Abstract
Aminoacyl-tRNA synthetases (aaRSs) are multidomain proteins that specifically attach amino acids to their cognate tRNAs. Their most conserved, and presumably evolutionarily oldest, domains are the catalytic cores, which activate amino acids and transfer them to the 3′ ends of tRNAs. Additional domains appended to or inserted in the body of aaRSs increase efficiency and specificity of the aminoacylation process, either by providing additional tRNA contacts, or by hydrolyzing noncognate amino acid products (cis-editing). Here, we report specific tRNA-dependent trans-editing by aaRS-like proteins that reciprocate the editing domains of aaRSs, but not the remainder of the corresponding enzyme. A freestanding homologue of the prolyl-tRNA synthetase-editing domain, the PrdX protein from Clostridium sticklandii, efficiently and specifically hydrolyzes Ala-tRNAPro. Similarly, autonomous alanyl-tRNA synthetase-editing domain homologues (AlaX proteins) from Methanosarcina barkeri and Sulfolobus solfataricus hydrolyze Ser-tRNAAla and Gly-tRNAAla substrates. The discovery of autonomous editing proteins efficient in hydrolyzing misacylated products provides a direct link between ancestral aaRSs consisting solely of the catalytic core and extant enzymes to which functionally independent modules are appended.
Accurate aminoacylation depends on successful discrimination between cognate and noncognate amino acid and tRNA substrates. Inaccuracies are more frequent in amino acid selection (1 in 104 to 105) than in the selection of tRNA (1 in 106) (1, 2), due to the larger surface area of the tRNA substrate and greater diversity of structural elements that serve as discriminating factors (3). It has been postulated that error rates of >1 in 3,000 in the initial amino acid selection require correction mechanisms to increase the accuracy of aminoacylation and thereby reduce errors in protein synthesis to a tolerable level (1). When error rates exceed this threshold, the incorrect products are hydrolyzed at the secondary amino acid binding sites (editing sites), either by pretransfer (hydrolysis of aminoacyl-adenylate) or posttransfer (hydrolysis of aminoacyl-tRNA) editing mechanisms (4).
Editing sites have hitherto been described in distinct domains (editing domains) corresponding to particular structural modules within the characteristic synthetase multidomain structure. In some synthetases [isoleucyl- or alanyl-tRNA synthetase (Il-eRS, AlaRS)], the occurrence of such domains seems to be universal (5). In contrast, although well described for some organisms (6–8), editing domains are absent in many archaeal threonyl-tRNA synthetases (ThrRS) (9) and are limited to only some bacterial groups of prolyl-tRNA synthetases (ProRS) (8). Moreover, the isolated editing domains of valyl-tRNA synthetase and IleRS were shown to perform some degree of in vitro deacylation of Thr-tRNAVal and Val-tRNAIle, respectively (10). Similarly, the isolated editing domain of Escherichia coli ProRS and the freestanding editing domain homologue Ybak from Haemophilus influenzae were recently shown to hydrolyze Ala-tRNAPro, although somewhat less efficiently than the full-length ProRS (11).
The ability of isolated editing domains and proteins reciprocating these domains to edit in trans led us to speculate that the apparently absent editing modules from ProRS and ThrRS might be functionally complemented by the occurrence of trans-acting paralogs. We found that naturally occurring paralogs of parts of ProRS, AlaRS, and ThrRS efficiently and specifically hydrolyze misacylated tRNA substrates. These results suggest that editing domains of aminoacyl-tRNA synthetases (aaRS) may have originated from similar autonomous editing modules.
