Abstract
Objective
Endothelial cells (ECs) lining arteries respond to laminar shear stress (LSS) by suppressing pro-inflammatory changes, in part through the activation of MEK5, ERK5 and induction of KLF4. We examined if this anti-inflammatory pathway operates in human ECs lining microvessels, the principal site of inflammatory responses.
Methods
We used immunofluorescence microscopy of human skin to assess ERK5 activation and KLF4 expression in human dermal microvascular (HDM)ECs in situ. We applied LSS to or overexpressed constitutively active MEK5 (MEK5/CA) in cultured HDMECs and assessed gene expression by microarrays and qRT-PCR and protein expression by western blotting. We assessed effects of MEK5/CA on TNF responses using qRT-PCR, FACS and measurements of HDMEC monolayer electrical resistance. We used siRNA knockdown to assess the role of ERK5 and KLF4 in these responses.
Results
ERK5 phosphorylation and KLF4 expression is observed in HDMECs in situ. LSS activates ERK5 and induces KLF4 in cultured HDMECs. MEK5/CA-transduced HDMECs show activated ERK5 and increased KLF4, thrombomodulin, eNOS, and ICAM-1 expression. MEK5 induction of KLF4 is mediated by ERK5. MEK5/CA-transduced HDMECs are less responsive to TNF, an effect partly mediated by KLF4.
Conclusions
MEK5 activation by LSS inhibits inflammatory responses in microvascular ECs, in part through ERK5-dependent induction of KLF4.
Keywords: inflammation, skin microvascualture, MAP kinases, vascular leak, adhesion molecules
INTRODUCTION
The microvasculature is lined by a monolayer of ECs that, under basal conditions, form a permselective barrier that is not interactive with circulating leukocytes or platelets and that inhibits coagulation [32]. Tumor necrosis factor (TNF) is the prototypical inflammatory cytokine and many of its actions in vivo are targeted at the ECs lining post-capillary venules [31,34,48]. In response to TNF, venular ECs selectively express new gene products that capture leukocytes, namely E-selectin (CD62E), intercellular adhesion molecule 1 (ICAM-1, also designated as CD54) and vascular cell adhesion molecule 1 (VCAM-1, also designated as CD106). TNF also reduces expression of molecules that normally inhibit leukocyte and platelet activation, such as endothelial nitric oxide synthase (eNOS), and that inhibit coagulation, such as thrombomodulin (TM). TNF-treated venules also become leaky to large plasma proteins, such as fibrinogen and fibronectin, which upon extravasation form a provisional matrix within the tissues necessary to support leukocyte migration and survival. In cultured human ECs both TNF induction of adhesion molecules and vascular leakiness depends on new gene transcription and protein synthesis [8] whereas TNF effects on TM and eNOS are thought to arise from destabilization of mRNAs encoding these proteins [27,45]. Cumulatively, these TNF changes result in leukocyte recruitment, thrombosis, and tissue induration characteristic of the inflammatory response.
The microvasculature is not homogeneous in its response to TNF. Specifically, arteriolar and capillary ECs within the same microvascular bed are generally much less responsive to TNF induction of E-selectin and VCAM-1 than are venular ECs although TNF induction of ICAM-1 does not show this same distinction [21,29]. Venular ECs are also more susceptible to induction of leakiness. The basis of the differential response to TNF among microvascular segments is not fully understood. Similar differences in TNF responsiveness have been observed between arterial (less) and venous ECs (more) [21], and at least part of the difference between arterial and venous ECs may be attributed to modulation of the ECs by laminar or oscillatory shear stress which is much higher in arteries than veins [9]. Levels of shear stress are also higher in arterioles and capillaries than in venules [26,35]. One mechanism by which shear stress can modulate TNF responsiveness is the activation of mitogen-activated protein (MAP) kinase pathways. MEK5 is a MAP kinase kinase that is constitutively expressed by ECs and is activated by LSS [19]. The principal downstream target of MEK5 is ERK5, also known as MAPK7 or BMK1 (Big Map Kinase) [15,44]. When ERK5 is activated by MEK5-catalyzed dual phosphorylation, it initiates responses that lead to activation of transcription factors such as MEF2A, MEF2C and MEF2D [16] which, in turn, leads to synthesis of other transcription factors such as KLF2 [37] and KLF4, although an obligatory role for ERK5 in the MEK5-dependent induction of KLF4 has recently been challenged [41]. MEK5 has also been implicated in the inhibition of inflammatory responses in cultured large vessel ECs [19,47] and both KLF2 and KLF4 have been identified as contributing to these effects through increased transcription of anti-inflammatory proteins while antagonizing TNF-induced expression of pro-inflammatory proteins [12,14,18,20,25,36]. In the present study, we address the question of whether the MEK5 signaling pathway can be activated by shear stress and play a similar role in human microvascular ECs.
MATERIALS AND METHODS
Reagents and Antibodies
TNF (also called TNFα) was obtained from R&D Systems (Minneapolis, MN). MEK5 inhibitors (BIX2189 and BIX2188) [38]were provided by Boehringer-Ingelheim Pharmaceuticals, Inc. (Ridgefield, CT) and JAK Inhibitor 1 was from CalBiochem (San Diego, CA). Human thrombin was obtained from Sigma (St. Louis, MO). Mouse monoclonal antibodies reactive with influenza hemagglutinin (HA), human TM, human VCAM-1, human KLF2 (H-60) were from Santa Cruz Biotechnology (Santa Cruz, CA), with human ERK5 from Upstate Biotechnologies (Temecula, CA), with human eNOS and MEK5 from BD Biosciences (Franklin Lakes, NJ), with human KLF4 and ICAM-1 from R&D Systems (Minneapolis, MN) and with β˜ actin from Sigma (St Louis, MO). For FACS analysis, mouse antibodies conjugated with fluorescein isothiocyanate (FITC) and reactive with human ICAM-1, human E-selectin, or human VCAM-1 (Immunotech), were used. Unless otherwise specified, all other reagents were from Sigma.
Immunofluorescence Analysis of Human Skin
Normal human skin from de-identified healthy donors was obtained as discarded tissue from cosmetic plastic surgery procedures under a protocol approved by the Yale Human Investigations Committee. Tissue was frozen in OCT and sectioned to a thickness of 5 μm. Tissue sections were fixed in 4% paraformaldehyde and incubated in blocking solution (5% BSA, 1X TBS, 5% Normal Donkey Serum, and 0.1% Triton X-100). Anti-KLF4 antibody (rabbit) from Santa Cruz (Cat No. SC-20691), anti-pERK5 antibody (rabbit) from Santa Cruz (Cat No. SC-16564R) and smooth muscle actin from Dako (Cat No. M0851) were diluted to final concentration of 1 μg/ml and Ulex Europaeus Agglutinin I Rhodamine from Vector (Cat No. RL-1062) were diluted to final concentration of 10 μg/ml in blocking solution. Sections were incubated with species-specific biotin-conjugated secondary antibodies from Jackson Labs (Cat. No. 705-065-147 & 711-065-152) diluted to a final concentration of 7 μg/ml in blocking solution.
Streptavidin Alexa Fluor 488 (Invitrogen ; Cat No. S-11223) and Goat anti-mouse Alexa 647 (Invitrogen ; Cat No. A21237) was next incubated with the sections diluted to a final concentration of 10 μg/ml in blocking solution. Control sections were stained in a similar manner using rabbit IgG1 isotype control from R&D systems (Cat No. AB105C). Peptide blocking control sections were prepared using either a pERK5 epitope (Specific) or KLF4 epitope peptide (Non-specific) from Santa Cruz (Cat No. SC-16564P & SC-12538P, respectively). The slides were mounted using ProLong Gold Anti-Fade Reagent with DAPI from Invitrogen (Cat. No. P3931). Sections were viewed and images were generated using a Zeiss Axiovert fluorescence microscope with a digital camera (Zeiss, Thornwood, NY). pERK5 and KLF4 expression in microvascular ECs in situ was quantified by identifying UEA I positive microvessels that were contained within or closely associated with epidermal rete [6,30]. Such vessels are capillaries that contain few perivascular mural cells and positive staining is readily assigned to ECs. Using antibody isotype control-stained sections of skin, thresholds were set and the percentage of vessels that displayed antigen-specific fluorescence above these levels were scored as positive. Data were pooled from three separate human skin specimens, each from a different donor. No significant differences were noted among these specimens.
Culture of HDMECs and Application of LSS
HDMEC cultures were prepared under protocols approved by the Yale Human Investigations Committee under the auspices of the Yale Skin Disease Research Center cell culture core. HDMECs were harvested from the superficial vascular plexus of the upper dermal layers of de-identified and discarded normal adult human skin of a single donor. Except where specified all experiments were conducted using at least three different serially cultured EC isolates, each from separate donors, to control for donor heterogeneity. ECs were prepared by digestion of the skin with Dispase (50 U/ml; BD Biosciences) for 30 minutes at 37° C and fine mincing as described previously [8,17]. Released cells were cultured on 10 μg/ml human plasma fibronectin-coated plastic in EGM2-MV growth medium (Cambrex; East Rutherford, N.J). Primary adherent cells were resuspended using trypsin and immunoselected on a mini-MACS column using anti-CD-31-biotin followed by streptavidin-magnetic beads (Miltenyi Biotec, Auburn, CA). Unmodified HDMECs were grown on Superfrost microscope slides coated with 10μg/ml human plasma fibronectin. Cells were seeded at 5×105 cells per slide and cultured in 500 cm2 culture dishes in EGM2-MV growth medium. For all experiments, unique primary HDMEC cultures were used for each independent experiment and the cultures were used before passage 6. Cells were grown on microscope slides and placed in a multislide parallel plate flow chamber (Streamer; FlexCell International, Hillsborough, NC, USA). EGM2-MV medium was circulated at 600ml/min to produce 12.6 dynes/cm2 laminar shear for 24 hours. Matched control (static) cells were cultured in EGM2-MV for the same period without shear. Sheared and static cells were collected and analyzed for protein expression by Western blot. Where indicated, medium conditioned by ECs subjected to LSS or to static conditions was collected after 24 hours and transferred to fresh ECs under static conditions.
