Abstract
We have improved upon the methodology and dramatically shortened the time required for accurate assembly of 5- to 6-kb segments of DNA from synthetic oligonucleotides. As a test of this methodology, we have established conditions for the rapid (14-day) assembly of the complete infectious genome of bacteriophage φX174 (5,386 bp) from a single pool of chemically synthesized oligonucleotides. The procedure involves three key steps: (i) gel purification of pooled oligonucleotides to reduce contamination with molecules of incorrect chain length, (ii) ligation of the oligonucleotides under stringent annealing conditions (55°C) to select against annealing of molecules with incorrect sequences, and (iii) assembly of ligation products into full-length genomes by polymerase cycling assembly, a nonexponential reaction in which each terminal oligonucleotide can be extended only once to produce a full-length molecule. We observed a discrete band of full-length assemblies upon gel analysis of the polymerase cycling assembly product, without any PCR amplification. PCR amplification was then used to obtain larger amounts of pure full-length genomes for circularization and infectivity measurements. The synthetic DNA had a lower infectivity than natural DNA, indicating approximately one lethal error per 500 bp. However, fully infectious φX174 virions were recovered after electroporation into Escherichia coli. Sequence analysis of several infectious isolates verified the accuracy of these synthetic genomes. One such isolate had exactly the intended sequence. We propose to assemble larger genomes by joining separately assembled 5- to 6-kb segments; ≈60 such segments would be required for a minimal cellular genome.
Chemical synthesis of life in the laboratory has been a standing challenge to synthetic organic chemistry since Wöhler's synthesis of urea in 1828 (1), and the doctrine of spontaneous generation was put to rest by an address by Louis Pasteur in 1864.§ With an understanding of the genetic role of DNA, much work has focused on the synthesis of oligonucleotides and genes. The synthesis of the 207-bp gene for tyrosine suppressor tRNA in 1979 by Khorana and 17 coworkers (2) was a monumental undertaking. Since then, the automated DNA synthesizer has been developed based on fundamental advances in synthetic methods from the laboratories of Letsinger (3, 4) and Caruthers (5, 6).
In 1999 we described a minimal prokaryotic genome based on results from random whole genome transposon mutagenesis that inactivated one gene per cell (7). By using this approach, ≈300 essential genes for self-replicating cellular life were described, and we proposed to make a synthetic chromosome to test the viability of this hypothesis (7). Before attempting synthesis of a microbial chromosome, we commissioned an independent bioethical review of our proposed scientific plan (8). After >1 year of deliberation, the reviewers concluded that we were taking a reasonable scientific approach to an important biological question. The broader implications of the creation of life in the laboratory can now be considered a realistic possibility. However, there are several technical barriers to the synthesis of microbial chromosome-sized stretches of DNA that are hundreds of thousands to millions of nucleotides long, the most notable being the contamination of the oligonucleotides by truncated species. Although such oligonucleotides are highly useful as primers for PCR amplification and DNA sequencing, only small (a few hundred base pairs) synthetic genes can generally be accurately and directly synthesized without multiple repair/selection steps. For example, the recent report (9) of the assembly of a partially active poliovirus from cloned synthetic segments of DNA from which polio genomic RNA (7,440 bases) could be transcribed was quite complex and took many months to accomplish. First, segments 400–600 bp long were individually assembled and cloned, and 5–15 isolates of each were sequenced to find one that was correct or readily correctable by oligonucleotide mutagenesis. These segments were then assembled into three larger segments of the polio genome, recloned, and finally assembled to produce a full-length product. This slow process would not be practical for synthesizing a 300,000-bp chromosome. We have now improved the methodology for synthesis of multigene segments of a genome as a step toward synthesis of a cellular genome. As a test of this methodology we have established conditions for global assembly of the infectious genome of bacteriophage φX174 (5,386 bp) from a single pool of chemically synthesized oligonucleotides. φX174 presents no known hazard because it infects only certain enteric bacteria and is not a human, plant, or animal pathogen. Therefore, its choice for synthesis serves to separate safety issues from ethical considerations and other potential risks associated with synthetic genomics.
φX174 (φX) is the prototypical minute icosahedral bacteriophage that was first characterized in the Sinsheimer laboratory beginning in the 1950s. The φX virion contains a circular single-stranded DNA that replicates through a double-stranded replicative form of DNA (RF). Its chromosome was the first DNA molecule purified to homogeneity (10), and, consequently, φX DNA has been used in many landmark experiments. It was the first viral DNA shown to be infectious in 1961, and in 1967 Goulian, Kornberg, and Sinsheimer (11) demonstrated that φX DNA synthesized with DNA polymerase, using the intact genome as a template, was infectious. This feat was hailed as “life in the test tube” (12). The φX174 genome was also the first DNA completely sequenced by Sanger et al. in 1978 (13). The sequence describes a remarkably compact gene organization with several cases of genes expressed by translating overlapping regions of the DNA in two different reading frames. Thus, φX provides a historic and demanding test bed for accurate synthetic chromosomes.
