Abstract
V(D)J recombination, the rearrangement of gene segments to assemble Ig and T cell receptor coding regions, is vital to B and T lymphocyte development. Here, we demonstrate that the V(D)J recombinase protein RAG1 undergoes ubiquitylation in cells. In vitro, the RING finger domain of RAG1 acts as a ubiquitin ligase that mediates its own ubiquitylation at a highly conserved K residue in the RAG1 amino-terminal region. Ubiquitylation is best supported by a specific ubiquitin-conjugating enzyme, UbcH3/CDC34, and requires an intact RAG1 RING finger motif. Disruption of the RING finger and certain RAG1 N-terminal truncations are associated with immunodeficiency in human patients, suggesting that RAG1's ubiquitin ligase is required for its biological role in lymphocyte development.
The development of B and T cells depends on the rearrangement of variable (V), diversity (D), and joining (J) gene segments to produce mature Ig and T cell receptor coding regions (1). This rearrangement process, known as V(D)J recombination, must occur only in the correct lymphoid lineages and at the appropriate loci and developmental stages to minimize extraneous damage to the genome. Many factors are known to contribute to regulation of V(D)J recombination, and new evidence suggests that ubiquitin conjugation may also play an important role (2, 3).
V(D)J recombination is initiated by the RAG1 and RAG2 gene products (4, 5). Together these proteins act as a recombinase that introduces double-stranded DNA breaks at recombination signal sequences (RSSs) flanking the coding DNA (6, 7). The breaks are then repaired by the nonhomologous DNA end-joining machinery, resulting in fusions between pairs of coding gene segments and pairs of RSSs. Deletion analysis has defined minimal “core” regions of RAG1 and RAG2 (amino acids 384-1008 of 1,040 in RAG1 and 1–383 of 527 in RAG2) (8–11). A recombinase composed of core RAG1 and RAG2 retains DNA binding and cleavage activity in vitro and supports recombination of extrachromosomal V(D)J substrates (8, 9, 12–14), although with some differences from the full-length recombinase (15–18). In vitro, core RAG1 and RAG2 are more active than their full-length counterparts in DNA transposition (15, 18), a potentially damaging reaction that could lead to RSS ends attacking other regions of the genome. The RAG2 core also displays a fundamental defect in recombination of the Ig heavy chain locus in the cell (19), apparently because of an inability to gain access to certain regions of the locus.
Mutations of the “dispensable” regions of RAG1 also affect its activity. The RAG1 core can support D to J rearrangement at the Ig heavy chain locus in RAG1-/- pro-B cells at a somewhat reduced level relative to the full-length protein, but deletions of smaller regions within the N terminus reduce chromosomal recombination considerably more (16). A naturally occurring RAG1 mutation that disrupts the conserved RING finger motif (C328Y) and two N-terminal truncation mutants have been found to be defective in B cell development while supporting limited T cell development (20–22), resulting in a fatal primary immunodeficiency called Omenn's syndrome. The core proteins lack many conserved regions, and these data indicate that the missing regions contribute to V(D)J recombination of the chromosomal loci, perhaps performing a regulatory function.
Modification of proteins by ubiquitin conjugation (ubiquitylation) plays a central regulatory role in all eukaryotic cells (23). Ubiquitin is a highly conserved 76-aa protein that can be covalently attached by its C terminus to various target proteins (24). Conjugation is initiated by the ubiquitin-activating enzyme (E1), which becomes linked to a ubiquitin moiety through a high-energy thiol-ester bond. The activated ubiquitin is then transferred to the active-site cysteine residue on one of several ubiquitin carrier proteins (E2s). With the help of a third class of proteins, ubiquitin ligases (E3s), an E2 recognizes its ubiquitylation target and transfers the activated ubiquitin to an internal lysine (K) residue therein (24). Ubiquitin can also be selfconjugated on internal K residues, most often K48. Through multiple iterations of this cascade, proteins become attached to long polyubiquitin chains, usually leading to their degradation by the 26S proteasome (23, 25). For other target proteins, monoubiquitylation or polyubiquitylation, sometimes at multiple sites, has been shown to regulate protein activity independent of degradation (26–28). Ubiquitin conjugation is already known to play at least one important role in V(D)J recombination. RAG2 undergoes ubiquitylation (2), which may be linked to its cell cycle-dependent destruction (29). However, the E2 and E3 proteins that target RAG2 for ubiquitylation have not been identified.
Much of the specificity of the ubiquitin cascade is provided by the E3 family (30). E3 activity is being recognized in an increasing number of proteins, often with very diverse functions outside of their role as E3s. Many of these proteins themselves become monoubiquitylated or polyubiquitylated with resultant modulation of their various functions (31–33). E3s can be grouped into two classes, the so-called HECT domain proteins and proteins including zinc binding domains of the RING finger configuration (C3HC4 or C3H2C2) (34, 35). The primary Zn binding region of RAG1 (amino acids 264–389) includes a C3HC4 RING finger motif closely associated with an adjacent C2H2 Zn finger (36, 37). Although the Zn binding domain lies mostly outside the RAG1 core, mutation of conserved residues within the RING finger in the context of an otherwise intact N terminus has been shown to severely reduce rearrangement of extrachromosomal V(D)J substrates (8, 16) and can cause immunodeficiency in humans (22).
