was >75%.
Composite Agarose/Acrylamide Gel-Mobility Shift Assays. Ribosomal subunits were purified from HeLa cells as described (6). 5′ end-labeled RNAs (0.5 nM final concentration) were incubated in buffer E as described (6). Buffer E contained 100 mM KCl instead of 100 mM KOAc. Gel shifts were performed in composite gels containing 2.75% acrylamide/bis (19:1), 25 mM Tris·OAc (pH 7.0), 6 mM KCl, 2 mM MgCl2, 1 mM DTT, 1% sucrose (wt/vol), 0.5% NuSieve GTG agarose, 0.45% 3-dimethylaminopropionitrile (DMAPN), and 0.045% ammonium persulfate (APS). RNAs were incubated with increasing equimolar amounts of 40S and 60S subunits. No difference was observed in the order of addition of 40S, 60S, and RNAs. Gel-mobility shifts were quantitated by PhosphorImager (Molecular Dynamics) analysis.
Analysis of Ribosome Translocation. Dicistronic RNAs (7.5 ng/μl) were first annealed with primer PrEJ69 (6) in 40 mM Tris·Cl, pH 7.5, and 0.2 mM EDTA by slow cooling from 65°C to 35°C. Annealed RNAs were incubated in Buffer E (containing 100 mM KCl) containing 40S (final concentration 80 nM); 60S (final concentration 160 nM); 1 mM ATP, 0.4 mM GTP, or 0.4 mM 5′-guanylylimidodiphosphate (GMP-PNP); 30 ng/μl yeast or rabbit eukaryotic elongation factor (eEF)-1A; 50 ng/μl yeast eEF2; and 230 ng/μl bulk aminoacylated tRNAs. 80S ribosomes were first assembled on the IGR IRES by incubating at 37°C for 5 min. After addition of elongation factors and aminoacylated tRNAs, the reaction was further incubated for 10 min. Toeprinting analysis was performed as described (6). Where noted, recombinant termination eukaryotic release factor (eRF)-1 was added to the reaction at a final concentration of 50 ng/μl.
Peptide Synthesis Analysis. For the peptide synthesis experiments, bulk tRNAs were aminoacylated by using a mixture of amino acids including [35S]Met. Reactions contained 30 nM dicistronic RNA, 130 nM 40S subunits, 330 nM 60S subunits, 1 mM ATP, 1 mM GTP, 80 ng/μl eEF1A, 100 ng/μl eEF2, and 380 ng/μl aminoacylated tRNAs. 80S ribosomes were assembled for 5 min at 37°C. After addition of elongation factors and aminoacylated tRNAs, reactions were incubated at 37°C for 30 min. Peptide synthesis reactions were loaded on a 16.5% Tris·N-tris-(hydroxymethyl)methylglycine (Tricine) gel and run at a constant 50 V overnight.
Results
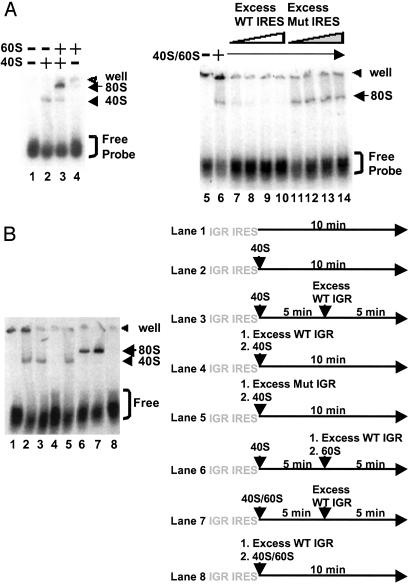
IRES-80S Ribosome Complex Formation by Joining 60S Subunit to Preformed IRES-40S Complexes. We have previously shown that the IGR IRES can bind 40S subunits directly and can assemble 80S complexes in reaction mixtures that contained only RNA and salt-washed 40S and 60S subunits (3). IRES-40S and IRES-80S complexes could be isolated in sucrose gradients, and complex formation was shown to be sensitive to EDTA, as expected for functional IRES-ribosome complexes (3, 11). To monitor the formation of IRES-ribosome complexes directly, we developed a composite agarose polyacrylamide gel system in which the kinetics of assembly of IRES-40S and IRES-80S complexes could be examined. Fig. 1 shows that binary IGR IRES-40S complexes migrated more slowly than free radiolabeled RNA in these composite gels (Fig. 1 A, lanes 1 and 2). So far, the only other RNA sequence that can assemble binary RNA-40S complexes without initiation factors is the hepatitis C virus IRES (12). Addition of purified 60S subunits to the reaction mixture resulted in the formation of putative IGR IRES-80S complexes that migrated even more slowly (Fig. 1 A, lane 3). In the absence of 40S subunits, 60S subunits did not bind efficiently to the IGR IRES (Fig. 1 A, lane 4). Formation of 80S complexes could be abolished by the addition of excess unlabeled WT IGR IRES RNA (Fig. 1 A, lanes 7–10) but not mutated IGR IRES molecules (Fig. 1 A, lanes 11–14), which harbored nucleotide changes in pseudoknots I and III that rendered the IRES nonfunctional in translation (6).