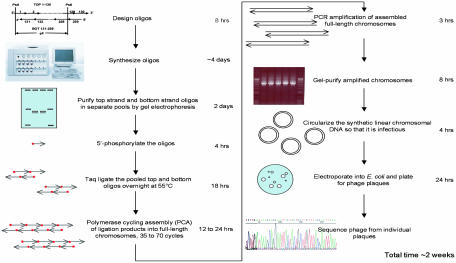
We set out to develop a procedure by which any 5- to 6-kb segment of DNA can be quickly and easily assembled from a single pool of synthetic oligonucleotides with high fidelity. In 1995, Stemmer et al. (14) used a polymerase cycling reaction to assemble a 2.7-kb plasmid from a single pool of oligonucleotides that was able to replicate in Escherichia coli. Synthesis of the larger φX genome, however, provides a more stringent test for optimizing oligonucleotide assembly methods. Its compact genetic organization makes it relatively intolerant to mutation compared with a plasmid, which may require only a replication origin and a drug resistance marker. We report here a procedure (Fig. 1) that utilizes sequential ligation and polymerase cycling reactions to accomplish the assembly of the φX genome 1 order of magnitude more rapidly than with previous methods (9) and with great savings in the sequence analysis of intermediate products and in human effort. This method should make it possible to assemble a minimal microbial genome from as few as 60 synthetic 5- to 6-kb segments.
Fig. 1.
Schematic diagram of the steps in the global synthesis of infectious φX174 bacteriophage from synthetic oligonucleotides.
Materials and Methods
Synthetic Oligonucleotides. Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) in 96-well format and normalized to 100 μM each. Based on the accompanying mass spectrometer data and information from the supplier, we ascertained that, on average, each 42-mer contained ≈50% truncated species. To purify the oligonucleotides, 10 μl of each of the top-strand (or bottom-strand) oligonucleotides were pooled in two separate pools (top-1 to -130 and bot-131 to -259), dried, and dissolved in 50 μl of water. Twenty microliters of concentrated pool plus 20 μl of formamide was heated to 95°C for 2 min, and 10-μl aliquots were loaded into 1.5-mm × 15-mm slots of a preparative 12% sequencing gel and electrophoresed at 1,300 V for ≈4 h. The bands, which migrated close to the xylene cyanol marker, were visualized with a handheld 254-nm UV lamp and excised. The gel was extruded through a tuberculin syringe into 0.7 ml of TE buffer, frozen at –20°C overnight, eluted at 37°C on a rotating wheel for 1 h, and filtered through a glass, wool-plugged, 1-ml Eppendorf pipette tip. The recovered oligonucleotides were ethanol-precipitated and dissolved in 50 μl of water.
Phosphorylation of Oligonucleotides. The oligonucleotides were phosphorylated before ligation. A 100-μl reaction mixture containing 20 μl of purified top (or bottom) oligonucleotides, 10 μl of 10× T4 polynucleotide kinase buffer (NEB, Beverly, MA), 1 mM ATP, and 40 units of T4 polynucleotide kinase (NEB) was incubated at 37°C for 1 h. The reaction was terminated by phenol-chloroform extraction and ethanol precipitation. The phosphorylation reaction was repeated a second time. After extraction and precipitation, the oligonucleotides were dissolved in 50 μl of water.
Ligation Reactions. The 100-μl ligation reaction mixture contained 10 μl of top 5′P oligonucleotides, 10 μl of bottom 5′P oligonucleotides, 10 μlof10× Taq ligation buffer (NEB), and 60 μl of water. The mixture was heated to 95°C for 2 min and slow-cooled over 30 min to 55°C. Ten microliters of Taq ligase (40 units/μl, NEB) was added, and incubation was continued at 55°C for 18 h. The reaction was terminated by phenol-chloroform extraction and ethanol precipitation. The products were dissolved in 90 μl of TE buffer.