Here, we demonstrate that RAG1 undergoes ubiquitylation in intact cells and that the RAG1 Zn binding domain possesses E3 activity that promotes autoubiquitylation in an upstream basic region. This modification occurs at a single highly conserved K residue and is best promoted by a specific E2 enzyme, UbcH3/CDC34. E3 activity of RAG1 has also been shown by Yurchenko et al. (3), although with some significant differences from the work presented here, as discussed later. In addition to promoting autoubiquitylation, RAG1's E3 activity may potentially target proteins such as RAG2. This activity could help to regulate DNA cleavage by RAG1/2 and other aspects of V(D)J recombination.
Methods
Plasmids and Proteins. Plasmids expressing murine RAG1 residues 218–389 (pJMJ029) and 264–389 (pJMJ030) were constructed by PCR using the pTrc-His-TOPO-TA vector (Invitrogen). Coding segments were amplified by PCR of pJH548 (8) by using the following primer pairs: 218–389, JMJ 056-TTACCCT T TAT TGATATGCACCA A AGTCTC and JMJ057-CGGGGACTCA AGAGGA AGAGACATCAG; 264–389, JMJ042-TCCAACTGCAGTAAGATTCATCTCAGTACC and JMJ056. This resulted in bacterial expression constructs linked to six-histidine tags and Xpress epitopes on their N termini. Mutations were introduced into pJMJ029 by using the QuikChange mutagenesis kit (Stratagene). Subcloning and mutations were confirmed by sequencing. Numbering is for murine RAG1.
Histidine-tagged PK-ubiquitin was expressed in bacteria from the plasmid pPK-HA-Ub, the kind gift of Yue Xiong (Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill) (38). PK-ubiquitin was labeled by using cAMP-dependent kinase (Sigma) and [γ-32P]ATP (DuPont/NEN).
For bacterial expression, plasmids were transformed into BL21- or STBL-competent Escherichia coli, which were grown at 37°C to midlog phase, then induced for 2 h by using 0.5 mM isopropyl β-d-thiogalactoside. Cell pellets were resuspended in lysis buffer [20 mM Tris, pH 7.4/0.5 M NaCl/10 mM 2-mercaptoethanol/0.1% Nonidet P-40/0.05 mM ZnCl2/10% glycerol for RAG1 constructs or 20 mM Tris, pH 7.4/0.25 M NaCl/10 mM 2-mercaptoethanol/0.1% Nonidet P-40/10% glycerol for ubiquitin] and incubated on ice for 30 min in the presence of 1 mg/ml lysozyme. Cell debris were collected by high-speed centrifugation, and histidine-tagged proteins in the soluble fraction were purified on Ni-NTA Superflow resin (Qiagen, Chatsworth, CA) by using standard techniques. Purified proteins were dialyzed overnight at 0°C into storage buffer (20 mM Tris, pH 7.4/0.15 M KCl/1 mM DTT/10% glycerol, plus 0.05 mM ZnCl2 for the RAG1 constructs) and stored at -80°C in small aliquots. To remove the Zn from RAG1-B/RING, the protein was dialyzed for 18 h at 0°C, with three buffer changes, against storage buffer (without Zn) supplemented with 5 mM diethylenetriaminepenta-acetic acid.
Full-length RAG1 and RAG2 tagged with nine-histidine and FLAG epitopes were expressed from vaccinia virus vectors, the kind gift of Marjorie Oettinger (Massachusetts General Hospital, Boston) (15). WT mammalian ubiquitin, CH3-ubiquitin, ubiquitin [K48R], leporine E1, and human E2 enzymes were purchased from Boston Biochem (Cambridge, MA). UbcH2, H5a, H5b, and H5c were tagged with glutathione-S-transferase, and UbcH3, H6, and H10 were tagged with six histidine residues. Apyrase was purchased from Sigma.
In Vitro Ubiquitylation Assay. E1 (45 nM), E2 (0.3 μM UbcH3, unless otherwise indicated), PK-ubiquitin (500 μM), and RAG1 (8 μM as dimer, unless otherwise indicated) were combined in reaction buffer (50 mM Tris, pH 7.4/0.001% Brij/2 mM Mg-ATP/50 mM NaCl/60 mM KCl/0.4 mM DTT/0.02 mM ZnCl2) and incubated for 1 h at 37°C. Reaction products were separated on 4–12% NuPAGE gels (Invitrogen). When radiolabeled PK-ubiquitin was used, gels were dried, exposed to phosphor-storage autoradiography, and analyzed on a Typhoon 8600 (Amersham Pharmacia Biotech) by using imagequant software (Amersham Pharmacia Biotech). When unlabeled ubiquitin was used, proteins were transferred to nitrocellulose and subjected to Western blot by using anti-Xpress antibody (Invitrogen) conjugated to horseradish peroxidase and SuperSignal West Dura Extended Duration Substrate (Pierce). Alternatively, unconjugated primary antibody was detected with Alexa 488-conjugated secondary antibody (Molecular Probes), and bands were analyzed on a Typhoon 8600 (Amersham Pharmacia Biotech) by using imagequant software (Amersham Pharmacia Biotech).
