Abstract
The targeting of molecular repertoires to complex systems rather than biochemically pure entities is an accessible approach that can identify proteins of biological interest. We have probed antigens presented by a monolayer of tumor cells for their ability to interact with a pool of aptamers. A glioblastoma-derived cell line, U251, was used as the target for systematic evolution of ligands by exponential enrichment by using a single-stranded DNA library. We isolated specifically interacting oligonucleotides, and biochemical strategies were used to identify the protein target for one of the aptamers. Here we characterize the interaction of the DNA aptamer, GBI-10, with tenascin-C, an extracellular protein found in the tumor matrix. Tenascin-C is believed to be involved in both embryogenesis and oncogenesis pathways. Systematic evolution of ligands by exponential enrichment appears to be a successful strategy for the a priori identification of targets of biological interest within complex systems.
The use of molecular repertoires is becoming increasingly important in the fields of drug discovery and biological research (1–3). These strategies involve the selection of combinatorially derived species. The most accessible techniques are based on the phage display of peptide or antibody libraries (4–8) and the use of libraries of oligonucleotides (9, 10). Systematic evolution of ligands by exponential enrichment (SELEX) is an iterative selection procedure used to identify oligonucleotides with desired properties, most often binding to a molecular target. The starting libraries (11, 12) are as large as 1015 unique sequences, some of which will be able to adopt secondary and tertiary structures (13). High-affinity oligonucleotide ligands to a plethora of high- and low-molecular-weight targets have been identified (3). However, the vast majority of these experiments have targeted biochemically pure entities.
The targeting of complex systems with SELEX lends itself to the concept of a priori identification of targets of biological interest and possibly to in vivo efficacy of such bioactive molecules. Here we demonstrate oligonucleotide targeting of the glioblastoma cell line U251. Glioblastomas are the most common of the human brain malignancies (14–16). Their aggressive nature is believed to be due to a combination of hypervascularity, focal necrosis, and rapid cellular proliferation. The glioblastoma remains refractory to therapy because of tumor heterogeneity, local invasion, and nonuniform vascular permeability to drugs. Our goal was to generate oligonucleotide ligands that recognize tumor-associated proteins on/within living cells, simultaneously identifying target proteins and DNA aptamers.
Experimental Procedures
Cell SELEX. Synthetic DNA template (10 pmol; Operon Technologies, Alameda, CA) containing 34 random nucleotides flanked by fixed regions 5′-GCCTGTTGTGAGCCTCCT-N34-CGCTTATTCTTGTCTCCC-3′ complementary to the primers 5′-BBB-GCCTGTTGTGAGCCTCCT-3′ and 5′-GGGAGACAAGA ATA AGCG-3, where BBB denotes three biotin phosphoramidite couplings, were amplified by PCR (Perkin-Elmer/Cetus thermal cycler). The PCR mixtures contained 50 mM KCl, 10 mM Tris·HCl (pH 8.6), 2.5 mM MgCl2, 170 μg/ml BSA, all four dNTPs at 1 μM each [except for dCTP (0.1 μM) and [α-32P]dCTP (1.25 μM)], primers (1 μM each), and 1,000 units/ml TaqDNA polymerase (Boehringer Mannheim). Twenty-five thermal cycles were conducted at 93°C for 30 sec, 52°C for 20 sec, and 72°C for 60 sec. The nonbiotinylated single-stranded DNA was then separated from the larger biotinylated DNA strand by gel electrophoresis (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3/8% acrylamide/8 M urea).
The SELEX process was performed on cell monolayers at 4°C. U251 glioblastoma cells (NCI) were plated at 107 per T175 flasks (Falcon) and allowed to form a monolayer at 37°C in RPMI medium 1640 (Sigma) containing 10% (vol/vol) FBS (Sigma). Cells were washed three times with cold serum-free RPMI medium 1640 (25 ml) before addition of competitor oligonucleotide. Yeast tRNA and salmon sperm DNA were denatured and sonicated to ≈200 nucleotides before use. Body-labeled DNA library (50–100 pmol) was incubated with the cell monolayer for 20 min before washing the cell monolayer (as described above except that washes were 25 min each). Cells were harvested by trypsinization before being heated (at 95°C for 15 min). DNA was recovered by phenol extraction, ethanol precipitation, and PCR amplification. To monitor aptamer pool binding at various rounds, 32P-labeled DNA was incubated with 2e5 U251 cells per well for 1 h, washing was performed as above, and cells were alkaline-lysed and 32P-quantified by scintillation counting. In this cell SELEX process, we attempted to derive specific and high-affinity aptamers by increasing the molar ratio of nonamplifiable competitor tRNA and DNA to aptamer pool at each round of selection. The early rounds contained a 50-fold molar excess, and the later rounds contained a 1,000-fold molar excess. A second feature of the process was designed to select DNA aptamers with slower off rates: cells were washed for long periods (three times for 25 min each) before bound aptamers were collected. Twenty-one rounds of cell SELEX were performed before cloning.