Materials and Methods
Cloning and Preparation of Enzymes. All DNA and predicted amino acid sequences were obtained from the available databases. Genomic DNA from Clostridium sticklandii was a gift from A. Pich [Universität Halle-Wittenberg, Halle (Saale), Germany], Plasmodium falciparum DNA was a gift from T. McCutchan (National Institutes of Health, Bethesda), Methanosarcina barkeri DNA was a gift from K. Sowers (University of Maryland Biotechnology Institute, Rockville), and Sulfolobus solfataricus DNA was purchased from American Type Culture Collection (ATCC) bioproducts (ATCC 35092D). Methanocaldococcus jannaschii cells were obtained from K. O. Stetter and M. Thomm (University of Regensburg, Regensburg, Germany). DNA was prepared by standard methods. E. coli AlaRS was a gift from A. Ambrogelly (Yale University, New Haven, CT). ProRS, ProX, and AlaX genes were identified from available genomic sequences by blast searches (12), amplified by PCR, and cloned into pET15b (Invitrogen). The M. jannaschii AlaRS gene has been described (13). The QuikChange system (Stratagene) was used for site-directed mutagenesis to obtain the 5′-truncated P. falciparum proS gene (nucleotides 670-2238) that encodes ΔPF-ProRS lacking the whole N-terminal extension, and the M. jannaschii AlaRS gene that codes for AlaRS with the single amino acid substitution C699A.
Expression was induced in E. coli BL21-Codon Plus (DE3)-RIL strain at A600 = 0.6 with 0.8 mM isopropyl β-d-thiogalactoside for 2 h at 37°C. N-terminally His-6-tagged proteins were purified on nickel-columns as described (8). Native M. jannaschii AlaRSs were purified as described (13) with an additional purification step over a DEAE-cellulose column (8). S. solfataricus AlaX was flocculated at 65°C for 45 min to minimize the amount of contaminating E. coli proteins before purification by affinity chromatography.
Aminoacylation Assay. To determine the amount of the active enzyme in the preparation, active site titration was performed as described (8). Unfractionated mature tRNA from M. jannaschii was prepared according to the published procedure (14). Aminoacylation with P. falciparum ProRSs was performed at 37°C in 50 mM Hepes-KOH (pH 7.2), 50 mM KCl, 15 mM MgCl2, 5 mM DTT, 10 mM ATP, 50 μM [3H]proline (200 cpm/pmol) or 300 μM [3H]alanine (200 cpm/pmol) (Amersham Pharmacia Biosciences), 2 mg/ml Saccharomyces cerevisiae unfractionated tRNA (Roche) in 120-μl reactions. Twenty-microliter aliquots were spotted on 3MM filter disks (Whatman) and washed in 10% trichloroacetic acid. The amount of radioactivity was determined by liquid scintillation counting.
Posttransfer Editing Assay. E. coli Ala-tRNAPro, Pro-tRNAPro, and Cys-tRNAPro were prepared as described (8). E. coli Ala-tRNAAla was prepared by using E. coli AlaRS. Ser-tRNAAla and Gly-tRNAAla were generated by charging unfractionated M. jannaschii or S. cerevisiae unfractionated tRNA with the M. jannaschii AlaRS mutant C699A, analogous to the procedure described for the editing-deficient E. coli AlaRS variant C666A (5). Additionally, M. jannaschii tRNA was charged with S. solfataricus ThrRS to produce Ser-tRNAThr (unpublished results), with seryl-tRNA synthetase (SerRS) from M. barkeri (D.K., unpublished results) to produce Ser-tRNASer, or with M. jannaschii AlaRS to produce Ala-tRNAAla. After aminoacylation, tRNA was extracted by phenol and chloroform, ethanol-precipitated and dried. It was subsequently used in deacylation assays as described (8).
Phylogenetic Inference. ProRS sequences were aligned by using clustalx and analyzed by the neighbor-joining method from the same package (15). Phylogenetic trees were viewed and edited with the treeview program (16).