Pull-Down Assays. HeLa cells were grown to 80% confluence and infected at a multiplicity of infection of 3 with vaccinia viruses expressing T7 RNA polymerase plus RAG1 or RAG1 and RAG2. After 16 h, clasto-lactacystin-β-lactone (2 μM; Boston Biochem) was added to plates as indicated, and incubation was continued for 8 h. Cells (5 × 106) were washed twice with PBS, then lifted with a scraper and collected by centrifugation. Cell pellets were resuspended in 1 ml of urea buffer (20 mM Tris, pH 8.0/0.5 M NaCl/6 M urea) and sonicated extensively to shear DNA. Lysate (0.2 ml) was combined with 20 μl of Ni-NTA Superflow resin (Qiagen) and incubated overnight at 4°C. Beads were collected in spin filters (Sigma) and washed four times with 0.7 ml of urea buffer (pH 7.4) and three times with 0.7 ml of the same buffer plus 50 mM imidazole. Proteins were eluted from the beads with 50 μl of urea buffer (pH 7.4) plus 0.3 M imidazole. Samples (10 μl) were separated on 4–12% NuPAGE gels (Invitrogen), and proteins were transferred to nitrocellulose. Blots were probed with antiubiquitin protein conjugate antibody (1:500; Affiniti Research Products, Exeter, U.K.) and secondary anti-rabbit-horseradish peroxidase conjugate (1:2,000; Vector Laboratories) and detected as described above. Blots were stripped overnight at room temperature with Restore buffer (Pierce), then reprobed with anti-FLAG M2 peroxidase conjugate (1:20,000; Sigma).
Results
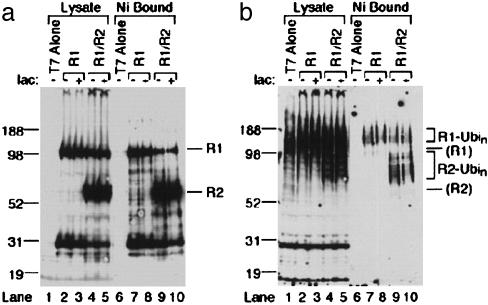
RAG1 Is Ubiquitylated in Cells. We used nickel (Ni) pull-down assays to examine ubiquitylation of RAG1. Full-length RAG1, tagged with nine histidines and a FLAG epitope, or RAG1 plus similarly tagged full-length RAG2 were expressed from vaccinia virus vectors in HeLa cells (15). In some cases, cells were treated with clasto-lactacystin-β-lactone to inhibit 26S proteasome activity. Expression of RAG1 and RAG2 was confirmed by Western blot with anti-FLAG antibody (Fig. 1a). Bands corresponding to full-length RAG1 (Fig. 1a, lanes 2–5) and full-length RAG2 (Fig. 1a, lanes 4–5) and some lower molecular weight bands were present in the appropriate cell lysates but were absent when virus expressing only T7 RNA polymerase was used (Fig. 1a, lane 1). The RAG1 and RAG2 bands were precipitated with Ni resin (Fig. 1a, lanes 7–10). Western blots with antiubiquitin protein conjugate antibody indicated that many ubiquitylated species were present in the lysate fractions regardless of the presence of RAG1 or RAG2 (Fig. 1b, lanes 1–5), but in the absence of RAG1 and RAG2, no ubiquitylated species were precipitated with Ni resin (Fig. 1b, lane 6). When RAG1 was present, a smear of ubiquitylated species of higher molecular weight than RAG1 was precipitated (Fig. 1b, lanes 7–10). When RAG2 was also present, an additional smear of ubiquitylated species of higher molecular weight than RAG2 was observed (Fig. 1b, lanes 9 and 10). These data indicate that full-length RAG1 can undergo ubiquitylation in intact cells, and that this reaction does not require nor is it inhibited by RAG2. Bands corresponding to ubiquitylated RAG1 and RAG2 were only barely detectable with the anti-FLAG antibody (Fig. 1a and data not shown), suggesting that only a small fraction of RAG1 and RAG2 was ubiquitylated under these conditions. The presence of proteasome inhibitor did not have a strong impact on the quantity of ubiquitylated species.
Fig. 1.
Full-length RAG1 is ubiquitylated in intact cells. Full-length RAG1 (R1; lanes 2–5 and 7–10) and RAG2 (R2; lanes 4, 5, 9, and 10) were expressed in HeLa cells, and pull-down assays were performed as described in Methods. Products were analyzed by Western blot probed with anti-FLAG antibody (a) and antiubiquitin protein conjugate (b).
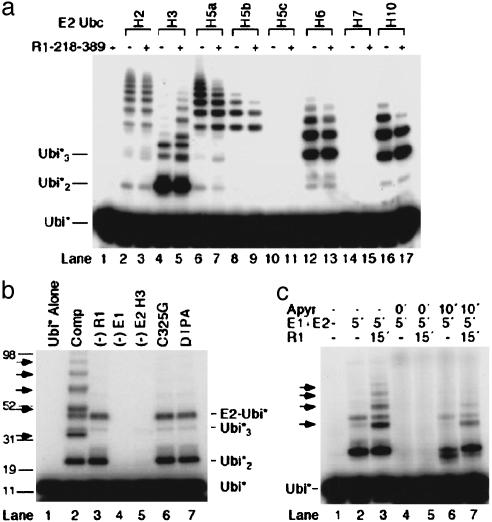
The RAG1 Zn Binding Domain Acts as a Ubiquitin Ligase. To test for E3 activity of the RAG1 N terminus, a fragment of murine RAG1 spanning the RING finger, adjoining zinc finger, and conserved upstream basic regions (amino acids 218–389, henceforth called RAG1-B/RING) was combined with leporine E1 enzyme, radiolabeled ubiquitin tagged with a protein kinase motif (PK-ubiquitin), and one of several recombinant human E2 enzymes. UbcH3 was the only E2 species that consistently supported robust RAG1-B/RING-dependent uptake of PK-ubiquitin. (Fig. 2a, compare lanes 4 and 5), although bands corresponding to autoubiquitylation of the E2 enzymes were present in many samples regardless of the presence of RAG1 (Fig. 2a, lanes 2–9, 12, 13, 16, and 17). For the E2 proteins tagged with glutathione-S-tranferase (UbcH2, H5a, H5b, and H5c), these products could be eliminated by pull-down with glutathione-agarose (data not shown). RAG1-independent bands representing diubiquitin and triubiquitin were also generated (Fig. 2a, lanes 2–7, 12, 13, 16, and 17). RAG1-B/RING-dependent products were present at very low levels in reactions including UbcH2 and H5a (e.g., Fig. 2a, lanes 3 and 7, and data not shown). Quantification of radiolabeled ubiquitin moieties incorporated into RAG1-B/RING-dependent bands demonstrated that the RAG1-B/RING fragment possessed a 5- to 10-fold preference for UbcH3 (Table 1).