Affinity Purification of Target Membrane Proteins with DNA. A monolayer of U251 glioblastoma cells (plated at 107 in a T175 flask) was washed three times at 4°C with PBS containing 1 mM CaCl2 (pH 7.4). One milliliter of hypotonic buffer [10 mM Tris·HCl (pH 7.5) containing 5 mM KCl and 0.5 mM MgCl2 with protease inhibitors (1.5 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μM PMSF)] was added to the cell monolayer (for 30 min at 4°C) before cells were harvested by scraping. Stripped cells were homogenized on ice with a Dounce tissue grinder in a sucrose solution (0.25 M, 10 ml). Whole cells and nuclei were removed by centrifugation (1,000 × g for 10 min at 4°C). The supernatant was then recentrifuged to pellet the membrane component (105,000 × g for 1 h at 4°C). The pellet was solubilized (for 4 h at 4°C with rotation) in extraction buffer [10 mM Tris·HCl (pH 7.5) containing 200 mM NaCl, 0.1% (vol/vol) Triton X-100, 1 mM MgCl2, 1 mM CaCl2, and protease inhibitors (described above)]. This cell-surface extract was incubated (at 4°C for 2 h) with biotinylated aptamer (GBI-10, Scb10-1, and Scb10-2) and streptavidin magnetic beads (Dynal) in affinity purification buffer [extraction buffer containing 0.5 μg/μl BSA and 4% (vol/vol) glycerol]. The beads were washed in extraction buffer (for 30 min at 4°C) before the bound protein was eluted [10 mM Tris·HCl (pH 7.5) containing 1 M NaCl, 5 mM EDTA, and 0.1% (vol/vol) Triton X-100). Aptamer-purified proteins were analyzed by SDS/PAGE and silver staining.
Liquid Chromatography Tandem MS (LC-MS/MS). After polyacrylamide gel electrophoresis (SDS/PAGE), the aptamer-purified protein band (250 kDa) was excised and digested in situ with trypsin according to a previously described protocol (17), except that Tween 20 was not added. The digestions were analyzed by automated microcapillary LC-MS/MS analysis. Online HPLC separation of peptides was performed by using 100-mm ID capillary columns (6–8 cm in length) packed with Vydac C18 reverse-phase support. The sample was analyzed by using a linear gradient from 2% to 92% buffer B [A, 0.1% trifluoroacetic acid (TFA) in water; B, 90% acetonitrile, 0.07% TFA in water (vol/vol)] at a flow rate of 100–200 nl/min. Postcolumn UV detection was performed at 200 nm by using a 759A UV/VIS spectrophotometer (Applied Biosystems) equipped with a capillary flow cell holder. MS was performed on a Finnigan MAT (San Jose, CA) TSQ-700 triple sector quadruple mass spectrometer equipped with a Finnigan MAT electrospray ion source modified for microelectrospray as described (18). The mass spectrometer and HPLC system were controlled by a DECStation 5000/240 computer (Digital Equipment, Merrimack, NH) running Finnigan icis data system software version 7.2. Programs for data acquisition and instrument control were developed by using Finnigan instrument control language, version 7.27. Precursor mass spectra and collision-induced dissociation (CID) product ion spectra were collected as described (19). A single LC-MS/MS analysis of the 250-kDa protein generated 63 CID analyses; however, 18 were of poor quality and thus were not included in the database search. Correlation analysis using sequest matched 34 of the remaining 45 CID spectra with peptide sequences derived from the database. The remaining 11 spectra could not be interpreted by sequest or by manual interpretation and could not be correlated to a known peptide with confidence.
ELISA of the Aptamer–Tenascin Interaction. In ELISA format A, wells of the ELISA plate (Corning) were coated overnight at 37°C with streptavidin (Pierce, 5 μg/ml in 300 μl of H2O). The following day, wells were washed four times at room temperature with 240 μl of binding buffer [10 mM Tris·HCl (pH 7.5) containing 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 0.1% (wt/vol) BSA, 0.1% (vol/vol) Triton X-100, and 0.1% (vol/vol) Tween 20] before blocking with 300 μl of the same buffer (for 2 h at 4°C). Wells were washed (as before) before the addition of biotinylated oligonucleotides (50 pmol) in binding buffer (for 4 h, at 21°C). Wells were washed again before the incubation of cell-surface extract (100 μg) diluted in the binding buffer (overnight at 4°C). After washing, bound tenascin was detected (for 1 h at 4°C) by mAb 1923 (Chemicon). After washing, bound antibody was detected by a goat-anti-mouse peroxidase (Pierce) diluted 1,000-fold before a 1-h incubation at 4°C. The reaction was developed and assayed as per described protocols (20). ELISA format B was performed similarly to format A with the exception that the anti-tenascin monoclonal antibody, mAb 1911, was first immobilized in ELISA wells (250 ng). Purified tenascin (2 μg; Sigma) was captured on the monoclonal antibodies and assayed for binding to biotinylated oligonucleotides (50 pmol per well). Bound biotinylated aptamers were detected by a streptavidin-conjugated peroxidase (1:1,000 in binding buffer, Pierce). All blocking, washing, and color development steps were performed as described (20).