Retroviral Construction and HDMEC Transduction
A cDNA encoding constitutively active MEK5 (MEK5/CA) was subcloned from pCDNA3-MEK5.7CA-1 [16] (provided by Boehringer-Ingelheim Pharmaceuticals, Inc.) using PCR primers that included a 9 amino acid influenza hemagglutinin (HA) tag inserted onto the 3’ end of the cDNA. The PCR product was ligated into the LTR region of the retroviral vector pLZRS immediately downstream of a CMV promoter. Retroviral supernatants were produced by pLZRS plasmid transfection of Phoenix packaging cells (gift of Dr. Garry Nolan, Stanford University) grown in DMEM supplemented with 10% FBS, L-glutamine, and antibiotics followed by selection for positive transfectants in the same media supplemented with 10 μg/ml puromycin. Drug-free viral culture supernatants were used to transduce low passage, primary HDMEC cultures as described [8]. Cells were routinely transduced with neat viral supernatants 4 times for 6 hours. Expression of MEK5/CA and KLF4 was verified by Western blot analysis (see below).
Western Blotting
Cultured cells to be analyzed by Western blot were lysed with 20mM Tris-HCl, 150mM NaCl, 1mM EDTA, 1mM EGTA, 1% Triton, 2.5mM Na pyrophosphate, 1mM β-glycerophosphate, 1mM orthovanadate, and 2% SDS (Cell Signaling Technology, Danvers, MA) supplemented with a protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Ten (10) μg of each lysate was resolved using SDS-PAGE, proteins were transferred to Immobilon-P PVDF (Millipore, Bedford, MA), and the membranes were blocked in 5% non-fat milk (Bio-Rad, Hercules, CA). Primary antibodies were diluted in 5% non-fat milk at 1:1000 and incubated with membranes overnight. Isotype appropriate secondary horseradish peroxidase (HRP)-conjugated antibodies (Jackson ImmunoResearch) were incubated for 2 hours and HRP reactivity was detected using Femto West Chemiluminescent substrate (Thermo Scientific). KLF2 could not be detected by this method and required further amplification of the signal using biotin-conjugated anti-rabbit secondary antibody (Jackson Labs) and HRP-conjugated anti-biotin tertiary antibody (Cell Signaling Technology). Where indicated, densitometric analysis of band intensity was performed using ImageJ software (http://rsbweb.nih.gov/ij/index.html) and is detailed in the figure legends.
Microarray Analyses of mRNA Expression
Total RNA was isolated from 8 separate paired (derived from same primary isolate) MEK5/CA and LacZ transduced HDMEC lines. Cells were grown to confluence in EGM2-MV growth medium on 75cm2 cultureware. RNA was purified with RNeasy RNA purification system from Qiagen (Valencia, CA) using manufacturers instructions. RNA concentration and quality was evaluated by spectrophotometric analysis at 260 and 280nM and gel electrophoresis. The purified RNA was converted to double-stranded complementary DNA (cDNA) using an Affymetrix one-cycle cDNA synthesis kit and an oligo-dT primer containing a T7 RNA polymerase promoter. Biotin-labeled complementary RNA (cRNA) was then generated from the cDNA samples by performing an in vitro transcription reaction with T7 RNA polymerase. The resulting labeled cRNA was then fragmented to an average size of 35 to 200 bases by incubation at 94oC for 35 min. Hybridization (16 hr), washing, and staining were conducted according to the manufacturer’s specifications. The Affymetrix GeneChip® Human Genome U133 Plus 2.0 Array was used to identify changes in messenger RNA (mRNA) levels in response to transfection. The signal value of each gene was calculated using the MAS5 algorithm and normalized using global scaling which set the average signal intensity of an array to 750. A 2-way analysis of variance (ANOVA) using Partek Genomics Suite 6.3 (Partek Incorporated, St. Louis, MO), was carried out on the log(2) transformed data using the gene (MEK5/CA and LacZ control), and each cell line paired with rounds of transduction as variables (factors) for the analysis. The resulting p-values were adjusted using the False Discovery Rate (FDR). A total of 670 genes showed differential expression of 2-fold or more up and down-regulated. A complete list of differentially expressed genes has been deposited at the public archive, Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
qRT-PCR Analyses of mRNA Expression
Total RNA was isolated from HDMEC cultures using the RNeasy column system (Qiagen) and was reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Briefly, cDNA, MgCl2, and primers were added to Platinum SYBR Green qPCR SuperMix (Invitrogen). Samples were amplified for 35 cycles on a iCycler thermocycler (Bio-Rad): 50°C for 2 min., 95°C for 2 min., 95°C for 15 sec, 62°C for 15 sec, 72°C for 15 sec, 72°C for 5 min. cDNA input levels for each sample were normalized to levels of β-actin generated with β-actin-specific primers. Threshold cycles for each sample were compared to control (untreated) samples after actin normalization. Primer design using PrimerBank (http://pga.mgh.harvard.edu/primerbank). Primers used in PCR reactions measuring gene expression and the efficacy of siRNA knockdown in Table 1.
Table 1.
qRT-PCR Primer Sequences
| Primer | Sequence |
|---|---|
| β-actin Forward: | 5′ATGGGTCAGAAGGATTCCTAAGTG3′ |
| β-actin Reverse: | 5′CTTCATGAGGTAGTCAGTCAGGTC3′ |
| MEK5 Forward: | 5′ATGCTGTGGCTAGCCCTTGG 3′ |
| MEK5 Reverse: | 5′GTAATATCTAGTAGTATGACC3′ |
| KLF4 Forward: | 5′GCCACCCACACTTGTGATTA 3′ |
| KLF4 Reverse: | 5′CCCCGTGTGTTTACGGTAGT3′ |
| KLF2 Forward: | 5′ GCCGTCCTTCTCCACTTTC3′ |
| KLF2 Reverse: | 5′CGGGTTCGGGGTAATAGAAC3′ |
| TM Forward: | 5′CACAGGTGCCAGATGTTTTG 3′ |
| TM Reverse: | 5′GAGTCACAGTCGGTGCCAAT3′ |
| CD132 Forward: | 5′ TTGGAAGCCGTGGTTATCTC3′ |
| CD132 Reverse: | 5′TTTGGGGGAATCTCACTGAC3′ |
| ESAM Forward: | 5′GACTTTCTTTGCACCAGCATT3′ |
| ESAM Reverse: | 5′ATTGTGGGCCTTGCAGAC3′ |
| STAT6 Forward: | 5′CCTTTTGGCAGTGGTTTGATG3′ |
| STAT6 Reverse: | 5′GTTTGCTGATGAAGCCAATGATC3′ |
siRNA Knockdown of Specific Proteins
HDMECs were plated on gelatin-coated 12-well plates at 50 % confluence. siRNA complexes of Oligofectamine (Invitrogen) at 20 μg/ml and siRNA (Dharmacon, Lafayette, CO or Qiagen, Germany) at 50 nM were prepared in Opti-MEM I Reduced Serum Medium (Invitrogen) and then diluted five-fold in Opti-MEM to yield a final siRNA concentration of 10 nM. HDMECs cultures were exposed to siRNA complexes for 6 hours at 37 ° C. Fresh medium was added overnight and cells were re-transfected 24 hours later. Cells were rested for 24 hours prior to further stimulation with cytokine or inhibitors. siRNA sequences used in this study are described in Table 2.
Table 2.
siRNA Sequences
| siRNA | AntisenseSequence |
|---|---|
| MEK5-1: | 5′PGAAAUGUACAAAUGGCUCCUU3′ |
| MEK5-2: | 5′PCAUAUAAGCAUUUGUUCCAUU3′ |
| MEK5-3: | 5PUUUAGCAUAUUGGAGGUU3′ |
| KLF4-1: | 5′CUAGUUGUAAAUACUGGAU3′ |
| KLF4-2: | 5′CCCAUAUUUAAUAUAGGCA3′ |
| KLF4-3: | 5′CCACUCAGAACCAAGAUUU3′ |
| LAMINA/C: | 5′UGUUCUUCUGGAAGUCCAGdTdT3′ |
FACS Analysis of Surface Protein Expression
Confluent HDMEC cultures, either unstimulated or treated with TNF following siRNA transfection, were collected using trypsin. E-selectin, ICAM-1, or VCAM-1 surface levels on HDMECs were determined by FACS analysis (FACaliber; BD Biosciences, San Jose, CA) after immunostaining with saturating concentrations of fluorochrome-labeled antigen-specific monoclonal antibodies or with isotype-matched control antibodies (see above). Corrected mean fluorescence intensity (specific MFI-isotype control MFI) was used to calculate changes in adhesion molecule expression.
Measurements of HDMEC Monolayer Resistance
Electrical resistance in HDMEC monolayers was monitored by electric cell-substrate impedance sensor (ECIS) (Applied BioPhysics, Troy, NY) [39]. MEK5/CA and LacZ HDMEC were cultured on 8W10E+, gold electrode 8-chamber slides coated with 10 μg/ml fibronectin. Individual wells were seeded with 1 × 105 cells/well in complete EGM2-MV media that serves as an electrolyte. HDMEC resistance was calculated from daily impedance readings to determine the point of maximum resistance, at which point monolayers were stimulated, in triplicate, with increasing doses of human TNF. Real-time changes in endothelial monolayer resistance were measured by application of a 1 μA constant AC current at 4000 Hz between a large and small electrode embedded in the chamber slide. The TNF and thrombin EC50 (Effective Concentration 50), the concentration of agent to produce a 50% fall in barrier resistance, were determined on LacZ and MEK5/CA HDMEC lines. All readings were controlled by computer and accompanying ECIS software supplied by manufacturer (Applied BioPhysics).