Fig. 1.
80S ribosome assembly on the IGR IRES using purified 40S and 60S subunits. (A)5′ end-labeled IGR IRES RNAs (0.5 nM) were incubated alone (lane 1), with 40S (14 nM, lane 2), with 60S (16 nM, lane 4), or with 40S and 60S (lane 3) ribosomal subunits in 5-μl reactions. Excess (150 nM, 300 nM, 600 nM, and 800 nM) unlabeled WT IGR IRES RNAs (lanes 7–10) and mutated (Mut) IRES RNAs (lanes 11–14) were added as indicated. Excess RNAs were added to reactions before addition of 40S and 60S subunits. Reactions were separated on composite agarose polyacrylamide gels, dried, and exposed to x-ray film. (B) Formation of IRES-80S complexes by joining 60S to preformed binary IRES-40S complexes. (Right) The sequential order of reagents is shown. (Left) Radiolabeled WT IGR IRES RNAs were incubated alone (lane 1) or with 40S subunits (lane 2). Excess unlabeled WT IGR IRES RNAs or mutated (Mut) IGR IRES RNAs were added after (lane 3) or before (lanes 4 and 5) 40S subunit incubation. Addition of 60S subunits to prebound WT IGR IRES-40S complexes assembled 80S complexes in the presence of excess unlabeled WT IGR IRES RNAs (lane 6). Excess unlabeled WT IGR IRES RNAs were added after (lane 7) or before (lane 8) addition of 40S and 60S subunits. Reactions were separated on composite agarose polyacrylamide gels, dried, and exposed to x-ray film.
To investigate the mechanism of 80S assembly, we tested whether preformed IGR IRES-40S complexes directly recruited 60S subunits to form IGR IRES-80S complexes. First, to determine the stability of preformed IGR IRES-40S complexes or whether bound 40S must first dissociate from the IRES, radio-labeled IGR IRES-40S complexes (Fig. 1B, lane 2) were challenged with excess unlabeled IRES and shown not to dissociate after 5 min (Fig. 1B, lane 3). Even prolonged incubation with excess free IRES RNA failed to trap any 40S complex that dissociated from preformed IGR IRES-40S complexes (data not shown). As expected, addition of excess unlabeled WT (Fig. 1B, lane 4), but not of mutated (Fig. 1B, lane 5), IGR IRES molecules before the addition of 40S subunits prohibited formation of binary complexes. Therefore, the off-rate of bound 40S subunits from IGR IRES RNAs was negligible under the conditions of this assay. Addition of excess unlabeled WT IGR IRES RNAs followed by addition of 60S subunits to prebound 40S-radiolabeled IGR IRES complexes resulted in the formation of 80S complexes (Fig. 1B, lane 6). Although this result does not exclude the possibility that preformed 80S complexes can in principle bind to the IGR IRES, it clearly demonstrates that this diverse RNA element can assemble 80S complexes by joining 60S subunits to preformed binary RNA-40S complexes via an unprecedented pathway that does not require canonical initiation factors.