Biosensor Analysis of the Aptamer–Tenascin Interaction. Biotinylated GBI-10, along with the sequence-scrambled oligonucleotide Scb10-2, were coupled to a streptavidin-coated carboxyl methyl dextran surface. Full-length tenascin-C (Chemicon) or three bacterially expressed fragments of tenascin-C, TNfbg, TNfn3–5, and TNfnA-D (graciously supplied by H. P. Erickson, Duke University, Durham, NC) (21) were passed across this surface in a BIACORE 2000, and binding was measured by surface plasmon resonance (22, 23). Data from the control surface containing Scb10-2 were subtracted.
Results
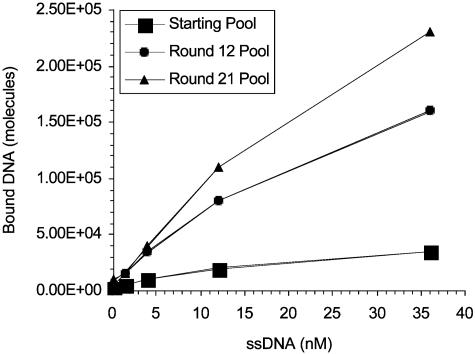
Targeting the U251 Glioblastoma with DNA Aptamers. The U251 cell line, derived from a human glioblastoma, was used as the target for in vitro selection of aptamers from a random pool of DNA molecules. The starting repertoire was composed of 70-mer single-stranded DNA molecules containing randomized 34-nucleotide inserts. This library was applied to a monolayer of cultured cells in the presence nonamplifiable competitor oligonucleotide, which minimized nonspecific interaction. In each round of selection, the concentration of competitor DNA was increased to further drive selection toward a high-affinity and high-specificity aptamer pool. After 21 rounds of selection, the U251-binding properties of the starting, intermediate, and final pools were compared (Fig. 1). The round-12 and -21 pools displayed increased binding to U251 cells compared with the starting pool. The U251-selected round-21 pool was then cloned and sequenced.
Fig. 1.
Relative cell binding of U251-evolved SELEX pools. SELEX pools from round 21, round 12, and the starting round were assayed for binding to U251 cells.
Sequences from 163 clones were obtained, and their inserts were analyzed and sorted into putative families by the alignment of consensus motifs. Motifs were identified by inspection with the aid of computer-assisted search engines (data not shown). On the basis of clonal dominance and demonstrable binding to U251 cells, one aptamer, GBI-10, was chosen for further characterization. GBI-10 and its homologues represented 10% of the sequenced pool. The DNA sequence of GBI-10 along with two sequence-scrambled versions, Scb10-1 (scrambled variable region) and Scb10-2 (scrambled variable and fixed region), are given in Table 1.
Table 1. Sequence of U251 affinity-selected aptamer GBI-10.
| GBI-10 | ggctgttgtgagcctcctCCCAGAGGGAAGACTTTAGGTTCGGTTCACGTCCcgcttattcttactccc |
| Scb 10-1 | ggctgttgtgagcctcctATCTTGAGCTTACGGCCAAGCAGTTTCCGCGGAGcgcttattcttactccc |
| Scb 10-2 | ccgttcagtctagtcgctATCTTGAGCTTACGGCCAAGCAGTTTCCGCGGAGttgtgcctcctcctttga |
The fixed regions are denoted in lowercase, and the U251 affinity-selected region is denoted in uppercase. The sequences of two nonbinding variants, Scb 10-1 and Scb 10-2, are shown for comparison.
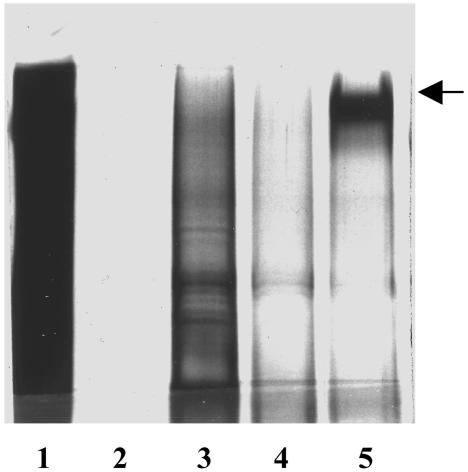
Identification of the GBI-10 Protein Target. An affinity purification procedure was used to identify the GBI-10 target on U251 cells. Cell-surface extracts were incubated with biotinylated oligonucleotides immobilized on streptavidin magnetic beads. The aptamer–bead complexes were washed extensively, and bound proteins were eluted and then resolved electrophoretically before silver staining (Fig. 2). The affinity purification profiles of unconjugated beads, Scb10-1, Scb10-2, and GBI-10, were compared with the cell-surface extracts. Strikingly, GBI-10 (Fig. 2, lane 5) exhibited a profile different from that of its scrambled peers. GBI-10 appeared to specifically interact with a high-molecular-weight polypeptide corresponding to ≈250 kDa. In contrast, there appeared to be less binding either to the beads alone or to the two control oligonucleotides.