Fig. 2.
RAG1-B/RING exhibits E3 activity. (a) In vitro ubiquitylation assays were performed as described in Methods with E1 enzyme, radiolabeled PK-ubiquitin (Ubi*), E2 enzymes (0.3 μM), and RAG1-B/RING as indicated. (b) In vitro ubiquitylation assays were performed as described with the complete (Comp) reaction including Ubi*, RAG1-B/RING (R1), E1, UbcH3 (E2). Components were omitted as indicated (lanes 3–5). In lanes 6 and 7, WT RAG1-B/RING was replaced with the C325G mutant form or with RAG1-B/RING from which Zn had been removed with diethylenetriaminepenta-acetic acid (DTPA). RAG1-B/RING-dependent products are indicated by arrows. (c) In vitro ubiquitylation assays including apyrase (0.1 mg/ml) were staged, with components added at the times (in min) indicated. Buffer components and Ubi* were present from time 0. Reactions were incubated for an additional 2 h after the addition of R1. RAG1-B/RING-dependent products are indicated by arrows.
Table 1. RAG1-B/RING ubiquitylation activity.
| E2 enzyme* | Ubiquitin incorporation† | Relative activity‡ |
|---|---|---|
| UbcH2 | 13 ± 6 | 0.1 |
| UbcH3 | 114 ± 51 | 1 |
| UbcH5a | 28 ± 17 | 0.2 |
RAG1-B/RING-dependent ubiquitylation activity supported by UbcH5b, H5c, H6, H7, and H10 was below the level of detectability (Fig. 2a).
Results represent three independent trials with two different preparations of E2 enzymes.
Activity with the most active E2 (UbcH3) was arbitrarily set to 1.
RAG1-B/RING E3 activity completely depended on the presence of E1 and E2 enzymes (Fig. 2b, lanes 1–5). In addition, an intact RING finger was necessary for RAG1 E3 activity. To test this requirement, a mutation was introduced at one of the conserved cysteine residues (C325) that coordinates Zn ions in the RING domain. The RAG1 RING finger has a high degree of homology to the BRCA1 RING finger, which also possesses E3 activity (32, 39). Mutation of the equivalent cysteine in BRCA1 (C61) to glycine abolishes ubiquitylation activity and has been associated with tumor predisposition (32, 39). As expected, RAG1-B/RING[C325G] lacked E3 activity (Fig. 2b, lane 6). Removal of Zn from WT RAG1-B/RING by extensive dialysis against diethylenetriaminepenta-acetic acid also abolished ubiquitylation (Fig. 2b, lane 7).
ATP was required for charging of the E1 enzyme, but not for subsequent steps in the reaction. When apyrase was added before the E1 and E2 components to deplete ATP from the solution, ubiquitylation was abolished (Fig. 2c, compare lanes 3 and 5). However, incubation of E1 and E2 with ATP for 5 min before the addition of apyrase resulted in some ubiquitylation activity, even though RAG1-B/RING was not added until after the apyrase (Fig. 2c, compare lanes 5 and 7).
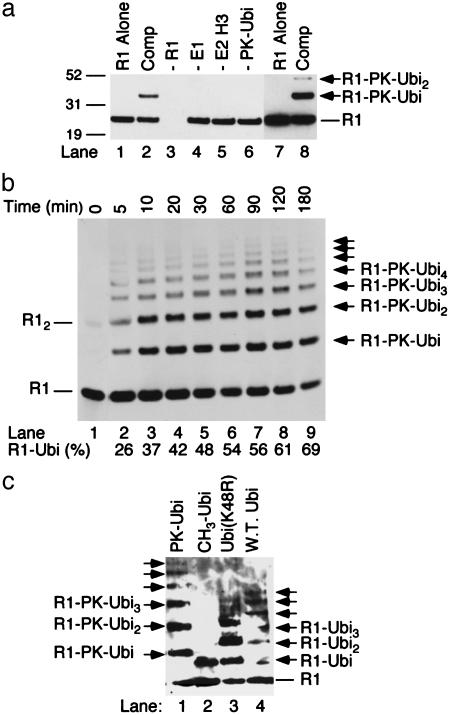
RAG1 Mediates Autoubiquitylation. The apparent size of the radiolabeled RAG1-B/RING-dependent ubiquitylated products suggested that the RAG1 fragment itself was becoming ubiquitylated, but this result was complicated by the similar mobilities of monoubiquitylated RAG1-B/RING and triubiquitin. Autoubiquitylation of RAG1 was confirmed by Western blot. Products from ubiquitylation reactions including RAG1-B/RING, E1, UbcH3, and unlabeled PK-ubiquitin were probed with an antibody specific for the RAG1 construct. In the complete reaction, higher molecular weight species corresponding in size to monoubiquitylation and diubiquitylation products of the RAG1-B/RING protein were present in addition to the unmodified protein (Fig. 3a, lanes 2 and 8). These products were absent when RAG1, E1, UbcH3, or PK-ubiquitin was omitted (Fig. 3a, lanes 3–6). For quantification of ubiquitylation, a fluorescent-conjugated antibody system was used. Approximately one-quarter of the RAG1-B/RING fragments had become conjugated to one or more ubiquitin moieties within the first 5 min of the reaction (Fig. 3b, lane 2), and by 30 min, nearly half of the RAG1-B/RING was ubiquitylated (Fig. 3b, lane 5). Products continued to accumulate over the course of a 3-h incubation (Fig. 3b). The primary products were monoubiquitylation and diubiquitylation species, but chains of up to seven ubiquitins conjugated to RAG1-B/RING were observed at later time points (Fig. 3b, lanes 7–9). The increased sensitivity of this system also revealed that a small amount of RAG1-B/RING (<1%) remained as a dimer even under the denaturing conditions of the gel (Fig. 3b, lane 1).