Polymerase Cycling Assembly (PCA) of Oligonucleotides. PCA was carried out in reaction mixtures (50 μl) containing 5 μl of 10× Advantage 2 buffer, 5 μl of 10× dNTP mixture, 1 μl of 50× HF polymerase mixture (Clontech Advantage HF 2 PCR Kit, catalog no. K1914-1), and 0.2, 0.5, or 1 μl of the Taq ligation product. The polymerase mixture contains both an N-terminal deletion mutant of TaqDNA polymerase that lacks 5′-exonuclease activity and Deep VentR polymerase (NEB) with 3′-exonuclease proofreading activity. Cycling parameters were 94°C for 15 sec, slow cool at –0.1°C/sec to 55°C, annealing at 55°C for 2 min, and extension at 72°C for 6 min. Thirty-five cycles were carried out followed by finishing at 72°C for 5 min.
PCR Amplification of Synthetic φX174 (synφX) DNA Molecules Produced by PCA. The 25-μl PCR mixtures contained 2.5 μl of 10× Advantage 2 buffer, 2.5 μlof10× dNTPs, 0.5 μl of HF polymerase mixture, 0.5 μl of each second-stage PCA product, and 1 μl each of 10 μM top-1 and bot-259 oligonucleotides (unpurified). PCR parameters were 94°C for 15 sec, 55°C for 30 sec, and 72°Cfor6min for 25 cycles, finishing with 72°C for 5 min. The PCR mixtures were pooled, phenol-chloroform-extracted, ethanol-precipitated, and redissolved in 10 μl of TE buffer.
Conversion of Linear synφX DNA Molecules to Infectious Circular Molecules. The pooled PCR-amplified synφX DNA was cleaved with PstI, and the linear DNA was gel-purified to yield ≈1 μg of linear synφX. The linear synφX molecules were circularized by ligation under dilute conditions (≈1 μg/ml) with T4 ligase (NEB) using recommended conditions. The ligation mixture was phenol-chloroform-extracted, ethanol-precipitated, and redissolved in 10 μl of TE/5 buffer in preparation for infectivity testing.
Assay of φX DNA Infectivity. One microliter of synφX ligation product (an estimated 3–5 ng of circular molecules based on ethidium bromide staining intensity) was electroporated into DH10B cells (Invitrogen), immediately diluted with 500 μl of SOC broth (Invitrogen), and then aliquoted into two screw-capped glass culture tubes (A and B) containing 2 ml of KC broth each (10 g of Bacto tryptone, 5 g of KCl, and 0.5 ml of 1 M CaCl2 per liter). The tubes were rotated at 37°C for ≈40min, and then 225 μl of 2 mg/ml lysozyme (Sigma) and 47.5 μl of 0.5 M EDTA (pH 8) were added, and incubation was continued on ice for 30 min. The tubes were freeze-thawed twice in dry ice-ethanol to release the synφX phage. Aliquots of the synφX-A and -B lysates were plated undiluted in 3 ml of top agar containing 0.3 ml of log phase HF4704 at 5 × 108 cells per ml on LB plates. Phage plaques were visualized after 6–18 h of incubation at 37°C.
DNA Sequencing. Plaques were picked directly into 50-μl PCRs with a 10-μl micropipette tip and subjected to 30 cycles of amplification consisting of 10 sec at 94°C, 30 sec at 55°C, and 6 min at 72°C. Fifty microliters of a mixture of 40 μl of shrimp alkaline phosphatase (1 unit/μl), 8 μl of exonuclease I (20 units/μl), and 752 μl of water was added to each PCR mixture, and digestion was at 37°C for 45 min and 45°C for 15 min, followed by 72°C for 15 min to inactivate the enzymes. The synφX DNA was gel-purified before carrying out standard sequencing reactions and sequencing on a 3730 XL sequencer (Applied Biosystems). Both strands were sequenced, and depth was generally >2-fold.
Rationale and Theoretical Considerations
The steps we used to accomplish the synthesis of infectious φX double-stranded RF DNA from a single pool of oligonucleotides are summarized in Fig. 1. In developing this strategy, we considered how impurities present in oligonucleotide preparations can result in assembly errors and mutations in the final product. Thus, we were led to purify the oligonucleotides before assembly. We also analyzed and determined the theoretical endpoints and limitations of the two basic assembly steps: ligation and PCA.
Oligonucleotide Purity. If automated DNA synthesizers produced pure oligonucleotides of the programmed sequence, then assembly of long double-stranded DNA molecules would be straightforward. In reality, only ≈50% of the molecules in preparations such as those used in our work have the correct chain length. The population of other molecules includes both truncated species capped at the growing end and uncapped molecules containing errors (mostly deletions). These incorrect molecules will either block assembly of the oligonucleotides or result in mutations in the assembled DNA. Because, on average, one of every two molecules is correct, the probability that a strand of our φX genome would completely assemble correctly by random selection from 130 unpurified oligonucleotides is (1/2)130 or ≈10–39. We estimated that, even with selection for infectivity, it was essential to reduce the number of incorrect oligonucleotides to ≤10% to allow detection of correct molecules. Because purification of each oligonucleotide would be time-consuming and laborious, and because all our synthetic oligonucleotides were of equal chain length, we chose to gel-purify pooled oligonucleotides. We pooled the nucleotides from each strand in two separate pools to minimize the likelihood of annealed structures that could interfere with gel purification on the basis of chain length.
