Abstract
Small molecule inhibitors of protein kinases are widely used in signal transduction research and are emerging as a major class of drugs. Although interpretation of biological results obtained with these reagents critically depends on their selectivity, efficient methods for proteome-wide assessment of kinase inhibitor selectivity have not yet been reported. Here, we address this important issue and describe a method for identifying targets of the widely used p38 kinase inhibitor SB 203580. Immobilization of a suitable SB 203580 analogue and thoroughly optimized biochemical conditions for affinity chromatography permitted the dramatic enrichment and identification of several previously unknown protein kinase targets of SB 203580. In vitro kinase assays showed that cyclin G-associated kinase (GAK) and CK1 were almost as potently inhibited as p38α whereas RICK [Rip-like interacting caspase-like apoptosis-regulatory protein (CLARP) kinase/Rip2/CARDIAK] was even more sensitive to inhibition by SB 203580. The cellular kinase activity of RICK, a known signal transducer of inflammatory responses, was already inhibited by submicromolar concentrations of SB 203580 in intact cells. Therefore, our results warrant a reevaluation of the vast amount of data obtained with SB 203580 and might have significant implications on the development of p38 inhibitors as antiinflammatory drugs. Based on the procedures described here, efficient affinity purification techniques can be developed for other protein kinase inhibitors, providing crucial information about their cellular modes of action.
Protein kinases are key regulators of cellular signaling and therefore represent attractive targets for therapeutic intervention in a variety of human diseases (1). Various small molecule inhibitors for target-selective inhibition of diseaserelevant protein kinases are currently in different stages of clinical testing, and the first drugs of this class have already received FDA approval (2, 3). Most of these inhibitors interact with the relatively conserved ATP binding site and are therefore likely to target several protein kinases, even when assumed to be highly specific based on currently available data. Because the knowledge about an inhibitor's true selectivity is a prerequisite for the correct interpretation of its biological effects, efficient methods to determine the cellular targets of protein kinase inhibitors are of central importance for both signal transduction research and drug development. Inhibitor selectivity is usually assessed in parallel enzymatic assays for a set of recombinant protein kinases (4, 5). Because even the largest of these currently available selectivity panels comprise <100 members of the protein kinase family, the great majority of the >500 human protein kinases are not tested, and, moreover, alternative protein targets such as different types of enzymes are not analyzed (6). Thus, efficient methods for proteome-wide assessment of kinase inhibitor selectivity are needed. Classical affinity chromatography employing immobilized protein kinase inhibitors has been occasionally used to address this important issue (7, 8). In terms of sensitivity and efficiency, limitations of these previously described affinity purifications become apparent when the results are seen in comparison with published data from selectivity panels comprising only ≈30 human protein kinases (5). However, because of the power of affinity chromatography combined with new advances in protein identification, a substantial optimization of the affinity approach is urgently required to gain new insights into the cellular modes of action of kinase inhibitors.
Antiinflammatory drugs, such as SB 203580, that belong to the pyridinyl imidazole class of compounds were originally recognized as protein kinase inhibitors when the mitogen-activated protein kinase p38 was identified as their major cellular target (9, 10). SB 203580 is deemed to be relatively specific for p38 with respect to protein kinase inhibition because it did not significantly inhibit a variety of other kinases in vitro (4). In addition to p38, SB 203580 potently inhibits hepatic cytochrome P450 enzymes and was further shown to affect cyclooxygenase and thromboxane synthase activities in vitro, although at higher concentrations than p38 kinase (11, 12). But these described side effects did not impair the use of this pharmacological inhibitor in studies resulting in >1,000 published articles to characterize and implicate p38 function in cellular signaling and a plethora of biological processes. Despite the widespread application of SB 203580, its true selectivity for p38 has not yet been assessed on a proteome-wide scale.
Here, we describe the immobilization of a suitable analogue of SB 203580 on chromatography beads. With this reagent, we establish an efficient proteomics method for the identification of cellular targets affected by a protein kinase inhibitor. To achieve this goal, various biochemical parameters had to be thoroughly optimized. Importantly, by using these conditions, we not only identify several previously unknown SB 203580 targets of high significance but also establish a robust procedure of general utility for the analysis of kinase inhibitor selectivity.
Materials and Methods
Reagents and Plasmids. Cell culture media and Lipofectamine were purchased from Invitrogen. Radiochemicals and epoxy-activated Sepharose 6B were from Amersham Biosciences. SB 203580 and histone H1 were from Merck. GST-activating transcription factor 2 (ATF2) was obtained from Upstate Biotechnology (Lake Placid, NY). All other reagents were from Sigma.
Antibodies used were rabbit polyclonal anti-p38 antibody (Cell Signaling Technology, Beverly, MA), polyclonal anti-Rip-like interacting caspase-like apoptosis-regulatory protein (CLARP) kinase (RICK) antibody (ABR), mouse monoclonal anti-glycogen synthase kinase 3α/β (GSK3α/β) antibody, goat polyclonal anti-CK1α and anti-CK1ε antibodies, rabbit polyclonal anti-c-jun N-terminal kinase (JNK) antibody (all from Santa Cruz Biotechnology), and mouse monoclonal anti-vesicular stomatitis virus G protein (VSV-G) antibody (Roche). Recombinant protein kinases purchased were human p38α, human JNK1α1, human JNK2α2 (Upstate Biotechnology), and rat CK1δ (New England Biolabs).