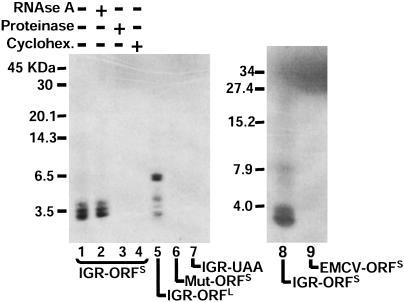
P-Site-Occupied IGR IRES Directs Peptide Synthesis in a Minimal Reconstituted System. To determine whether RNA-80S complexes assembled in vitro can support translation elongation, we examined the translation products synthesized from mRNAs containing the IGR IRES linked to an ORFS. In summary, total tRNAs were aminoacylated with an amino acid mixture that contained radiolabeled [35S]Met. Incubation of the reporter mRNA with this tRNA pool, ribosomal subunits, and elongation factors eEF1A and eEF2 resulted in the synthesis of a radiolabeled product that migrated as a heterogeneous band in the 3.5-kDa range (Fig. 2, lanes 1 and 8), the predicted size of the ORF. In a control reaction, the EMCV IRES, which requires canonical initiation factors to start translation initiation (13), did not direct the synthesis of small peptides (Fig. 2, lane 9); however, the EMCV IRES was active in translation-competent lysates (data not shown). Appearance of these bands was insensitive to RNase A treatment but sensitive to proteinase treatment (Fig. 2, lanes 2 and 3), indicating that these bands represent proteins. Inclusion of cycloheximide in the reaction prevented the appearance of the proteins, indicating that these peptides were synthesized by ribosomes (Fig. 2, lane 4). Moreover, a template mRNA that contained a UAA stop codon after the Ala GCU codon in the A-site inhibited synthesis of the peptides, further demonstrating that synthesis of the peptide required elongating ribosomes (Fig. 2, lane 7). Synthesis of the peptides was mediated by the IGR IRES, because a mutated, nonfunctional IRES (6) failed to direct the synthesis of peptides (Fig. 2, lane 6). To examine whether this reconstituted system allowed the synthesis of longer peptides, the IGR IRES was linked to an ORF that should direct the synthesis of a 6.5-kDa protein. Indeed, peptides with a molecular mass of 6,500 Da could be synthesized in the reconstituted system (Fig. 2, lane 5). Therefore, the IGR IRES can recruit 40S and 60S subunits to assemble 80S ribosomes that can perform several elongation steps in which peptide bonds form. However, the mechanism of the first elongation step, in which a portion of the IRES is present in the P-site instead of Met-tRNAi, remained to be investigated.
Fig. 2.
Peptide synthesis directed by the IGR IRES in a reconstituted system. Dicistronic constructs containing the IGR IRES were engineered to synthesize ≈3-kDa (IGR-ORFS) or 6.5-kDa (IGR-ORFL) peptides. RNAs were incubated with 40S and 60S subunits for 5 min at 37°C, followed by addition of elongation factors and aminoacylated tRNAs and further incubation for 25 min at 37°C. Translation products from WT or mutated (Mut) IRES-containing RNAs are shown as indicated. Reactions were treated with 0.7 mg/ml RNase A (lane 2) or 2 mg/ml proteinase (lane 3) (Pronase, Boehringer Mannheim). Cycloheximide (Cyclohex., 500 μg/ml, lane 4) was added to the reactions before the addition of elongation factors. Mutated IGR IRES (Mut-ORFS) contains the mutation CC6214-5GG, which disrupts pseudoknot I (6). Lane 9, reaction containing the EMCV IRES (EMCV-ORFS) is shown. All reactions were loaded on 16.5% Tris·Tricine gels, which were dried and exposed to x-ray films. Autoradiographs are shown.