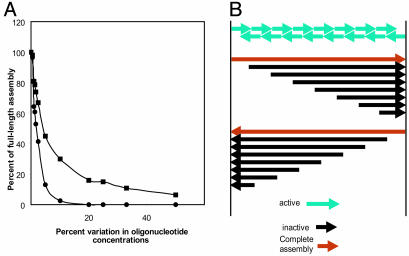
Ligation. We chose Taq ligation as the first step in our φX assembly, because ligation under stringent annealing conditions (55°C) would diminish the possibility of incorrect pairing and might also select against ligation of oligonucleotides containing mutations. In principle, it should have been possible to obtain full-length φX genomes by ligation. A major reason why this was not attained is that the concentrations of all of the oligonucleotides in the ligation mixture are not equal. Growing assemblies terminate prematurely when particular oligonucleotides become exhausted. Fig. 2A shows the results of a computer simulation of the ligation reaction. Both the fraction of full-length product and the average chain length of assemblies drop rapidly as the percentage of random variation in the oligonucleotide concentrations increases. For example, if the oligonucleotide concentrations vary by as little as 20%, essentially no full-length assemblies are made. Other contributing factors are incomplete oligonucleotide phosphorylation and sequence errors that interfere with efficient ligation. For these reasons we should not expect complete φX genomes to assemble in our ligation reactions, and subsequent assembly of the ligation products by polymerase cycling is needed.
Fig. 2.
(A) Computer simulation of the ligation reaction as a function of variation in the concentrations of the input oligonucleotides. Computer simulation parameters are 130 top oligonucleotides (with bottom oligonucleotides saturating) and 2,000 molecules of each top oligonucleotide. Percent variation of oligonucleotide concentrations is determined by the randomly chosen number of molecules of each oligonucleotide. For example, at 10% variation in oligonucleotide concentrations, the number of each oligonucleotide is randomly chosen between 1,800 and 2,000. At each iteration of the program, a random pair of assemblies is selected and the pairs are joined together to form a larger assembly if the end coordinate of one assembly is one less than the beginning of the other assembly. The process is iterated until the number of assemblies no longer changes during 1 million pairings. -▪-▪-, average percent length of assemblies; -•-•-, percent of full-length assemblies. (B) The final products of a theoretical PCA reaction that starts with oligonucleotides of uniform size. Only the two terminal oligonucleotides can be extended to full length because polymerization is only in the 5′ to 3′ direction. An assembly is considered “active” if it can be extended by overlapping with another assembly followed by fill-in synthesis. An assembly is “inactive” if it cannot be extended further. Mass increase factor = final mass/beginning mass = sn (n + 2)/2sn = (n + 2)/2 ≈ n/2. Fraction of final mass full length = [2 (sn + s/2)]/[sn (n + 2)] = [2 (n + 1/2)]/[n (n + 2)]≈ 2/n. s = nucleotide length of oligonucleotide; n = number of oligonucleotides on one strand.
PCA. The PCA reaction is a thermocycling polymerase reaction, similar to a PCR but without a pair of primers present in excess compared with template (14). At each cycle, DNA is melted and overlapping single strands reanneal. If the 3′ ends of reannealed strands are such that they can be extended by using the opposite strand as template, then polymerase extends the strands to form duplex molecules. DNA strands continue to elongate at each cycle until they either are full-length or can no longer be extended. It should be noted that PCA is not an amplification reaction. Only limited total synthesis can occur, and, because the polymerase mixture we used contains no 5′ exonuclease, no DNA is degraded. It is difficult to analyze the kinetics of the process; however, the final end products are simple to describe (Fig. 2B). Molecules present in the starting mixture can be extended in the 5′→3′ direction to the end of the genome just once, over a number of cycles. Only the molecules containing the 5′ ends of each strand can be extended to full length. The total increase in mass of the DNA is limited to ≈n/2, and the fraction of final mass that can achieve full length is ≈2/n, where n is the number of oligonucleotides on one strand (Fig. 2B). We assumed it would be unlikely that either the ligation or the PCA reactions are a major source of errors, because no synthesis occurs in the former and synthesis is limited in the latter (although some errors could occur by deamination or depurination during these steps). Most errors can be attributed to incorrect synthesis of the oligonucleotides. If our oligonucleotides are 90% pure, then each synthetic chain would, on average, contain ≈13 mutations. The fraction of correct synφX molecules would be (0.9)130 or ≈10–6.