Results
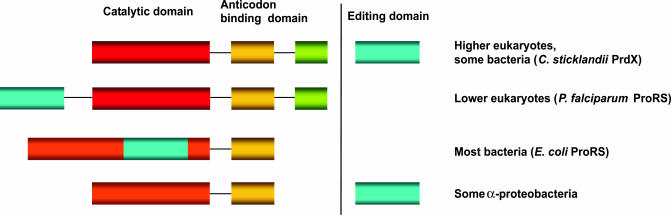
Identification of Editing Domain Paralogs. We used the E. coli ProRS insertion domain sequence (from amino acids 224 to 399) to search available genomic databases for homologues of the previously described ProRS editing domains, while also tracking hypothetical autonomous editing modules. Our search revealed domains with some degree of similarity (the typical P values ranged from 0.0001 to 1) and of corresponding length to the editing domain of ProRS (Fig. 1), found in three different genomic contexts (Fig. 2). In addition to the described editing domain inserted in the catalytic core of most bacterial enzymes, a fairly homologous domain was identified in lower eukaryotes appended to the N terminus of ProRS. Furthermore, autonomous proteins resembling the insertion (editing) domain of ProRS were identified in many bacteria, eukarya, and archaea. They were designated ProX. Additionally, some proteobacterial genomes (E. coli and many α-proteobacteria) encode two putative ProX paralogs, implying that the ProX protein family may have diverged into several subtypes. The phylogenetic tree in Fig. 3 indicates separation of insertion domain paralogs into three major groups: the first includes ProRS insertion domains, the second group comprises a subfamily of ProX (represented by the ProX protein from C. sticklandii annotated in genome databases as PrdX) and N-terminal extensions in ProRSs of lower eukaryotes, whereas the third group is constituted solely by autonomous ProX proteins, including the recently described H. influenzae Ybak protein (17).
Fig. 1.
Multiple alignment of the ProRS editing domain homologues. Asterisks represent residues that were shown important for editing function of the E. coli ProRS enzyme. “>OXH<” defines the position of the oxyanion hole. The last two sequences represent Ybak proteins [the crystal structure has been solved for the H. influenzae protein (17)]. M.mus, Mus musculus (NM_026465); M.mag, Magnetospirillum magnetotacticum (ZP_00053959); R.rub, Rhodospirillum rubrum (ZP_00016210); C.sti, C. sticklandii (CAB71308); P.fal, P. falciparum (AE014846); T.bru, Trypanosoma brucei (www.sanger.ac.uk/cgi-bin/blast/getseq?id=115XH77A6700k03xeEJ;db=tryppub/contigs;acc=tryp_x-97f05.q1k); E.col, E. coli (ProRS AAB08622, Ybak AAC73583); B.sub, Bacillus subtilis (NP_389539); C.ace, Clostridium acetobutylicum (NP_349775); A.aeo, Aquifex aeolicus (NP_213250); H.inf, H. influenzae (P45202); Ins, insertion domain of bacterial-type ProRS; Nterm, N-terminal extension in ProRSs from lower eukaryotes. GenBank accession numbers are in parentheses.
Fig. 2.
Different architectures of the Prolyl-tRNA synthetases and their editing domains. In blue are shown editing domains (note that they can be present in cis or trans; as an N-terminal domain or an insertion). In red are shown catalytic domains (orange is used for bacterial-type ProRS). Anticodon-binding domains are yellow, and archaeal-type ProRS-specific C termini are green. Representatives of the analyzed editing domain homologues are shown in parentheses.
Fig. 3.
Unrooted phylogenetic tree implied by the neighbor-joining method showing possible relationships between ProRS insertion domain homologues. If not specifically labeled in genomic databases, the free-standing ProRS editing domain homologues are denoted ProX. GenBank accession numbers are in parentheses: C. sticklandii PrdX (CAB71308), Pseudomonas aeruginosa ProX (AAG05230), M. magnetotacticum ProX (ZP_00053959), Agrobacterium tumefaciens ProX1 (NP_356919), M. musculus ProX (NM_026465), Arabidopsis thaliana ProX (AAO63892), P. falciparum ProRS (AE014846), Chlamydia trachomatis ProRS (NP_219903), E. coli ProRS (AAB08622), Thermotoga maritima ProRS (NP_228324), C. acetobutylicum ProRS (NP_349775), E. coli YeaK (AAC74857), Chlorobium tepidum ProX (NP_662760), Aeropyrum pernix ProX (NP_148681), A. tumefaciens ProX2 (AAK85924), Enterococcus faecalis EbsC (AAC36853), Streptomyces coelicolor ProX (NP_626318), E. coli YbaK (AAC73583), and H. influenzae YbaK (P45202).