A partial cDNA encoding amino acids 24–646 of cyclin G-associated kinase (GAK) was PCR-amplified from human lung cDNA and inserted into vector pcDNA3 (Invitrogen) modified to attach a C-terminal VSV-G epitope (13, 14). The GAK sequence encoding amino acids 26 to 392 was cloned into pGEX-4T1 for expression of recombinant GST fusion protein in Escherichia coli.
The full-length RICK coding sequence fused to a C-terminal hemagglutinin epitope tag was cloned into pPM7 expression vector (13, 15). Kinase-inactive K47R and inhibitor-insensitive T95M mutants were generated by using a mutagenesis kit (Stratagene). Plasmids pPM7-RICK-dCst and pPM7-RICK-KRdCst express the first 353-aa residues of wild-type or kinase-inactive RICK fused to a C-terminal streptag epitope. The expression cassette from pPM7-RICK-dCst was inserted into an adenovirus genome by recombination in bacteria (13). Recombinant RICK enzyme production using adenovirus-directed expression is described elsewhere (16).
Compound Synthesis and Immobilization. Pyridinyl imidazole (PI) 51 and PI 51peg were prepared by and purchased from Evotec-OAI (Abingdon, Oxon, U.K.). PI 51 was synthesized as described (17). PI 51peg was prepared by dissolving 0.59 mM PI 51, 0.59 mM diisopropylamine, and 0.59 mM 1-bromo-2-[2-(2-ethoxy-ethoxy)-ethoxy]-ethane in 10 ml of 1,4-dioxane, followed by refluxing the mixture for 16 h until complete consumption of PI 51. The mixture was cooled to room temperature, and the solvent was removed under vacuum. The residue was purified by column chromatography [silica gel; gradient dichlorometane/methanol (95:5) to dichlorometane/methanol/25% aq. ammonia (90:10:1)].
For immobilization, drained epoxy-activated Sepharose 6B was resuspended in 2 vol of 20 mM PI 51 dissolved in 50% dimethylformamide (DMF)/0.1 M Na2CO3. After adding of 10 mM NaOH, coupling was performed overnight at 30°C in the dark. After three washes with 50% DMF/0.1 M Na2CO3, remaining reactive groups were blocked with 1 M ethanolamine. Subsequent washing steps were performed according to the manufacturer's instructions. To generate the control matrix, epoxy-activated Sepharose 6B was incubated with 1 M ethanol-amine and equally treated as described above. The beads were stored at 4°C in the dark.
Cell Culture and Transfections. COS-7 and HeLa cells were cultured in DMEM supplemented with 10% FBS. COS-7 cells were transiently transfected as described (13). On the second day after transfection, cells were either lysed or phosphate-starved for a further 2 h in phosphate-free medium containing 10% dialysed FBS. Cells were then treated with inhibitor for 15 min and subsequently metabolically labeled with 70 μCi (1 Ci = 37 GBq) [32P]orthophosphate per ml for 30 min before cell lysis.
Cell Lysis and in Vitro Association Experiments. HeLa cells or transfected COS-7 cells were lysed in buffer containing 50 mM Hepes (pH 7.5), 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM EDTA, 10 mM sodium pyrophosphate plus additives (10 mM sodium fluoride/1 mM orthovanadate/10 μg/ml aprotinin/10 μg/ml leupeptin/1 mM phenylmethylsulfonyl fluoride/0.2 mM DTT). Lysates were precleared by centrifugation and equilibrated to 1 M NaCl for in vitro association experiments. Twenty-five microliters of drained PI 51 matrix or control matrix was incubated with 250 μl of high-salt lysate for 3 h at 4°C. Optionally, 2 mM free PI 51 was added to the lysate. After washing with 500 μl of 2× lysis buffer without additives containing 1 M NaCl (high salt) and with 500 μl of 1× lysis buffer without additives containing 150 mM NaCl (low salt), the beads were eluted with 1.5× SDS sample buffer. To test different elution conditions for bound p38, beads were incubated in 100 μl of low salt lysis buffer supplemented with 1 mM PI 51 or 10 mM ATP/20 mM MgCl2 as indicated. For precipitation of strep-tagged proteins, 250 μl of lysate containing 150 mM NaCl was incubated with StrepTactin-MacroPrep beads (IBA, Göttingen, Germany) for 3 h at 4°C. Beads were then washed three times with the same buffer without additives. After SDS/PAGE, proteins were transferred to nitrocellulose membrane and immunoblotted with the indicated antibodies. Radioactively labeled RICK-KRdC was visualized by autoradiography before detection with StrepTactin-horseradish peroxidase (IBA).
