Abstract
Fusion peptides mediate viral and host cell membrane fusion during viral entry. The monomeric form of the internal fusion peptide from Ebola virus was studied in membrane bilayer and water environments with computer simulations using replica exchange sampling and an implicit solvent description of the environment. Wild type Ebola fusion peptide, the W8A mutant form, and an extended construct with flanking residues were examined. It was found that the monomeric form of wild type Ebola fusion peptide adopts coil-helix-coil structure with a short helix from residue 8 to 11 mostly sampling orientations parallel to the membrane surface. W8A mutation disrupts the helicity in the N-terminal region of the peptide, and leads to a preference for slightly oblique orientation relative to the membrane surface. The addition of flanking residues also alters the fusion peptide conformation with either a helix-break-helix structure or extended N and C-termini and reduced membrane insertion. In water, the fusion peptide is found to adopt structures with low helicity.
Keywords: molecular dynamics, implicit membrane, generalized Born, replica exchange, membrane fusion
Introduction
Ebola virus belongs to the Filoviridae family and causes severe hemorrhagic fever in primates1. Viral infection requires the fusion between viral and host cell membranes. The membrane fusion process is mediated by fusion proteins that extrude from the viral membrane2–5. Fusion peptides that are part of the fusion proteins are the key components that are in contact with the host cell membrane. In case of Ebola virus, Ebola glycoprotein (GP) is responsible for both receptor binding and membrane fusion6. This protein is composed of two sub-domains, GP1 and GP2, which are connected via a disulfide bond. Ebola GP shares many common features with other membrane fusion proteins such as HA1 and HA2 in influenza and gp120 and gp41 in HIV type 1. In all of these fusion proteins, the first subunit binds to the cell receptor while the second subunit mediates membrane fusion7. The role of the fusion peptide during the fusion process is further illustrated in Fig. 1. The crystal structure of soluble GP2 reveals a trimer where a long coiled coil structure is surrounded by C-terminal helices. A key part of GP2 is the fusion peptide which interacts with the host cell membrane to induce fusion. In the soluble GP2 construct, the fusion peptide is missing8. Based on structures of influenza virus fusion protein, it is assumed that the fusion peptide is buried within the hydrophobic core of fusion protein in its inactive form, but becomes solvent exposed and able to insert into the host cell membrane once the fusion protein undergoes a structural change to its fusogenic form. Ebola fusion peptide (EFP) (G524AAIGLAWIPYFGPAA539) is thought to be in direct contact with the host cell membrane and is conserved within the virus family9. Unlike the N-terminal influenza and HIV fusion peptides5, EFP is an internal fusion peptide that is located 22 residues from the N-terminus of GP2. In the following, EFP residues will be referred to with a more convenient numbering scheme that starts at 1 for the first residue of the fusion peptide (G524 in GP2).
Figure 1.
Schematic representation of Ebola fusion protein in fusiogenic state. The globular domain GP1 is initially responsible for binding to the host cell receptor. The GP2 domain contains a helical bundle with the fusion peptide near the N-terminus.
Circular dichroism (CD) and infrared (IR) spectroscopy studies show that EFPE (wild type EFP with one additional glutamic acid residue at the C terminus) has three states10: random coil in solution and either an α-helix or a β-sheet when bound to the membrane10. The secondary structure of the membrane-bound peptide depends on the presence of Ca2+11. In the presence of Ca2+ a β-sheet structure is preferred while in the absence of Ca2+ helical structures are dominant11. In a different nuclear magnetic resonance (NMR) study of EFP it was also observed that the peptide adopts a random coil structure in aqueous buffers and more defined structure in the presence of sodium dodecyl sulfate (SDS) micelles12. Furthermore, tryptophan fluorescent emission data suggests that W8 enters the hydrophobic core of SDS micelles and according to chemical shifts and distance constraints according to nuclear Overhauser effect (NOE) measurements obtained from 1H NMR there is a short 310 helix form I9 to F12 in the middle of the peptide while N- and C-termini appear to be less structured12. The presence of a short helix in the middle of the peptide suggests that this region is in contact with the membrane core whereas the presumably flexible N and C termini may interact more favorably with the solvent. The flexibility of the EFP structure is presumably related to the presence of glycines13, but the glycines are also proposed to facilitate favorable insertion of EFP at the membrane head group-tail interface12. The secondary structure of EFP is apparently stabilized by an aromatic-aromatic interaction between W8 and F12 since the W8A mutation leads to a loss of helicity around I9 and a tendency to form helical structures between I4 and A812. Another key residue is P14 which is believed to be essential for destabilizing the membrane and promoting fusion since the P14R mutant disrupts membrane destabilization but does not affect peptide membrane association14. Since proline residues are known to promote kinked helical structures15, it was hypothesized that the mutation would disrupt a bent structure of EFP and result in a more linear, less destabilizing helical structure in the membrane14. The sequence context of the N- and C-termini is also essential for fusion activity. Addition of glutamic acid residues to N-and C-termini was found to reduce fusion activity10. In particular, addition of glutamate to the N-terminus of the peptide severely impairs peptide-vesicle interaction while addition of glutamate to the C-terminus of the peptide appears to only affect peptide association to membranes that lack phosphatidyl-inositol10.