Fig. 3.
RAG1-B/RING undergoes autoubiquitylation. (a) In vitro ubiquitylation assays were performed as described in Methods with the complete reaction including E1, E2, RAG1-B/RING (R1), and unlabeled PK-ubiquitin (PK-Ubi). Components were omitted as indicated (lanes 3–6). RAG1-B/RING was detected by using anti-Xpress epitope antibody conjugated to horseradish peroxidase; lanes 7 and 8 are a longer exposure of lanes 1 and 2. (b) In vitro ubiquitylation assays were performed as described in Methods for the incubation times indicated. RAG1-B/RING was detected as in a except that unconjugated anti-Xpress antibody was detected with secondary antibody conjugated to Alexa 488 fluorescent probe. (c) In vitro ubiquitylation assays were performed by using PK-Ubi, untagged, reductively methylated ubiquitin (CH3-Ubi), ubiquitin K48R, or WT ubiquitin.
RAG1 Is Autoubiquitylated Primarily at K233 and Can Form Ubiquitin Chains with Residues Other Than K48. The presence of multiubiquitylated products raises the possibility that RAG1-B/RING is ubiquitylated at several sites. Polyubiquitylation at one site can be distinguished from monoubiquitylation at multiple sites by using ubiquitin whose K residues have been blocked by methylation (CH3-ubiquitin). CH3-ubiquitin can be covalently attached to the K residues on target proteins through its carboxyl terminus, but it is unable to form polyubiquitin chains. When RAG1-B/RING was ubiquitylated in a reaction including WT, untagged ubiquitin, both monoubiquitylation and polyubiquitylation products were present (Fig. 3c, lane 4; these products are smaller than those generated by ubiquitylation with the 11.3-kDa PK-ubiquitin). Ubiquitylation with CH3-ubiquitin produced only monoubiquitylated products (Fig. 3c, lane 2), indicating that RAG1-B/RING was ubiquitylated primarily on one K residue. Polyubiquitylation most often occurs through ubiquitylation of ubiquitin K48 but can also occur through other K resides. RAG1-B/RING could be polyubiquitylated with a ubiquitin K48R mutant (Fig. 3, lane 3), indicating that it does not depend on K48 to form polyubiquitin chains.
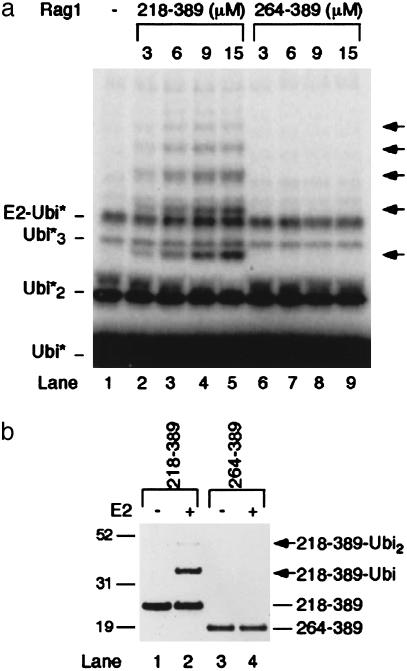
The basic region upstream of the RING finger motif was required for autoubiquitylation. The RING finger and adjoining Zn finger form a single domain (36, 37). A fragment spanning only this domain, RAG1[264–389], did not demonstrate ubiquitylation activity in an assay with radiolabeled PK-ubiquitin (Fig. 4a, compare lanes 2–5 to 6–9), and no species consistent with autoubiquitylated RAG1[264–389] were observed when products were analyzed by Western blot (Fig. 4b, compare lanes 2 and 4).
Fig. 4.
Conserved basic domains are required for RAG1-B/RING E3 activity. In vitro ubiquitylation assays were performed as described in Methods with radiolabeled PK-ubiquitin (a) or unlabeled PK-ubiquitin (b) and RAG1 B/RING (218–389) or RING (264–389) at the concentration indicated (a) or 8 μM (b). (a) Radiolabeled products were detected by phosphor-storage autoradiography. (b) RAG1 218–389 or 264–389 was detected by using anti-Xpress epitope antibody.