Fig. 2.
A GBI-10 affinity matrix reveals a high-molecular-weight protein target. Shown are silver-stained gels depicting proteins affinity-purified by GBI-10 (lane 5), Scb10-1 (lane 3), Scb10-2 (lane 4), and beads alone (lane 2) from U251 membrane extracts (lane 1).
To identify the GBI-10 protein target, affinity purification was performed with 12 nmol of GBI-10 and 12 × 107 U251 cells. The eluted protein was further resolved on a 4–20% gradient-reducing SDS gel. The 250-kDa polypeptide band was excised before being digested with trypsin, and a fraction of the resulting digestion mixture was analyzed by automated LC-MS/MS. Intriguingly, 34 of the identified peptides localized to 24 regions of a protein sequence in the GenBank database, tenascin-C (Table 2) (24). The results show the range of peptide sequences identified after GBI-10 affinity purification. These results identify the GBI-10 protein target as tenascin-C. Furthermore, the ability of GBI-10 to purify tenascin-C from among the array of DNA- and RNA-binding proteins in the cell extract indicates that this aptamer specifically binds tenascin-C.
Table 2. GBI-10 interacts with tenascin.
| LEELENLVSSLR | 126-138 |
| VTEYLVVYTPTHEGGLEMQFR | 648-668 |
| SIPVSARVATYLPAPEGLK | 700-718 |
| NMNKEDEGEITK | 747-758 |
| RPETSYRQTGLAPGQEYEISLHIVKNNTR | 762-790 |
| LDAPSQIEVKD | 802-812 |
| AKETFTTGLDAPR | 884-896 |
| VSQTDNSITL EWR | 901-913 |
| TLTGLRPGTEY GIGVSAVK | 949-968 |
| GLEPGQEYNVLLTAEK | 1,041-1,056 |
| AATPYTVSIYGVIQGY | 1,135-1,152 |
| STDLPGLK | 1,218-1,125 |
| AVDIPGLEAATPYR | 1,400-1,413 |
| TAHISGLPPSTDFIVYLSGL APSIR | 1,491-1,515 |
| LSWTADEGVFDNFVLKI | 1,637-1,653 |
| KQSEPLEITLLAPER | 1,658-1,672 |
| RSQTVSAIATTAMGSPK | 1,697-1,713 |
| ITYVPITGGTPSMVTVDGTK | 1,741-1,760 |
| WQPAIATVDSYVI SYTGEKVPEITR | 1,818-1,842 |
| FTTDLDSPR | 1,882-1,890 |
| GRENFYQNWK | 2,031-2,040 |
| REEFWLGLDNL NKITAQGQYELRV | 2,050-2,073 |
| DHGETAFAVYDKFSVGDAK | 2,077-2,095 |
| YGDNNHSQGVNWFHWKGHEHSIQFAEMK | 2,156-2,083 |
GBI-10 affinity-purified peptides identified by LC-MS/MS (column 1) along with their location within tenascin-C (column 2).
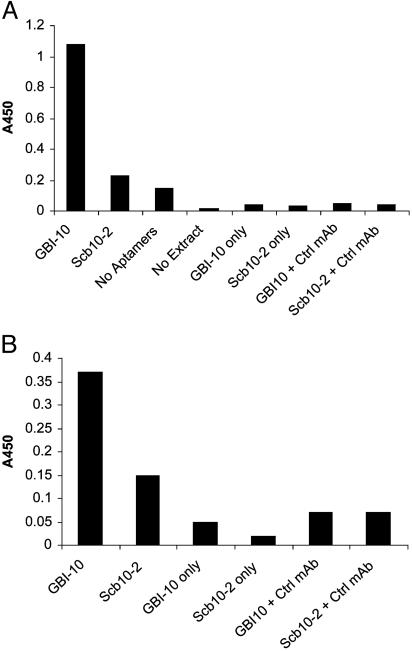
The GBI-10–Tenascin Interaction. To verify that GBI-10 binds tenascin-C from cell-surface extracts, the ability of biotinylated GBI-10 to interact with tenascin-C was compared with the background interaction of biotinylated Scb10-2. Biotinylated aptamers were captured on streptavidin before being incubated with cell-surface extracts. Bound tenascin-C was then detected by a tenascin-C monoclonal antibody (mAb 1923) and a peroxidase-conjugated secondary antibody (Fig. 3A). The results show that tenascin-C contained within the extract selectively interacted with GBI-10 whereas there was little nonspecific interaction with Scb10-2. To ascertain whether GBI-10 binds purified tenascin-C, purified tenascin was captured upon a tenascin-C-specific monoclonal antibody (mAb 1911) before the incubation of either GBI-10 or Scb10-2. Bound aptamer was detected by a streptavidin-linked peroxidase. (Fig. 3B). The results revealed that GBI-10 bound purified tenascin-C. As before, there was little nonspecific binding to Scb10-2. We therefore concluded that GBI-10 specifically interacted with tenascin-C in cell-surface extracts and as a purified protein.