Computational studies of EFP have offered a more detailed view of EFP structure and dynamics. Molecular dynamics (MD) simulations of EFP in SDS micelles and water-hexane using explicit solvent show that during 10 ns of simulation, the NMR structure (Protein data bank (PDB) code 2RLJ12) is largely maintained in SDS micelles, especially the short central helix, while larger structural deviations were found at the water-hexane interface12. Another MD simulation of EFP with a relatively simple implicit membrane model16 suggests that EFP adopts an α-helical structure with an interfacial membrane orientation at the center of the peptide while both termini are exposed to high- dielectric solvent17.
Here, we report results from extensive replica exchange simulations of wild-type EFP and the W8A mutant with a more realistic implicit membrane model. Wild-type EFP is studied both as the 16-residue construct and with three additional residues from the GP2 sequence at either terminus in order to examine the effect of the sequence context on EFP structure and dynamics. The resulting conformational sampling is in overall good agreement with the recent MD studies12,17 and data from NMR12 but also offers new insights into the effect of the flanking sequence on EFP structure and dynamics.
In the following, the computational methodology is described before results are presented and discussed.
Methods
Replica exchange MD simulations of EFP were carried out. Three systems were studied: wild type EFP with the sequence G1AAIGLAWIPYFGPAA (G1 corresponds to G524 in the GP2 fusion protein), the W8A mutant with the sequence GAAIGLAAIPYFGPAA, and a longer construct (EFP-X) with three extra residues from Ebola fusion protein on either side: QDE-G1AAIGLAWIPYFGPAA-EGI. Wild-type EFP was studied in an implicit membrane environment as well as a homogeneous dielectric to represent aqueous solvent. The W8A mutant and EFP-X were only simulated in the membrane environment. All simulations were carried out with the CHARMM program18 version 34a2 in conjunction with the MMTSB Tool Set19. The lowest energy structure from the NMR ensemble of EFP in SDS micelles (PDB code 2RLJ12) was chosen as the initial structure for the EFP simulations. The initial structure for EFP-X was built by adding the extra residues with a random coil secondary structure using pyMol20. Both N- and C-termini were assumed to be neutral and modeled as NH2 and CONH2, respectively. To describe the peptide interactions, the CHARMM2221 force field was used including the CMAP correction22. To implicitly model the membrane, we used the heterogeneous dielectric generalized Born model (HDGB)23 which has been developed in our group and applied previously in the simulation of membrane-bound proteins and peptides24–26. The HDGB model describes the phospholipid bilayer with a continuously varying dielectric constant and surface tension along the membrane normal (z-axis). The thickness of the membrane measured as the distance between the center of the membrane and water with dielectric constant equal to 80 was set to 25 Å. The dielectric and non-polar profiles used in this paper were modified from the initial HDGB model23 as described elsewhere26. Here, we also used a modified periodic dielectric and non-polar profiles to achieve a finite peptide concentration in z-direction as described earlier25. Other implicit solvent parameters were set as described previously27. The simulation of EFP in implicit water was performed with GBMV28,29 using a solvent dielectric constant of 80 and the following parameters to obtain numerically stable simulations30: β=−12, S0=0.65, C0=0.3225, C1=1.076 and D=−0.1418. The surface tension coefficient in the implicit water simulations was set to 0.015 kcal/Å2. Since the peptide is small, no cutoff was used for the calculation of non-bonded interactions. In all simulations, SHAKE31 was applied to constrain bonds involving hydrogen atoms and an integration time step of 1.5 fs was used for all simulations to maintain stable simulations with the implicit solvent model32. Simulations were run in the canonical ensemble with Langevin dynamics33 using a friction constant of 1ps−1 to enhance sampling34. Each system was minimized for 50 steps of steepest decent method followed by 500 steps with the adapted-basis Newton-Raphson method18. The minimized structures were then used as initial structures to run temperature replica exchange molecular dynamics simulations35,36. Eight replicas were run with temperatures spaced exponentially between 300 and 500 K. Exchanges between neighboring replicas were attempted after every 200 steps (0.3 ps). The acceptance ratio for successful exchanges was 23–37%. Total simulation times for different peptides ranged from 23–45 ns/replica (see Table I) but the first 9 ns of each simulation were considered equilibration and discarded during subsequent analysis.
Table I.
