Abstract
The analysis of completely sequenced genomes uncovers an astonishing variability between species in terms of gene content and order. During genome history, the genes are frequently rear-ranged, duplicated, lost, or transferred horizontally between genomes. These events appear to be stochastic, yet they are under selective constraints resulting from the functional interactions between genes. These genomic constraints form the basis for a variety of techniques that employ systematic genome comparisons to predict functional associations among genes. The most powerful techniques to date are based on conserved gene neighborhood, gene fusion events, and common phylogenetic distributions of gene families. Here we show that these techniques, if integrated quantitatively and applied to a sufficiently large number of genomes, have reached a resolution which allows the characterization of function at a higher level than that of the individual gene: global modularity becomes detectable in a functional protein network. In Escherichia coli, the predicted modules can be bench-marked by comparison to known metabolic pathways. We found as many as 74% of the known metabolic enzymes clustering together in modules, with an average pathway specificity of at least 84%. The modules extend beyond metabolism, and have led to hundreds of reliable functional predictions both at the protein and pathway level. The results indicate that modularity in protein networks is intrinsically encoded in present-day genomes.
The combined history of genomes provides a glimpse at past evolutionary events, revealing selective forces that acted at all levels of cellular and organismal function. Although the individual gene and its immediate regulatory elements form the primary unit of selection, evolution does not stop there (1). Instead, selection can also act on entire groups of genes, leading to joint transfers of genes between genomes (2, 3), concerted gene loss (4), gene fusion events (5), coregulation of genes through common regulatory elements (6), and the creation and maintenance of operons containing nonhomologous but cotranscribed genes (7, 8).
All of the above genomic events define evolutionarily selected (and thereby, functional) connections between genes, an invaluable resource in annotating gene function (9–11) and in understanding how gene products interact globally to support cellular systems. Through systematic comparison of extant genomes, many of the relevant genomic events can be inferred. This, in turn, allows the objective and unbiased prediction of thousands of functional associations among genes (or proteins) from genome sequences alone, albeit in some instances with considerable error rates (refs. 12–17, for reviews, see refs. 11 and 18–22).
Here, we apply a rigorously benchmarked and quantitatively integrated combination of the three main prediction techniques (12, 15, 16) to 89 completely sequenced genomes, and construct a global network of functionally interacting proteins. We use this network to quantitatively study functional modularity in protein networks. We find that functional modules are detectable by using unsupervised clustering, and without any use of prior knowledge about protein function; however, the predicted modules agree remarkably well with previously annotated metabolic pathways. Functional modularity is an important feature of the topology of many real-world complex systems (23), and has recently been suggested to exist in biological systems as well (24, 25). Here we show that functional modules correspond to well characterized cellular systems, based on analysis of an objective, unbiased, and highly specific interaction network. Importantly, we find that the signal is quite robust, being detectable by using a variety of prediction methods and parameters. Additionally, we show that the predicted modules help in annotating previously uncharacterized proteins and cellular systems.
Data Sources and Procedures
Input Data. Functional associations between orthologous groups of proteins were predicted using string (ref. 26, version 3.0). We considered only groups containing at least one protein from the target organism (Escherichia coli K12), and we excluded groups containing on average more than four distinct genes per species (these fail to resolve orthology with sufficient detail). We additionally processed a limited number of large groups manually, enhancing the orthology resolution by splitting the groups into two or more smaller groups. The splitting was done before the analysis presented in this study, and was guided solely by inherent sequence information. In total, we split 28 groups; these were selected based on the appearance of phylogenetic trees, the availability of operon information, and their relevance to metabolism.
Clustering. The predicted functional associations define a network with undirected, weighted edges connecting proteins. To identify functional modules in this network, we used three algorithmically distinct unsupervised clustering techniques: unweighted pair group method with arithmetic mean (UPGMA) clustering, single-linkage clustering (both as implemented in the oc package, www.compbio.dundee.ac.uk/manuals/oc/oc_manual.txt) as well as Markov clustering (27). We explored parameter space by testing several different cutoff values (for single-linkage and mean clustering) or inflation values (for Markov clustering).