Fig. 3.
ELISA analysis of the tenascin–GBI-10 interaction. (A) The interaction of GBI-10 and Scb10-2 with tenascin derived from cell-surface extract. Biotinylated aptamers were adsorbed to a streptavidin surface, incubated with U251 cell extract, washed, and further incubated with anti-tenascin mAb 1923 and a secondary peroxidase-conjugated antibody. (B) The interaction of pure tenascin with GBI-10 and Scb10-2. Anti-tenascin monoclonal antibody mAb 1911 was adsorbed to the surface, and then purified tenascin was captured by the adsorbed antibody. Biotinylated oligonucleotides were then incubated with the surface, followed by washing and detection with a peroxidase-conjugated streptavidin.
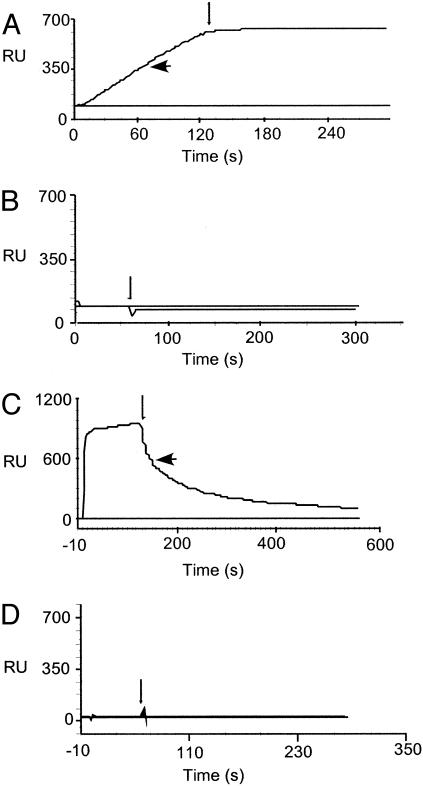
The GBI-10–tenascin-C interaction was further characterized by using surface plasmon resonance (BIACORE 2000). Biotinylated GBI-10 and Scb10-2 were coupled to a streptavidincoated carboxyl methyl dextran surface, and tenascin-C was passed across the surface (Fig. 4A). The results show that tenascin-C interacts with GBI-10 but not with the control aptamer Scb10-2. Interestingly, the protein does not display measurable dissociation from the DNA-containing surface. In this experiment the low level of tenascin-C that bound to the control aptamer surface (<100 response units) also did not display dissociation. It is possible that the lack of dissociation could be an artifact, whereby the large size of protein slows dissociation from the surface to a point where rebinding dominates. To identify the domain of tenascin-C that is targeted by GBI-10, three protein fragments were assayed for binding (Fig. 4 B–D). These fragments correspond to the C-terminal fibrinogen-like globule (TNfbg), fibronectin type III repeats 3–5 (TNfn3–5), and fibronectin type III repeats A–D (TNfnA–D) (21). GBI-10 specifically targets the TNfn3–5 domain of tenascin-C (Fig. 4 B–D). The affinity and temperature sensitivity of GBI-10 binding to TNfn3–5 were also measured under analytical conditions (23) leading to an equilibrium binding constant, Kd, of 150 nM at 4°C. The Kd was at least 10-fold weaker at 22°C and 37°C (data not shown). These results are consistent with the fact that the U251 SELEX was conducted at 4°C. The direct demonstration that GBI-10 targets domain fn3–5 of tenascin is not unexpected in the light of the fact that this domain binds to another polyanion, heparin (25).
Fig. 4.
Biosensor analysis of the tenascin–GBI-10 interaction. Binding of tenascin-C and subdomains of tenascin-C to aptamer GBI-10 is shown. Biotinylated GBI-10 (large arrow) or a scrambled-sequence control aptamer, Scb10-2 (no arrow), was bound to a streptavidin-coupled carboxyl methlyl dextran surface. Proteins were passed over the surface at the indicated concentration, followed by buffer only (small arrow). (A) TN-C (50 nM). (B) TNfbg (200 nM). (C) TNfn3–5 (200 nM). (D) TNfnA-D (200 nM). Responses to the control aptamer Scb10-2, always <100 response units, were subtracted from responses to GBI-10, generating the flat lines observed for Scb10-2 in A–D.
Discussion
Complex Target SELEX Identifies Protein Targets of Biological Interest. The a priori identification of proteins within complex targets promises to open doors to the discovery of proteins not previously associated with the disease state under study. Red blood cell membranes have recently been used to demonstrate the concept of complex target SELEX and the deconvolution strategies necessary for the rapid isolation of oligonucleotide ligands to multiple targets (26). In addition, SELEX experiments using live trypanosomes identified RNA aptamers specific for a 42-kDa protein located in the parasite's flagellar pocket, and the authors have elegantly defined the target protein by photo-crosslinking (27) and demonstrated that the aptamer is internalized into lysosomes (28). However, the idea of specifically targeting subpopulations of cells is not a novel one. Antibody fragments with binding specificity for the blood group antigens of the ABO, I, Rh, and Kell blood systems have been successfully isolated from phage-display libraries (29). In a similar experiment, de Kruif et al. (30) isolated phage-displayed antibody fragments specific for subsets of blood leukocytes. In contrast to both of these studies, Goodson et al. (31) used a strategy of alternate screening of receptor-positive and -negative cell lines to target a specific cell-surface receptor with phage-displayed peptides. It is notable that all three approaches used cell suspension selection.