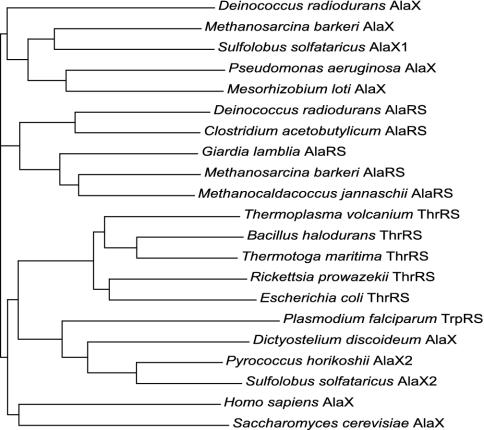
Similar patterns of duplication are evident for both AlaRS and ThrRS. The editing domains of these two synthetases share a notable sequence similarity and are evolutionarily related (5). Our query for the autonomous homologues of AlaRS and ThrRS editing domains using the E. coli AlaRS editing domain sequence (amino acids 547–675) yielded proteins of considerable similarity in all of the living kingdoms. Some of these proteins were previously identified as AlaX proteins, but their incidence was not linked to a specific function (18). As seen for ProRSs, a systematic analysis revealed several groups of freestanding proteins, all of which are partially related to the editing modules found in the two aaRSs of interest (Fig. 4). The phylogenetic tree in Fig. 5 includes various representatives of AlaX proteins, as well as the editing domains of different AlaRSs and ThrRSs. AlaX proteins were scattered throughout our tree, which may be suggestive of different substrate specificities for the identified paralogs.
Fig. 4.
Multiple alignments of the editing modules of AlaRS and ThrRS. Asterisks represent residues that were shown to be important for the editing function of the E. coli ThrRS or the E. coli AlaRS enzymes. M.jan, M. jannaschii (Q57984); P.aby, Pyrococcus abyssi (Q9UY36); E.col, E. coli (Q8XE27); B.sub, B. subtilis (P18256); M.bar, M. barkeri (ZP_00076591); A.per, A. pernix (NP_147589); S.sol, S. solfataricus (AlaX1, NP_341986; AlaX2, NP_342718); D.rad, Deinococcus radiodurans (NP_294225). GenBank accession numbers are in parentheses. S. solfataricus possesses two different AlaX.
Fig. 5.
Unrooted phylogenetic tree implied by the neighbor-joining method, showing relationships between the editing domains of AlaRS and ThrRS and their free-standing homologues found elsewhere. Note that the homologous module is found as an insert (AlaRS), an N-terminal extension (ThrRS), and a freestanding protein (AlaX). A homologous domain was also found at the N terminus of Plasmodium TrpRS. GenBank accession numbers are in parentheses: D. radiodurans AlaX (NP_294225), M. barkeri AlaX (ZP_00076591), S. solfataricus AlaX1 (NP_341986), P. aeruginosa AlaX (NP_250796), Mesorhizobium loti AlaX (NP_102167), D. radiodurans AlaRS (Q9RS27), C. acetobutylicum AlaRS (Q97IG3), Giardia lamblia AlaRS (AAG23137), M. barkeri AlaRS (ZP_00077929), M. jannaschii AlaRS (Q57984), Thermoplasma volcanium ThrRS (Q978W0), Bacillus halodurans ThrRS (Q9K866), T. maritima ThrRS (Q9WZJ9), Rickettsia prowazekii ThrRS (O05947), E. coli ThrRS (Q8XE27), P. falciparum TrpRS (NP_705269), Dictyostelium discoideum AlaX (AAO52108), Pyrococcus horikoshii AlaX2 (NP_142539), S. solfataricus AlaX2 (NP_342718), Homo sapiens AlaX (NP_079543), S. cerevisiae AlaX (NP_014358).
Ala-tRNAPro Is Hydrolyzed by Cis- and Trans-Editing Factors. The available crystal structure of the H. influenzae Ybak protein (17) provided a structural framework from which to infer a potential function for ProRS editing domain paralogs. Our analysis was accordingly focused on the presence of the oxyanion hole, a standard element used by proteins catalyzing ester bond hydrolysis [including serine proteases and d-Tyr-tRNA hydrolases (19, 20)]. Such a structural motif comprised by GXXXP is clearly distinguishable in Ybak. Given the conservation of this element among the analyzed sequences of ProRS editing domain paralogs (Fig. 1), we surmised that ProX proteins possess a hydrolytic function. Conservation of several amino acids known to be crucial for the function of ProRS (7) additionally supported this hypothesis. tRNA-dependent deacylation was accordingly tested with various members of the ProRS editing domain paralog family identified in different genomic contexts (Fig. 2).