Overview of EFP simulations
| System | Environment | Abbreviation | Simulation time/replica |
|---|---|---|---|
| GAAIGLAWIPYFGPAA | Membrane | EFP | 37ns |
| Membrane | W8A | 36ns | |
| GAAIGLAWIPYFGPAA | Water | EFP-AQ | 23ns |
| Membrane | EFP-X | 45ns |
Potentials of mean force (PMF) at 300K were calculated from the replica exchange simulations using weighted histogram analysis (WHAM)37,38 to include weighted sampling from higher temperature replicas in the overall free energy surface. For membrane-interacting peptides, the peptide insertion depth and angle were used as the primary reaction coordinates in the construction of the PMFs. For the wild-type EFP and W8A mutant, the insertion depth was calculated as the distance of the center of mass (COM) of the entire peptide from the center of the membrane (z=0) along the z-direction. For EFP-X, the center of mass was calculated as above but without considering the three additional residues at each terminus. Peptides interacting with a different implicit membrane due to periodic replication or with opposite leaflets of the same membrane after spontaneous crossing of the bilayer were mapped onto a common scale by calculating the distance to the closest membrane for all structures. The insertion angle was calculated as the angle between membrane normal (z-axis) and the vector that connects the N(i+4) and O(i) atoms of the backbone for residues i from 3 to 6 for wild type and W8A mutant and residues 7 to 10 for EFP-X corresponding to the same sequence as in EFP. The conformation of the peptides was further analyzed by calculating α-helicity as a function of residue number. Two criteria were used to define the presence of α-helical structure: 1) An O(i) – H-N(i+4) distance of less than 2.6 Å39 and 2) backbone torsion values in the range of −70≤φ≤−50 and −35≤ψ≤−55. An O(i) – H-N(i+3) distance of less than 2.6 Å was also considered as the criterion for defining 310 helicity.
The PMF for EFP in water was projected onto two reaction coordinates: helicity based on hydrogen bond formation between O(i) – H-N(i+4) as described above and radius of gyration. Representative structures for each system were obtained through clustering based on mutual similarities as measured by root mean square deviations (RMSDs).
For analysis of EFP structures at the membrane interface, the superposition scheme used during the RMSD calculation was modified to preserve peptide orientations with respect to the z-axis and distinguish between different membrane insertion depths. This was achieved by allowing only transitions along x and y directions and rotation around the z-axis25. Using the modified superposition scheme, the K-means method was then used to cluster conformations based on RMSD with a radius of 3 Å and representative structures were extracted based on proximity to the center of each cluster. The standard superposition procedure was applied for clustering conformations from the simulation in implicit aqueous solvent.
Results
Replica exchange simulations were used to study the dynamics of EFP in implicit membrane/solvent environments. The wild-type EFP, a W8A mutant, and the extended sequence EFP-X were simulated in a membrane environment. Wild-type EFP was also simulated in aqueous solvent. The simulations are summarized in Table I.
Conformational sampling of EFP, W8A, and EFP-X in membrane bilayer
All of the systems exhibited significant dynamics involving different peptide conformations and different interactions with the model membrane but most of the sampled structures displayed at least some degree of helicity. Conformational sampling was characterized in more detail based on a 2D-PMF as a function of the N-terminal insertion angle and the distance of the center of mass from the membrane center (see Fig.2 A–C). Conformations with a center of mass distance of less than 25 Å were considered to be associated with the membrane. The corresponding structures were clustered and representative structures near the centers of the most populated clusters are shown in Fig. 2. Furthermore, average properties for membrane-associated peptides are reported in Table II.
Figure 2.
Left panel: Potentials of mean force (PMFs) for EFP (A), W8A (B), and EFP-X (C) from weighted-histogram analysis (WHAM), projected onto insertion angle (in degrees) and distance of the peptide center of mass from the membrane center (in Å). Different colors reflect relative free energies as indicated by the color bar (in kcal/mol). Right panel: Dominant structures for simulations with EFP (A1–A4), W8A (B1–B4) and EFP-X(C1–C4) from clustering. The backbone is shown as a ribbon and side chains are shown as stick representation. Charged residues are colored in red, glycines in green, prolines pale cyan, aromatic residues in white and hydrophobic residues in gray. The N-terminus is marked by a green sphere. Residue 8 is shown in ball and stick representation and colored magenta (W8) and orange (A8 in mutant). The z-axis is pointing up in all figures. All figures were generated with pyMol20.
Table II.