Affinity Chromatography and Preparative Gel Electrophoresis. Frozen HeLa cells (2.5 × 109, 4C Biotech, Seneffe, Belgium) were lysed in 30 ml of buffer containing 20 mM Hepes (pH 7.5), 150 mM NaCl, 0.25% Triton X-100, 1 mM EDTA, 1 mM EGTA, 1 mM DTT plus additives (10 mM sodium fluoride/1 mM orthovanadate/10 μg/ml aprotinin/10 μg/ml leupeptin/1 mM phenylmethylsulfonylfluoride/10% glycerol), cleared by centrifugation, and adjusted to 1 M NaCl. The filtrated lysate was loaded with a flow rate of 100 μl/min on an HR 5/2 chromatography column (Amersham Biosciences) containing 600 μl of PI 51 matrix equilibrated to lysis buffer without additives containing 1 M NaCl. The column was washed with 15 column volumes and equilibrated to lysis buffer without additives containing 150 mM NaCl, and bound proteins were eluted in the same buffer containing 1 mM PI 51, 10 mM ATP, and 20 mM MgCl2 with a flow rate of 50 μl/min. The volume of protein-containing elution fractions was reduced to 1/10 in a Centrivap concentrator (Labonco, Kansas City, MO) before precipitation according to Wessel and Flügge (18). Precipitated proteins were dissolved in 16-benzyldimethyl-n-hexadecylammonium chloride (16-BAC) sample buffer and after reduction/alkylation separated by two-dimensional 16-BAC/SDS/PAGE (13). Coomassie-stained spots were picked and subjected to analysis by mass spectrometry.
Mass Spectrometry. Picked samples were destained in 30% ethanol/10% acetic acid overnight. Destained samples were washed twice in 0.1 M ammonium bicarbonate (NH4HCO3) and reduced with 10 mM DTT in 0.1 M NH4HCO3 for 30 min at 56°C. Samples were then dehydrated with acetonitrile, rehydrated and alkylated with 55 mM iodoacetamide in 0.1 M NH4HCO3 for 30 min in the dark, and washed twice with 0.1 M NH4HCO3. Dried samples were reswollen in trypsin (Promega) solution containing 50 mM NH4HCO3/10% acetonitrile and digested overnight at 37°C. Peptides were washed out once with 50 mM NH4HCO3 and twice with 5% formic acid. Guanidination for matrix-assisted laser desorption ionization (MALDI) mass mapping was performed as described (19). Sample cleanup was performed with ZipTips by using the manufacturer's procedures (Millipore).
MALDI spectra were acquired by using a Bruker (Billerica, MA) Ultraflex time-of-flight (TOF)/TOF mass spectrometer with LIFT technology and anchor chip targets. Data analysis was performed by using Bruker's Biotools and the mascot program. Searches were done against the National Center for Biotechnology Information database.
In Vitro Kinase Assays. Kinase reactions were performed for either 10 min (p38α, JNK1, JNK2, and CK1δ) or 30 min (RICK and GAK) at 30°C in a total volume of 50 μl. All kinases were assayed in 50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 0.1 mM EGTA, 100 μM ATP, and 1 μCi [γ-32P]ATP in the presence of indicated SB 203580 concentrations. In addition, JNK1 and JNK2 assays were performed in the presence of 2 μM ATP. Kinase substrate proteins included were 0.4 mg/ml myelin basic protein (p38α and RICK), 0.4 mg/ml casein (CK1δ), 0.2 mg/ml histone H1 (GAK), and 0.1 mg/ml GST-activating transcription factor 2 (ATF2) (JNK1 and JNK2). Reactions were stopped by addition of 3× SDS sample buffer. After gel electrophoresis, phosphorylated substrate proteins were visualized by autoradiography and quantified by phosphorimaging. Determination of IC50 (0–100%) values was performed by using grafit (Erithacus, Horley, Surrey, U.K.).