Metabolic Pathways. For reference, we used pathways defined for small-molecule metabolism of E. coli, as annotated in the database EcoCyc (version 6.5) (28, 29). The pathways group 583 proteins (enzymes or enzyme subunits) into 144 partially over-lapping metabolic units.
Benchmarking. Before benchmarking, we removed from the predicted modules all proteins not present in the EcoCyc pathways. Modules remaining with zero or only one annotated enzyme (singletons) were not considered further.
For each of the remaining modules, the best-matching metabolic pathway was selected for comparison and the following measures were computed: specificity, defined as Tp/(Tp+Fp); sensitivity, defined as Tp/(Tp+Fn); and overlap function, defined as Tp/(Tp+Fp+Fn), where Tp denotes true positives, Fp denotes false positives, and Fn denotes false negatives. Another measure was “total coverage,” defined as the fraction of enzymes found clustered in predicted modules together with at least one other enzyme. For all measures, counting was done on the level of proteins; i.e., orthologous groups containing several E. coli proteins generated multiple counts. Enzyme subunits were counted as separate entities. The choice as to which pathway a predicted module should be compared to (“best matching pathway”), was by selecting the pathway with maximal overlap function.
Random Background. We compared predicted modules with random expectations at two different levels of randomization. One very conservative random model was to (i) keep the module size distribution as predicted, (ii) keep the number of enzymes within each module fixed, and (iii) only swap enzyme identities across those fixed modules. On average, this led to a 2.2-fold reduction in observed specificity and to a 2.4-fold reduction in observed overlap.
A more realistic comparison to random expectation was to ask how many modules can be expected, by chance, to consist entirely of enzymes only. Given the known numbers of enzymes and nonenzymes in E. coli, this expectation was computed by using the hypergeometric distribution (sampling without replacement). When comparing the actual predictions against the expectation, we observed a strong deviation from randomness (especially for the larger modules); the predicted modules differed by more than one order of magnitude from the random expectation for all module sizes larger than two (Table 2, which is published as supporting information on the PNAS web site).
Functional Categories. To functionally classify proteins, we used gene ontology (GO) categories (30), as assigned to proteins in E. coli (31). We considered only the subcategory “biological processes,” and further reduced the number of terms by grouping related terms as follows: first, we checked the distribution of all E. coli proteins over the whole GO hierarchy, by traversing from assigned leaf terms through all possible paths up to the root term. Throughout this procedure, we marked all nodes visited at least 100 times as terms of sufficiently high generality. For any protein of interest, we then traversed from its assigned terms up the hierarchy, and stopped at the first encountered “marked” node, thereby objectively grouping functional assignments at a medium level of detail. For Fig. 4, GO annotations proved impractical, and we instead chose the high-level categories defined for orthologous groups in the COG database (32).
Fig. 4.
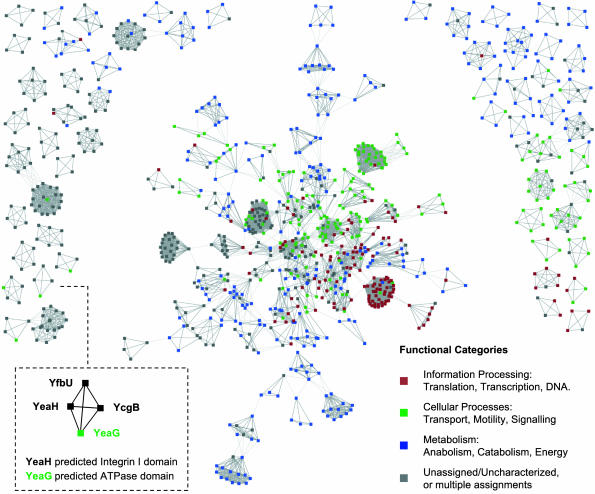
A network of predicted functional modules in E. coli. Only modules of size four or larger are shown. Nodes represent single proteins or groups of highly similar proteins as defined in the COG database. Genomic context links within predicted modules are shown in dark gray, and those across modules are shown in light gray. For clarity, the latter links are limited to those with an association score of 0.650 or higher (on a scale from zero to one; ref. 26). Functional categories are as defined in the COG database. (Inset) A typical example of a largely uncharacterized pathway.