Here, we have used SELEX to target the glioblastoma cell line U251 with a single-stranded random DNA library. Twenty-one rounds of SELEX, in the presence of nonamplifiable competitor nucleic acid, were performed against a monolayer of cells. Many of the selection rounds were performed without benefit of knowing how rapidly the aptamer pool complexity was decreased. Second-generation U251 cell SELEX experiments (32) monitored SELEX progress by quantitating pool complexity by using classical Cot analysis (33), a useful method for monitoring aptamer pool convergence from 1013 sequences toward a small number (<104) of sequences. We performed these selections at 4°C to minimize loss of any aptamers bound to cell-surface receptors that undergo receptor-mediated internalization. Cloning and sequencing of the final pool revealed one DNA ligand, GBI-10 and its homologues, to be prevalent at 10% of the total clones. Affinity purification using the aptamer, coupled to LC-MS/MS analysis, identified GBI-10's binding partner as the extracellular matrix protein tenascin-C. It is likely that the abundance and accessibility of tenascin-C as an extracellular matrix protein drove the selection of GBI-10 toward its clonal dominance within the final pool.
The discovery that tenascin presents a dominant epitope in this glioblastoma SELEX is exciting and intriguing (a review of tenascin-C is beyond the scope of this article; see refs. 34–37). Tenascin was first demonstrated to be an integral part of the dense mesenchyme surrounding the growing epithelium of embryonic and neoplastic tissues (38). However, on closer inspection, it appeared that tenascin may, in fact, be a stromal marker for epithelial malignancy. Immunohistochemical studies on the mammary gland tumors of human, rat, and mouse revealed the intensity of tenascin staining to correlate with the progression of the tumor. That is, malignant tumors expressed high levels of tenascin in contrast to benign tumors (39). Earlier, Bourdon et al. (40) identified a monoclonal antibody to the human glioma cell line U251MG, a relative of the U251 cell line used in this SELEX. Characterization of the antibody 81C6 revealed strong staining patterns of what the authors termed GMEM (glioma mesenchymal extracellular matrix) in glioblastomas but not in benign tumors or normal brain tissue (40). Later studies deduced that GMEM and tenascin were one and the same. Notably, anti-tenascin monoclonal antibodies are now in phase II trials for the treatment of patients with recurrent gliomas (41, 42). A study conducted by Wikstrand and Bigner (43) may be seen as a striking parallel to this study. Both experiments challenged large naïve libraries (109 for the murine immune system vs. 1013 for the initial SELEX pool) with a complex target, the U251 cell line, resulting in the identification of ligands specific for tenascin-C.
During the last two decades a great deal of effort has been aimed at the identification of tumor-associated antigens with the aid of monoclonal antibodies through the immunization of mice with tumor tissue or cell lines established from human tumor (44). However, transition from animal model to human tumor therapy is complicated by the need to humanize murine antibodies and, more importantly, by the slow rate of tumor penetration of full-length antibodies (40, 44, 45). In comparison with their antibody counterparts, SELEX-derived aptamers may be less immunogenic, and their smaller size may allow for increased tumor penetration rate and more rapid removal from blood (reviewed in ref. 45). The DNA aptamer described here does not bind well at 37°C; this is because of selection for binding at 4°C to minimize receptor-mediated endocytosis. In addition, the DNA ligand is subject to nuclease activity in vivo. To obtain an aptamer suitable for testing as a tumor-targeting agent, we are developing (nuclease-resistant) modified RNA aptamers that bind with high affinity to tenascin-C at physiological temperatures (32).
Complex Target SELEX and the Dissection of Biological Systems. Aside from the prediction that complex target SELEX will identify proteins of biological interest, a second assumption is that an array of protein targets should be selected for. The composition of an evolved SELEX pool (such as the one we cloned) will be dictated by the individual selective pressures driven by the characteristics of the mixture of protein targets. Pool composition is based on a combination of relative concentrations of accessible target proteins, relative native affinities of targets for aptamers, and arbitrary SELEX conditions. A recent study by Vant-Hull et al. (46) systematically addresses the issues pertaining to the mathematics of SELEX against complex targets. A computer program simulating SELEX against multiple targets was developed. On the basis of protein concentration, partitioning efficiency, and relative protein affinity, the authors model the dynamics of SELEX against heterogeneous mixtures of a small number of protein targets (46).