Results
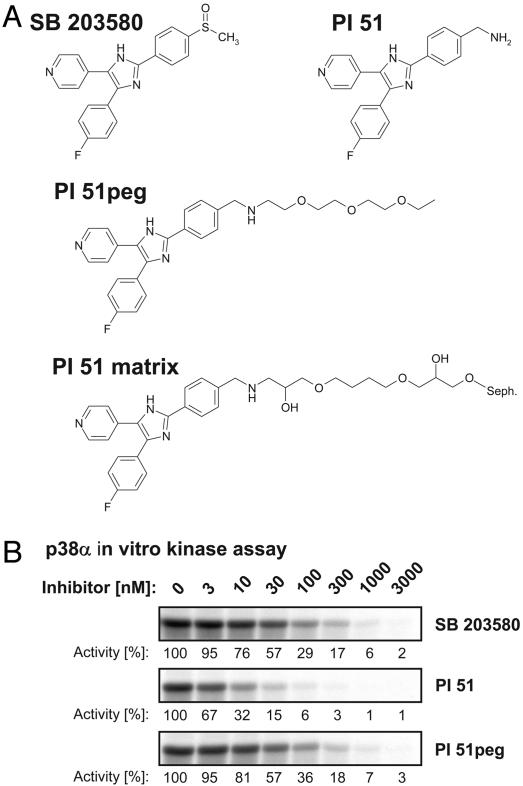
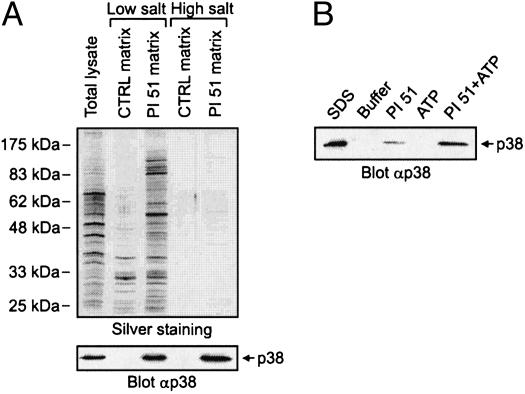
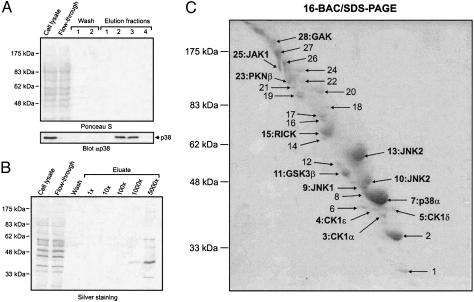
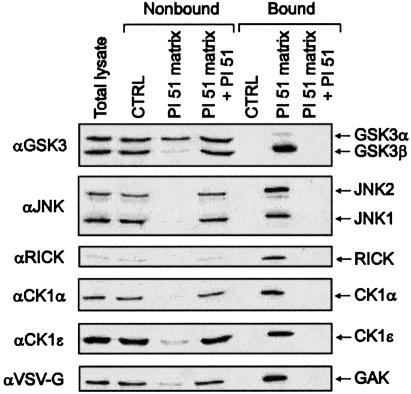
The proteome-wide selectivity of SB 203580 has not been investigated yet, an analysis which might be achieved with affinity matrices possessing essentially the same binding properties as soluble SB 203580 itself. The crystal structure of p38 in complex with SB 203580 shows exposure of the inhibitor's sulfoxide moiety at the protein surface, suggesting a suitable site for the attachment of linkers extending from solid support materials (20). For the purpose of immobilization, we selected a closely related derivative of SB 203580, possessing a primary methylamine function instead of the sulfoxide moiety at the accessible position. This p38 inhibitor has previously been described as compound 51 of a series of pyridinyl imidazoles by Gallagher et al. (17) and is therefore referred to as PI 51 in this study (Fig. 1A). PI 51 was pegylated to yield PI 51peg, resulting in a structure similar to PI 51 covalently coupled to epoxy-activated Sepharose (Fig. 1 A). SB 203580, PI 51, and PI 51peg were then tested in kinase assays using recombinant p38α as enzyme and myelin basic protein (MBP) as substrate (Fig. 1B). In agreement with published data, SB 203580 and PI 51 inhibited p38 activity with IC50 values of ≈40 nM and 4 nM, respectively (4, 17). Importantly, extension of PI 51 by pegylation reduced its inhibitory effect on p38 by one order of magnitude, but, even so, PI 51peg was as effective in inhibiting p38 as SB 203580 (Fig. 1B). We therefore concluded that the interaction of PI 51 with p38 should be retained after immobilization of PI 51 on epoxy-activated Sepharose to generate the PI 51 matrix (Fig. 1 A). This assumption could be confirmed by the in vitro association experiment shown in Fig. 2A, in which p38 from HeLa total cell lysate specifically bound to the PI 51 matrix, but not to control beads devoid of covalently coupled PI 51. However, when the batch purification was performed at a physiological ionic strength of 150 mM NaCl, parallel silver staining showed a variety of bound proteins, indicating only moderate p38 enrichment. Testing of different buffer compositions led to the identification of high salt concentrations of 1 M NaCl as critical for efficient retention and separation of p38 from the vast majority of cellular proteins (Fig. 2 A). Furthermore, elution under nondenaturing conditions turned out to be far more efficient when both PI 51 and ATP were included in the elution buffer rather than either component alone (Fig. 2B). Employing these optimized adsorption and elution conditions, we established an affinity chromatography method on PI 51 matrix-containing columns by using total cell extracts from 2.5 × 109 HeLa cells as starting material. As shown by immunoblotting, the mitogen-activated protein kinase p38 quantitatively bound to the PI 51 matrix whereas the general protein pattern detectable in the initial extract reappeared in the flow-through. After extensive washing, p38 was efficiently eluted in the presence of both ATP and PI 51 (Fig. 3A Lower). Remarkably, at this level of sensitivity, no protein bands could be visualized in the elution fraction (Fig. 3 A Upper), demonstrating a substantial enrichment, which was at least 5,000-fold as shown by comparison of sample and flow-through with increasing aliquots of the fraction containing the proteins specifically eluted from the column (Fig. 3B). Silver staining revealed several bands, indicating the purification of several other proteins in addition to p38 on the PI 51 affinity matrix. To identify these proteins, they were first resolved by preparative gel electrophoresis in the presence of the cationic detergent 16-benzyldimethyl-n-hexadecylammonium chloride (16-BAC) followed by standard SDS/PAGE in the second dimension (13). Coomassie staining of 16-BAC/SDS gels visualized ≈30 protein spots, which were excised and analyzed by mass spectrometry (Fig. 3C). In addition to p38, which was represented by one of the most prominent spots, a variety of additional protein kinases could be identified. These included JNK isoforms and GSK3β, kinases known to be affected by micromolar concentrations of SB 203580 (4, 21). Strikingly, in addition to these known interactors of SB 203580, we