Results
Delineation of Functional Modules. We integrated all three major comparative genomics (genomic context) techniques currently capable of inferring functional associations between proteins, based on common phylogenetic distribution (16), conserved gene neighborhood (14, 15), and gene fusions (12, 13). The methods were quantitatively combined by using a benchmarked, unified scoring scheme (26). Functional associations detected this way may correspond to physically interacting proteins such as those involved in protein complexes, biochemically related proteins such as those involved in the same metabolic pathways, or genetically interacting proteins such as transcriptional regulators and their target genes (for review, see refs. 11 and 18–22). All these binary associations can be seen as edges (with weights provided by the score) that connect groups of orthologous proteins (32) (nodes) in a network of functional associations.
We derived a complete network of associated genes for 89 species, connecting 19,473 orthologous groups (26, 32) corresponding to 260,023 proteins participating in a total of 1,908,210 binary links. When projecting this network to the Escherichia coli K12 genome with 4,290 annotated genes (33), 3,256 of these are connected through 113,864 links.
In this E. coli K12 network, we were able to detect inherent modularity in an objective and reproducible manner, by applying three algorithmically different clustering approaches (34), namely unweighted pair group method with arithmetic mean (UPGMA) clustering, single linkage, and Markov clustering (27).
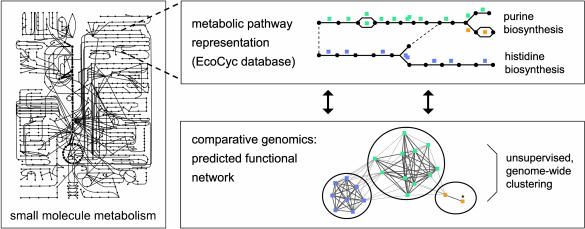
Validation of Predicted Functional Modules. To show that the resulting tight clusters indeed correspond to functional modules, we benchmarked the automatic analysis against manually curated annotations. The probably best-understood functional subnetwork in E. coli is that of small molecule metabolism. Hence, we compared the obtained clusters to the EcoCyc knowledge base of metabolic pathways (28), which encompasses the entire known metabolic complement of E. coli (Fig. 1).
Fig. 1.
Correlation between metabolic pathways and genomic context predictions. Metabolic databases such as EcoCyc describe metabolites and enzymes, and subjectively group them into metabolic “pathways.” In contrast, comparative genomics can reveal selective pressures shared by groups of enzymes, thereby defining functional modularity objectively. Surprisingly, a good agreement between both is observed. Note that the purine biosynthesis pathway is covered by two predicted modules, which are separated by a branching point in the pathway. The node marked by an asterisk consists of two enzymes (GuaC and ImdH), which are too closely related to be resolved into separate orthologous groups (32). Both enzymes are involved in purine metabolism, but only ImdH is part of the biosynthesis pathway, so GuaC is counted here as a false positive. [The schematic overview of metabolism is reproduced with permission from ref. 42 (Copyright 1994, Garland Publishing, New York).]
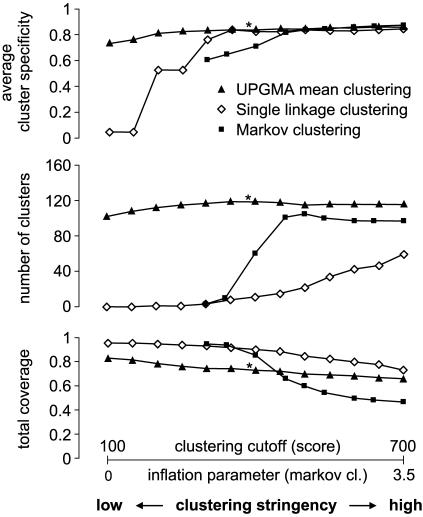
We first explored the parameter space of the clustering methods and their performance with respect to how well the resulting clusters matched pathway definitions (Fig. 2; see Data Sources and Procedures for the benchmarking procedure). As expected, we observed a tradeoff between the accuracy in reconstructing known pathways and the total coverage (the latter being the fraction of pathway proteins which are found clustered in groups of at least two). However, we observed some tolerance to the parameter choice, suggesting an implicit signal toward functional modularity (Fig. 2). When demanding a high specificity, all methods achieved a remarkably high total coverage, >70% (Fig. 2 and Table 3, which is published as supporting information on the PNAS web site).