We believe the enrichment of GBI-10 is not a unique event; this aptamer may represent one of many in the final evolved pool that may specifically target U251 cells. Indeed, preliminary analysis of the affinity purification profiles of a subset of U251-selected aptamers identifies polypeptides of distinct sizes as well as tenascin-C (data not shown). We envisage that a number of parallel ligand-mediated target purifications would be required to decipher the full range of epitopes targeted in this multiple-target SELEX. A recent complex target SELEX study from this laboratory presents methodologies for grouping aptamers according to their protein target. The authors then go on to show how “deconvolution SELEX” can be used to partition aptamer pools that have been evolved against multiple targets (26). Ligand-directed (presented here; see also ref. 27) and target-mediated (26) deconvolution strategies will be invaluable to the dissection of pools evolved by complex target SELEX against biological systems.
An obvious advantage of complex SELEX is the ability to target a disease state without prior knowledge of molecular changes associated with the new condition. For example, a recent study has targeted atherosclerosis and injured vasculature (Andrew Stephens, personal communication). The tandem use of ex vivo and in vivo complex target SELEX in balloon-injured rat carotid arteries evolved an RNA ligand capable of specifically interacting with injured arteries in preference to normal arteries; the target of this aptamer is not yet known. This tumor cell SELEX demonstrates the ability to identify known tumor markers, such as tenascin-C. It is likely that complex–target SELEX experiments can discover previously uncharacterized markers for tumors, pathologies, development, and differentiation. Such dissection of complex biological systems by the SELEX process may generate aptamers for diagnostic and therapeutic uses.
Acknowledgments
We thank Carol Bell for cell culture expertise, Tim Fitzwater for help with sequence analysis, and past members of NeXstar Pharmaceuticals for inspiration and support. This work was supported by National Institutes of Health grants (to L.G.).
Abbreviations: LC-MS/MS, liquid chromatography tandem MS; SELEX, systematic evolution of ligands by exponential enrichment.
References
- 1.Janda, K. D. (1994) Proc. Natl. Acad. Sci. USA 91, 10779–10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gold, L. (1995) Harvey Lect. 91, 47–57. [PubMed] [Google Scholar]
- 3.Gold, L., Polisky, B., Uhlenbeck, O. & Yarus, M. (1995) Annu. Rev. Biochem. 64, 763–797. [DOI] [PubMed] [Google Scholar]
- 4.Cwirla, S. E., Peters, E. A., Barrett, R. W. & Dower, W. J. (1990) Proc. Natl. Acad. Sci. USA 87, 6378–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott, J. K. & Smith, G. P. (1990) Science 249, 386–390. [DOI] [PubMed] [Google Scholar]
- 6.Daniels, D. A. & Lane, D. P. (1995) Exp. Opin. Ther. Pat. 5, 901–912. [Google Scholar]
- 7.Marks, J. D., Hoogenboom, H. R., Bonnert, T. P., McCafferty, J. Griffiths, A. D. & Winter, G. (1991) J. Mol. Biol. 222, 581–597. [DOI] [PubMed] [Google Scholar]
- 8.Winter, G., Griffiths, A. D., Hawkins, R. E. & Hoogenboom, H. R. (1994) Ann. Rev. Immunol. 12, 433–455. [DOI] [PubMed] [Google Scholar]
- 9.Ellington, A. D. & Szostak, J. W. (1990) Nature 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 10.Tuerk, C. & Gold, L. (1990) Science 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 11.Fitzwater, T. & Polisky, B. (1996) Methods Enzymol. 267, 275–301. [DOI] [PubMed] [Google Scholar]
- 12.Tuerk, C. (1997) Methods Mol. Biol. 67, 219–230. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann, G. R., Jenison, R. D., Wick, C. L., Simorre, J. P. & Pardi, A. (1997) Nat. Struct. Biol. 4, 644–649. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro, W. R. & Shapiro, J. R. (1998) Oncology 12, 233–240. [PubMed] [Google Scholar]
- 15.Damek, D. M. & Hochberg, F. H. (1997) Curr. Opin. Neurol. 10, 452–458. [DOI] [PubMed] [Google Scholar]
- 16.Freeman, C. R. & Farmer, J. P. (1998) Int. J. Radiat. Oncol. Biol. Phys. 40, 265–271. [DOI] [PubMed] [Google Scholar]