ProRSs from lower eukaryotes with an N-terminal putative editing domain were represented by P. falciparum ProRS (PF-ProRS). As shown in Fig. 6A, the PFProRS efficiently hydrolyzed Ala-tRNAPro whereas a deletion mutant lacking the N-terminal domain (ΔPFProRS) was unable to do so. On the other hand, the efficiency of ΔPFProRS in aminoacylation was only three times decreased compared with the wild-type enzyme (data not shown). Furthermore, ΔPFProRS showed misacylation of tRNAPro with alanine, unlike the wild-type PFProRS (Fig. 6B). Interestingly, the ProRS from S. cerevisiae was unable to catalyze editing (Fig. 6A) and accordingly showed very low, but detectable, alanylation of tRNAPro (Fig. 6B), in correlation with the absence of the oxyanion hole and significant truncations in its N-terminal extension (data not shown).
Fig. 6.
(A) Specific deacylation of E. coli Ala-tRNAPro. Shown are 0.3 μM CSPrdX (•), 0.3 μM CSPrdX plus 1 μM CSProRS (▴), 0.3 μM ECYbaK (▪), 0.5 μM PFProRS (○), 0.5 μM ΔPFProRS (□), 0.5 μM S. cerevisiae ProRS (▵), 0.1 μM E. coli ProRS (▾), and control without enzyme (▿). (Inset) Pro-tRNAPro and Ala-tRNAAla deacylation [0.3 μM CSPrdX (• and ○, respectively), 0.5 μM PFProRS (▪ and □, respectively)]. (B) Mischarging of alanine onto S. cerevisiae tRNAPro. Shown are 6 μM PFProRS (•), 6 μM ΔPFProRS (▪), and 6 μM S. cerevisiae ProRS (▵).
The autonomous editing domain paralogs of ProRS from C. sticklandii (CSPrdX) and E. coli (ECYbak), drawn from each of the two main groupings in the phylogenetic tree (Fig. 3), were investigated. We considered CSPrdX a particularly promising candidate, due to the operon-like arrangement of the proS and prdX genes in the C. sticklandii genome. As shown in Fig. 6A, Ala-tRNAPro is efficiently hydrolyzed by CSPrdX but not by ECYbak. Because the addition of C. sticklandii ProRS did not enhance the deacylation activity of CSPrdX (Fig. 6A), CSPrdX represents an example of an efficient editing protein autonomous of the corresponding synthetase. These results are in agreement with the molecular phylogeny, as CSPrdX groups together with the N-terminal extension of PFProRS, and possesses a conserved histidine residue shown to be important for the amino acid specificity of editing in E. coli ProRS (Fig. 1 and ref. 7). Furthermore, occurrence of the prdX gene in particular organisms correlates with the absence of the editing domain in the respective ProRS (Fig. 2). In contrast, ECYbak lacks the conserved histidine residue, and, because E. coli ProRS possesses an intact insertion domain, ECYbak may have altered its substrate specificity in the genomic context of a more efficient editing enzyme.
In addition to confirming the inferred editing function, we analyzed the substrate specificity of PFProRS and CSPrdX enzymes. Their editing is specific toward both amino acid and tRNA, because neither PFProRS nor CSPrdX showed notable hydrolysis of Pro-tRNAPro and Ala-tRNAAla substrates (Fig. 6A Inset). In addition, Cys-tRNAPro was also not a substrate for these enzymes (data not shown).