Fig. 2.
Clustering genomic context associations: parameter exploration and benchmarking. Shown are graphs summarizing the benchmarking performance of the clustering, considering only clusters containing at least two enzymes. The achievable pathway specificity quickly reaches a plateau at a high level of prediction accuracy, independent of the clustering algorithm used. In contrast, the observed number of the predicted functional modules and the fraction of total metabolism they cover are both somewhat more sensitive to clustering algorithms and cutoffs. The data set marked with an asterisk was chosen for detailed manual analysis and is the basis of all subsequent figures.
Mean clustering demonstrated the best overall performance (Fig. 2), grouping as many as 74% of the possible 583 proteins into 119 clusters; these matched 89 EcoCyc pathway definitions with 84% average specificity and 49% average sensitivity. In other words, more than half of the metabolic network is recovered with very high specificity, solely by objective, comparative genome analysis. These measurements are several times higher than expected when compared to random models (see Data Sources and Procedures for details).
Deviations from Current Knowledge: Limitations and Biological Discovery. Following parameter exploration, one representative set of predicted modules (unweighted pair group method with arithmetic mean clustering; score cutoff, 0.400; see Supporting Text, which is published as supporting information on the PNAS web site) was chosen for further analysis. Of particular interest were any deviations from the current knowledge of metabolic pathways, because such discrepancies might represent areas of discovery and indicate key biological properties of the metabolic web. For example, the relatively low average sensitivity of predicted modules (49%) is partly due to coverage of a single EcoCyc pathway by several distinct modules (e.g., Fig. 1). We often observed that such “submodules” are separated by branching points in the metabolic web, i.e., by metabolites participating in several reactions and leading to distinct products. The branching points separate subpathways that may be used in different contexts by serving different physiological roles, and may be subject to distinct regulation, thereby justifying the representation as individual modules (e.g., Fig. 1). In support of this notion, we globally observe that the quantitative strength of genomic context associations correlates well with the structure of the metabolic web: the average association scores are markedly higher for nonbranching, linear sections of metabolism (Fig. 3B).
Fig. 3.
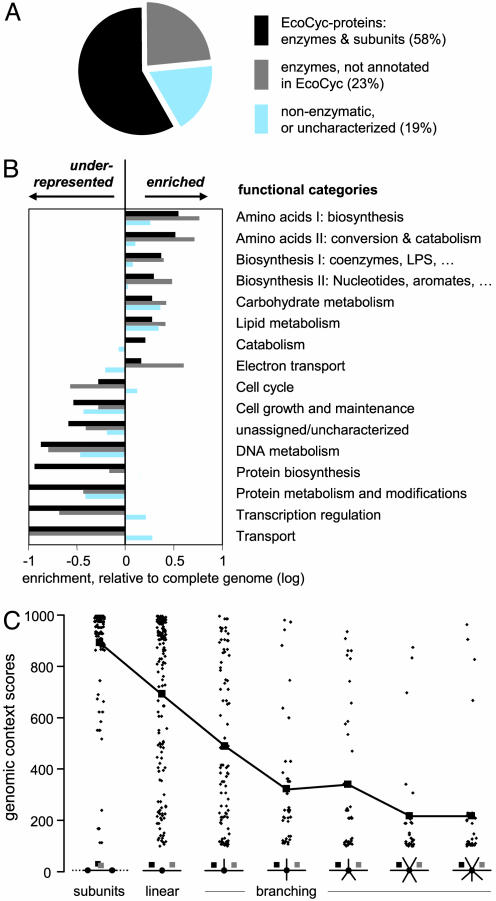
Global properties of the predicted metabolic modules. (A) Functional composition. In addition to the annotated enzymes, the predicted modules often contain putative enzymes that are not yet assigned to pathways, as well as proteins from other functional categories. (B) As expected, metabolic modules are strongly enriched in enzymatic functions, but they also contain other functions, most notably transport and transcription regulation. The categories shown are a subset of the gene-ontology (30) subtree “biological process” (see Data Sources and Procedures for details). (C) Pathway topology and genomic context. The graph shows the scores of all genomic context associations between enzymes that are direct neighbors in metabolism of E. coli. The pathway topology is defined by the number of enzymes metabolizing the same substrate (not considering frequent substrates such as water or ATP, frequency cutoff is 8). Any substrate metabolized by more than two enzymes constitutes a branching point.