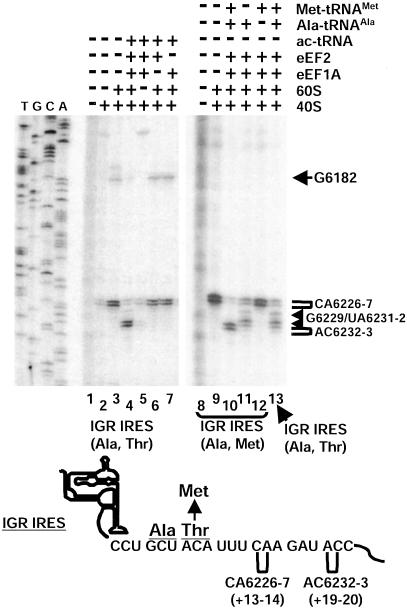
Requirements for Ribosome Translocation on the IGR IRES. To address the factor requirement for translation elongation of IRES-80S complexes assembled in vitro, we monitored ribosome movement by toeprinting analysis. In this technique, an oligodeoxynucleotide primer is annealed 3′ to the ribosome binding site. Addition of reverse transcriptase produces primer-initiated cDNA whose synthesis is arrested when the enzyme encounters an RNA-bound ribosome. The arrested cDNA product is usually 15–16 nt from the first nucleotide of the triplet in the ribosomal P-site occupied by a tRNA (13–15). IGR IRES-80S (Ala, Thr) complexes assembled from purified 40S and 60S subunits produced strong toeprints at CA6226-7 (Fig. 3, lane 3), which is +13–14 nt from the first cytidine (C6213) residue of the CCU triplet in the P-site of the ribosome (3). Addition of purified elongation factors eEF1A and eEF2 and aminoacylated tRNAs to assembled IGR IRES-80S complexes in the presence of cycloheximide produced toeprints at AC6232-3 (Fig. 3, lane 4), which is 6 nt downstream of CA6226-7, indicating a rearrangement or movement of the IGR IRES-80S complex similar to that observed after two cycles of translocation, with the deacylated tRNAAla in the ribosomal E-site. This toeprint change did not occur when elongation factors, aminoacylated tRNAs, or 60S subunits were omitted from the reaction (Figs. 3, lanes 5–7, and Fig. 5, which is published as supporting information on the PNAS web site). The change in toeprint most likely represented movement of the ribosome, because addition of nonhydrolyzable GTP (GMP-PNP) or edeine, which inhibits the delivery of an aminoacylated tRNA to the ribosomal A-site, inhibited the shift in the toeprint (Fig. 5). To further analyze IGR IRES-80S movement, we monitored the shift in toeprint in the presence of individual tRNAs, which are synthesized and aminoacylated in vitro, and purified eEF1A and eEF2. Normally, the first two codons in the ORF after the IGR IRES are alanine-encoding GCU and threonine-encoding ACA (9). Because we were unable to aminoacylate tRNAThr, we generated an ORF that contained the alanine-encoding GCU codon followed by a methionine-encoding AUG. Addition of elongation factors, Ala-tRNAAla, and Met-tRNAMet resulted in toeprints +6 nt downstream of toeprint CA6226-7, consistent with a ribosome positioned after two cycles of translocation (Fig. 3, lane 10). Addition of only Ala-tRNAAla to the IGR (Ala, Met) IRES-80S (Fig. 3, lane 11) or addition of both Ala-tRNAAla and Met-tRNAMet to the IGR (Ala, Thr) IRES-80S (Fig. 3, lane 13) resulted in a toeprint at an intermediate position, suggesting that the ribosomes had performed one cycle of translocation.
Fig. 3.
Ribosome rearrangement in IGR IRES–ribosome complexes in a reconstituted system. Movement of the ribosome on the IGR IRES was monitored by toeprinting analysis. Ribosomes assembled on dicistronic RNAs containing the WT IGR IRES (Ala, Thr) (lanes 2–7 and 13) or a WT IGR IRES (Ala, Met) with a Ala GCU codon followed by a Met AUG codon (lanes 8–12). Purified 40S, 60S, eEF2, eEF1A, and bulk aminoacylated tRNAs (ac-tRNA) were added in combinations to reactions with the dicistronic RNAs and analyzed by primer extension analysis (lanes 1–7). All reactions shown were in the presence of cycloheximide. Ala-tRNAs synthesized in vitro and elongator Met-tRNAs were used in lanes 8–12 instead of bulk aminoacylated tRNAs. Reactions were displayed on denaturing polyacrylamide gels and quantitated by PhosphorImager analysis. Toeprints of ribosome translocation on the IGR IRES are marked on the right. Numbering of the nucleotides refers to the nucleotide's position in the CrPV genome. A summary of the toeprints on the dicistronic RNAs is shown below the gel.