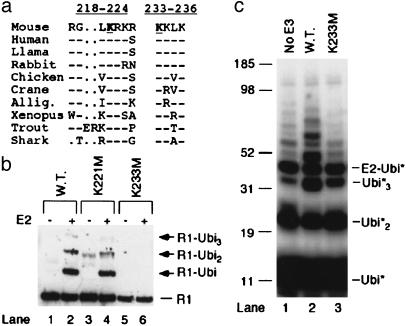
McMahan et al. (40) found that highly conserved basic regions contributed to rearrangement of extrachromosomal V(D)J substrates. Within these domains, we examined two K residues (K221 and K233 in murine RAG1) that are conserved in all species examined from a wide variety of vertebrate classes (Fig. 5a). Mutation of K221 to methionine had no effect on RAG1-B/RING ubiquitylation activity (Fig. 5b, compare lanes 2 and 4), although this mutation did increase the amount of dimer present on the denaturing gel (Fig. 5b, lane 3). Mutation of K233 abolished autoubiquitylation (Fig. 5b, lane 6), suggesting that this residue was the primary position of ubiquitylation. This was also true for the lower level of RAG1-B/RING autoubiquitylation supported by the E2 enzymes UbcH2 and UbcH5a (data not shown). RAG1-B/RING[K233M] mildly stimulated formation of triubiquitin relative to no E3 (Fig. 5c, compare lanes 1 to 2 and 3), suggesting that mutation of K233 does not disrupt RAG1 interaction with ubiquitin or UbcH3. After long incubation (>12 h), a very low level of ubiquitylation of RAG1-B/RING[K233M] was observed (data not shown), suggesting that other K residues can act as ubiquitin acceptors, but very inefficiently. These data indicate that K233 is the primary position of autoubiquitylation and that it is not generally required for RAG1-B/RING E3 activity. It is possible that full-length RAG1 may be modified at additional K residues.
Fig. 5.
RAG1-B/RING autoubiquitylation occurs primarily on K233. (a) Alignments of RAG1 segments from various species were conducted by using the National Center for Biotechnology Information blast resource. (b) In vitro ubiquitylation assays were carried out as described in Methods with unlabeled PK-ubiquitin and B/RING WT, K221M, or K233M. RAG1 B/RING was detected by using anti-Xpress epitope antibody. (c) In vitro ubiquitylation assays were carried out by using radiolabeled PK-ubiquitin and RAG1-B/RING WT or K233M.
Discussion
Ubiquitin conjugation plays vital regulatory roles in a wide variety of cellular processes. The previous finding that RAG2 is ubiquitylated before its destruction suggests that ubiquitin conjugation participates in regulation of V(D)J recombination (2). We find that the Zn binding domain of RAG1 and upstream basic region (amino acids 218–389) with the assistance of UbcH3/CDC34 can promote ubiquitylation at a single highly conserved RAG1 K residue, K233, with as much as 70% of RAG1 undergoing modification in vitro. In addition, full-length RAG1 undergoes polyubiquitylation in intact cells. RAG1's ability to act as an E3 protein, described here and elsewhere (3), suggests that promotion of ubiquitin conjugation is intimately linked to the other biochemical activities of the recombinase.
The structure of the RAG1 RING finger is unusual in several respects. Typical RING fingers such as cCBL and BRCA1 coordinate two Zn ions in a so-called cross-brace motif (41, 42). The RAG1 RING also adopts the cross-brace structure but binds three Zn ions, with two included in a binuclear cluster coordinated in part by additional cysteines and histidines upstream of the canonical RING finger residues (36). RING proteins like Rbx1 also exhibit an atypical RING structure including coordination sites for a third zinc ion (43), and this family of RING proteins is frequently associated with UbcH3/CDC34 or its homologues. However, in Rbx1 the additional coordination sites are contributed by an extra loop within the canonical RING domain instead of by residues upstream, suggesting that the RAG1 RING is not closely related evolutionarily to Rbx1. Unlike other RING proteins, the RAG1 RING motif forms a single, integrated domain with a downstream C2H2 Zn finger, which coordinates a fourth Zn ion (36, 37). Finally, the shallow groove within the cross-brace that has been demonstrated to play an important role in other RING-E2 interactions (42) is nearly absent in RAG1 (36), being completely filled by amino acid side chains. Additional study will be necessary to determine how these factors influence RAG1-promoted ubiquitylation.
Yurchenko et al. (3) reported that the N-terminal region of RAG1 (amino acids 1–383) attached to a short S-peptide linker possesses weak E3 monoubiquitylation activity directed toward S protein bound to the linker. This activity was supported by the E2 enzymes UbcH4 and UbcH10 but by no other E2s. This group also found low levels of substrate-independent ubiquitin polymerization by the E2s UbcH5b and UbcH5c in the presence of the RAG1 RING alone (amino acids 250–350) without the attached Zn finger (3). The robust autoubiquitylation activity of the RAG1-B/RING (amino acids 218–389) presented here was best promoted by UbcH3/CDC34, but was also observed to a lesser extent with UbcH2/Rad6 and UbcH5a. The different RAG1 constructs used by the two laboratories (see above) are likely to have contributed to the different observations. It is also possible that RAG1 is active with different E2 enzymes in the cell under various circumstances.
There are numerous potential roles for RAG1-promoted ubiquitylation in V(D)J recombination. The RING finger lies outside of the core domain, which includes the minimal residues necessary for DNA cleavage. However, the conservation of the RING and the upstream basic domains among RAG1 sequences from many species suggests that they play some important role. In addition, mutation of the RING finger and truncations of the N terminus are associated with a fatal immunodeficiency syndrome (20–22), indicating that this region is essential for normal lymphocyte development in humans. Deletion of more than a third of the protein to form the core may remove multiple positive and negative regulatory regions, making it difficult to assess their true roles in normal RAG1 biology by examining activity of the core alone. In addition, if the role of the RING finger is to down-regulate RAG1 activity, this may not be apparent in the extrachromosomal V(D)J recombination assays typically used to determine mutant activity, particularly if RAG1 is not the limiting factor. Nevertheless, there is some genetic data that these domains do influence RAG1 activity and expression (8, 16, 17, 40). RAG1 has a short half-life (8), and various truncations of the N terminus lead to higher steady-state levels of RAG1 expression (40). Moreover, mutation of the basic region including K233 increases RAG1 expression to a greater extent than mutation of other nearby basic domains (40). Deletion of the RING finger in the context of an otherwise full-length N terminus can also increase expression of the protein (17, 40). These data suggest that one function of the RAG1 RING may be to modulate its own turnover, potentially through autoubiquitylation of K233.