AlaX Hydrolyzes Misacylated tRNAAla. Our efforts to define a functional link between AlaX and editing domains in AlaRSs and ThrRSs was facilitated by identification of the HXXXH and CXXXH motifs in a vast majority of AlaX proteins (Fig. 4). Because these residues have been shown to be important for editing capacity in both the AlaRS and ThrRS systems (5, 6), we inferred a hydrolytic function for AlaX homologues. We were primarily interested in AlaX representatives from archaea because most of the corresponding genomes do not encode ThrRSs with recognizable editing domains (9), and it was plausible to assume that the “missing” editing domain might be complemented by a trans-acting factor.
tRNA-dependent deacylation was tested with AlaX proteins from M. barkeri and S. solfataricus. As shown in Fig. 7, these proteins were unable to deacylate Ser-tRNAThr but were efficient in Ser-tRNAAla and Gly-tRNAAla hydrolysis. Furthermore, hydrolysis of Ala-tRNAAla or Ser-tRNASer could not be detected (data not shown), which demonstrated that the editing reaction catalyzed by AlaX is specific toward misacylated tRNAAla substrates. We also tested the S. cerevisiae AlaX homologue with the same set of substrates used for archaeal AlaXs but could not observe efficient hydrolysis (Fig. 7 and data not shown).
Fig. 7.
Specific deacylations by AlaXs. Shown are 100 nM M. barkeri AlaX and Ser-tRNAAla (•), 100 nM S. solfataricus AlaX and Ser-tRNAAla (▪), 100 nM M. jannaschii AlaRS and Ser-tRNAAla (▴), 100 nM S. cerevisiae AlaX and Ser-tRNAAla (□), 100 nM M. barkeri AlaX and Gly-tRNAAla (○), 100 nM M. barkeri AlaX and Ser-tRNAThr (▾), and control without enzyme (▵).
Discussion
Origin of Editing Domains in aaRSs. Although a matter of speculation, it is generally believed that ancient aaRSs existed as single-domain proteins resembling the catalytic core domains of modern aaRSs (21, 22). However, even the simplest architectures of contemporary synthetases contain additional domains involved in functions such as editing or tRNA binding (23). Because they are not universally distributed among aaRSs, editing domains represent promising systems for evaluation of the hypothesis that the additional domains in contemporary aaRSs originated from autonomous freestanding proteins. The diversity of editing modules found inserted or appended to the body of aaRSs supports this notion, but genomic and experimental evidence for extant trans-acting editing proteins has proved elusive (24). The only exception is a recent report describing Ala-tRNAPro hydrolysis by H. influenzae Ybak. Our demonstration of efficient and specific freestanding editing domains strongly suggests that editing domains of aaRSs may have previously existed as autonomous proteins that were incorporated into the respective aaRS genes in a later evolutionary event. This scenario is corroborated by the observation that the editing domains of isoleucyl-tRNA synthetase, valyl-tRNA synthetase, and LeuRS (25) are flanked by CXXC motifs, which characterize many mobile domains found inserted in larger proteins (26). Interestingly, examination of the bacterial ProRSs reveals that many of them also contain conserved CXXC motifs flanking the insert (Fig. 1), providing additional evidence for its external origin.
Our findings also imply that domain acquisitions and rearrangements must have occurred frequently during the evolution of some aaRSs. In the ProRS family, the contexts in which editing modules are found are rather diverse and include insertions, N-terminal additions, and autonomous protein forms. This finding implies that the acquisition of editing modules by ProRSs occurred independently at least twice. The situation for AlaRS seems analogous because the editing domains of AlaRS and ThrRS share notable sequence similarity, and apparently evolutionary origin (5). However, in AlaRS, this domain is found inserted in the enzyme body whereas the homologous domain found in ThrRS is positioned at its N terminus. Considering that the editing domain is not found in most archaeal ThrRSs, which are deep rooted in the AlaRS tree (5), it is plausible that acquisition of the editing module in AlaRS preceded the analogous event in ThrRS. These examples illustrate that the same type of module could have entered different aaRSs at different positions in their structures and at different points in evolution, hence substantiating the proposed origin of editing domains.