Population of inserted peptides, average center of mass (COM) insertion depth, insertion angles, and helicity fractions from simulated EFP structures at 300 K for membrane-associated conformations (defined as having COM insertion depths of less than 25 Å). Analysis of EFP-X was only applied to the core EFP sequence. Standard deviations are reported in parentheses. The standard errors based on block averaging are about 0.3 Å for COM, 10.5 degrees for insertion angles and 0.03 for helical fractions.
| Simulation | Population of inserted peptides |
COM insertion[Å] |
Insertion Angle [deg] |
Helicity fraction (from H-bonds) |
Helicity fraction (from φ/ψ sampling) |
|---|---|---|---|---|---|
| EFP | 72.5% | 19.05 (2.87) | 85.64 (25.74) | 0.345 (0.14) | 0.214(0.10) |
| W8A | 25.6% | 20.84 (2.45) | 91.80 (41.09) | 0.229 (0.12) | 0.161(0.09) |
| EFP-AQ | - | - | - | 0.154(0.14) | 0.047(0.07) |
| EFP-X | 2.2% | 22.49 (2.18) | 92.49 (31.27) | 0.362 (0.13) | 0.262(0.11) |
For wild-type EFP, there is a deep minimum at insertion depths of 15–22 Å and insertion angles of 60–100° (Fig. 2A) indicating that the peptide tends to lie parallel to membrane surface and strongly prefers interfacial association. The most dominant structure is A1, which consists of a bent helix with a kink at P10 and a less-ordered C-terminus. A second highly populated cluster (A2) adopts a straight helix for residues 3 to 12 also followed by a disordered C-terminus. A V-shaped orientation of EFP with the N- and C-termini pointing towards the membrane was observed as well (A3). In this structure, there is a short helix from residues 3 to 7 followed by a kink with a helix from residues 9 to 15. Interestingly, there was also a small population of trans-membrane (TM) orientations with an insertion depth of 0–10 Å and insertion angles of 0–40° (A4 in Fig. 2). The TM structures are mostly helical with an oblique orientation and with the N-terminus facing away from the membrane. The sampling of predominantly helical structures with a kink near P10 is consistent with experiments14 and a previous simulation17. The observation of spontaneous TM insertion may suggest a mechanism by which the host cell membrane is disrupted and membrane fusion is facilitated but this point will be discussed in more detail below.
The conformational sampling of W8A was significantly different from wild-type EFP. First, W8A did not associate as strongly with the membrane as the wild-type peptide. Secondly, membrane-associated structures were overall less helical (see B1–B3). However, helical structures with parallel membrane orientation and insertion depths of 15 to 20 Å similar to the dominant wild-type conformation were also observed in the mutant (B4) but with a lower probability. The loss of helicity in the middle of the dominant structures (B1–B3) as a result of the W8A mutation is in agreement with earlier experimental studies12 and is presumed to be a consequence of the lost interactions between W8 and F12. The loss of helicity in turn results in a more flexible peptide and broader sampling of different conformations. The relative populations indicated in Fig. 2 suggest that the mutated peptide samples a broader range of different structures with similar frequency.
The sampling of EFP-X with three additional residues at the N and C-termini also differs significantly from the shorter EFP construct (see Fig. 2C). Membrane association is even less favorable than in the W8A mutant, presumably as a result of the added negatively charged aspartate and glutamate residues at the N-terminus and another glutamate at the C-terminus. The structures that are associated with the membrane appear to cover a broader ensemble consisting of oblique and parallel orientations relative to the membrane. The representative structure from the most populated cluster has an insertion angle of 110° and an insertion depth of 24 Å (see C1 in Fig. 2) It is mostly helical with kinks at residues P10 and P14. The second most populated membrane-associated conformation (C2) is more deeply inserted and consists of a short helix at residues 6 to 12 but has disordered N- and C-termini. The short helix lies parallel to the membrane surface as in A1 but the helix does not extend as far in C2 as in A1. C3 is similar to C1 but it is turned around with respect to the membrane allowing the turn region to interact with the membrane and the termini to be exposed towards the aqueous solvent. Finally, C4 has a reduced helical structure but lies parallel to the membrane surface similar to the A1 conformation for EFP.
EFP in aqueous solvent
Conformational sampling of EFP in aqueous solvent was also investigated to better understand the effect of the membrane. A PMF based on the average α-helicity and radius of gyration is shown in Fig. 3. There is a broad minimum region that reaches from helical fractions of 0 to 0.7 with two preferred states: compact structures with partial helicity (D2–D4 in Fig. 3) and fully extended conformations with no helical structure (D1). The helical structures have only short helices either from A3 to A7 (D2) from I9 to Y11 (D3) (a short 310 helix) or from I4 to Y11 (D4). A loss of secondary structure in aqueous buffer was previously observed12 and our simulation results confirm that the membrane stabilizes the secondary structure of EFP.
Figure 3.
Left panel: Potential of mean force (PMF) of EFP-AQ from weighted-histogram analysis (WHAM), projected onto average helicity based on hydrogen bond criterion and radius of gyration (in Å). Different colors reflect relative free energies as indicated by the color bar (in kcal/mol). Right panel: Dominant structures for simulations with EFP-AQ from clustering as in Fig. 2.