identified a set of protein kinases that had not been implicated as targets of p38 inhibitors before. These included RICK, different CK1 isoforms, GAK, protein kinase Nβ (PKNβ) and Janus kinase 1 (JAK1). Most of the other species were identified as relatively abundant proteins, and their presence could be accounted for by their abundance in the extract combined with a weak affinity for the PI 51 matrix (Table 1, which is published as supporting information on the PNAS web site). To confirm the mass spectrometry results, total lysates from HeLa cells or COS-7 cells expressing an epitope-tagged GAK (24–646) fragment were subjected to in vitro association with either control beads or PI 51 beads with or without free PI 51 present in the lysate. Both unbound proteins from the supernatants and proteins retained on the beads were immunoblotted with specific antibodies for JNK, GSK3, RICK, CK1α, CK1ε, or for the epitope tag of GAK (24–646). All endogenous kinases from HeLa cells and the truncated GAK specifically interacted with the PI 51 matrix, thus verifying the identifications by mass spectrometry (Fig. 4). Interestingly, GSK3 binding was specific for the β isoform, with no significant interaction observed for GSK3α.
Fig. 1.
Identification of a p38 inhibitor analogue suitable for immobilization. (A) Chemical structure of SB 203580 compared with PI 51 in its free, pegylated, and immobilized form. (B) In vitro kinase reactions were performed with recombinant p38α enzyme by using MBP as a substrate. The pyridinyl imidazoles SB 203580, PI 51, and PI 51peg were added at the indicated concentrations. After SDS/PAGE, MBP phosphorylation was detected by autoradiography and quantified by phosphorimaging. Kinase activity in the absence of inhibitor was set to 100%, and remaining activities at different inhibitor concentrations are expressed relative to this value.
Fig. 2.
Optimization of adsorption and elution conditions for a functional p38 inhibitor matrix. (A) Total lysates from HeLa cells were subjected to in vitro associations with either control or PI 51 matrix. Incubations were performed in the presence of either low or high NaCl concentrations of 150 mM or 1 M, respectively. Relative to the total cell lysate, 100× aliquots of the bound protein fractions were resolved by gel electrophoresis for subsequent silver staining (Upper), and, in parallel, 5× aliquots were analyzed by immunoblotting with p38-specific antibody (Lower). (B) Cellular p38 bound to PI 51 beads was eluted with lysis buffer supplemented with 1 mM PI 51 or 10 mM ATP/20 mM MgCl2 as indicated. For comparison, quantitative p38 elution was performed with SDS sample buffer. After gel electrophoresis, eluted p38 was detected by immunoblotting.
Fig. 3.
Efficient affinity purification of protein kinases specifically targeted by immobilized p38 inhibitor. HeLa whole cell lysate was subjected to PI 51 affinity chromatography, and the bound proteins were eluted with a combination of ATP and free PI 51. (A) After gel electrophoresis and transfer onto nitrocellulose membrane, cell lysate, flow-through, wash, and elution fractions were analyzed by Ponceau S staining and subsequent immunoblotting using specific anti-p38 antibody. (B) Increasing aliquots of pooled fractions containing protein specifically eluted from the PI 51 column were visualized in comparison with cell lysate, flow-through, and wash fractions. The silver-stained SDS/polyacrylamide gel demonstrates at least 5,000-fold enrichment of retained proteins. (C) The large remainder of the proteins purified by affinity chromatography on the PI 51 matrix was separated by 16-BAC/SDS/PAGE and stained with Coomassie blue. The indicated spots (arrows) were analyzed by mass spectrometry. Spots containing identified protein kinases are marked by their names. See a full list of identified proteins in Table 1.
Fig. 4.
Confirmation of results from mass spectrometry analysis. In vitro association of total lysates from HeLa cells or COS-7 cells expressing truncated GAK (24–646) protein fused to a VSV-G epitope with either control matrix or PI 51 matrix. Free PI 51 was present in the lysate where indicated. Both nonbound proteins from the supernatants and bound proteins eluted from the matrix were immunoblotted with specific antibodies for GSK3α/β, JNK, RICK, CK1α, and CK1ε and for the epitope tag of GAK (24–646)-VSV-G.