Statistical analyses
Significance of differences between groups was tested by an unpaired Ratio t-test or paired, two-tailed t-test and among more than two groups, by one-way analysis of variance (ANOVA) followed by the Bonferroni post-test. P values < 0.05 were accepted as significant. A sigmoid curve non-linear regression curve fit analysis was used to describe the concentration dependence of TNF- and thrombin-induced changes in EC50 for HDMEC lines. All results were computed using Prism version 4.0b (GraphPad Software,Inc, La Jolla, CA).
RESULTS
Analyses of HDMECs in situ
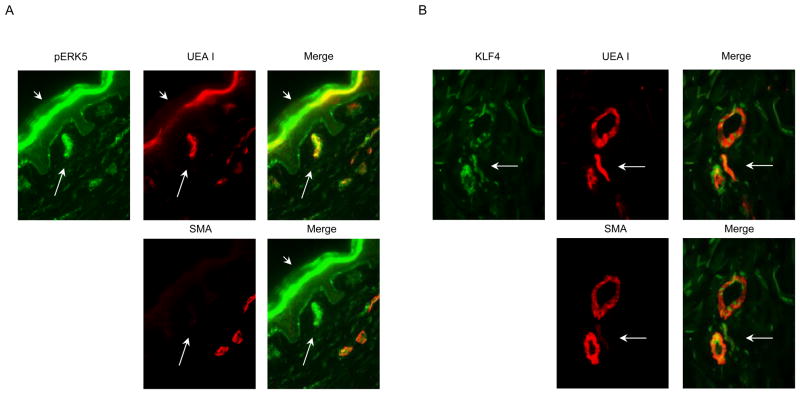
MEK5 is activated in cultured human large vessel ECs by shear stress and the microvasculature is subjected to shear stress in situ. To assess if MEK5 is activated in microvascular ECs in situ, samples of healthy human skin were analyzed by immunofluorescence microscopy as described in the Materials and Methods. Microvessel structures stained positively for phospho-ERK5 (pERK5), an indicator of MEK5 activity, and for KLF4, a gene known to be induced in large vessel ECs by arterial shear stress in a MEK5-dependent manner. Such staining could be associated with EC and/or with mural pericytes or smooth muscle cells. We therefore double stained the same sections with Ulex Europeus Agglutinin I (UEA I), a lectin that detects the H blood group on human ECs as well as human epithelia but that is not expressed on vascular mural cells, and with smooth muscle α-actin (SMA), a marker for pericytes and smooth muscle cells that is absent from ECs. Within the dermal papillae, we identified positive UEA I staining of capillary ECs that were minimally and only focally covered by SMA-stained pericytes. pERK5 and KLF-4 both co-localize to these UEA I positive, SMA-negative ECs (Figures 1A and 1B, respectively). pERK5 staining was specifically inhibited with a pERK5 epitope blocking peptide, but was unaffected by an irrelevant peptide (data not shown). A similar analysis was not feasible for KLF4 staining as a blocking peptide used to generate the antibody used for tissue staining is unavailable. We quantitated the frequency of expression for both pERK5 and KLF4 in capillary EC (which we identified by UEA I positivity and inclusion in or juxtaposition to the epidermal rete) and observed that 84% (of 195) and 63% (of 198) dermal capillaries, counted in three different human skin specimens, were positive for pERK5 and KLF4, respectively in data pooled from three separate donors. Expression of pERK5 and KLF4 was also observed on ECs lining larger microvessels as well as in some microvessel SMA-positive mural cells located below the level of the epidermal rete, i.e. the location of the arterioles and venules of the superficial vascular plexus. These data are consistent with the idea that MEK5 is activated, albeit not exclusively, within ECs of the microvasculature in human skin.
Figure 1. pERK5 and KLF4 Immunofluorescent Staining of Human Skin.
Normal human skin was snap frozen, sectioned on a cryostat, and fixed in 4% paraformaldehyde. A) pERK5-specific antibody was used to determine the presence of pERK5. B) KLF4-specific antibody (KLF4) was used to detect KLF4. pERK5 and KLF4 antigen specific staining was detected by Alexa 488 (green) fluorescence. All sections were stained with Ulex Europaeus Agglutinin I-Rhodamine (UEA I) or smooth muscle α-actin (SMA) to identify the location of human microvascular ECs and vascular mural cells, respectively. Capillaries are identified by long arrows and epithelium by short arrows. One of three independent experiments with similar results are depicted in the photomicrographs.
Effects of LSS on Cultured HDMECs
Microvascular ECs in situ have been calculated to undergo shear stress that is greatest in the arterioles, intermediate in the capillaries and lowest in the venules [35]. The effects of shear stress are well characterized in cultured large vessel ECs, but have not been extensively studied on cultured microvascular ECs. We first subjected cultured HDMECs to levels of LSS intermediate between that of arteries and of veins, namely 12.6 dynes/cm2 for 24 hours to investigate ERK5 activation and associated changes in protein expression. Western blotting showed a shift in position of ERK5, consistent with its phosphorylation and activation (Figure 2A) (The antibody used to detect pERK5 by immunofluorescent staining was not effective for detection in Western blots). This effect was not a result of stable soluble factors released by HDMEC exposed to LSS since shear conditioned media was insufficient to activate ERK5 (Figure 2A). We also observed strong induction of KLF4 protein in response to LSS but not in response to medium conditioned by cells exposed to LSS. We further investigated the role of MEK5 in transducing shear stress stimuli leading to ERK5 activation and KLF4 induction by pre-treating EC with MEK5 siRNA to knock down MEK5 protein. ERK5 activation, again detected by a shift in the position of ERK5 in Western blotting, was strongly inhibited and KLF4 induction was substantially inhibited by MEK5 knockdown confirming that both ERK5 activation and KLF4 induction by shear stress are MEK5-dependent in human microvascular EC (Figures 2B & 2C). KLF2, which is strongly induced in large vessel EC by LSS, was detected in some, but not all instances, under the conditions of shear stress applied in this experiment and detection of KLF2 required an additional amplification of the signal not required to detect KLF4. However, when shear-induced KLF2 was detected, induction was inhibited by MEK5 knockdown in the same manner as KLF4 (Figure 2B). These experiments show that microvascular ECs, like large vessel ECs, are responsive to shear stress and that MEK5 and ERK5 are activated in this setting, leading to significant KLF4 induction and a variable and lesser degree of KLF2 induction.
Figure 2. ERK5 and KLF4 Activation and Inhibition by MEK5 siRNA in Shear Stressed Primary HDMEC.
A) Wild type HDMEC or B) wild type HDMEC transfected with Lamin control or MEK5 siRNA (as described in Methods) were grown on fibronectin coated slides and subjected to shear stress of 12.6 dynes/cm2 in a flow chamber (Shear) or maintained in static (Static) culture for 24 hours. SDS whole cell lysates from each of the slides were prepared and analyzed by SDS-PAGE and Western blotting as described in the Methods. To assess the effect of shear-conditioned media on ERK5 activation, static HDMEC cultures were exposed to fresh media (EGM-2MV), to media from static HDMEC cultures (Static Cond. Media), or to media collected from the flow chamber (Shear Cond. Media) for 24 hours and analyzed by Western blotting. One of three independent experiments with similar results. B) Wild type HDMEC transfected with Lamin control or MEK5 siRNA (as described in Methods) were subjected to shear stress as above and analyzed by SDS-PAGE and Western blotting. Phosphorylated ERK5 (pERK5) and ERK5 (ERK5) are indicated with arrows. One of three independent experiments with similar results. C) KLF4 band intensity was compared to a loading control (KLF4:Control) using ImageJ software. One-way ANOVA and Bonferroni post-tests were used to assess statistical significance.
Development and Characterization of HDMECs Expressing a Constitutively Active Form of Human MEK5
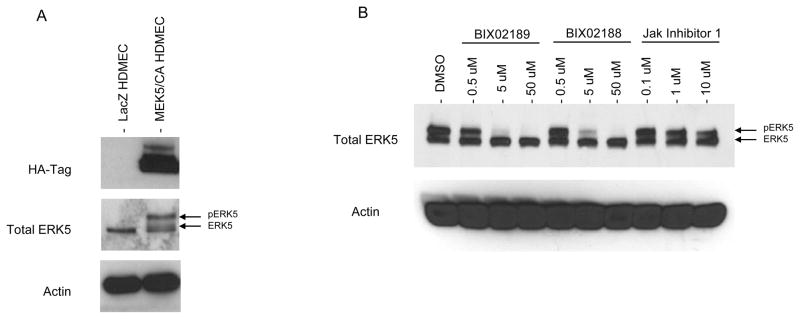
To examine more directly the effects of MEK5 signaling in microvascular ECs independent of other changes that may be induced by shear stress, HDMECs were transduced with retroviral vectors encoding either HA-tagged, constitutively active MEK5 (MEK5/CA HDMECs) or, as a transduction control, LacZ (LacZ HDMECs). Western blotting with an HA-specific antibody demonstrated that HA-tagged MEK5/CA was expressed in MEK5/CA HDMECs and, as expected, was absent from control LacZ transduced HDMECs (Figure 3A). MEK5/CA overexpression appeared functional since ERK5 was strongly phosphorylated and displayed the band shift characteristic of phosphorylated MAPKs (Figure 3A). This change was not observed in control LacZ HDMECs. We confirmed that ERK5 activation was induced through MEK5 because two separate MEK5-selective kinase inhibitors, designated BIX2189 and BIX2188 [38], led to a dose-dependent ERK5 band-shift diminution, indicating ERK5 inactivation. JAK Inhibitor 1, used as a specificity control, had no effect on ERK5 status (Figure 3B).
Figure 3. Generation of MEK5/CA Overexpressing Primary HDMEC.