- 17.Hellman, U., Werstedt, C., Gonez, J. & Heldin, C. H. (1995) Anal. Biochem. 224, 451–455. [DOI] [PubMed] [Google Scholar]
- 18.Davis, M. T., Stahl, D. C., Hefta, S. A. & Lee, T. D. (1995) Anal. Chem. 67, 4549–4556. [DOI] [PubMed] [Google Scholar]
- 19.Blyn, L. B., Swiderek, K. M., Richards, O., Stahl, D. C., Semler, B. L. & Ehrenfeld, E. (1996) Proc. Natl. Acad. Sci. USA 93, 11115–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniels, D. A. & Lane, D. P. (1994) J. Mol. Biol. 243, 639–652. [DOI] [PubMed] [Google Scholar]
- 21.Aukhil, I., Joshi, P., Yan, Y. & Erickson, H. P. (1993) J. Biol. Chem. 268, 2542–2553. [PubMed] [Google Scholar]
- 22.Karlsson, R., Roos, H., Fägerstam, B. & Persson, B. (1994) Methods Companion Methods Enzymol. 6, 99–110. [Google Scholar]
- 23.Karlsson, R. & Fält, A. (1997) J. Immunol. Methods 200, 121–133. [DOI] [PubMed] [Google Scholar]
- 24.Gherzi, R., Caremolla, B., Siri, A., Ponassi, M., Balza, E. & Zardi, L. (1995) J. Biol. Chem. 270, 3429–3434. [DOI] [PubMed] [Google Scholar]
- 25.Leahy, D. J., Hendrickson, W. A., Aukhil, I. & Erickson, H. P. (1992) Science 258, 987–991. [DOI] [PubMed] [Google Scholar]
- 26.Morris, K. N., Jensen, K. B., Julin, C. M., Weil, M. & Gold, L. (1998) Proc. Natl. Acad. Sci. USA 95, 2902–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homann, M. & Goringer, H. U. (1999) Nucleic Acids Res. 27, 2006–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homann, M. & Goringer, H. U. (2001) Bioorgan. Med. Chem. 9, 2571–2580. [DOI] [PubMed] [Google Scholar]
- 29.Marks, J. D., Ouwehand, W. H., Bye, J. M., Finnern, R., Gorick, B. D., Voak, D., Thorpe, S. J., Hughes-Jones, N. C. & Winter, G. (1993) Biotechnology 11, 1145–1149. [DOI] [PubMed] [Google Scholar]
- 30.de Kruif, J., Terstappen, L., Boel, E. & Logtenberg, T. (1995) Proc. Natl. Acad. Sci. USA 92, 3938–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodson, R. J., Doyle, M. V., Kaufman, S. E. & Rosenberg, S. (1994) Proc. Natl. Acad. Sci. USA 91, 7129–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hicke, B. J., Marion, C., Chang, Y-F., Gould, T., Lynott, C. K., Parma, D., Schmidt, P. G. & Warren, S. (2001) J. Biol. Chem. 276, 48644–48654. [DOI] [PubMed] [Google Scholar]
- 33.Charlton, J. & Smith, D. (1999) RNA 5, 1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erickson, H. P. (1993) Curr. Opin. Cell Biol. 5, 869–876. [DOI] [PubMed] [Google Scholar]
- 35.Leprini, A., Querze, G. & Zardi, L. (1994) Pers. Dev. Neurobiol. 2, 117–123. [PubMed] [Google Scholar]
- 36.Kurpad, S. N., Zhao, X. G., Wikstrand, C. J., Batra, S. K., McLendon, R. E. & Bigner, D. D. (1995) Glia 15, 244–256. [DOI] [PubMed] [Google Scholar]
- 37.Mackie, E. J. (1997) Int. J. Biochem. Cell Biol. 29, 1133–1137. [DOI] [PubMed] [Google Scholar]
- 38.Chiquet-Ehrismann, R., Mackie, E. J., Pearson, C. A. & Sakakura, T. (1986) Cell 47, 131–139. [DOI] [PubMed] [Google Scholar]
- 39.Mackie, E. J., Chiquet-Ehrismann, R., Pearson, C. A., Inaguma, Y., Taya, K., Kwarada, Y. & Sakakura, T. (1989) Proc. Natl. Acad. Sci. USA 84, 4621–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bourdon, M. A., Wikstrand, C. J., Furthmayr, H., Matthews, T. J. & Bigner, D. D. (1983) Cancer Res. 43, 2796–2805. [PubMed] [Google Scholar]
- 41.Bigner, D. D., Brown, M. T., Friedman, A. H., Coleman, R. E., Akabani, G., Friedman, H. S., Thorstad, W. L., McLendon, R. E., Bigner, S. H., Zhao, X. G., et al. (1998) J. Clin. Oncol. 16, 2202–2212. [DOI] [PubMed] [Google Scholar]
- 42.Paganelli, G., Grana, C., Chinol, M., Cremonesi, M., De Cicco, C., F., Robertson, C., Zurrida, S., Casadio, C., Zoboli, S., Siccardi, A. G. & Veronesi, U. (1999) Eur. J. Nucl. Med. 26, 348–357. [DOI] [PubMed] [Google Scholar]
- 43.Wikstrand, C. J. & Bigner, D. D. (1982) Cancer Res. 42, 267–275. [PubMed] [Google Scholar]
- 44.Khawli, L. A. & Epstein, A. L. (1997) Q. J. Nucl. Med. 41, 25–35. [PubMed] [Google Scholar]
- 45.Hicke, B. J. & Stephens, A. W. (2000) J. Clin. Invest. 106, 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vant-Hull, B., Payano-Baez, A., Davis, R. H. & Gold, L. (1998) J. Mol. Biol. 279, 579–597. [DOI] [PubMed] [Google Scholar]