Average insertion depths, insertion angles, and helicity based on hydrogen bonding and dihedral angles are reported in Table II. For membrane simulations, only membrane associated structures were considered. Again, it can be seen that EFP is inserted most and EFP-X is inserted least according to the average insertion depths of the peptide centers of mass. The average N-terminal insertion angle is near 90° for all peptides. However, while the average reflects the dominant structures for EFP and W8A it is a result of an average over populations with different insertion angles for EFP-X (see also Fig. 2).
Helical propensity in EFP
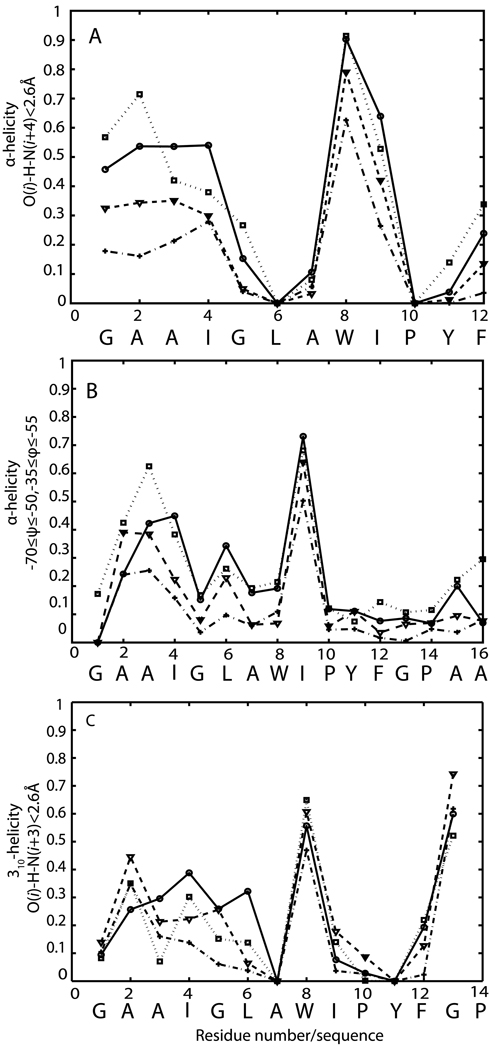
Experimental data suggests that EFP has a tendency to form helical structures in the presence of the membrane12. From our simulation data, helicity was calculated according to two criteria: based on hydrogen bonding and backbone ϕ/ψ-sampling (see Methods section). Overall, average helicity is largest for EFP-X and slightly lower for EFP (see Table II). Helicity is significantly reduced in the W8A mutant and even further for EFP in aqueous solvent. In order to examine helical propensity in more detail, we have calculated helicity as a function of residue along the EFP sequence for all of the peptides studied here. The results shown in Fig. 4 indicate that all peptides have a strong tendency to form helical structures in the central region of the peptide (residues 8–10) consistent with NMR chemical shift measurements12. Furthermore, there is significant helical propensity at the N-terminus. Mutating W8 to alanine reduces the helicity in the N-terminal domain but does not cause any significant change in helicity in the C-terminal part of the peptide. This observation is also consistent with experiment12. There are two breaks in helicity close to the proline residues (P10 and P14). Since there is no amide hydrogen on P10 and P14 these two residues are not able to form hydrogen bonds with L6 and P10. The role of proline in inducing helical breaks was also observed experimentally and is hypothesized to be important for fusion activity in Ebola14 as well as influenza fusion peptide40. Including extra residues at the N- and C-termini of EFP changes the overall structure and orientation of EFP as described above but does not appear to significantly change the α-helicity profiles.
Figure 4.
Average fraction of helical conformations as a function of residue for EFP (circle), W8A (triangle), EFP-X (square) and EFP-AQ (cross). A:Helicity is defined by O(i) – H-N(i+4) hydrogen bonding (< 2.6 Å distance); B: Helicity is defined by −70≤ψ≤−50 and −35≤φ≤−55; C: 310 helicity is defined by O(i) – H-N(i+3) hydrogen bonding (< 2.6 Å distance)
Formation of 310-helices was also investigated based on an i,i+3 hydrogen bond criterion (see Methods section). In all four simulations, the peptide has a high propensity to form 310-helix at W8. It should be noted that a residue may be in both α- and 310-helical form if bifurcated hydrogen bonds are present. Formation of a 310-helix in the central region of the peptide was also observed experimentally12. In addition to W8, G13 also shows a high propensity to form 310-helices. According to Fig. 4C, EFP appears to prefer α-helical structures from G1 to I4 310-helices. This preference for α-helices is even more pronounced for EPF-X whereas W8A and EFP-AQ have similar propensities towards either helical form.