A) HDMEC were transduced with retroviral vectors encoding the consitutively active HA-tagged MEK5 (MEK5/CA) or LacZ transgenes. Western blot analysis of LacZ and MEK5/CA HDMEC using HA-, ERK5-, and actin-specific antibodies. Representative of one of five results. B) MEK5/CA HDMEC were treated with increasing amounts of MEK5 inhibitor (BIX02189 and BIX02188) or JAK Inhibitor I for 1.5 hrs followed by Western blot analysis using an ERK5-specific antibody. One of two independent experiments with similar results.
Microarray mRNA Expression profiling of MEK5/CA vs. Control Transduced HDMECs
To more fully characterize the downstream effects of MEK5 activity, we conducted a global gene expression profiling analysis on MEK5/CA HDMECs using microarray technology. A total of 8 separate paired mRNA samples were prepared from LacZ HDMEC and MEK5/CA HDMEC lines from 5 independently transduced cultures. Included were 3 duplicate cell line pairs that differed by having been subjected to either 3 or 4 rounds of retroviral transduction. In all cases enhanced MEK5 expression in MEK5/CA cells was confirmed by qRT-PCR using MEK5-specific primers and the rates of induction were between 8.5 and 48 fold (data not shown). All transcripts displaying analysis of variance (ANOVA) p-Values <0.05 (see Methods) and Fold Changes greater than 2, or less than −2, were considered statistically valid. We screened our validated list of genes preliminarily on the basis of EC association using the Pubmed Biomedical database (www.ncbi.nlm.nih.gov/sites/entrez). A secondary screen was then performed on the basis of biological function, including Inflammation, Vascular Permeability, Hemodynamic Shear Stress, and Coagulation and Thrombolysis and a selected list of genes is presented in Table 3. A complete list of differentially expressed genes has been deposited at the online searchable public archive, Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/).
Table 3.
Response of Selected Genes to the Expression of MEK5/CA in HDMEC (MEK5JCA HDMEC: LacZ HDMEC)
| Accession No. | Gene Name | Gene Symbol | Fold Change | Function |
|---|---|---|---|---|
| NM_002036 | Duffy blood group, chemokine receptor | DARC | 106.2 | Endothelial Inflammation |
| AK023795 | ADAM metallopeptidase | ADAMTS1 | 99.0 | Vascular Permeability/Hemodynamic Shear Stress |
| AW157548 | Insulin-like growth factor binding protein 5 | IGFBP5 | 46.0 | Vascular Permeability |
| NM_000361 | Thrombomodulin | THBD | 21.9 | Coagulation and Thrombolysis |
| NM_004591 | Chemokine (C-C motif) ligand 20 | CCL20 | 18.9 | Inflammation/Hemodynamic Shear Stress |
| NM_002933 | Ribonuclease, RNase A family, 1 | RNASE1 | 18.1 | Hemodynamic Shear Stress |
| NM_004235 | Kruppel-like factor 4 | KLF4 | 15.5 | Inflammation/Hemodynamic Shear Stress |
| X90579 | Cytochrome P450, family 3, subfamily A | CYP3A5 | 12.9 | Hemodynamic Shear Stress |
| AK001855 | Apelin | APLN | 10.5 | Coagulation and Thrombolysis |
| NM_002276 | Junction plakoglobin | JUP | 10.4 | Vascular Permeability |
| BC005939 | Prostaglandin D2 synthase 21 kDa | PGD2 | 8.3 | Hemodynamic Shear Stress |
| Al091047 | Solute carrier family 2, member 1 | SLC2A1 | 7.1 | Vascular Permeability |
| U58828 | G protein-coupled estrogen receptor 1 | GPER | 6.9 | Hemodynamic Shear Stress |
| NM_000050 | Argininosuccinate synthetase 1 | ASS1 | 6.7 | Hemodynamic Shear Stress |
| AL518391 | Aquaporin 1 | AOP1 | 6.5 | Vascular Permeability |
| BF594294 | TEK tyrosine kinase | TIE2 | 6.3 | Vascular Permeability/Angiogenesis |
| AK002203 | Nitric oxide synthase trafficker | NOSTRIN | 6.2 | Endothelial Inflammation |
| NM_005114 | Heparan sulfate 3-O-sulfotransferase 1 | HS3ST1 | 6.0 | Coagulation and Thrombolysis |
| U43522 | PTK2B protein tyrosine kinase 2 beta | PTK2B | 5.6 | Vascular Permeability/Hemodynamic Shear Stress |
| NM_000104 | Cytochrome P450, (1B1) | CYP1B1 | 5.6 | Hemodynamic Shear Stress |
| NM_016270 | Kruppel-like factor 2 | KLF2 | 5.6 | Inflammation/Hemodynamic Shear Stress |
| AB024518 | Interleukin 33 | IL33 | 5.2 | Endothelial Inflammation |
| MM_002658 | Plasminogen activator, urokinase | uPA | 5.2 | Coagulation and Thrombolysis |
| NM_003823 | Decoy Receptor 3 | DCR3 | 5.0 | Endothelial Inflammation |
| AL033377 | G protein-coupled receptor 126 | VIGR | 4.8 | Coagulation and Thrombolysis |
| AF089868 | CD146 (melanoma cell adhesion molecule) | CD 146 | 4.7 | Inflammation/Vascular Permeability |
| NM_000906 | Natriuretic peptide receptor A | NPR1 | 4.5 | Vascular Permeability |
| NM_000591 | CD14 | CD14 | 3.9 | Endothelial Inflammation |
| BF305661 | Integrin, beta 4 | ITGB4 | 3.8 | Coagulation and Thrombolysis |
| AK000667 | ADAM metallopeptidase domain 15 | ADAM15 | 3.7 | Hemodynamic Shear Stress |
| BE903880 | CD44 | CD44 | 3.6 | Inflammation/Vascular Permeability |
| AA812232 | Thioredoxin interacting protein | TXNIP | 3.4 | Inflammation/Hemodynamic Shear Stress |
| Al653117 | CD59 | CD59 | 3.3 | Shear Stress/Coagulation and Thrombolysis |
| NM_000930 | Plasminogen activator, tissue | PLAT | 3.2 | Coagulation and Thrombolysis |
| AL573851 | Endothelial cell adhesion molecule | ESAM | 3.0 | Inflammation/Vascular Permeability |
| NM_000201 | Intercellular adhesion molecule 1 | ICAM1 | 2.2 | Inflammation/Vascular Permeability |
| NM_000963 | COX-2 | PTGS2 | −2.3 | Inflammation |
| NM_007036 | Endocan | ESM1 | −2.7 | Inflammation |
| NM_001993 | Tissue Factor | TF | −2.7 | Coagulation and Thrombolysis |
| Al986120 | Latent TGF beta binding protein 1 | LTBP1 | −2.7 | Hemodynamic Shear Stress |
| AF187858 | Angiopoietin 2 | ANGPT2 | −2.9 | Inflammation/Permeability/Angiogenesis |
| AA669336 | Coagulation factor C homolog, cochlin | COCH | −3.4 | Coagulation and Thrombolysis |
| AL574096 | Tissue factor pathway inhibitor 2 | TFPI-2 | −3.8 | Coagulation and Thrombolysis |
| AJ224869 | Chemokine (C-X-C motif) receptor 4 | CXCR4 | −64 | Inflammation/Permeability/Shear Stress |
| NM_013409 | Follistatin | FST | −7.4 | Vascular Permeability |
ANOVA and False Discovery Rate were used to generate p-Value and the Fold Change (log2). All transcripts displaying p-Values <0.05 and Fold Changes greater than 2, or less than −2, were considered statistically valid. Selected list of valid induced and repressed gene transcripts resulting from MEK5/CA transduction in HDMEC grouped according to biological function.
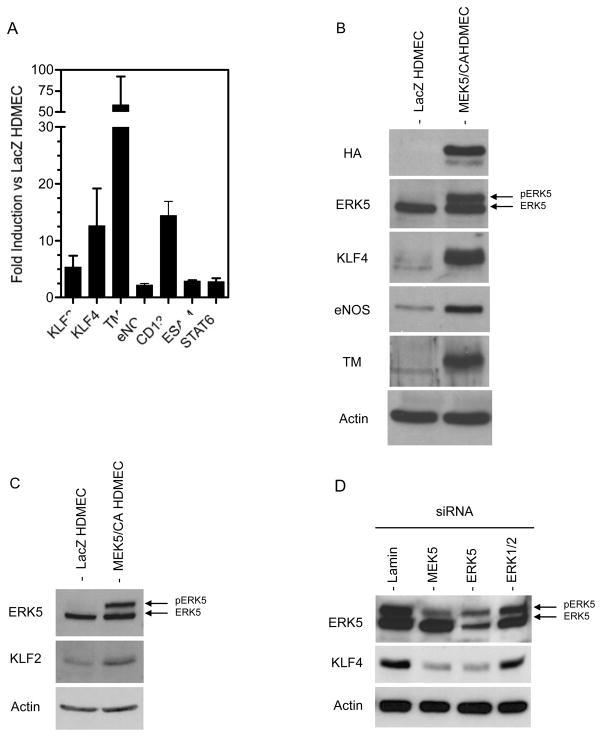
To confirm the results of the microarray analysis we selected 7 mRNAs for analysis by qRT-PCR based upon their known relevance to EC regulation and function. These were KLF2, KLF4, TM, eNOS, CD132, ESAM, and STAT6, of which all 7 transcripts were elevated in MEK5/CA HDMECs compared to LacZ HDMECs (Figure 4A). We then extended our microarray analysis of mRNA to examine protein levels by Western blotting. Both KLF4 and TM were strongly induced in all MEK5/CA HDMEC lines tested (Figure 4B) and eNOS, present at low levels in LacZ HDMECs, was also substantially increased in MEK5/CA cells (Figure 4B). KLF2 protein, absent in LacZ HDMECs, was induced in three separate lines following MEK5/CA transduction, but could be detected only after signal amplification (Figure 4C).