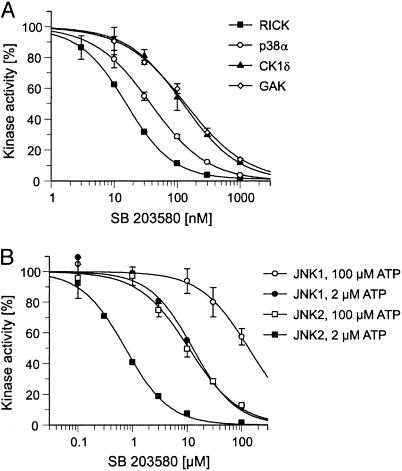
Association with the PI 51 matrix did not provide a quantitative measure of the inhibitory potency of pyridinyl imidazoles such as SB 203580 toward the specifically bound protein kinases. To test how binding might translate into inhibitor sensitivity, we analyzed the effect of SB 203580 on the in vitro kinase activities of p38α, RICK, GAK, and CK1δ in the presence of 100 μM cold ATP, a concentration that allows comparisons of these results with previously published data (4). SB 203580 inhibited recombinant p38α in our assays with an IC50 value of 38 nM, in good agreement with published data (Fig. 5A) (4). Strikingly, RICK was even more potently inhibited by SB 203580 than p38α, with an IC50 value of only 16 nM. Moreover, the inhibitor concentrations required to inhibit CK1δ and GAK kinase activities by 50% were only ≈3-fold higher than that determined for p38α. Thus, their respective IC50 values were still significantly below the 500 nM reported for the less sensitive p38β isoform (Fig. 5A) (4). Although the specificity of SB 203580 has been examined against >25 protein kinases previously, the most potently inhibited kinase other than p38 was c-Raf, with an IC50 value of 2 μM (4, 21–23). Thus, SB 203580 was previously deemed to be relatively specific for p38. Our data demonstrate, in stark contrast, that low concentrations of SB 203580 inhibit several protein kinases. These results establish the proteomics approach presented here as a powerful tool to investigate the selectivity of protein kinase inhibitors.
Fig. 5.
In vitro characterization of protein kinases inhibited by SB 203580. (A) The concentration-dependent inhibition of p38α, RICK, CK1δ, and GAK by SB 203580 was determined as described in Materials and Methods. Kinase activities in the absence of inhibitor were set to 100%, and remaining activities at different SB 203580 concentrations are expressed relative to this value. (B) Effect of different SB 203580 concentrations on the activities of JNK1 and JNK2 in the presence of 2 or 100 μM ATP.
Surprisingly, JNK1 was also efficiently retained by the PI 51 matrix although this kinase is not inhibited by 10 μM SB 203580 in vitro (4, 21). To resolve this issue, we performed kinase assays with both JNK1 and JNK2 at either 100 μM ATP, the standard concentration used, or at only 2 μM ATP, which resembles the conditions during affinity purification under which cell-derived Mg2+-ATP cannot compete for binding due to the presence of EDTA as a chelating reagent. As shown in Fig. 5B, the SB 203580 concentrations required for half maximal kinase inhibition significantly dropped from >100 μM to 13 μM for JNK1 and from 11 μM to 0.7 μM for JNK2 on reduction of the ATP concentration to 2 μM, thereby providing a plausible explanation for the specific binding to the PI 51 matrix observed.
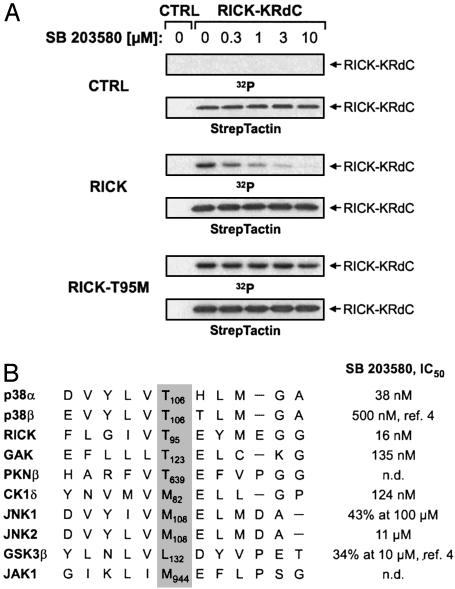
Finally, we chose the highly SB 203580-sensitive serine/threonine kinase RICK to analyze how the enzymatic activity of one of the new inhibitor targets is affected on SB 203580 treatment in intact cells. As a cellular kinase substrate, we expressed a kinase-inactive fragment of RICK (RICK-KRdC), which was heavily phosphorylated on cotransfection of the catalytically active, full-length kinase (Fig. 6A). RICK-mediated substrate phosphorylation was inhibited by SB 203580 in a dose-dependent manner, with 0.3 μM compound already conferring ≈50% inhibition. Interestingly, RICK possesses a conserved threonine residue equivalent to Thr-106 of p38, which is critical for inhibitor binding and was shown to render p38 resistant to SB 203580 when mutated to a larger amino acid (20, 23). This structural determinant is equally important for RICK because mutation of the corresponding Thr-95 to methionine rendered RICK resistant to SB 203580 at all concentrations tested, further demonstrating that the inhibitor directly affected RICK kinase activity in intact cells (Fig. 6A). With the notable exception of CK1, the critical threonine residue is conserved in all kinases found to be potently inhibited by SB 203580 although ≈75% of all protein kinases possess a larger hydrophobic residue in this position (Fig. 6B).