Despite the high overall specificity of 84%, there remains a sizeable fraction of apparent “false positive” assignments (40 of the predicted modules cannot fully be matched by a single metabolic pathway). About 40% of these cases result from the limited resolution of orthology assignments (32), i.e., two or more very similar E. coli proteins belong to one orthologous group, but have been assigned to different pathways in EcoCyc. For such proteins, the comparative genomics methods used here currently cannot resolve which protein participates in which predicted association. This is clearly a current technical rather than a fundamental limitation. Remarkably, about half of the measured false positives may in fact represent genuine functional connections: 33% of the assignments correspond to true metabolic connections, because they are linking pathways known to be connected through a common metabolite, and another 17% correspond to pathways previously connected through various types of experimental or genetic evidence, as reported in recent literature. The remaining 10% of false positive assignments predict pathway links that were discovered in this study (Table 1 and Supporting Text).
Table 1. Examples of predictions derived from the functional modules.
| Type of prediction | Confirmed through literature | Novel prediction |
|---|---|---|
| Extensions to existing pathways (enzymatic) | IspG and IspH; two additional enzymes in the nonmevalonate isoprenoid biosynthesis pathway. Known, but not yet in EcoCyc | YqiA; an α—β hydrolase predicted to be associated to ubiquinone metabolism |
| Extensions to existing pathways (nonenzymatic) | NarU and NarK; two nitrate/nitrite transporters linked with the nitrate reductase complex | YbaD; a transcription factor possibly regulating riboflavin biosynthesis enzymes |
| Functional links between pathways | A predicted link between the metabolism of selenocysteine, and formate dehydrogenase; the latter is known to contain selenocysteine | A predicted link between coenzyme A biosynthesis and the metabolism of nucleotides. Supported by multiple observations |
| A functional link between nonmevalonate isoprenoid biosynthesis and the subsequent biosynthesis of polyisoprenoids | A predicted link between phospholipid biosynthesis and thiamine biosynthesis: conserved neighborhood between pgpA and thiL | |
| Entirely novel functional systems/pathways | A large, conserved module consisting of enzymes needed to use ethanolamine as a carbon/nitrogen source. Not yet annotated in EcoCyc | An uncharacterized, conserved functional module, containing the domains integrin I and AAA—ATPase. This combination of domains is known to occur in metal chelatases. |
Taken together, the differences to the manually annotated EcoCyc database generally cannot be seen as misassignments of an automatic prediction method, but reflect a more fine-grained functional clustering and the retrieval of known associations that have not yet been annotated in the knowledge base. When including the latter associations, the actual specificity of the automatic approach is >90%. Thus, the predicted functional modules should be a rich source for reliable biological discovery at the protein and pathway-annotation level (see Table 1 for examples).
Prediction of Extensions to Known Pathways. Many of the predicted metabolic modules contain proteins that are not annotated in EcoCyc (Fig. 3A). These additional proteins (>300 in total) are predicted to represent functional pathway extensions, and their placement is expected to be as accurate as that of the annotated proteins as the clustering procedure is based on objective genome data. In the average metabolic module, annotated pathway enzymes in EcoCyc constitute the largest fraction (58%), whereas proteins identified as putative enzymes in other databases (32, 35) contribute another 23% (Fig. 3A) and can thus be associated at the pathway level. Only 19% of the proteins in the modules appear to be hypothetical or noncatalytic.
The hypothetical proteins are excellent candidates for enzymes that can “fill” gaps in our current pathway knowledge (10), and we found several such examples in the predicted modules, some of which have been proven correct by recent literature (Table 1 and Supporting Text and Figs. 5–7, which are published as supporting information on the PNAS web site). Although the predicted metabolic modules are usually enriched in enzymes, they also contain links to many other cellular processes. Of those, certain functional categories such as transport and transcriptional regulation are overrepresented (Fig. 3B). This is in accordance with individual observations that such proteins are coregulated in metabolic operons. The modules recover several known links, such as the transcriptional regulator HycA, which is coupled to formate hydrogenlyase subunits (36), and also imply previously undescribed associations. For example, the hypothetical transporter system YliA/B/C/D can be tentatively linked to asparagine import because the genes encoding the transporter are in a predicted module together with the asparaginase gene asgX.