Discussion and Conclusion
In this study we describe results from simulation of EFP in the presence of membrane and water environments. An implicit description of the environment allowed us to reach much longer time scales than what would be possible with explicit lipid/water simulations. In particular, the implicit membrane model avoids the need for relatively slow lipid relaxation in response to different peptide conformations. Further sampling enhancement is gained from using replica exchange sampling and a reduced friction coefficient in Langevin dynamics. While the exact time scales covered in our simulations are difficult to determine, we estimate that the µs-ms range is reached despite a nominal length of the simulations of only tens of nanoseconds.
On the other hand, the use of an implicit membrane model introduces certain approximations and in particular focuses on the mean-field effect of the membrane environment while neglecting specific molecular details of peptide-membrane interactions. While this may appear to be a serious limitation, we have used the same approach previously to study membrane-bound influenza virus fusion peptide25 and phospholamban26. In both cases, we found very good agreement with experimental data. In fact, the agreement appears to be better than with previous explicit lipid simulations presumably because of the much longer time scales that could be covered with the implicit solvent model. In particular, the dominant conformation predicted for membrane-bound influenza virus fusion peptide predicted by our simulations was recently confirmed experimentally41. Furthermore, similar but simpler implicit membrane models than the HDGB model used here, have also been very successful in describing the conformational ensemble of other membrane-bound peptides in good agreement with experiment42–45. This suggests that the approach taken here is reasonable for obtaining realistic conformational ensembles of membrane-bound EFP. However, it is clear that a full understanding of fusion peptide-membrane interactions will eventually also have to include simulations with explicit lipids.
In our simulations, EFP samples a variety of conformations with different degrees of partial helicity. Sampling of helical structures is reduced in the W8A mutant and in water while extending EFP by three residues at the N- and C-termini slightly enhances helicity. EFP by itself mostly prefers a central helix that is oriented parallel to the membrane. In contrast, the addition of extra residues from the GP2 sequence context leads to significant sampling of helix-break-helix motifs where N- and C-terminal helices are oriented more obliquely with respect to the membrane normal. A conformation with a central helix parallel to the membrane is also observed, but the helix is shorter in the EFP-X construct than in the EFP sequence (see C2 in Fig. 2). The extra residues also significantly reduce the affinity of the peptide for the peptide as a result of charged residues at both ends. These results indicate that the sequence context is important in determining the conformation of membrane-bound EFP. Similar conclusions were found for influenza virus fusion peptide (IFP) where the addition of an experimentally used C-terminal tag also appeared to affect conformational sampling and orientation relative to the membrane25. Our findings therefore strongly suggest that implications based on structural studies of just the fusion peptide alone should be treated with caution and that it may be necessary to study fusion peptides in the context of the fusion protein to fully understand their interactions with membranes and ultimately the mechanism by which they exert their fusogenic activity.
EFP is an internal fusion protein with extra residues from the GP2 fusion protein on both the N- and C-termini. Under the assumption that neither the N- or C-terminal part of GP2 inserts deeply into the host cell membrane the type of EFP conformations that are possible in the context of GP2 are constrained. Conformations where both N- and C-termini are inserted into the membrane (like A1 or C1) appear to be unlikely. Furthermore, the trans-membrane (TM) structures observed for the short EFP construct would be ruled out because it would require the 22 N-terminal residues of GP2 to either cross the membrane or be inserted into the membrane as well. Membrane insertion or crossing is unlikely due to the presence of charged and polar residues at the N-terminus of GP2. More likely conformations of EFP in the context of GP2 appear to be C2, C3, or C4 all of which would allow N- and C-terminal residues to connect to the peptide outside the membrane or just at the membrane-water interface.
It is also interesting to compare the structures observed for EFP with earlier results for IFP. In our simulations, IFP samples largely helix-break-helix structures as confirmed by recent new experimental data25,41,46. Those structures resemble the structures C1 and C3 seen for EFP-X but in the IFP simulations, those conformations where found to lie mostly parallel to the membrane while the EFP helix-break-helix structures appear to be oriented perpendicular to the membrane. It therefore seems from this study and our previous work that there are common structural elements shared between viral fusion peptides. This would suggest a common fusogenic mechanism, but more work is clearly needed to come to definite conclusions in this regard.
Another issue that has been so far neglected is that viral fusion proteins and in particular their fusion peptides are known to oligomerize. It will therefore be critical to expand future studies to fusion peptide oligomers to better understand the membrane fusion process. The implicit membrane methodology applied here could also be used to study fusion peptide oligomers albeit with more extended conformational sampling because of the additional degrees of freedom.