Figure 4. Gene Expression Confirmation for Microarray Analysis of MEK5/CA HDMEC.
A) Total RNA from four separate HDMEC cell lines, each of which were used for the initial microarray analysis, were tested for expression of selected transcripts to confirm 42 array gene expression results. RNA was reverse transcribed and analyzed by qPCR using gene-specific primers for KLF2, KLF4, TM, eNOS, CD132, ESAM, and STAT6. Actin-specific primers were used to normalize for cDNA input and gene-specific fold increase for each MEK5/CA HDMEC line was calculated by comparison to it’s paired LacZ HDMEC control. B&C) Whole cell lysates from confluent LacZ and MEK5/CA transduced HDMEC were prepared. Lysates were analyzed by SDS-PAGE and Western blot. Antigen-specific antibodies for HA, ERK5, KLF4, eNOS, TM and actin (B) and ERK5, KLF2 and HSP90 (C) were used to detect EC proteins as described in Methods. One of three independent experiments with similar results.
To confirm the role of MEK5 and ERK5 in the pathway leading to KLF4 induction and maintenance in HDMEC, we treated MEK5/CA HDMEC, with MEK5, ERK5, ERK1/2 or control siRNA. Control siRNA and ERK1/2 treated HDMEC showed no change in ERK5 phosphorylation status and maintained a high level of KLF4 protein following transfection (Figure 4D). In contrast, MEK5 knockdown, which limited phosphorylation and activation of the downstream substrate, ERK5, lead to a significant reduction of KLF4 levels. In addition, ERK5 knockdown also lead to lower KLF4 levels, suggesting that MEK5 utilizes primarily ERK5, and not ERK1/2, to transduce signals that lead to KLF4 induction in HDMEC.
Alteration of TNF-induced Adhesion Molecule Expression in MEK5/CA HDMECs
TNF is a major endogenous mediator of inflammation and acts principally by changing the expression of genes in microvascular ECs to create a state that promotes leukocyte adhesion and activation, activates the coagulation system, and produces vascular leakiness. These responses to TNF are antagonized by arterial levels of shear stress, in part through MEK5/ERK5 activation. We tested our MEK5/CA HDMECs to determine if MEK5 activation could modulate the expression levels of EC surface adhesion molecules on unstimulated MEK5/CA-transduced HDMECs by direct immunofluorescent staining and FACS analysis. Expression of E-selectin and VCAM-1 were essentially absent in both MEK5/CA HDMECs and LacZ transduced control HDMECs, but ICAM-1 was substantially increased in MEK5/CA expressing cells and displayed an electrophoretic mobility shift as determined by Western blot (data not shown). The basis of this difference is unknown.
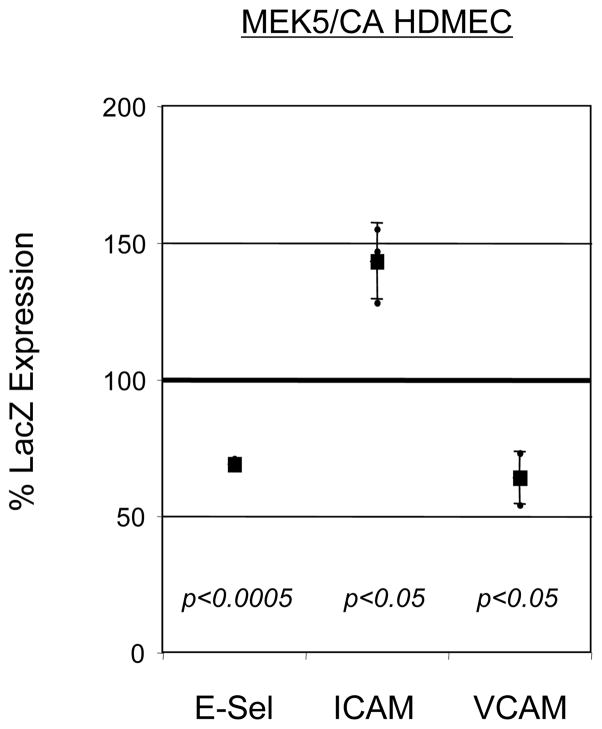
We next measured the induction of cell surface E-selectin, ICAM-1, and VCAM-1 in TNF-treated MEK5/CA and LacZ HDMECs. Compared to control LacZ HDMECs, both E-Selectin and VCAM-1 expression levels were suppressed in MEK5/CA HDMECs, exhibiting 69% and 64% of control cells, respectively (Figure 5). In contrast, ICAM-1 cell surface expression was elevated on TNF-treated MEK5/CA cells to 143% of that on LacZ HDMEC controls.
Figure 5. Alteration of TNF-induced Adhesion Molecule Expression in MEK5/CA HDMEC Cell Line.
LacZ and MEK5/CA HDMEC were stimulated with 2 ng/ml human TNF for either 4 (E-selectin) or 24 (ICAM-1 and VCAM-1) hours, when optimal protein expression is seen. Cells were collected and immunostained with FITC-conjugated, antigen-specific monoclonal antibodies and analyzed by flow cytometry. Percent MEK5/CA surface adhesion molecule expression compared to LacZ control (“% LacZ Expression”) was calculated as follows: ((Corrected MFI(MEK5/CA HDMEC)/Corrected MFI(LacZ HDMEC))*100), +/− SD. Statistical comparison of the means of the Corrected MFI for LacZ HDMEC vs. MEK5/CA HDMEC was done using a Ratio Paired t-test. Data represents pooled results from three independent trials.
To confirm that MEK5 signalling was responsible for the altered response to TNF we used MEK5-specific siRNA to ablate MEK5 activity and measured TNF-induced adhesion molecule expression in LacZ and MEK5/CA HDMECs. MEK5 knockdown effectively reversed TNF-induced adhesion molecule expression profiles. Specifically TNF-induced E-selectin was increased by 51% following MEK5 knockdown compared with a control siRNA while VCAM-1 was increased by 75% (Table 4). Conversely, MEK5 siRNA knockdown in TNF-treated MEK5/CA HDMECs led to a 23% reduction (or diminished induction) of ICAM-1 expression. These results support the conclusion that MEK5 activation can regulate both basal and TNF-responsive adhesion molecule expression in HDMECs.
Table 4.
TNF-induced Adhesion Molecule Expression Due to MEK5 or KLF4 siRNA Knockdown in MEK5/CA HDMEC: Percent of ‘Control’ siRNA Expression
| Adhesion Molecules | |||
|---|---|---|---|
| siRNA | E-Selectin | ICAM-1 | VCAM-1 |
| MEK5 | 151 (+/− 45)a (p<0.001)b | 77 (+/− 12) (p<0.0005) | 175 (+/− 42) (p<0.0001) |
| KLF4 | 150 (+/− 33) (p<0.0005) | 106 (+/− 9) (ns) | 123 (+/− 24) (p<0.05) |
Percent of Control siRNA surface adhesion molecule expression due to siRNA treatment.
Comparison of the means of Corrected MFI for Control vs. Specific siRNA.
MEK5/CA HDMEC were transfected with a cocktail of either 3 MEK5- or 3 KLF4-specific siRNA (‘Specific’) or an irrellevant Control siRNA. siRNA-treated cells were stimulated with 2 ng/ml human TNF for 24 hours and cells were immunostained with FITC-conjugated, antigen-specific monoclonal antibodies and analyzed by flow cytometry. Results are presented as percent adhesion molecule expression following siRNA treatment compared to an irrelevant, ‘Control’ siRNA (“% Control siRNA”) and are calculated as follows: ((Corrected MFI(Specific siRNA)/Corrected MFI (Control siRNA))*100), +/− SD.. Statistical comparison of the means of corrected MFI for Control vs. Specific siRNA was performed using Ratio Paired t-test. Values represent aggregate data from 6 or more trials for each condition
Role of KLF4 in Altered Gene Expression in MEK5/CA HDMECs
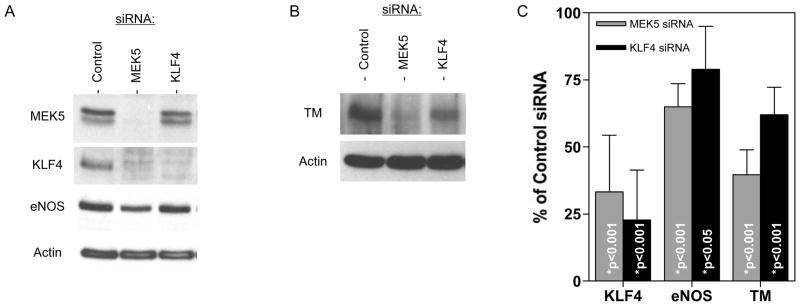
To determine the extent to which KLF4 was a mediator of MEK5-induced responses in HDMECs, we compared the effects of MEK5 and KLF4 knockdown using siRNA and Western blot analysis (Figures 6A & 6B). Knockdown of MEK5 protein (including MEK5/CA) in MEK5/CA cells resulted in a 67% reduction of KLF4 expression (Figures 6A & 6C) and a 35% reduction of eNOS (Figures 6A & 6C) as determined by densitometric analysis of band intensity. Knockdown of KLF4 was more effective using a KLF4-specific siRNA and this treatment also led to a reduction of eNOS, but not to the same extent as did MEK5 knockdown, 21% vs 35% inhibition, respectively (Figure 6C). We also observed a decrease in TM expression following either MEK5 or KLF4 knockdown (Figure 6B), but MEK5 knockdown had a more pronounced negative effect on TM expression than did KLF4, 60.3% vs. 38%, respectively (Figure 6C).