Results
The oligonucleotides were designed to synthesize a φX genome with exactly the sequence reported by Sanger et al. in 1978 (13) [several database entries have exactly this sequence, NC_001422 (GenBank database), J02482, and V01128]. In designing the oligonucleotide set, we adopted the strategy of appending sequence to either end of the φX sequence to make a slightly larger molecule that could be cleaved to size with PstI and then circularized to produce infectious DNA. We appended 5′-TAACGCTGCA to the left end and one G plus a randomly generated 73-nucleotide sequence to the right end, thereby restoring a PstI site at each end. Starting at the left end, 130 oligonucleotides (top-1 to top-130), each 42 bases long, were consecutively generated. Similarly, starting on the bottom strand at position 22, oligonucleotides numbered bot-131 to bot-259 were generated (see Fig. 1, upper left). These oligonucleotides were pooled and gel-purified as described in Materials and Methods.
We phosphorylated the oligonucleotides and mixed the top and bottom pools. To improve the ligation fidelity we used Taq ligase at 55°C for 18 h. A sample of the ligation reaction was analyzed on an Invitrogen 2% E-gel (Fig. 3E). The average size of the double-stranded products was ≈700 bp, with a small fraction extending to 2 or 3 kb (Fig. 3E, lane N). Fig. 3E, lane D, shows a sample denatured in formamide before loading on the gel. The single-stranded products ranged in size, with the largest being ≈1 kb.
Fig. 3.
PCA of full-length synφX molecules. The first stage of PCA (50-μl reaction volume) was carried out for 35 cycles with 0.2, 0.5, or 1 μlofthe Taq ligation product. PCA products were analyzed on a 0.8% E-gel. (A) Lanes 1 and 6, 1-kb ladder; lane 2, 0.5 μlof Taq ligation product; lane 3, 2 μl of the 0.2-μl PCA; lane 4, 2 μl of the 0.5-μl PCA; lane 5, 2 μl of the 1-μl PCA. The second stage of PCA was for an additional 35 cycles in five new 50-μl reactions. For reaction 1, the 0.2-μl first-stage reaction was continued without change for another 35 cycles with the addition of 0.5 μl of fresh HF polymerase mixture. For reactions 2 and 3, 10 and 20 μl of the 0.5-μl first-stage PCA product was used. For reactions 4 and 5, 5 and 10 μl of the 1-μl first-stage PCA product was used. Analysis was on 0.8% E-gels. (B) Formamide-denatured DNA. (C) Native DNA. (B and C) Lanes 1 and 7, 1-kb ladder; lane 2, 2 μl of reaction 1; lanes 3 and 4, 2 μl of reactions 2 and 3; lanes 5 and 6, 2 μl of reactions 4 and 5. (D) PCR amplification of the products of the second set of PCA products as shown in B and C.(E) Taq ligase assembly of 259 oligonucleotides. A 0.5-μl sample of the ligation products was analyzed on a 2% E-gel (Invitrogen) in duplex form (lane N). One microliter of the ligation products was mixed with 20 μl of formamide, heated to 95°C for 2 min, and then analyzed (lane D). Denatured standards run approximately the same as native standards, based on other experiments (data not shown).
To obtain full-length φX DNA molecules, we diluted 0.2-, 0.5-, and 1-μl samples of the ligation product to 50 μl and subjected them to 35 cycles of PCA as shown in Fig. 3A. With the 0.2- and 0.5-μl samples, single-stranded assemblies approaching full length or nearly full length were produced, whereas assemblies from the 1-μl sample were shorter. PCA of ligation samples exceeding 1 μl yielded even shorter PCA products (data not shown). Apparently, dilute conditions favor the annealing of only two partially overlapping strands at a time followed by fill-in synthesis, whereas concentrated conditions favor multibranched, annealed structures that interfere with fill-in synthesis.
To improve the yield of full-length φX, diluted samples of the first stage of PCA assembly were subjected to an additional 35 cycles of PCA. A small fraction of the products was now visible as full-length single strands (Fig. 3B) and as full-length duplexes (Fig. 3C). The presence of products of size apparently greater than full-length suggests either that some incorrect assemblies had accumulated during the later PCA cycles or that branched structures might have formed by reannealing either during electrophoresis (Fig. 3B) or during the final cycle of PCA (Fig. 3C).