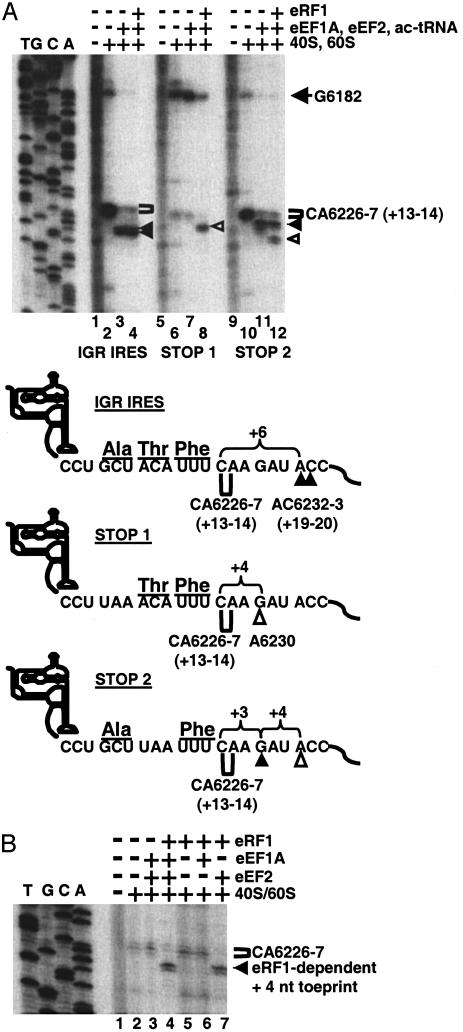
P-Site-Occupied IGR IRES Mediates Translation Termination. Because the IGR IRES can mimic the function of a Met-tRNAi in the P-site and direct translation initiation and translocation from the A-site, we examined whether the P-site-located IRES could mimic the function of a peptidyl tRNA by directing translation termination if a stop codon occupied the following A-site. During termination of eukaryotic protein synthesis, eRF1 recognizes and binds to all three stop codons (UAA, UAG, and UGA) in the ribosomal A-site and induces peptidyl tRNA hydrolysis, thereby releasing the nascent polypeptide chain (16). In prokaryotes, release factor (RF)-3 and GTP then mediate the release of RF1 from the ribosome, resulting in a posttermination complex with a deacylated tRNA in the P-site of the ribosome (17–19). It is known from cell-free reconstitution experiments that eRF1 can contact a stop codon located in the ribosomal A-site if the P-site is occupied by tRNA (20–22). To test whether eRF1 could contact a stop codon in the A-site if the IGR IRES was in the P-site, the movement of mRNA-80S complexes assembled in vitro was analyzed by toeprint analyses. Incubation of IGR IRES-80S complexes with eEF1A, eEF2, and aminoacylated tRNAs, in the presence of cycloheximide, changed the IGR IRES-80S toeprint by 6 nt, indicating that 80S ribosomes have translocated (Fig. 4A, lanes 2 and 3). Addition of purified eRF1 did not change the toeprint pattern (Fig. 4A, lane 4). In IGR IRES-80S complexes that contained a UAA stop codon instead of the alanine-encoding GCU codon in the A-site (Fig. 4A, STOP 1), addition of eRF1 resulted in a shift of the toeprint by 4 nt (Fig. 4A, lane 8), suggesting that eRF1 altered the structure of the IGR IRES-80S complex, likely by interacting with the stop codon in the A-site. To test this prediction, we exchanged the next codon, a threonine-encoding ACA codon, for a UAA stop codon (Fig. 4A, STOP 2). Assembled STOP 2–80S complexes displayed the CA6226-7 toeprints (Fig. 4A, lane 10). Addition of elongation factors and aminoacylated tRNAs shifted the toeprint by 3 nt (Fig. 4A, lane 11), suggesting that the ribosome had performed one translocation step and that the GCU codon had moved from the A- to the P-site. Addition of eRF1 produced the +3-nt toeprint and an additional toeprint of 4 nt downstream of the observed toeprint in STOP 2–80S complexes (Fig. 4A, lane 12). Similar toeprints were observed of ribosomes bound to STOP 1 and STOP 2 RNAs in rabbit reticulocyte lysates (Fig. 6, which is published as supporting information on the PNAS web site). Although we do not know whether the eRF1-elicited +4-nt toeprints, observed for both STOP 1 and STOP 2 mRNA-80S complexes, reflect ribosome movement, ribosome rearrangement, or both, they clearly depended on the presence of a stop codon and eRF1.