Prediction of Functional Links Between Pathways. At the pathway level, the modules also suggest previously undescribed connections. We identified 12 links between pathways that are not connected in EcoCyc, and for which the links cannot be explained easily by shared metabolites or orthology assignment artifacts. One such link is a previously unsuspected connection between CoA biosynthesis and nucleotide metabolism. Two distinct enzymes in CoA biosynthesis (PanC and CoaA/B) both have independent links to nucleotide metabolism, with evidence stemming from all three genomic context methods. Intriguingly, mutants in CoaA/B have been reported to have defects in DNA-synthesis (37). Conceivably, the two pathways could be functionally coupled because the nucleotide adenine is functioning as a structural component in CoA or because the CoA/B enzyme uses the nucleotide CTP as an unusual energy source. More speculatively, one of the enzymes in CoA biosynthesis could have a second, additional function in nucleotide metabolism (Fig. 8, which is published as supporting information on the PNAS web site).
Prediction of Pathways and Cellular Systems. Although the bench-marking can only demonstrate high accuracy within metabolism, numerous modules were also predicted for other functional categories. Of the total of 508 modules predicted for E. coli, 375 are dominated by one of four broad categories as defined in the COG database (32) (see also Fig. 4). Of these, 114 are metabolic (in good agreement with the 119 modules matching metabolism as defined in EcoCyc). About 60 modules are related to key processes such as cell motility, division, and signaling; and another 38 are related to translation, transcription, recombination, replication, and repair. Although hypothetical (uncharacterized) proteins are underrepresented in the metabolic modules (Fig. 3A), there are as many as 167 modules containing 677 proteins that are dominated by hypothetical proteins, indicating the existence of many undiscovered cellular processes or systems. In total, 1,636 proteins in E. coli were annotated as “hypothetical” at the time of analysis. Of those, 841 were associated to at least one partner through the modules predicted here (the remainder were either in modules of size one, or not sufficiently conserved in other organisms; 235 and 560 proteins, respectively). For 247 of those 841 hypothetical proteins, however, all predicted partners were hypothetical as well.
A typical example for a previously undescribed cellular system is a predicted module with four proteins, of which only two have vague annotations (Fig. 4). Three of these proteins form a very well supported and evolutionarily widespread unit of unknown function (Fig. 9, which is published as supporting information on the PNAS web site), whereas the fourth protein is more loosely connected and may be dispensable. Mutants in two of the proteins have been described as having defects in endospore formation in Bacillus subtilis (38, 39); however, the module occurs also in many non-spore-forming organisms, suggesting a broader functional role. Detailed homology analysis and structure prediction reveals remote similarity of two of the proteins to subunits of a Mg-chelatase (40), leading to the hypothesis that this module may represent a chelatase with unknown metal specificity. Candidate metals include calcium, because calcium is required in endospore formation (41), and mutants lacking one of the proteins are reported to have low levels of calcium-dipicolinate (38).
Discussion
The approach presented here relies only on genome sequences, thus providing an objective and unbiased view on functional modularity in a variety of organisms. The procedure is based on a combination of methods to predict functional associations from whole genomes. For full coverage and accuracy, this combination is crucial: an analysis of the relative contributions of the methods (conserved gene neighborhood, common phylogenetic distribution of genes, and gene fusions) showed that only 56% of the pathways were detected by all three methods, 20% of pathways were predicted by any two of these methods, whereas 24% of pathways were identified only on the basis of a single method (Table 4, which is published as supporting information on the PNAS web site). Gene neighborhood was found to be the major contributor, recovering the highest number of pathways (89% of pathways detected by any methods). Because each methodology relies on a different measure of the driving forces in microbial evolution, the integration of all three methods provides for a robust and extensive coverage of the events that have shaped genome organization, revealing the underlying functional modularity.