After viewing the intensity of the bands and realizing that only 1/25th of the PCA reaction mixture was analyzed on the gel, we estimated that several nanograms of full-length product were produced. Because 1 ng of φX RF is ≈1.7 × 108 molecules, we must have generated >109 independent synφX molecules, and we can infer that some of these would have the correct sequence.
A small sample (0.5 μl) of each second-stage PCA reaction was amplified by PCR by using top-1 and bot-259 oligonucleotides as primers (Fig. 3D). The PCRs were pooled and cleaved with PstI, and the linear DNA was gel-purified to yield ≈1 μg of linear synφX. The linear synφX molecules were circularized by ligation under dilute conditions (<1 μg/ml) in preparation for infectivity testing. Gel analysis showed that ≈50% of the linear synφX were converted to circular molecules (data not shown).
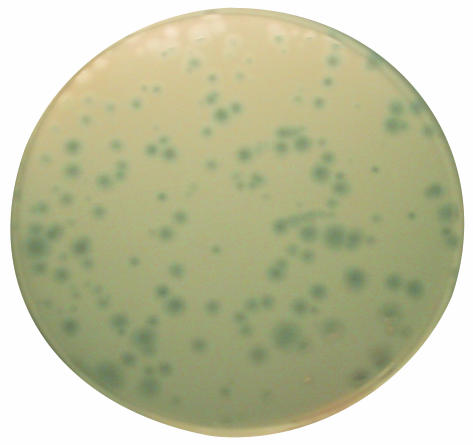
One microliter of synφX ligation product (an estimated 3–5ng of circular molecules based on ethidium bromide staining intensity) was electroporated into DH10B cells (Invitrogen), divided into two aliquots (A and B), and plated for infectious particles as described in Materials and Methods. The synφX-A lysate yielded 194 plaques (Fig. 4), and the synφX-B lysate yielded 181 plaques per 100 μl plated. Some variation in plaque size was observed. The phage yield from 3 to 5 ng of the synφX product was 2 × 10–4 of the yield from 1 ng of commercial NEB RF DNA; therefore, we estimate that the synφX product is ≈5 × 10–5 of that for natural φX, leading us to conclude that there are 9–10 inactivating mutations per synthetic genome. This is in reasonable agreement with our estimate of 90% purity for the gel-purified oligonucleotides.
Fig. 4.
Plaques of synφX-A. There appear to be several plaque morphologies: small plaques with sharp borders, medium-sized plaques, and large plaques with fuzzy borders.
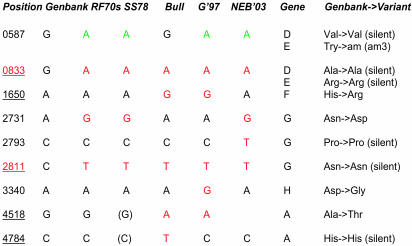
Several plaques were picked from each plate directly into PCR mixtures for amplification and sequencing. The A plaques are independent of the B plaques, but individual plaques from A, or from B, are not necessarily independent, because they may have arisen from the same infected cell. Representative sequences from four plaques were compared in Fig. 5. Phage DNA from plaque B3 gave a sequence that was identical to GenBank accession no. J02482. The B1 DNA sequence contained one silent T→C transition, two silent G→A transitions, one G→A transition at position 4170 (resulting in a Gly-to-Ser amino acid change in the gene A protein), and one T→G transversion at position 3606 (resulting in a Ser-to-Ala change in the gene H protein). DNA from plaque A4 contained a T→C silent mutation at position 1045 and A8 contained a G→A silent mutation at position 446, a C→T mutation at position 4399 (resulting in an Ala→Val amino acid change in gene A protein), and a C→A change at position 5144 (resulting in a Gln-to-Lys change in gene B and silent changes in gene A and A*). Infectious titers of synthetic phage from plaque B3 were indistinguishable from commercially available phage.
Fig. 5.
Sequence comparisons of natural φX and synφX genomes. Differences from the Sanger sequence (13) are indicated. A4, A8, B1, and B3 are the synφX described in the text. NEB, φX RF I DNA supplied by NEB (catalog no. N3021S); RF70s, DNA prepared in the late 1970s and stored since then by C.A.H.