Figure 6. Effect of MEK5 and KLF4 Knockdown on Gene Expression in MEK5/CA.
siRNA were used to knock down MEK5 and KLF4 expression in MEK5/CA HDMEC. Western blot analysis was performed to measure the effect on A) KLF4 and eNOS and B) TM protein levels. C) For quantitation purposes, band intensity from independent experiments was measured using ImageJ software and the mean +/− SD of the % protein remaining relative to Control siRNA was determined from n=4, 5, and 4 trials for KLF4, eNOS, and TM, respectively. One-way ANOVA and Bonferroni post-tests were used to assess statistical significance relative to Control.
We next examined the role of KLF-4 in the modulation of TNF responses in HDMECs. Using FACS analysis to quantify adhesion molecule expression, we found that TNF-induced E-selectin was increased by 50% and VCAM-1 by 23% following KLF4 knockdown (Table 4), confirming a role for KLF4 in E-selectin and VCAM-1 suppression in microvascular ECs. ICAM-1 levels did not decrease following KLF4 knockdown in the same manner as they did following MEK5 ablation, suggesting that KLF4 was not involved in this part of the response. VCAM-1 expression, previously shown to be suppressed in MEK5/CA overexpressing cells by Western blotting (data not shown), was strongly induced following MEK5 knockdown in TNF-treated MEK5/CA. KLF4 knockdown also resulted in higher VCAM-1 expression, but again, to a lesser extent than for MEK5 knockdown (Table 4). Taken together, these results suggest a role for KLF4 in regulating (suppressing) E-selectin and VCAM-1 in TNF-treated HDMECs and inducing eNOS and TM in MEK5/CA cells. However, the greater overall effect of MEK5 knockdown than of KLF4 knockdown on eNOS, TM, and VCAM-1, further suggests that additional elements other than KLF4 contribute to the regulation of these important EC regulatory molecules in MEK5/CA transduced cells.
Role of MEK5 /CA Overexpression on TNF-Induced Vascular Leak
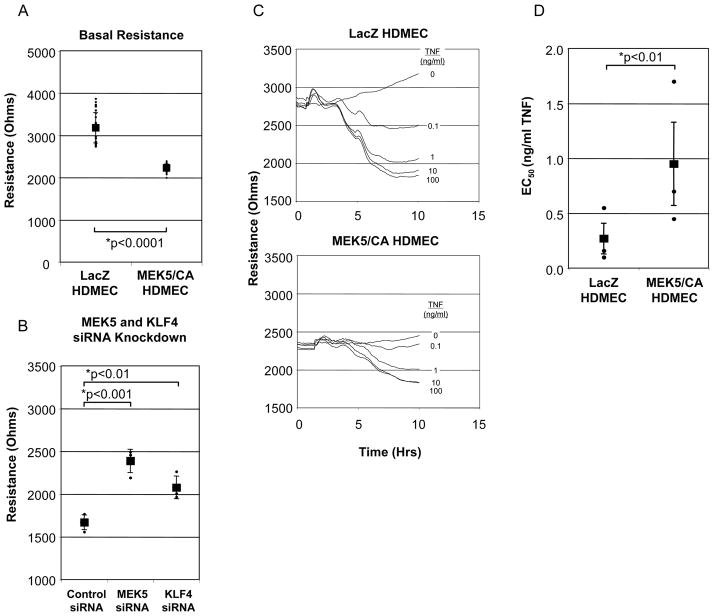
A critical function of the microvasculature is to control permselectivity across capillaries and to permit macromolecule extravasation across venules at sites of inflammation. In a final series of experiments, we examined the effects of MEK5 overexpression on EC barrier formation and its regulation by TNF. We compared the basal levels of monolayer electrical resistance, a measure of barrier integrity, in both MEK5/CA and LacZ HDMEC monolayers. Analysis of 3 separate cell lines revealed that MEK5/CA cells consistently produced less basal barrier resistance (2234 +/− 89 Ohms (measured at 4000 Hz)) when compared to LacZ HDMECs (3179 +/− 356 Ohms) (Figure 7A). The level of basal barrier resistance was significantly increased by either MEK5 or KLF4 knockdown using MEK5- or KLF4-specific siRNA in MEK5/CA HDMEC monolayers, although MEK5 knockdown had a somewhat larger effect (Figure 7B). Neither siRNA knockdown of MEK5 or KLF4 had any significant effect on basal barrier resistance in LacZ control HDMEC (data not shown). These results were consistent whether a control siRNA or an Oligofectamine (lipid) alone (without siRNA) comparison was used (data not shown). These results suggest that KLF4, alone or in combination with MEK5 signaling, plays a key role in regulating basal barrier function in microvascular EC.
Figure 7. Role of MEK5/CA on Barrier Function and TNF-induced Vascular Leak.
A) LacZ and MEK5/CA HDMEC were cultured on fibronectin coated, 8-well ECIS chamber slides. Basal resistance of LacZ and MEK5/CA HDMECs were measured at maximal resistance. Individual points and Mean (+/− Std Dev.) presented. B) siRNA reversal of MEK5/CA barrier reduction. MEK5/CA cells were transfected with either Control, MEK5-, or KLF4-specific siRNA. Electrical resistance was monitored until maximum resistance was achieved in the Control group. Data are from one of two independent experiments with similar results and are presented as mean +/− SD; Statistical significance was determined by ANOVA. C) LacZ and MEK5/CA HDMEC were cultured on ECIS chamber slides. Electrical resistance (Ohms), was monitored to determine point of maximal monolayer resistance. Increasing doses of TNF (0.1, 1, 10, and 100 ng/ml) were added in triplicate to culture wells and electrical resistance was measured for a period of 10 hours. Curves represent means of triplicate wells. Error bars were excluded for clarity. D) Results from (C) were used to calculate EC50. The untreated control (0 ng/ml TNF) was designated as 0% vascular leak and 100 ng/ml TNF dose as 100% leak and used to calculate a maximum change in resistance. The maximum resistance difference (RD) between 0 and 100 % vascular leak was used to calculate percentage of leak for each experimental condition: [(RD (TNF dose)/RD (maximum)*100)]. Non-linear regression curve and EC50 was calculated. The results represent the means and SEM of three independent experiments. Statistical significance was determined by paired t-test.
We next compared the effects of TNF on the barrier functions of MEK5/CA and LacZ HDMECs (Figure 7C). Because basal barrier functions were different and this could affect the magnitude of the change, we determined the dose of TNF required to produce a 50% fall in resistance (Effective Concentration 50; EC50). In three separate trials, using three different transduced cell lines, control LacZ HDMECs displayed an average EC50 of 0.27 ng/ml TNF (SEM=0.14ng/ml) (Figure 7D), similar to the EC50 previously observed in untransduced HDMECs using porated membrane inserts and an Ussing chamber to measure electrical resistance [8]. In contrast, MEK5/CA HDMECs displayed an elevated mean EC50 of 0.95 ng/ml TNF (SEM=0.38ng/ml). This effect did not appear specific for TNF in that thrombin responses also showed a shift in the EC50 even though thrombin works by a different mechanism that is protein synthesis-independent.
DISCUSSION
In this report we describe an interplay between shear stress, MEK5, KLF4 and inhibition of inflammation in microvascular ECs. As previously observed in large vessel ECs [19,28,44], we find that LSS at levels between those experienced by arteries and those experienced by veins is sufficient to activate MEK5 in cultured HDMECs as shown by phosphorylation of ERK5 by Western blot (Figure 2). Staining of human skin with an antibody to pERK5 indicates that this pathway is activated in HDMECs in situ (Figure 1). A potential complication of the in situ staining is that the anti-pERK5 reagent used in this study may cross react with pERK1/2 and we cannot be certain that the signal detected in situ as immunoreactive pERK5 is truly pERK5. However, the mobility shift of ERK-5 induced by shear stress or by overexpression of MEK5/CA in cultured HDMECs, interpreted to indicate phosphorylation, can be readily distinguished from the phosphorylation of ERK1/2 by its markedly different apparent size in SDS-PAGE. Moreover, while ERK1/2 can be activated by shear stress in large vessel ECs, this effect is quite transient compared to sustained activation of ERK513. We also observe strong induction by LSS of the transcription factor KLF4 (Figure 2), and a much weaker effect on KLF2. KLF4 expression was also observed in HDMECs in situ. Prior reports using large vessel ECs have linked MEK5, KLF4 and KLF2 to inhibition of inflammatory changes, such as those induced by TNFa [10,14,18,21,25]. In the current report we have explored this question in microvascular ECs by transducing HDMECs to overexpress a constitutively active form of MEK5 (Figure 3). As expected, these cells show a phosphorylation –dependent “gel shift” of total ERK5 protein indicative of activation of ERK5. We then compared these cells to control, LacZ transductants by microarrays. Among the genes that are induced, we found prominent increases in KLF4, TM and eNOS, all of which were confirmed by qRT-PCR and Western blotting for protein (Figure 4). ICAM-1 was also increased at the mRNA and protein level. At the same time as showing increased basal expression of ICAM-1 in MEK5/CA HDMEC transductants, the same cultures show enhanced TNF-mediated induction of ICAM-1 but inhibition of TNF-induced expression of E-selectin and VCAM-1 (Figure 5). These changes are reversed upon siRNA-mediated knockdown of MEK5 and, to a lesser extent, knockdown of KLF4 (Table 4). MEK5/CA HDMECs formed a less resistant barrier to passage of electrical current, a measure of vascular leakiness, than did control transduced HDMECs. Effects on basal changes in electrical resistance were reversed by siRNA knockdown of MEK5 (Figure 7A). Importantly, MEK5/CA HDMECs showed a blunted response to TNF that was measured as a shift in the EC50 in response to TNF (Figure 7D).
KLF2 mRNA was increased and readily detectable in MEK5/CA HDMEC, although expression levels were consistently lower than KLF4 (Figure 4). Furthermore, KLF2 protein was significantly more difficult to detect than KLF4, requiring an additional step of amplification. A different anti-KLF2 antibody from a second source completely failed to detect this protein in HDMEC (unpublished results, PC). These observations suggest that KLF4 serves as the dominant shear-responsive KLF family member in microvascular EC.