Fig. 4.
P-site-located IGR IRES modulates the movement of A-site-located stop codon–eRF1 complexes. (A) Ribosome translocation on a dicistronic RNA containing the WT IGR IRES or the WT IGR IRES with a UAA stop codon in the first (STOP 1) or second (STOP 2) triplet of the downstream ORF. IGR IRES, STOP 1, or STOP 2 dicistronic RNAs were incubated alone (lanes 1, 5, and 9); with 40S and 60S subunits (lanes 2, 6, and 10); with 40S, 60S, elongation factors, and aminoacylated tRNAs (lanes 3, 7, and 11); or with 40S, 60S, elongation factors, aminoacylated tRNAs, and purified eRF1 (lanes 4, 8, and 12). All reactions were incubated in the presence of cycloheximide. Reactions were analyzed by toeprinting assays, displayed on denaturing polyacrylamide gels, and quantitated by PhosphorImager analysis. A summary of the toeprinting analyses is shown below the gel. Brackets denote the location of toeprints induced by ribosome that had not translocated. Filled arrowheads point to toeprints of ribosomes that had translocated. Open arrowheads indicate the +4-nt toeprint observed in the presence of eRF1. (B) eRF1-induced toeprint depends on the presence of eEF2. STOP 1 dicistronic RNAs were incubated alone (lane 1), with 40S and 60S subunits (lane 2), with 40S, 60S, eEF1A, and eEF2 (lane 3), or with 40S, 60S, eEF1A, eEF2, and eRF1 (lane 4). eEF1A, eEF2, and eRF1 were each omitted from the reaction (lanes 5–7). Reactions were analyzed by toeprinting assay and displayed on a denaturing polyacrylamide gel as above.
To determine the requirements for the eRF1-dependent +4-nt toeprint on the STOP 1 RNA, we performed toeprint analyses in reconstituted systems in which specific translation factors were omitted. eRF1 alone (Fig. 4B, lane 5) or in combination with eEF1A (Fig. 4B, lane 6) did not change the CA6226-7 toeprint on STOP 1–80S complexes. Surprisingly, the eRF1-dependent +4-nt toeprint required eEF2 (Fig. 4B, lane 7). Ribosome translocation is known to be catalyzed by eEF2 and GTP hydrolysis; these experiments suggest that the eRF1-induced ribosome rearrangements or translocations that occur during termination require eEF2, interacting with eRF1 bound in the ribosomal A-site.
Discussion
Overall, these findings describe unique properties of a divergent RNA molecule that can function as an IRES and recruit ribosomes in the absence of initiation factors and Met-tRNAi. The IRES occupies the ribosomal P-site to set the correct translation ORF (3). These unusual features raised the question of whether the IRES mimics the functions of Met-tRNAi during translation initiation. We have shown here that the IRES can indeed direct the synthesis of peptides by triggering the ribosome to perform the first translocation step without formation of a peptide bond, without initiation factors, and aided only by eEF1A, eEF2, and aminoacylated tRNAs. A similar conclusion has been reached very recently by Pestova and Hellen (23), who showed that the IRES can synthesize peptidylpuromycin dipeptides. However, we measured the synthesis of oligopeptides in our reconstituted system, suggesting that the IRES can indeed mediate the synthesis of proteins under conditions when initiation factors are limiting. Furthermore, we showed that the IGR IRES can function as both a Met-tRNAi and an elongator tRNA in the ribosomal P-site. The latter observation opens the possibility of studying distinct steps involved in peptide chain termination in a reconstituted system. These findings underscore the biological impact IGR IRES-like RNA molecules have in translation initiation. First, translation initiation directed by such IRESs is predicted not to be inhibited during nutritionally, hypoxically, apoptotically, stress-, or virus-induced activation of eIF2 kinases such as PKR, GCN2, or PERK (8, 24–26). Thus, the synthesis of proteins that aid cells in recovery from a variety of stress responses may be regulated by an IGR IRES-type mechanism. By a similar mechanism, infectious agents such as CrPV may use IGR IRES-like RNAs to counteract the phosphorylation of eIF2 by the innate arm of the cellular immune response. Second, IRES molecules with Met-tRNAi-like properties may have preceded the evolution of the Met-tRNAi, thereby assembling ribosomes and setting correct reading frames themselves.