Discussion
Synthesis of a molecule with the same properties as a naturally occurring compound has traditionally been used as evidence for correctness of a proposed molecular structure. Therefore, it is interesting to consider whether synthesis of an infectious φX genome proves the correctness of the Sanger sequence (13) on which our synthesis was based. Clearly, the sequence is accurate enough to specify an infectious phage. One synφX isolate (B3) with wild-type plaque morphology is identical in sequence to the Sanger sequence. In a study of convergent evolution using φX, Bull et al. (15) determined that the wild-type φX used in their experiments differed at five positions from the Sanger sequence (13). In the present work we have also sequenced four different preparations of natural φX. It is interesting that none of these sequences is identical to the Sanger sequence (Fig. 6), differing from it by three to five single base substitutions in addition to the single base change associated with the am3 mutation, which is present in our natural DNA preparations but absent from the Sanger sequence as presented in the GenBank database (13). SynφX-A4, A8, and B3 are consequently closer to the Sanger sequence (13) than any of the natural φX DNA preparations (10) we have sequenced. It should be noted that the commercial φX DNA present in the laboratory while synthesis was ongoing differs at five positions (including the am3 site) from the Sanger sequence. None of those differences are found in our synφX sequences, demonstrating that this isolate is synthetic rather than a contaminant. The natural sequences closest to the Sanger sequence are from DNA preparations contemporaneous with the original sequencing (Fig. 6). Our sequences were determined by using template DNA prepared in the late 1970s from φX brought back from a sabbatical in the Sanger laboratory by one of the authors (C.A.H.). It is difficult to conclude whether any of the three differences between the Sanger sequence and contemporaneous φX preparations represents sequencing errors. Only two of these differences are shared by all four of the natural φX sequences and the sequence of Bull et al. (15), and these are both silent changes. synφX-B3 is identical to the Sanger sequence (13) at both positions and is fully infectious. The quality of a synthetic genome sequence depends in part on the accuracy of the original DNA sequence from which it is derived. It is truly remarkable how well the first sequence of a DNA genome holds up to close scrutiny a quarter of a century later.
Fig. 6.
Sequence differences between the Sanger sequence and more recent sequencing of natural φX DNAs. RF70s, a preparation of φX double-stranded RF from the late 1970s; SS78, a preparation of φX virion single-stranded DNA from 1978; Bull, the sequence of wild-type φX used by Bull et al. (15); G'97, φX RF DNA from 1997; NEB'03, φX RF DNA from NEB in use at the Institute for Biological Energy Alternatives during the φX genome synthesis.
We have considered the source of mutations limiting the accuracy of assembled synthetic DNA. The predominant impurities in oligonucleotide preparations are molecules shorter than the desired product that are expected to result in deletions after assembly. In our experiments, selection for viable φX strongly selects against such frameshift mutations, and none were seen in the viable synφX isolates. The observed mutations are all single-base substitutions, predominantly transitions [changes from one pyrimidine to the other (C↔T) or from one purine to the other (A↔G)]. The two obvious sources of these substitutions are enzymatic replication errors and preexisting base substitutions in the synthetic oligonucleotide preparations. The mutation frequency expected from enzymatic errors is simply due to the 25 cycles of PCR carried out before circularization of the genome. PCA reactions extend molecules present in the original ligation mixture to full length only a single time, and so are not expected to contribute significantly to the mutation frequency. The error rate for PCR using Taq polymerase has been measured to be ≈10–5 (mutation frequency/bp/duplication), and the HF polymerase mixture used in these experiments is reported to have significantly higher fidelity. This leads us to estimate an average of approximately one PCR-induced mutation per genome. The observed rate appears to be somewhat higher, leading us to speculate about the possible origin of base substitutions in the oligonucleotide preparations. The predominant transition type observed (G.C to A.T, which results in G→A and C→T changes) could arise by deamination of C to produce U. PCR amplification would then copy U in DNA as T to produce normal DNA that is insensitive to the uracil N-glycosylase of E. coli. The phosphoramidite of U might exist as a contaminant of the C phosphoramidite used in oligonucleotide synthesis, or deamination might occur at any stage before PCR amplification of the full-length φX genome.