A central aspect of our report is its focus on microvascular EC. Shear stress has largely been considered as a modulator of the arterial wall where laminar or regularly oscillating shear is believed to inhibit proinflammatory responses while disturbed flow does not, contributing to the predispostion of atheromas to arise in regions of disturbed flow [2,9,11,13,40]. In fact, inflammation in the arterial tree is relatively rare and typically chronic. The intense inflammatory infiltrates and vascular leak that are the hallmarks of acute inflammation develop principally in the microvasculature, especially around the post-capillary venules [1,32]. The ECs that line these segments are those that are most prone to express TNF-induced E-selectin and VCAM-1 and to allow plasma proteins to extravasate [24,29,33]. ECs lining the microvasculature are also exposed to LSS, and calculations suggest that shear is highest in the arterioles, intermediate in the capillaries and lowest in the venules [26,35]. It is appealing to think that differences in shear, mediated through a MEK5/ERK5/KLF4 pathway may contribute to the differential responses of these ECs to TNF.
The mechanism(s) by which shear force activates MEK5 is not known. MEK5 is a MAP kinase kinase and is thought to be downstream of MEKK2 or MEKK3 [7]. Developmentally, knock out of MEKK3, MEK5 or ERK5 all give a similar phenotype, characterized by vascular instability and embryonic death [15,46]. This phenotype is also mimicked by knock out of the ERK5 activated transcription factor MEF2C [4]. It is not clear if KLF4 or KLF2 levels are reduced in such knock out cells or if they contribute to the phenotype.
We used transduced HDMECs and gene expression profiling to determine more fully the effects of MEK5 on gene expression in microvascular ECs. A survey of 47,000 human gene transcripts resulted in the identification of 498 induced and 68 repressed validated transcripts in MEK5/CA HDMECs, which suggests that MEK5 transduction affects cell function primarily by gene transactivation (as opposed to repression) since 88% of all “valid” transcripts were induced rather than repressed. We are able to compare the results of our microarray data to a number of microarray studies that have explored the genetic changes in ECs due to arterial levels of shear stress. Indeed, we have identified a number of transcripts that are induced both in MEK5/CA HDMECs and in HUVECs subjected to arterial levels of shear stress based on the reports of McCormick et al and Dekker et al. [11,23]. Along with TM, KLF2 and KLF4, CytochromeP450 1B1 (Accession NM_000104), TEK (TIE-2) (NM_000459.3), Argininosuccinate synthase (ASS) (NM_000050), Plasminogen activator inhibitor type 1 (PAI-1) (NM_000602), and RNase A Family, 1 (RNASE1) (NM_002933) are all up-regulated by either shear stress or MEK5 activation. Tissue Factor (TF/F3) (NM_001993) is down-regulated in both cases. There are also some notable differences. In large vessel ECs in culture, KLF2 is strongly induced by arterial levels of shear force [11,36] and KLF4 appears to be somewhat less responsive. The ratio appears reversed in HDMECs. KLF2 overexpression, like MEK5 overexpression, increases basal levels of ICAM-1 and blunts TNF induction of E-selectin and VCAM-1 [21,36]. The mechanism of this effect has been attributed to inhibition of the activation of AP-1 by SAPK/JNK [5,12] or to competition of activated transcription factors, such as NF-kB or AP-1, for recrutiment of histone acetylases like CBP or p300 [18,36]. Our recent study using chromatin immunoprecipitation more strongly supports the latter interpretation [21]. Our current experiments using siRNA suggest that KLF4 has a similar effect on adhesion molecule expression in microvascular ECs and therefore may act through a similar mechanism(s). KLF4 may directly repress gene expression, but it has not been shown to directly bind to the promotor of either E-selectin or VCAM-1. The failure of our KLF4 knockdown to completely phenocopy our MEK5 knockdown (Table 4 and Figure 6) may suggest some functional redundancy of KLF proteins in this regard. While uncertainty persists in how KLF proteins antagonize TNF induction of adhesion molecules, the role of KLF4 regarding TM and eNOS appears to be more straightforward. In this instance, KLF4 is a transcriptional activator of these two genes [14] while TNF acts to post-transcriptionally destabilize their mRNAs [27,45]. The net effect is that KLF4 antagonizes inflammation and thrombosis while TNF promotes these two responses.
Recently, Villareal et al. [41] published a microarray gene transcription profiling study in which a constitutively active form of MEK5 was overexpressed in cultured human umbilical vein EC using an adenovirus expression system. Comparisons of our two sets of data reveal that many genes are affected by MEK5/CA whether overexpressed in micro- or large vessel EC, including KLF4 and TM and e-NOS. However, these authors also see a consistent increase in KLF2 and do not observe effects on ICAM-1. These differences may reflect intrinsic differences between micro- and large vessel ECs, but also may relate to differences in the manner in which MEK5/CA is introduced; typically, retroviral transduction leads to lower, more physiological levels of expression that adenoviral transduction. In addition, the report by Villarreal et al suggests that KLF4 induction by chemical stimulation with resveratrol can be MEK5-dependent, but ERK5-independent. Our experiments with ERK5 siRNA show an obligatory role for this enzyme in the coupling of MEK5 to KLF4 induction in HDMEC.
Vascular leak is an essential component of the inflammatory response, creating a provisional matrix within the inflamed tissue that can support the migration of extravasated leukocytes [32,33]. Leak initially starts in the post-capillary venules and spreads at later time to the capillaries. Both TNF and thrombin can induce leakiness. The effect of thrombin is transient, resolving after 10–15 minutes, and is mediated by a Rho kinase pathway that stabilizes myosin light chain phsophorylation [22]. Although some investigators have reported similar actions by TNF in HUVECs [42,43], our experience with cultured HDMECs has instead shown a slower response that requires NF-κB-dependent new protein synthesis [3] and that appears independent of Rho kinase (MSK, unpublished observations). It is interesting that this TNF effect can also be inhibited by MEK5. This does not simply appear to be a result of stabilizing the cell junctions in some non-specific fashion in that basal leakiness is actually higher in MEK5/CA. Both MEK5 and KLF4 appear to play a specific role in regulating this basal barrier function (Figure 7B).
In summary, we have shown that microvascular ECs respond to shear stress by activating MEK5 and then this response leads to KLF4 induction. The effects of MEK5 activation in microvascular ECs are similar but not identical to those reported previously for large vessel ECs.
Acknowledgments
Supported by a grant from the Yale-Boehringer Ingelheim Pharmaceuticals, Inc. Research Alliance and from the National Institutes of Health (NIH) grant RO1-HL036003 to J.S.P.). Isolation and culture of HDMECs is supported by the Yale Skin Diseases Research Center (NIH grant P30-AR041942).
Footnotes
Disclosures: PRC, TJ, MSK, and JSP have no financial conflict of interest; AH and ZQ are employed by Boehringer-Ingelheim Pharmaceuticals Inc. and MM and RJT were employed by Boehringer-Ingelheim Pharamceuticals Inc. when this study was performed.