Fig. 6.
Structural determinants of SB 203580 sensitivity. (A) Effect of SB 203580 on cellular RICK kinase activity. COS-7 cells were transiently transfected with pPM7-RICK-KRdCst expression plasmid (1.3 μg per well) plus either control vector or pPM7 plasmids encoding RICK or RICK-T95M (0.2 μg per well). After metabolic labeling with [32P]orthophosphate and precipitation using StrepTactin beads, radioactively labeled RICK-KRdC was detected by autoradiography (blots 1, 3, and 5) before immunodetection with StrepTactin-horseradish peroxidase (blots 2, 4, and 6). (B) Amino acid residues surrounding Thr-106 in p38 were aligned with the corresponding sequences of various protein kinases binding to the PI 51 matrix. With the exception of CK1δ, all kinases possessing a larger amino acid residue at the indicated position (gray background) show significantly higher IC50 values than those having a threonine residue at this site.
Discussion
Given the lack of selectivity of SB203580 for p38 kinase as revealed by our experimental approach, incorrect conclusions might have been drawn from numerous experiments in which this particular compound was used as a “specific” p38 inhibitor. The pyridinyl imidazoles were initially characterized as inhibitors of lipopolysaccharide (LPS)-induced cytokine release (9). But, remarkably, recent genetic evidence implicated RICK as critical regulator of these inflammatory responses and further revealed a role of RICK upstream of p38 activation in LPS-triggered signaling in macrophages (24, 25). Reduced p38 activation might account for the decreased cytokine production observed in RICK-deficient cells, consistent with results showing that genetic ablation of the p38 downstream mediator mitogen-activated protein kinase-activated protein kinase 2 (MAPKAP kinase 2) also interferes with LPS-induced synthesis of several cytokines (26).
Functional characterization of the cellular SB 203580 targets should be feasible employing drug-resistant protein kinase mutants for in vivo validation of inhibitor specificity. Previous data by Cohen and colleagues (27) showed that some cellular effects of SB 203580 are indeed mediated by p38 whereas others were not. Because RICK acquires SB 203580 resistance by the corresponding threonine to methionine substitution, as previously shown for p38, the same experimental strategy might be useful to study cellular SB 203580 effects not related to p38 function and possibly mediated through RICK kinase. It is also noteworthy that overexpression of inhibitor-resistant p38 kinase was recently shown to revert some inhibitory effects of SB 203580 on lipopolysaccharide-induced cytokine production, which were in part overlapping with those reductions in cytokine release observed in RICK-deficient cells (25, 28). Although these results seem to be conflicting, they could mean that overexpression of a drug-resistant p38 also compensates for the inhibition of the second SB 203580 target, RICK, lying in the same pathway. Obviously, further experimental work is required to address these issues. But, based on the evidence already available, it is likely that the combined inhibition of both p38 and RICK activities by antiinflammatory drugs such as SB 203580 determine the physiological outcome, a conclusion of potential significance for drug development in this area.
In addition, parallel screening of the SB 203580-sensitive protein kinases identified in this study might lead to the identification of more selective pyridinyl imidazole derivatives and therefore better inhibitors for cell-based assays. Furthermore, initial results with protein kinase inhibitors structurally unrelated to the pyridinyl imidazoles show that the affinity purification methods established here for an SB 203580 derivative work equally well with other compound scaffolds and should therefore be of general utility to define the selectivity and molecular mode of action of small-molecule kinase inhibitors.
Supplementary Material
Acknowledgments
We thank Katrin Herzberger and Kerstin Stegmüller for excellent technical assistance and Dirk Brehmer for help with the figures. This work was supported by a grant from the German Bundesministerium für Bildung und Forschung.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VSV-G, vesicular stomatitis virus G protein; JNK, c-jun N-terminal kinase; GAK, cyclin G-associated kinase; PI, pyridinyl imidazole; GSK, glycogen synthase kinase; RICK, rip-like interacting caspase-like apoptosis-regulatory protein (CLARP) kinase.