It has been proposed that interactions of the E- and P-site-occupied deacylated tRNAs with the ribosome are important for mediating elongation factor G (EF-G)-dependent translocation (27, 28). The mechanism by which this process occurs is poorly understood. The IGR IRES occupies the P-site of the ribosome to set the correct translational frame for the first aminoacylated tRNA to be delivered to the ribosomal A-site. Moreover, a recent structural analysis of IGR IRES-40S complexes by cryoelectron microscopy reconstructions revealed that the IGR IRES occupies the E- and P-sites of the ribosome (C. Spahn, E.J., A. Mulder, R. A. Grassucci, P.S., and J. Frank, unpublished data), leading to the hypothesis that, like in the prokaryotic system, eukaryotic E- and P-site-occupied tRNA–ribosome interactions are also important for translocation. In this study, the development of a minimal translation system driven by the IGR IRES allows for future analysis of translocation-dependent RNA–ribosome interactions.
Although the CrPV IGR IRES itself does not carry out elongation functions in a physiological setting, we show here that the IRES can mimic functions of a peptidyl tRNA in the ribosomal P-site. By monitoring structural changes in IRES-80S complexes that contain the IRES in the P-site and a stop codon in the A-site, it was observed that eRF1 induced IRES-80S rearrangements that depended on the presence of eEF2. At present, we do not know whether eEF2 interacts directly or indirectly with eRF1. In prokaryotes, the ribosome recycling factor (RRF) and EF-G, the prokaryotic homologue of eEF2, catalyze the dissociation of the ribosome, mRNA, and deacylated tRNA (29–31). One model suggests that EF-G induces translocation with the RRF in the ribosomal A-site. Curiously, no RRF has been identified in eukaryotic cells. Therefore, it has been suggested that eukaryotic eRF3 can recycle both eRF1 and ribosomes (32). Our result, that the eRF1-induced +4-nt toeprint required eRF1 and eEF2, suggests that eEF2 is involved in ribosome translocation with eRF1 in the ribosomal A-site.
In conclusion, the development of a minimal reconstituted system that synthesizes peptides directed by the IGR IRES provides a framework for understanding not only the mechanism of tRNA-like elements such as the IGR IRES but also translation in general, initiation, elongation, and termination.
Supplementary Material
Acknowledgments
We thank Bill Merrick for the rabbit eEF1A; Adam Geballe for the recombinant eukaryotic eRF1; Dan Herschlag, Magdalena Dorywalska, Ruben Gonzalez, and Geoff Otto for helpful discussions and protocols; and Karla Kirkegaard and Jody Puglisi for critical reading of the manuscript. This work was supported by National Institutes of Health Grants GM55979 (to P.S.) and GM57483 (to T.G.K.) and by Damon Runyon Cancer Research Foundation Grant DRG-1630 (to E.J.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: eIF, eukaryotic initiation factor; Met-tRNAi; initiator tRNA; IRES, internal ribosome entry site; CrPV, cricket paralysis virus; IGR, intergenic; ORFS, small ORF; ORFL, large ORF; EMCV, encephalomyocarditis virus; eEf, eukaryotic elongation factor; eRF, eukaryotic release factor.