Our key observation is the surprisingly high accuracy with which gene context analysis defines functional modules. The rigorous testing of the approach with the small-molecule metabolism from E. coli, the best characterized functional system to date, reveals that using a large number of genomes and a combination of techniques increases the amount of information retrievable from whole genomes to the point where it can probably rival that of large scale experimental approaches. We have specifically tested the performance in relation to metabolic pathways, but we expect a roughly similar performance for other functional systems, because enzymes are only slightly above average in terms of sequence conservation and species coverage, when compared to other functional systems (Fig. 10, which is published as supporting information on the PNAS web site). We have also tested the performance in a separate organism, and found it to be comparable (Fig. 11 and Table 5, which are published as supporting information on the PNAS web site).
Nevertheless, although the accuracy seems high throughout, it is not entirely uniform. When analyzing the predicted network and the functional modules, we observed that several functional aspects of metabolism influence the predictive power of genomic context methods. For example, the observed coverage of biosynthesis pathways is considerably higher than that of degradation pathways, possibly because anabolic pathways tend to be more linear, consume more energy, and are more tightly regulated, thus presumably enforcing more constraints on genome evolution. We also found that enzymes consisting of several subunits have an extraordinarily high chance of having all subunits correctly represented in one predicted module, showing that physical dependencies are particularly well reflected in genome context.
Because genome sequences are an objective and quickly growing resource, automatic pathway definitions based on comparative analysis may become feasible. We show here that genome structure and evolution are intricately intertwined through biochemical function and interaction networks. We have also proven functional modularity within these networks and have developed a reliable tool for the prediction of these modules. Further work will be needed to incorporate gene expression, localization, and regulation information to increase the resolution within the functional modules and to better understand their interactions within cells.
Supplementary Material
Acknowledgments
We thank members of the Bork group for helpful discussions, in particular Jan Korbel and Tobias Doerks for sharing expertise in specific predictions. This work was supported by the U.K. Medical Research Council (S.T.), the Portuguese Foundation for Science and Technology (J.B.P.-L.), the German Federal Ministry for Education and Science (C.V.M. and F.C.), and the European Union (E.M.Z.). C.O. acknowledges support from the European Molecular Biology Laboratory, the British Council, and IBM Research.
References
- 1.Wolfe, K. H. & Li, W. H. (2003) Nat. Genet. 33, Suppl., 255–265. [DOI] [PubMed] [Google Scholar]
- 2.Koonin, E. V., Makarova, K. S. & Aravind, L. (2001) Annu. Rev. Microbiol. 55, 709–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence, J. G. (1997) Trends Microbiol. 5, 355–359. [DOI] [PubMed] [Google Scholar]
- 4.Aravind, L., Watanabe, H., Lipman, D. J. & Koonin, E. V. (2000) Proc. Natl. Acad. Sci. USA 97, 11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snel, B., Bork, P. & Huynen, M. (2000) Trends Genet. 16, 9–11. [DOI] [PubMed] [Google Scholar]
- 6.Pilpel, Y., Sudarsanam, P. & Church, G. M. (2001) Nat. Genet. 29, 153–159. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence, J. G. (2002) Cell 110, 407–413. [DOI] [PubMed] [Google Scholar]
- 8.Lathe, W. C., III, Snel, B. & Bork, P. (2000) Trends Biochem. Sci. 25, 474–479. [DOI] [PubMed] [Google Scholar]
- 9.Marcotte, E. M., Pellegrini, M., Thompson, M. J., Yeates, T. O. & Eisenberg, D. (1999) Nature 402, 83–86. [DOI] [PubMed] [Google Scholar]