Potential Advantages of Incorporating Editing Domains into aaRSs. The ability of autonomous editing domains to efficiently function in trans raises the question of why they have been acquired by aaRSs in some systems but not others. Presumably, the free-standing modules exhibit lesser tRNA specificity compared with the complete aaRSs, which can exploit more intricate tRNA contacts. Indeed, when using higher concentrations (up to 6 μM) of the freestanding proteins ProX and AlaX in deacylation assays, we observed some hydrolysis of Ala-tRNAAla and Ser-tRNASer, respectively (data not shown). In contrast, this effect was not observed with comparable concentrations of E. coli ProRS, P. falciparum ProRS, or M. jannaschii AlaRS. Furthermore, the efficiency and specificity of the freestanding modules were extremely sensitive to changes in the tRNA acceptor stem sequence (I.A., unpublished observation). We thus speculate that association of the editing module and the core of the aaRS may be advantageous in terms of specificity and efficiency of the editing reaction; additionally, such an arrangement provides a facile system for transcriptional coregulation, another potential advantage for the maintenance of the editing module within the multidomain synthetase structure.
The advantages of incorporating editing domains into certain aaRSs notwithstanding, the reverse process, deletion or mutational inactivation of an editing domain, might also be advantageous for some organisms. In support of this idea, some ProRS sequences indicate secondary loss of editing function, as observed in yeast, the aphidian intracellular symbionts Buchnera sp., and quite possibly mitochondria (data not shown). In yeast, editing domain sequences show significant truncations and a loss of the oxyanion hole and were accordingly shown to be hydrolytically impotent (Fig. 6A). The same scenario could be proposed for mitochondrial ProRSs, which have recognizable CXXC motifs and a very short stretch with some homology to the insertion domain of bacterial-type ProRSs but lack the majority of the insert. If true, this theory implies that the insertion domain existed in ProRSs even before the endosymbiotic event (27) and that incorporation of the editing modules by bacterial-type ProRSs is of an ancient origin. In any case, it seems that editing of Ala-tRNAPro is superfluous in certain organisms, which could be a consequence of improved amino acid selection at the level of activation. Accordingly, yeast ProRSs show almost no detectable alanine misacylation (Fig. 6B), and their discrimination against alanine at the level of aminoacylation was estimated to be adequate to negate the need to hydrolyze Ala-tRNAPro.
Flexibility of Editing Modules to Adopt New Functions. Our results show that archaeal AlaX proteins hydrolyze misacylated tRNAAla, but not tRNAThr substrates. Because the canonical AlaRS enzymes from M. barkeri and S. solfataricus are expected to perform editing in vivo themselves, the rationale underlying the apparent duplication of the editing function is unclear. Although there are no data on the expression of AlaX genes in archaea, it is plausible to speculate that these genes are differentially expressed or that they are not expressed at all. On the other hand, available proteomic data for S. cerevisiae AlaX suggest that, although this protein is nonessential, it is expressed at different stages of growth (www.yeastgenome.org). Nonetheless, the S. cerevisiae AlaX and E. coli Ybak proteins were unable to hydrolyze any of the tested misacylated substrates, which implies that these proteins may have developed rather different substrate specificities and functions, perhaps different to the tRNA-dependent deacylations studied here. The paralogous editing domains of AlaRS and ThrRS clearly demonstrate that such an alteration of specificity is both evolutionarily feasible and beneficial. Moreover, P. falciparum and Plasmodium yoelii tryptophanyl-tRNA synthetases contain AlaX homologs at their N termini (Fig. 5), possibly providing them with the ability to edit mischarged tRNATrp. In this respect, the families of ProX and AlaX proteins described here may represent a diverse pool of hydrolytically potent proteins with various specificities resulting from duplications and evolutionary divergence; if so, similar domains could be responsible for various types of tRNA-dependent deacylations, providing extant cells with an enhanced capacity to deal with incorrect products of aminoacylation.
Acknowledgments
We thank A. Pich, T. McCutchan, K. Sowers, K. O. Stetter, M. Thomm, and A. Ambrogelly for the gifts of DNAs, cells, and enzymes. This work was supported by grants from the National Institute of General Medical Sciences (to D.S. and M.I.) and the Department of Energy (to D.S.).
Abbreviations: AlaRS, alanyl-tRNA synthetase; ThrRS, threonyl-tRNA synthetase; ProRS, prolyl-tRNA synthetase; PFProRS, Plasmodium falciparum ProRS; aaRS, aminoacyl-tRNA synthetase.