Other data suggest that the RING motif plays a role specifically in promoting signal joint formation. Mutation of the RING region drastically reduces signal joint formation on extrachromosomal V(D)J recombination substrates (8, 16). Likewise, the RAG1 core is associated with an increase in overall levels of cleaved signal ends along with a decrease in signal joints, with coding joint levels remaining relatively unaffected (17). After cleavage, the RAG1/2 recombinase remains tightly bound to the recombination signal sequence ends and sequesters them from the repair machinery (44, 45). The ends are joined together to form signal joints at the G1/S transition (46), and it has been hypothesized that specific factors remodel or disassemble the complex at that time, allowing joining to proceed. The E2 enzyme with which we find RAG1 to be most active, UbcH3/CDC34, enables the transition from G1 to S phase by promoting degradation of p27KIP (47). UbcH3/CDC34-promoted RAG1 E3 activity occurring at the G1/S transition could also assist in remodeling of the postcleavage complex through ubiquitylation of RAG1, RAG2, or another target. The small percentage of RAG1 that was ubiquitylated in our unsynchronized, nearly confluent cell sample relative to the robust ubiquitylation we observed in vitro may be explained by RAG1 ubiquitylation occurring in only a fraction of the cells. This would be consistent with a cell-cycle dependent process.
RAG1 belongs to a family of proteins that includes many recombinases, integrases, and transposases characterized by a conserved trio of acidic amino acids, the so-called DDE motif (48–50). Several of these proteins, such as HIV integrase and RAG1, include traditional C2H2 zinc fingers, but to our knowledge RAG1 is the only DDE motif protein that also includes a RING finger and E3 activity. The RAG1/2 recombinase is hypothesized to have evolved from an ancient transposon that entered the genome at the level of the common ancestor to all jawed vertebrates (51). The absence of E3 activity in all other known transposases suggests that either (i) possession of this activity made it especially well suited to “domestication” by the immune system or (ii) RAG1 acquired this activity after entering the vertebrate genome as it was adapted for use by the cell. Thus, further research into the role of RAG1's E3 activity could well shed light on the evolution of the diversified immune system as a whole, in addition to answering vital questions regarding regulation of this developmentally important DNA rearrangement.
Acknowledgments
We thank Y. Xiong, M. Oettinger, and S. Elkin for their gifts of materials and advice. J.M.J. was supported by the Damon Runyon Cancer Research Foundation (Grant DRG1582).
References
- 1.Gellert, M. (2002) Annu. Rev. Biochem. 71, 101–132. [DOI] [PubMed] [Google Scholar]
- 2.Mizuta, R., Mizuta, M., Araki, S. & Kitamura, D. (2002) J. Biol. Chem. 277, 41423–41427. [DOI] [PubMed] [Google Scholar]
- 3.Yurchenko, V., Xue, Z. & Sadofsky, M. (2003) Genes Dev. 17, 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oettinger, M. A., Schatz, D. G., Gorka, C. & Baltimore, D. (1990) Science 248, 1517–1523. [DOI] [PubMed] [Google Scholar]
- 5.Schatz, D. G., Oettinger, M. A. & Baltimore, D. (1989) Cell 59, 1035–1048. [DOI] [PubMed] [Google Scholar]
- 6.McBlane, J. F., van Gent, D. C., Ramsden, D. A., Romeo, C., Cuomo, C. A., Gellert, M. & Oettinger, M. A. (1995) Cell 83, 387–395. [DOI] [PubMed] [Google Scholar]
- 7.van Gent, D. C., McBlane, J. F., Ramsden, D. A., Sadofsky, M. J., Hesse, J. E. & Gellert, M. (1995) Cell 81, 925–934. [DOI] [PubMed] [Google Scholar]
- 8.Sadofsky, M. J., Hesse, J. E., McBlane, J. F. & Gellert, M. (1993) Nucleic Acids Res. 21, 5644–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadofsky, M. J., Hesse, J. E. & Gellert, M. (1994) Nucleic Acids Res. 22, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuomo, C. A. & Oettinger, M. A. (1994) Nucleic Acids Res. 22, 1810–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silver, D. P., Spanopoulou, E., Mulligan, R. C. & Baltimore, D. (1993) Proc. Natl. Acad. Sci. USA 90, 6100–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiom, K. & Gellert, M. (1997) Cell 88, 65–72. [DOI] [PubMed] [Google Scholar]
- 13.Hiom, K. & Gellert, M. (1998) Mol. Cell 1, 1011–1019. [DOI] [PubMed] [Google Scholar]
- 14.van Gent, D. C., Ramsden, D. A. & Gellert, M. (1996) Cell 85, 107–113. [DOI] [PubMed] [Google Scholar]
- 15.Elkin, S. K., Matthews, A. & Oettinger, M. (2003) EMBO J. 22, 1931–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roman, C. A., Cherry, S. R. & Baltimore, D. (1997) Immunity 7, 13–24. [DOI] [PubMed] [Google Scholar]