References
- 1.Dever, T. E. (2002) Cell 108, 545–556. [DOI] [PubMed] [Google Scholar]
- 2.Hellen, C. U. & Sarnow, P. (2001) Genes Dev. 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 3.Wilson, J. E., Pestova, T. V., Hellen, C. U. & Sarnow, P. (2000) Cell 102, 511–520. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki, J. & Nakashima, N. (1999) J. Virol. 73, 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki, J. & Nakashima, N. (2000) Proc. Natl. Acad. Sci. USA 97, 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jan, E. & Sarnow, P. (2002) J. Mol. Biol. 324, 889–902. [DOI] [PubMed] [Google Scholar]
- 7.Kanamori, Y. & Nakashima, N. (2001) RNA 7, 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez, J., Yaman, I., Sarnow, P., Snider, M. D. & Hatzoglou, M. (2002) J. Biol. Chem. 277, 19198–19205. [DOI] [PubMed] [Google Scholar]
- 9.Wilson, J. E., Powell, M. J., Hoover, S. E. & Sarnow, P. (2000) Mol. Cell. Biol. 20, 4990–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestova, T. V. & Hellen, C. U. (2001) RNA 7, 1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jan, E., Thompson, S. R., Wilson, J. E., Pestova, T. V., Hellen, C. U. & Sarnow, P. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 285–292. [DOI] [PubMed] [Google Scholar]
- 12.Pestova, T. V., Shatsky, I. N., Fletcher, S. P., Jackson, R. J. & Hellen, C. U. (1998) Genes Dev. 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova, T. V., Hellen, C. U. & Shatsky, I. N. (1996) Mol. Cell. Biol. 16, 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anthony, D. D. & Merrick, W. C. (1992) J. Biol. Chem. 267, 1554–1562. [PubMed] [Google Scholar]
- 15.Hartz, D., McPheeters, D. S. & Gold, L. (1989) Genes Dev. 3, 1899–1912. [DOI] [PubMed] [Google Scholar]
- 16.Kisselev, L., Ehrenberg, M. & Frolova, L. (2003) EMBO J. 22, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kisselev, L. L. & Buckingham, R. H. (2000) Trends Biochem. Sci. 25, 561–566. [DOI] [PubMed] [Google Scholar]
- 18.Bertram, G., Innes, S., Minella, O., Richardson, J. & Stansfield, I. (2001) Microbiology 147, 255–269. [DOI] [PubMed] [Google Scholar]
- 19.Poole, E. & Tate, W. (2000) Biochim. Biophys. Acta 1493, 1–11. [DOI] [PubMed] [Google Scholar]
- 20.Chavatte, L., Frolova, L., Kisselev, L. & Favre, A. (2001) Eur. J. Biochem. 268, 2896–2904. [DOI] [PubMed] [Google Scholar]
- 21.Bulygin, K. N., Repkova, M. N., Ven'yaminova, A. G., Graifer, D. M., Karpova, G. G., Frolova, L. Y. & Kisselev, L. L. (2002) FEBS Lett. 514, 96–101. [DOI] [PubMed] [Google Scholar]
- 22.Chavatte, L., Seit-Nebi, A., Dubovaya, V. & Favre, A. (2002) EMBO J. 21, 5302–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestova, T. V. & Hellen, C. U. (2003) Genes Dev. 17, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufman, R. J., Scheuner, D., Schroder, M., Shen, X., Lee, K., Liu, C. Y. & Arnold, S. M. (2002) Nat. Rev. Mol. Cell. Biol. 3, 411–421. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman, R. J. (1999) Genes Dev. 13, 1211–1233. [DOI] [PubMed] [Google Scholar]
- 26.Harding, H. P., Calfon, M., Urano, F., Novoa, I. & Ron, D. (2002) Annu. Rev. Cell. Dev. Biol. 18, 575–599. [DOI] [PubMed] [Google Scholar]
- 27.Valle, M., Zavialov, A., Sengupta, J., Rawat, U., Ehrenberg, M. & Frank, J. (2003) Cell 114, 123–134. [DOI] [PubMed] [Google Scholar]
- 28.Zavialov, A. V. & Ehrenberg, M. (2003) Cell 114, 113–122. [DOI] [PubMed] [Google Scholar]
- 29.Inokuchi, Y., Hirashima, A., Sekine, Y., Janosi, L. & Kaji, A. (2000) EMBO J. 19, 3788–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janosi, L., Hara, H., Zhang, S. & Kaji, A. (1996) Adv. Biophys. 32, 121–201. [DOI] [PubMed] [Google Scholar]
- 31.Karimi, R., Pavlov, M. Y., Buckingham, R. H. & Ehrenberg, M. (1999) Mol. Cell 3, 601–609. [DOI] [PubMed] [Google Scholar]
- 32.Buckingham, R. H., Grentzmann, G. & Kisselev, L. (1997) Mol. Microbiol. 24, 449–456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.