We have demonstrated the rapid, accurate synthesis of a large DNA molecule based only on its published genetic code. The accuracy of our final product was demonstrated by DNA sequencing and phage infectivity. Our methods will permit serial synthesis of gene cassettes containing four to seven genes in a highly robust manner. However, without selectivity, these cassettes will contain mutations (≈2 per kb) derived from errors contained in the oligonucleotide pool. Thus, without error correction, they would currently be unsuitable for assembly into a chromosome for an entire organism. Selection for infectivity, such as we have used, or for ORFs in single genes provides advantages for synthesizing viruses or short sequences. However, when our method of synthesis is coupled with DNA sequencing and repair by site-directed mutagenesis, it will enable rapid production of accurate cassettes that can be assembled into larger genomes. The capabilities of DNA synthesis have lagged far behind our ability to determine sequences during the past 30 years. If this gap can be closed, then limitless possibilities for the application of synthetic methods to the study and practical application of genomics will emerge. There are many reasons to synthesize DNA chemically, rather than clone natural sequences, one of which is to prove correctness (or incorrectness) of a sequence, because many published sequences contain errors. The natural DNA may be unavailable to the experimenter for various reasons, including an uncooperative laboratory, an environmental sample that has been used up, an archaeological sample in short supply, or a sequence from badly degraded DNA or from an extinct organism. Also, the sequence could be deduced (rather than experimental) from an ancestral sequence, a designer protein, or a fusion of domains from different proteins. There may be a hazard associated with the source of the natural sequence, or the target sequence may be RNA or a protein sequence rather than a DNA sequence. The sequence may need to be reengineered to alter the codon usage (or the code) for a particular host; to alter closely spaced regulatory signals or protein initiation (ribosome binding sites), promoters, or transcription terminators; to introduce restriction sites; or to allow convenient construction of a family of related (but different) constructs. Synthesis may become the easiest way to get a sequence as methods are refined. A desired construct may require the assembly of many pieces from different sources. Synthesis obviates the need to develop a special strategy for each construct, providing complete flexibility of design. The combination of improved oligonucleotide synthesis combined with the methods described here will enable rapid, accurate synthesis of genomes of self-replicating organisms that will serve as a basis for understanding minimal cellular life. Synthetic genomics will become commonplace and will provide the potential for a vast array of new and complex chemistries altering our approaches to production of energy, pharmaceuticals, and textiles.
Acknowledgments
This work was supported by grants from the U.S. Department of Energy and the J. Craig Venter Science Foundation.
Abbreviations: RF, replicative form of DNA; PCA, polymerase cycling assembly; synφX, synthetic φX174.
Footnotes
Pasteur, L., Sorbonne Scientific Soiree, Apr. 7, 1864, Paris.
References
- 1.Wöhler, F. (1828) Ann. Phys. Chem. 88, 253–256. [Google Scholar]
- 2.Sekiya, T., Takeya, T., Brown, E. L., Belagaje, R., Contreras, R., Fritz, H. J., Gait, M. J., Lees, R. G., Ryan, M. J., Khorana, H. G., et al. (1979) J. Biol. Chem. 254, 5787–5801. [PubMed] [Google Scholar]
- 3.Letsinger, R. L. & Mahadevan, V. (1965) J. Am. Chem. Soc. 87, 3526–3527. [DOI] [PubMed] [Google Scholar]
- 4.Letsinger, R. L., Ogillvie, K. K. & Miller, P. S. (1969) J. Am. Chem. Soc. 91, 3360–3365. [Google Scholar]
- 5.Matteucci, M. D. & Caruthers, M. H. (1981) J. Am. Chem. Soc. 103, 3185–3191. [Google Scholar]
- 6.McBride, L. J. & Caruthers, M. H. (1983) Tetrahedron Lett. 24, 245–248. [Google Scholar]
- 7.Hutchison, C. A., III, Peterson, S. N., Gill, S. R., Cline, R. T., White, O., Fraser, C. M., Smith, H. O. & Venter, J. C. (1999) Science 286, 2165–2169. [DOI] [PubMed] [Google Scholar]
- 8.Cho, M. K., Magnus, D., Caplan, A. L. & McGee, D. (1999) Science 286, 2087–2090. [DOI] [PubMed] [Google Scholar]
- 9.Cello, J., Paul, A. V. & Wimmer, E. (2002) Science 297, 1016–1018. [DOI] [PubMed] [Google Scholar]
- 10.Sinsheimer, R. L. (1959) J. Mol. Biol. 1, 43-53. [Google Scholar]
- 11.Goulian, M., Kornberg, A. & Sinsheimer, R. L. (1967) Proc. Natl. Acad. Sci. USA 58, 2321–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornberg, A. (2000) J. Bacteriol. 182, 3613–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanger, F., Coulson, A. R., Friedmann, T., Air, G. M., Barrell, B. G., Brown, N. L., Fiddes, J. C., Hutchison, C. A., III, Slocombe, P. M. & Smith, M. (1978) J. Mol. Biol. 125, 225–246. [DOI] [PubMed] [Google Scholar]
- 14.Stemmer, W. P. C., Crameri, A., Ha, K. D., Brennan, T. M. & Heyneker, H. L. (1995) Gene 164, 49–53. [DOI] [PubMed] [Google Scholar]
- 15.Bull, J. J., Badgett, M. R., Wichman, H. A., Huelsenbeck, Hillis, D. M., Gulati, A., Ho, C. & Molineux, J. (1997) Genetics 147, 1497–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]