It is our hope that the present study motivates not just further computational studies of viral fusion studies but also stimulates a closer experimental look at the effect of the fusion protein context on the conformational sampling of the fusion peptide in Ebola and other viruses.
Acknowledgements
This work was supported in part through NSF grant MCB 0447799 and an Alfred P. Sloan fellowship (to MF). Computer resources at the High Performance Computer Center at Michigan State University as well as an NSF TeraGrid computer allocation (TG-MCB090003) are acknowledged.
References
- 1.Jahring PB, Kiley MP, Klenk HD, Peters CJ, A S, Swanepoel R. Arch Virol. 1995;10:289–292. [Google Scholar]
- 2.Malashkevich VN, Singh M, Kim PS. The trimer-of-hairpins motif in membrane fusion: Visna virus. Proc Natl Acad Sci U S A. 2001;98:8502–8506. doi: 10.1073/pnas.151254798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Rey FA. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 4.Yao Y, Ghosh K, Epand RF, Epand RM, Ghosh HP. Membrane fusion activity of vesicular stomatitis virus glycoprotein G is induced by low pH but not by heat or denaturant. Virology. 2003;310:319–332. doi: 10.1016/s0042-6822(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 5.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 6.Feldmann H, Klenk HD, Sanchez A. Arch Virol. 1993;7:81–100. doi: 10.1007/978-3-7091-9300-6_8. [DOI] [PubMed] [Google Scholar]
- 7.Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Strauss SE. Fields Virology. 3 ed. philadelphia: Lippincott; 1996. [Google Scholar]
- 8.Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci U S A. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito H, Watanabe S, Sanchez A, Whitt MA, Kawaoka Y. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol. 1999;73:8907–8912. doi: 10.1128/jvi.73.10.8907-8912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz-Arguello MB, Goni FM, Pereira FB, Nieva JL. Phosphatidylinositol-dependent membrane fusion induced by a putative fusogenic sequence of Ebola virus. J Virol. 1998;72:1775–1781. doi: 10.1128/jvi.72.3.1775-1781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suarez T, Gomara MJ, Goni FM, Mingarro I, Muga A, Perez-Paya E, Nieva JL. Calcium-dependent conformational changes of membrane-bound Ebola fusion peptide drive vesicle fusion. FEBS Lett. 2003;535:23–28. doi: 10.1016/s0014-5793(02)03847-4. [DOI] [PubMed] [Google Scholar]
- 12.Freitas MS, Gaspar LP, Lorenzoni M, Almeida FC, Tinoco LW, Almeida MS, Maia LF, Degreve L, Valente AP, Silva JL. Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts. J Biol Chem. 2007;282:27306–27314. doi: 10.1074/jbc.M611864200. [DOI] [PubMed] [Google Scholar]
- 13.Wong TC. Biochim Biophys Acta. 2003;1609:45–54. doi: 10.1016/s0005-2736(02)00652-1. [DOI] [PubMed] [Google Scholar]
- 14.Gomara MJ, Mora P, Mingarro I, Nieva JL. Roles of a conserved proline in the internal fusion peptide of Ebola glycoprotein. FEBS Lett. 2004;569:261–266. doi: 10.1016/j.febslet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Orzaez M, Salgado J, Gimenez-Giner A, Perez-Paya E, Mingarro I. Influence of proline residues in transmembrane helix packing. J Mol Biol. 2004;335:631–640. doi: 10.1016/j.jmb.2003.10.062. [DOI] [PubMed] [Google Scholar]
- 16.Im W, Feig M, Brooks CLI. An implicit membrane generalized Born theory for the study of structure stability and interactions of membrane proteins. Biophys J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson MA, Yeh I-C, Lee MS. Molecular Dynamics Simulations of Folding and Insertion of the Ebola virus Fusion Peptide into a Membrane Bilayer. Proceedings of the 2008 International Conference on Bioinformatics and Computational Biology; Las Vegas, NV; 2008. pp. 14–17. [Google Scholar]
- 18.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feig M, Karanicolas J, Brooks CL., 3rd MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J Mol Graph Model. 2004;22:377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Delano WL. The PyMol Molecular Graphics System. 2002. [Google Scholar]
- 21.Foloppe N, MacKerell AD., Jr All atom empirical force field for nucleic acids: I. parameter optimization based on small molecule and condensed phase macromolecular target data. J Comp Chem. 2000;21:86–104. [Google Scholar]
- 22.MacKerell AD, Jr, Feig M, Brooks CL., 3rd Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 23.Tanizaki S, Feig M. A generalized Born formalism for heterogeneous dielectric environments: application to the implicit modeling of biological membranes. J Chem Phys. 2005;122:124706. doi: 10.1063/1.1865992. [DOI] [PubMed] [Google Scholar]