References
- 1.Fersht, A. R. (1981) Proc. R. Soc. London Ser. B. 212, 351–379. [DOI] [PubMed] [Google Scholar]
- 2.Jakubowski, H. & Goldman, E. (1992) Microbiol. Rev. 56, 412–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giegé, R., Sissler, M. & Florentz, C. (1998) Nucleic Acids Res. 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakubowski, H. & Fersht, A. R. (1981) Nucleic Acids Res. 9, 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beebe, K., Ribas De Pouplana, L. & Schimmel, P. (2003) EMBO J. 22, 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dock-Bregeon, A., Sankaranarayanan, R., Romby, P., Caillet, J., Springer, M., Rees, B., Francklyn, C. S., Ehresmann, C. & Moras, D. (2000) Cell 103, 877–884. [DOI] [PubMed] [Google Scholar]
- 7.Wong, F. C., Beuning, P. J., Nagan, M., Shiba, K. & Musier-Forsyth, K. (2002) Biochemistry 41, 7108–7115. [DOI] [PubMed] [Google Scholar]
- 8.Ahel, I., Stathopoulos, C., Ambrogelly, A., Sauerwald, A., Toogood, H., Hartsch, T. & Söll, D. (2002) J. Biol. Chem. 277, 34743–34748. [DOI] [PubMed] [Google Scholar]
- 9.Sankaranarayanan, R., Dock-Bregeon, A. C., Romby, P., Caillet, J., Springer, M., Rees, B., Ehresmann, C., Ehresmann, B. & Moras, D. (1999) Cell 97, 371–381. [DOI] [PubMed] [Google Scholar]
- 10.Lin, L., Hale, S. P. & Schimmel, P. (1996) Nature 384, 33–34. [DOI] [PubMed] [Google Scholar]
- 11.Wong, F. C., Beuning, P. J., Silvers, C. & Musier-Forsyth, K. (October 6, 2003) J. Biol. Chem., 10.1074/jbc.M309627200.
- 12.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 13.Waters, E., Hohn, M. J., Ahel, I., Graham, D. E., Adams, M. D., Barnstead, M., Beeson, K. Y., Bibbs, L., Bolanos, R., Keller, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12984–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curnow, A. W., Tumbula, D. L., Pelaschier, J. T., Min, B. & Söll, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12838–12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Page, R. D. (1996) Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, H., Huang, K., Li, Z., Banerjei, L., Fisher, K. E., Grishin, N. V., Eisenstein, E. & Herzberg, O. (2000) Proteins 40, 86–97. [DOI] [PubMed] [Google Scholar]
- 18.Schimmel, P. & Ribas De Pouplana, L. (2000) Trends Biochem. Sci. 25, 207–209. [DOI] [PubMed] [Google Scholar]
- 19.Wilmouth, R. C., Edman, K., Neutze, R., Wright, P. A., Clifton, I. J., Schneider, T. R., Schofield, C. J. & Hajdu, J. (2001) Nat. Struct. Biol. 8, 689–694. [DOI] [PubMed] [Google Scholar]
- 20.Ferri-Fioni, M. L., Schmitt, E., Soutourina, J., Plateau, P., Mechulam, Y. & Blanquet, S. (2001) J. Biol. Chem. 276, 47285–47290. [DOI] [PubMed] [Google Scholar]
- 21.Rould, M. A., Perona, J. J., Söll, D. & Steitz, T. A. (1989) Science 246, 1135–1142. [DOI] [PubMed] [Google Scholar]
- 22.Ribas de Pouplana, L. & Schimmel, P. (2000) Cell. Mol. Life Sci. 57, 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander, R. W. & Schimmel, P. (2001) Prog. Nucleic Acid Res. Mol. Biol. 69, 317–349. [DOI] [PubMed] [Google Scholar]
- 24.Bishop, A. C., Beebe, K. & Schimmel, P. R. (2003) Proc. Natl. Acad. Sci. USA 100, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cusack, S., Yaremchuk, A. & Tukalo, M. (2000) EMBO J. 19, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grishin, N. V. (2001) Nucleic Acids Res. 29, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray, M. W., Burger, G. & Lang, B. F. (1999) Science 283, 1476–1481. [DOI] [PubMed] [Google Scholar]