References
- 1.Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 2.Berk BC. Atheroprotective signaling mechanisms activated by steady laminar flow in endothelial cells. Circulation. 2008;117:1082–1089. doi: 10.1161/CIRCULATIONAHA.107.720730. [DOI] [PubMed] [Google Scholar]
- 3.Bevilacqua MP, Pober JS, Mendrick DL, Cotran RS, Gimbrone MA., Jr Identification of an inducible endothelial-leukocyte adhesion molecule. Proc Natl Acad Sci U S A. 1987;84:9238–9242. doi: 10.1073/pnas.84.24.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi W, Drake CJ, Schwarz JJ. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev Biol. 1999;211:255–267. doi: 10.1006/dbio.1999.9307. [DOI] [PubMed] [Google Scholar]
- 5.Boon RA, Fledderus JO, Volger OL, van Wanrooij EJ, Pardali E, Weesie F, Kuiper J, Pannekoek H, ten Dijke P, Horrevoets AJ. KLF2 suppresses TGF-beta signaling in endothelium through induction of Smad7 and inhibition of AP-1. Arterioscler Thromb Vasc Biol. 2007;27:532–539. doi: 10.1161/01.ATV.0000256466.65450.ce. [DOI] [PubMed] [Google Scholar]
- 6.Braverman IMYA. Ultrastructure of the humandermal microcirculation. II. The capillary loops of the dermal papillae. J Invest Dermatol. 1977;68:44–52. doi: 10.1111/1523-1747.ep12485165. [DOI] [PubMed] [Google Scholar]
- 7.Chao TH, Hayashi M, Tapping RI, Kato Y, Lee JD. MEKK3 directly regulates MEK5 activity as part of the big mitogen-activated protein kinase 1 (BMK1) signaling pathway. J Biol Chem. 1999;274:36035–36038. doi: 10.1074/jbc.274.51.36035. [DOI] [PubMed] [Google Scholar]
- 8.Clark PR, Manes TD, Pober JS, Kluger MS. Increased ICAM-1 expression causes endothelial cell leakiness, cytoskeletal reorganization and junctional alterations. J Invest Dermatol. 2007;127:762–774. doi: 10.1038/sj.jid.5700670. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham KS, Gotlieb AI. The role of shear stress in the pathogenesis of atherosclerosis. Lab Invest. 2005;85:9–23. doi: 10.1038/labinvest.3700215. [DOI] [PubMed] [Google Scholar]
- 10.Dekker RJ, Boon RA, Rondaij MG, Kragt A, Volger OL, Elderkamp YW, Meijers JC, Voorberg J, Pannekoek H, Horrevoets AJ. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]
- 11.Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 12.Fledderus JO, van Thiene JV, Boon RA, Dekker RJ, Rohlena J, Volger OL, Bijnens AP, Daemen MJ, Kuiper J, van Berkel TJ, Pannekoek H, Horrevoets AJ. Prolonged shear stress and KLF2 suppress constitutive proinflammatory transcription through inhibition of ATF2. Blood. 2007;109:4249–4257. doi: 10.1182/blood-2006-07-036020. [DOI] [PubMed] [Google Scholar]
- 13.Gimbrone MA, Jr, Topper JN, Nagel T, Anderson KR, Garcia-Cardena G. Endothelial dysfunction, hemodynamic forces, and atherogenesis. Ann N Y Acad Sci. 2000;902:230–239. doi: 10.1111/j.1749-6632.2000.tb06318.x. discussion 239–240. [DOI] [PubMed] [Google Scholar]
- 14.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi M, Lee JD. Role of the BMK1/ERK5 signaling pathway: lessons from knockout mice. J Mol Med. 2004;82:800–808. doi: 10.1007/s00109-004-0602-8. [DOI] [PubMed] [Google Scholar]
- 16.Kato Y, Kravchenko VV, Tapping RI, Han J, Ulevitch RJ, Lee JD. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. Embo J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kluger MS, Johnson DR, Pober JS. Mechanism of sustained E-selectin expression in cultured human dermal microvascular endothelial cells. J Immunol. 1997;158:887–896. [PubMed] [Google Scholar]
- 18.Kumar A, Lin Z, SenBanerjee S, Jain MK. Tumor necrosis factor alpha-mediated reduction of KLF2 is due to inhibition of MEF2 by NF-kappaB and histone deacetylases. Mol Cell Biol. 2005;25:5893–5903. doi: 10.1128/MCB.25.14.5893-5903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Tatake RJ, Natarajan K, Taba Y, Garin G, Tai C, Leung E, Surapisitchat J, Yoshizumi M, Yan C, Abe J, Berk BC. Fluid shear stress inhibits TNF-mediated JNK activation via MEK5-BMK1 in endothelial cells. Biochem Biophys Res Commun. 2008;370:159–163. doi: 10.1016/j.bbrc.2008.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Z, Hamik A, Jain R, Kumar A, Jain MK. Kruppel-like factor 2 inhibits protease activated receptor-1 expression and thrombin-mediated endothelial activation. Arterioscler Thromb Vasc Biol. 2006;26:1185–1189. doi: 10.1161/01.ATV.0000215638.53414.99. [DOI] [PubMed] [Google Scholar]
- 21.Liu M, Kluger MS, D’Alessio A, Garcia-Cardena G, Pober JS. Regulation of arterial-venous differences in tumor necrosis factor responsiveness of endothelial cells by anatomic context. Am J Pathol. 2008;172:1088–1099. doi: 10.2353/ajpath.2008.070603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martorell L, Martinez-Gonzalez J, Rodriguez C, Gentile M, Calvayrac O, Badimon L. Thrombin and protease-activated receptors (PARs) in atherothrombosis. Thromb Haemost. 2008;99:305–315. doi: 10.1160/TH07-08-0481. [DOI] [PubMed] [Google Scholar]
- 23.McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, Chittur KK. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci U S A. 2001;98:8955–8960. doi: 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messadi DV, Pober JS, Fiers W, Gimbrone MA, Jr, Murphy GF. Induction of an activation antigen on postcapillary venular endothelium in human skin organ culture. J Immunol. 1987;139:1557–1562. [PubMed] [Google Scholar]
- 25.Methe H, Balcells M, Alegret Mdel C, Santacana M, Molins B, Hamik A, Jain MK, Edelman ER. Vascular bed origin dictates flow pattern regulation of endothelial adhesion molecule expression. Am J Physiol Heart Circ Physiol. 2007;292:H2167–2175. doi: 10.1152/ajpheart.00403.2006. [DOI] [PubMed] [Google Scholar]
- 26.Nagaoka T, Yoshida A. Noninvasive evaluation of wall shear stress on retinal microcirculation in humans. Invest Ophthalmol Vis Sci. 2006;47:1113–1119. doi: 10.1167/iovs.05-0218. [DOI] [PubMed] [Google Scholar]
- 27.Nan B, Lin P, Lumsden AB, Yao Q, Chen C. Effects of TNF-alpha and curcumin on the expression of thrombomodulin and endothelial protein C receptor in human endothelial cells. Thromb Res. 2005;115:417–426. doi: 10.1016/j.thromres.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA, Jr, Garcia-Cardena G. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petzelbauer P, Bender JR, Wilson J, Pober JS. Heterogeneity of dermal microvascular endothelial cell antigen expression and cytokine responsiveness in situ and in cell culture. J Immunol. 1993;151:5062–5072. [PubMed] [Google Scholar]
- 30.Petzelbauer P, Pober JS, Keh A, Braverman IM. Inducibility and expression of microvascular endothelial adhesion molecules in lesional, perilesional, and uninvolved skin of psoriatic patients. J Invest Dermatol. 1994;103:300–305. doi: 10.1111/1523-1747.ep12394720. [DOI] [PubMed] [Google Scholar]
- 31.Pober JS. Endothelial activation: intracellular signaling pathways. Arthritis Res. 2002;4(Suppl 3):S109–116. doi: 10.1186/ar576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pober JS. Physiology and Pathobiology of Microvascular Endothelium. In: Tuma RF, Duran WN, Ley K, editors. Handbook of Physiology: Microcirculation. 2. Amsterdam: Acedemic Press; 2008. pp. 37–55. [Google Scholar]
- 33.Pober JS, Cotran RS. Immunologic interactions of T lymphocytes with vascular endothelium. Adv Immunol. 1991;50:261–302. doi: 10.1016/s0065-2776(08)60827-5. [DOI] [PubMed] [Google Scholar]
- 34.Pober JS, Min W. Endothelial cell dysfunction, injury and death. Handb Exp Pharmacol. 2006:135–156. doi: 10.1007/3-540-36028-x_5. [DOI] [PubMed] [Google Scholar]
- 35.Pries ARSTW. Blood Flow in Microvascular Networks. In: Tuma RF, Duran WN, Ley K, editors. Handbook of Physiology: Microcirculation. Amsterdam: Acedemic Press; 2008. pp. 3–36. [Google Scholar]
- 36.SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA, Jr, Garcia-Cardena G, Jain MK. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sohn SJ, Li D, Lee LK, Winoto A. Transcriptional regulation of tissue-specific genes by the ERK5 mitogen-activated protein kinase. Mol Cell Biol. 2005;25:8553–8566. doi: 10.1128/MCB.25.19.8553-8566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatake RJ, O’Neill MM, Kennedy CA, Wayne AL, Jakes S, Wu D, Kugler SZ, Jr, Kashem MA, Kaplita P, Snow RJ. Identification of pharmacological inhibitors of the MEK5/ERK5 pathway. Biochem Biophys Res Commun. 2008;377:120–125. doi: 10.1016/j.bbrc.2008.09.087. [DOI] [PubMed] [Google Scholar]
- 39.Tiruppathi C, Malik AB, Del Vecchio PJ, Keese CR, Giaever I. Electrical method for detection of endothelial cell shape change in real time: assessment of endothelial barrier function. Proc Natl Acad Sci U S A. 1992;89:7919–7923. doi: 10.1073/pnas.89.17.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traub O, Berk BC. Laminar shear stress: mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler Thromb Vasc Biol. 1998;18:677–685. doi: 10.1161/01.atv.18.5.677. [DOI] [PubMed] [Google Scholar]
- 41.Villarreal G, Zhang Y, Larman HB, Garcia-Sancho J, Koo A, Garcia-Cardena G. Defining the regulation of KLF4 expression and its downstream transcriptional targets in vascular endothelial cells. Biochemical and Biophysical Research Communications. 2009;391:984–989. doi: 10.1016/j.bbrc.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wojciak-Stothard B, Entwistle A, Garg R, Ridley AJ. Regulation of TNF-alpha-induced reorganization of the actin cytoskeleton and cell-cell junctions by Rho, Rac, and Cdc42 in human endothelial cells. J Cell Physiol. 1998;176:150–165. doi: 10.1002/(SICI)1097-4652(199807)176:1<150::AID-JCP17>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 43.Wojciak-Stothard B, Williams L, Ridley AJ. Monocyte adhesion and spreading on human endothelial cells is dependent on Rho-regulated receptor clustering. J Cell Biol. 1999;145:1293–1307. doi: 10.1083/jcb.145.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan C, Takahashi M, Okuda M, Lee JD, Berk BC. Fluid shear stress stimulates big mitogen-activated protein kinase 1 (BMK1) activity in endothelial cells. Dependence on tyrosine kinases and intracellular calcium. J Biol Chem. 1999;274:143–150. doi: 10.1074/jbc.274.1.143. [DOI] [PubMed] [Google Scholar]
- 45.Yan G, You B, Chen SP, Liao JK, Sun J. Tumor necrosis factor-alpha downregulates endothelial nitric oxide synthase mRNA stability via translation elongation factor 1-alpha 1. Circ Res. 2008;103:591–597. doi: 10.1161/CIRCRESAHA.108.173963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Boerm M, McCarty M, Bucana C, Fidler IJ, Zhuang Y, Su B. Mekk3 is essential for early embryonic cardiovascular development. Nat Genet. 2000;24:309–313. doi: 10.1038/73550. [DOI] [PubMed] [Google Scholar]
- 47.Yoshizumi M, Abe J, Tsuchiya K, Berk BC, Tamaki T. Stress and vascular responses: atheroprotective effect of laminar fluid shear stress in endothelial cells: possible role of mitogen-activated protein kinases. J Pharmacol Sci. 2003;91:172–176. doi: 10.1254/jphs.91.172. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Park Y, Wu J, Chen X, Lee S, Yang J, Dellsperger KC, Zhang C. Role of TNF-alpha in vascular dysfunction. Clin Sci (Lond) 2009;116:219–230. doi: 10.1042/CS20080196. [DOI] [PMC free article] [PubMed] [Google Scholar]