- 17.Steen, S. B., Han, J. O., Mundy, C., Oettinger, M. A. & Roth, D. B. (1999) Mol. Cell. Biol. 19, 3010–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai, C. & Schatz, D. G. (2003) EMBO J. 22, 1922–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirch, S. A., Rathbun, G. A. & Oettinger, M. A. (1998) EMBO J. 17, 4881–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noordzij, J. G., Verkaik, N. S., Hartwig, N. G., de Groot, R., van Gent, D. C. & Van Dongen, J. J. M. (2000) Blood 96, 203–209. [PubMed] [Google Scholar]
- 21.Santagata, S., Gomez, C. A., Sobacchi, C., Bozzi, F., Abinun, M., Pasic, S., Cortes, P., Vezzoni, P. & Villa, A. (2000) Proc. Natl. Acad. Sci. USA 97, 14572–14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villa, A., Sobacchi, C., Notarangelo, L. D., Bozzi, F., Abinun, M., Abrahamsen, T. G., Arkwright, P. D., Baniyash, M., Brooks, E. G., Conley, M. E., et al. (2001) Blood 97, 81–88. [DOI] [PubMed] [Google Scholar]
- 23.Pickart, C. M. (2001) Annu. Rev. Biochem. 70, 503–533. [DOI] [PubMed] [Google Scholar]
- 24.Hershko, A., Heller, H., Elias, S. & Ciechanover, A. (1983) J. Biol. Chem. 258, 8206–8214. [PubMed] [Google Scholar]
- 25.Chau, V., Tobias, J. W., Bachmair, A., Marriott, D., Ecker, D. J., Gonda, D. K. & Varshavsky, A. (1989) Science 24, 1576–1583. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser, P., Flick, K., Wittenberg, C. & Reed, S. I. (2000) Cell 102, 303–314. [DOI] [PubMed] [Google Scholar]
- 27.Spence, J., Sadis, S., Haas, A. L. & Finley, D. (1995) Mol. Cell. Biol. 15, 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spence, J., Gali, R. R., Dittmar, G., Sherman, F., Karin, M. & Finley, D. (2000) Cell 102, 67–76. [DOI] [PubMed] [Google Scholar]
- 29.Lin, W. C. & Desiderio, S. (1994) Proc. Natl. Acad. Sci. USA 91, 2733–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laney, J. D. & Hochstrasser, M. (1999) Cell 97, 427–430. [DOI] [PubMed] [Google Scholar]
- 31.Honda, R. & Yasuda, H. (2000) Oncogene 19, 1473–1476. [DOI] [PubMed] [Google Scholar]
- 32.Mallery, D. L., Vandenberg, C. J. & Hiom, K. (2002) EMBO J. 21, 6755–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, Y., Fang, S., Jensen, J. P., Weissman, A. M. & Ashwell, J. D. (2000) Science 288, 874–877. [DOI] [PubMed] [Google Scholar]
- 34.Huibregste, J. M., Scheffner, M., Beaudenon, S. & Howley, P. M. (1995) Proc. Natl. Acad. Sci. USA 92, 2563–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S. & Weissman, A. M. (1999) Proc. Natl. Acad. Sci. USA 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellon, S. F., Rodgers, K. K., Schatz, D. G., Coleman, J. E. & Steitz, T. A. (1997) Nat. Struct. Biol. 4, 586–591. [DOI] [PubMed] [Google Scholar]
- 37.Rodgers, K. K., Bu, Z., Fleming, K. G., Schatz, D. G., Engelman, D. M. & Coleman, J. E. (1996) J. Mol. Biol. 260, 70–84. [DOI] [PubMed] [Google Scholar]
- 38.Furukawa, M., Ohta, T. & Xiong, Y. (2002) J. Biol. Chem. 277, 15758–5765. [DOI] [PubMed] [Google Scholar]
- 39.Hashizume, R., Fukuda, M., Maeda, I., Nishikawa, H., Oyake, D., Yabuki, Y., Ogata, H. & Ohta, T. (2001) J. Biol. Chem. 276, 14537–14540. [DOI] [PubMed] [Google Scholar]
- 40.McMahan, C. J., Difilippantonio, M. J., Rao, N., Spanopoulou, E. & Schatz, D. G. (1997) Mol. Cell. Biol. 17, 4544–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brzovic, P. S., Rajagopal, P., Hoyt, D. W., King, M. & Klevit, R. E. (2001) Nat. Struct. Biol. 8, 833–837. [DOI] [PubMed] [Google Scholar]
- 42.Zheng, N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. (2000) Cell 102, 533–539. [DOI] [PubMed] [Google Scholar]
- 43.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., et al. (2002) Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
- 44.Agrawal, A. & Schatz, D. G. (1997) Cell 89, 43–53. [DOI] [PubMed] [Google Scholar]
- 45.Jones, J. M. & Gellert, M. (2001) Proc. Nat. Acad. Sci. USA 98, 12926–12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsden, C. A. & Gellert, M. (1995) Genes Dev. 9, 2409–2420. [DOI] [PubMed] [Google Scholar]
- 47.Deshaies, R. J. (1999) Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- 48.Fugmann, S. D., Villey, I. J., Ptaszek, L. M. & Schatz, D. G. (2000) Mol. Cell 5, 97–107. [DOI] [PubMed] [Google Scholar]
- 49.Landree, M. A., Wibbenmeyer, J. A. & Roth, D. B. (1999) Genes Dev. 13, 3059–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim, D. R., Dai, Y., Mundy, C. L., Yang, W. & Oettinger, M. A. (1999) Genes Dev. 13, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gellert, M., Hesse, J. E., Hiom, K., Melek, M., Modesti, M., Paull, T. T., Ramsden, D. A. & van Gent, D. C. (1999) Cold Spring Harbor Symp. Quant. Biol. 64, 161–167. [DOI] [PubMed] [Google Scholar]