- 24.Tanizaki S, Feig M. Molecular dynamics simulations of large integral membrane proteins with an implicit membrane model. J Phys Chem B. 2006;110:548–556. doi: 10.1021/jp054694f. [DOI] [PubMed] [Google Scholar]
- 25.Panahi A, Feig M. Conformational sampling of influenza fusion peptide in membrane bilayers as a function of termini and protonation states. J Phys Chem B. 2010;114:1407–1416. doi: 10.1021/jp907366g. [DOI] [PubMed] [Google Scholar]
- 26.Sayadi M, Tanizaki S, Feig M. Effect of membrane thickness on conformational sampling of phospholamban from computer simulations. Biophys J. 2010;98:805–814. doi: 10.1016/j.bpj.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feig M, Im W, Brooks CL., 3rd Implicit solvation based on generalized Born theory in different dielectric environments. J Chem Phys. 2004;120:903–911. doi: 10.1063/1.1631258. [DOI] [PubMed] [Google Scholar]
- 28.Lee MS, Salsbury FR, Brooks CLI. Novel generalized Born Methods. J Chem phys. 2002;116:10606. [Google Scholar]
- 29.Lee MS, Feig M, Salsbury FR, Jr, Brooks CL., 3rd New analytic approximation to the standard molecular volume definition and its application to generalized Born calculations. J Comput Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 30.Chocholousova J, Feig M. Balancing an accurate representation of the molecular surface in generalized born formalisms with integrator stability in molecular dynamics simulations. J Comput Chem. 2006;27:719–729. doi: 10.1002/jcc.20387. [DOI] [PubMed] [Google Scholar]
- 31.Ryckaert J, Ciccotti G, Berensdsen H. Numerical integration of cartesin equations of motions of a system with constraints-molecular dynamics of N-alkanes. J Comp Chem. 1977;23:327. [Google Scholar]
- 32.Chocholousova J, Feig M. Implicit solvent simulations of DNA and DNA-protein complexes: agreement with explicit solvent vs experiment. J Phys Chem B. 2006;110:17240–17251. doi: 10.1021/jp0627675. [DOI] [PubMed] [Google Scholar]
- 33.Brooks CL, Berkowits M, Adelman S. Generalized Langevin theory for many-body problems in chemical dynamics gas surface collisions, vibrational energy relaxation in solids and recombination reactions in liquids. J Chem Phys. 1980;73:4353. [Google Scholar]
- 34.Feig M. Kinetics from Implicit Solvent Simulations of Biomolecules as a Function of Viscosity. J Chem Theory Comput. 2007;3:1734–1748. doi: 10.1021/ct7000705. [DOI] [PubMed] [Google Scholar]
- 35.Hansmann UH, Okamoto Y. New Monte Carlo algorithms for protein folding. Curr Opin Struct Biol. 1999;9:177–183. doi: 10.1016/S0959-440X(99)80025-6. [DOI] [PubMed] [Google Scholar]
- 36.Sugita Y, Okamoto Y. Replica exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141. [Google Scholar]
- 37.Kumar S, Rosenberg J, Bouzida D, Swedsen R, Kollman P. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comp Chem. 1992;13:1011–1021. [Google Scholar]
- 38.Kumar S, Rosenberg J, Bouzida D, Swedsen R, Kollman P. Multidimentional free-energy calculation using the weighted histogram analysis method. J Comp Chem. 1995;16:1339–1350. [Google Scholar]
- 39.Chen J, Im W, Brooks CL., 3rd Balancing solvation and intramolecular interactions: toward a consistent generalized Born force field. J Am Chem Soc. 2006;128:3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai AL, Park H, White JM, Tamm LK. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J Biol Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 41.Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc Natl Acad Sci U S A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Im W, Feig M, Brooks CL., 3rd An implicit membrane generalized born theory for the study of structure, stability, and interactions of membrane proteins. Biophys J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sammalkorpi M, Lazaridis T. Configuration of influenza hemagglutinin fusion peptide monomers and oligomers in membranes. Biochim Biophys Acta. 2007;1768:30–38. doi: 10.1016/j.bbamem.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Spassov VZ, Yan L, Szalma S. Introducing an implicit membrane in generalized born/solvent accessibility continuum solvent models. J Phys Chem B. 2002;106:8726–8738. [Google Scholar]
- 45.Bechor D, Ben-Tal N. Implicit solvent model studies of the interactions of the influenza hemagglutinin fusion peptide with lipid bilayers. Biophys J. 2001;80:643–655. doi: 10.1016/S0006-3495(01)76045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y, Weliky DP. 13C-13C correlation spectroscopy of membrane-associated influenza virus fusion peptide strongly supports a helix-turn-helix motif and two turn conformations. J Am Chem Soc. 2009;131:13228–13229. doi: 10.1021/ja905198q. [DOI] [PMC free article] [PubMed] [Google Scholar]