References
- 1.Cohen, P. (2002) Nat. Rev. Drug Discov. 1, 309–315. [DOI] [PubMed] [Google Scholar]
- 2.Capdeville, R., Buchdunger, E., Zimmermann, J. & Matter, A. (2002) Nat. Rev. Drug Discov. 1, 493–502. [DOI] [PubMed] [Google Scholar]
- 3.Muhsin, M., Graham, J. & Kirkpatrick, P. (2003) Nat. Rev. Drug Discov. 2, 515–516. [DOI] [PubMed] [Google Scholar]
- 4.Davies, S. P., Reddy, H., Caivano, M. & Cohen, P. (2000) Biochem. J. 351, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bain, J., McLauchlan, H., Elliott, M. & Cohen, P. (2003) Biochem. J. 371, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. (2002) Science 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- 7.Knockaert, M., Gray, N., Damiens, E., Chang, Y.-T., Grellier, P., Grant, K., Fergusson, D., Mottram, J., Soete, M., Dubremetz, K.-F., et al. (2000) Chem. Biol. 7, 411–422. [DOI] [PubMed] [Google Scholar]
- 8.Knockaert, M., Wieking, K., Schmitt, S., Leost, M., Grant, K. M., Mottram, J. C., Kunick, C. & Meijer, L. (2002) J. Biol. Chem. 277, 25493–25501. [DOI] [PubMed] [Google Scholar]
- 9.Lee, J. C., Laydon, J. T., McDonell, P. C., Gallagher, T. F., Kumar, S., Green, D., McNulty, D., Blumenthal, M. J., Heys, J. R., Landvatter, S. W., et al. (1994) Nature 372, 739–746. [DOI] [PubMed] [Google Scholar]
- 10.Cuenda, A., Rouse, J., Doza, Y. N., Meier, R., Cohen, P., Gallagher, T. F., Young, P. R. & Lee, J. C. (1995) FEBS Lett. 364, 229–233. [DOI] [PubMed] [Google Scholar]
- 11.Adams, J. L., Boehm, J. C., Kassis, S., Goeycki, P. D., Webb, W. F., Hall, R., Sorenson, M., Lee, J. C., Ayrton, A., Griswold, D. E., et al. (1998) Bioorg. Med. Chem. Lett. 8, 3111–3116. [DOI] [PubMed] [Google Scholar]
- 12.Börsch-Haubold, A. G., Pasquett, S. & Watson, S. P. (1998) J. Biol. Chem. 273, 28766–28772. [DOI] [PubMed] [Google Scholar]
- 13.Daub, H., Blencke, S., Habenberger, P., Kurtenbach, A., Dennenmoser, J., Wissing, J., Ullrich, A. & Cotten, M. (2002) J. Virol. 76, 8124–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura, S. H., Tsuruga, H., Yabuta, N., Endo, Y. & Nojima, H. (1997) Genomics 44, 179–187. [DOI] [PubMed] [Google Scholar]
- 15.Inohara, N., del Peso, L., Koseki, T., Chen, S. & Nunez, G. (1998) J. Biol. Chem. 273, 12296–12300. [DOI] [PubMed] [Google Scholar]
- 16.Cotten, M., Stegmüller, K., Eickhoff, J., Hanke, M., Herzberger, K., Herget, T., Choidas, A., Daub, H. & Godl, K. (2003) Nucleic Acids Res. 31, e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher, T. F., Seibel, G. L., Kassis, S., Laydon, J. T., Blumenthal, M. J., Lee, J. C., Lee, D., Boehm, J. C., Fier-Thompson, S. M., Abt, J. W., et al. (1997) Bioorg. Med. Chem. 5, 49–64. [DOI] [PubMed] [Google Scholar]
- 18.Wessel, D. & Flügge, U. I. (1984) Anal. Biochem. 138, 141–143. [DOI] [PubMed] [Google Scholar]
- 19.Beardsley, R. L. & Reilly, J. P. (2002) Anal. Chem. 74, 1884–1890. [DOI] [PubMed] [Google Scholar]
- 20.Tong, L., Pav, S., White, D. M., Rogers, S., Crane, K. M., Cywin, C. L., Brown, M. L. & Pargellis, C. A. (1997) Nat. Struct. Biol. 4, 311–316. [DOI] [PubMed] [Google Scholar]
- 21.Whitmarsh, A. J., Yang, S.-H., Su, M. S., Sharrocks, A. D. & Davis, R. J. (1997) Mol. Cell. Biol. 17, 2360–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall-Jackson, C. A., Goedert, M., Hedge, P. & Cohen, P. (1999) Oncogene 18, 2047–2054. [DOI] [PubMed] [Google Scholar]
- 23.Eyers, P. A., Craxton, M., Morrice, N., Cohen, P. & Goedert, M. (1998) Chem. Biol. 5, 321–328. [DOI] [PubMed] [Google Scholar]
- 24.Chin, A. I., Dempsey, P. W., Bruhn, K., Miller J. F., Xu, Y. & Cheng, G. (2002) Nature 416, 190–194. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, K., Inohara, N., Hernandez, L. D., Galan, J. E., Nunez, G., Janeway, C. A., Medzhitov, R. & Flavell, R. A. (2002) Nature 416, 194–199. [DOI] [PubMed] [Google Scholar]
- 26.Kotlyarov, A., Neininger, A., Schubert, C., Eckert, R., Birchmeier, C., Volk, H.-D. & Gaestel, M. (1999) Nat. Cell Biol. 1, 94–97. [DOI] [PubMed] [Google Scholar]
- 27.Eyers, P. A., van den Ijssel, P., Quinlan, R. A., Goedert, M. & Cohen, P. (1999) FEBS Lett. 451, 191–196. [DOI] [PubMed] [Google Scholar]
- 28.Guo, X., Gerl, R. E. & Schrader, J. W. (2003) J. Biol. Chem. 278, 22237–22242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.