- 10.Osterman, A. & Overbeek, R. (2003) Curr. Opin. Chem. Biol. 7, 238–251. [DOI] [PubMed] [Google Scholar]
- 11.Bork, P., Dandekar, T., Diaz-Lazcoz, Y., Eisenhaber, F., Huynen, M. & Yuan, Y. (1998) J. Mol. Biol. 283, 707–725. [DOI] [PubMed] [Google Scholar]
- 12.Enright, A. J., Iliopoulos, I., Kyrpides, N. C. & Ouzounis, C. A. (1999) Nature 402, 86–90. [DOI] [PubMed] [Google Scholar]
- 13.Marcotte, E. M., Pellegrini, M., Ng, H. L., Rice, D. W., Yeates, T. O. & Eisenberg, D. (1999) Science 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 14.Overbeek, R., Fonstein, M., D'Souza, M., Pusch, G. D. & Maltsev, N. (1999) Proc. Natl. Acad. Sci. USA 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dandekar, T., Snel, B., Huynen, M. A. & Bork, P. (1998) Trends Biochem. Sci. 23, 324–328. [DOI] [PubMed] [Google Scholar]
- 16.Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. (1999) Proc. Natl. Acad. Sci. USA 96, 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellor, J. C., Yanai, I., Clodfelter, K. H., Mintseris, J. & DeLisi, C. (2002) Nucleic Acids Res. 30, 306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doolittle, R. F. (1999) Nat. Genet. 23, 6–8. [DOI] [PubMed] [Google Scholar]
- 19.Eisenberg, D., Marcotte, E. M., Xenarios, I. & Yeates, T. O. (2000) Nature 405, 823–826. [DOI] [PubMed] [Google Scholar]
- 20.Marcotte, E. M. (2000) Curr. Opin. Struct. Biol. 10, 359–365. [DOI] [PubMed] [Google Scholar]
- 21.Sali, A. (1999) Nature 402, 23–26. [DOI] [PubMed] [Google Scholar]
- 22.Tsoka, S. & Ouzounis, C. A. (2000) FEBS Lett. 480, 42–48. [DOI] [PubMed] [Google Scholar]
- 23.Hartwell, L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. (1999) Nature 402, C47–C52. [DOI] [PubMed] [Google Scholar]
- 24.Snel, B., Bork, P. & Huynen, M. A. (2002) Proc. Natl. Acad. Sci. USA 99, 5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravasz, E., Somera, A. L., Mongru, D. A., Oltvai, Z. N. & Barabasi, A. L. (2002) Science 297, 1551–1555. [DOI] [PubMed] [Google Scholar]
- 26.von Mering, C., Huynen, M. A., Jaeggi, D., Schmidt, S., Bork, P. & Snel, B. (2003) Nucleic Acids Res. 31, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enright, A. J., Van Dongen, S. & Ouzounis, C. A. (2002) Nucleic Acids Res. 30, 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karp, P. D., Riley, M., Saier, M., Paulsen, I. T., Collado-Vides, J., Paley, S. M., Pellegrini-Toole, A., Bonavides, C. & Gama-Castro, S. (2002) Nucleic Acids Res. 30, 56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riley, M. (1993) Microbiol. Rev. 57, 862–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camon, E., Magrane, M., Barrell, D., Binns, D., Fleischmann, W., Kersey, P., Mulder, N., Oinn, T., Maslen, J., Cox, A. & Apweiler, R. (2003) Genome Res. 13, 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 34.Webb, A. (2002) Statistical Pattern Recognition (Wiley, Chichester, U.K.).
- 35.Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., Martin, M. J., Michoud, K., O'Donovan, C., Phan, I., et al. (2003) Nucleic Acids Res. 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauter, M., Bohm, R. & Bock, A. (1992) Mol. Microbiol. 6, 1523–1532. [DOI] [PubMed] [Google Scholar]
- 37.Spitzer, E. D. & Weiss, B. (1985) J. Bacteriol. 164, 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beall, B. & Moran, C. P., Jr. (1994) J. Bacteriol. 176, 2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eichenberger, P., Jensen, S. T., Conlon, E. M., van Ooij, C., Silvaggi, J., Gonzalez-Pastor, J. E., Fujita, M., Ben-Yehuda, S., Stragier, P., Liu, J. S. & Losick, R. (2003) J. Mol. Biol. 327, 945–972. [DOI] [PubMed] [Google Scholar]
- 40.Fodje, M. N., Hansson, A., Hansson, M., Olsen, J. G., Gough, S., Willows, R. D. & Al-Karadaghi, S. (2001) J. Mol. Biol. 311, 111–122. [DOI] [PubMed] [Google Scholar]
- 41.O'Hara, M. B. & Hageman, J. H. (1990) J. Bacteriol. 172, 4161–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K. & Watson, J. D. (1994) Molecular Biology of the Cell (Garland, New York).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.