Abstract
Robust high-throughput synthesis methods are needed to expand the repertoire of repetitive protein-polymers for different applications. To address this need, we developed a new method, overlap-extension rolling circle amplification (OERCA), for the highly parallel synthesis of genes encoding repetitive protein-polymers. OERCA involves a single PCR-type reaction for the rolling circle amplification of a circular DNA template and simultaneous overlap extension by thermal cycling. We characterized the variables that control OERCA and demonstrated its superiority over existing methods, its robustness, throughput and versatility by synthesizing variants of elastin-like polypeptides (ELPs) and protease-responsive polymers of a glucagon-like peptide-1 analog. Despite the GC-rich, highly repetitive sequences of ELPs, we synthesized remarkably large genes without recursive ligation. OERCA also enabled us to discover “smart” biopolymers that exhibit fully reversible thermally responsive behavior. This powerful strategy generates libraries of repetitive genes over a wide and tunable range of molecular weights in a “one-pot” parallel format.
Artificial repetitive polypeptides – also termed protein-polymers – derived from short peptide motifs found in elastin, collagen, silk and other structural proteins exhibit unique mechanical, structural and biological properties. These attributes have led to their application in biotechnology, tissue engineering, drug delivery, and biosensing1–5. Recombinant DNA technology is attractive for the synthesis of protein-polymers because it enables precise control of their length (number of repeats), composition and stereochemistry. This level of control is especially important for the in vivo applications of these biopolymers, because the polymer molecular weight controls their pharmacokinetics and biodistribution, while the amino acid sequence imparts biological activity to the biopolymer and affects their route, rate and mechanism of biodegradation. Recombinant DNA technology is also of interest for the synthesis of tandem repeats of naturally occurring peptides, as a strategy for the high-yield synthesis of peptide drugs and antigens 6–10. Furthermore, polymerization of peptide drugs with intervening protease cleavable sequences has the potential to improve their pharmacokinetics and drug efficacy11,12. However, current methods for the polymerization of DNA suffer from one or more critical limitations: they (1) require numerous steps, (2) are difficult to parallelize, and (3) do not provide tunable control over a range of molecular weights, all of which greatly limit the ability to simultaneously synthesize multiple variants with a range of repeat units and compositions.
Motivated by these limitations, we report a rapid, one-step, high-throughput and robust method for the recombinant polymerization of “monomer” DNA sequences with tunable control over the number of repeats. This method, which we term overlap-extension rolling circle amplification (OERCA), uses rolling circle amplification (RCA) to produce linear repeats of a circularized gene, followed by thermally cycled overlap-extension (OE) to yield a library of polymers of the monomer DNA, all in a single PCR reaction.
Here we show the utility of OERCA and its advantages over existing methods by the synthesis of two classes of protein-polymers. First, we demonstrate the parallel synthesis of genes that encode elastin-like polypeptides (ELPs), a family of thermally responsive protein-polymers derived from a recurring VPGVG pentapeptide found in elastin 13. We used OERCA to rapidly synthesize 9 different variants of ELPs, by substituting or inserting Alanine residues along the VPGVG motif. These studies revealed an unexpected degree of sequence promiscuity in the parent peptide motif and yielded new families of “smart” protein-polymers that display fully reversible thermo-responsive behavior, which will provide a new set of stimulus responsive motifs for biomedical and biotechnological applications. In a second example, we use OERCA to rapidly synthesize protease-responsive polymers of a glucagon-like peptide-1 (GLP-1) analog, with intervening protease sites of variable potency for the optimization of in vivo pharmacokinetics and release of GLP-1 from the polymer.
OERCA uses a circular ssDNA sequence that encodes for a “monomer” target gene. This ssDNA circle is then thermally cycled with primers that are complementary to the 5’- and 3’-ends of the linear monomer. During the annealing step in the first cycle, the antisense primer binds to the circular ssDNA template and forms linear oligomers as the polymerase rolls around the circle (Fig. 1). This is the only reaction in the first cycle, and is presumably the predominant extension reaction that produces sense strands of variable lengths during the initial cycles (Supplementary Fig S1). From the second cycle onward, the sense primer binds to the newly synthesized linear strands to produce double-stranded products of variable lengths. As these products accumulate, overlap extension – in which the DNA strands self-prime by binding asymmetrically within the repetitive regions – begins to dominate the reaction, producing longer products, while residual primers simultaneously synthesize complementary DNA of the same or shorter size from existing templates by binding internally at complementary sites on the repeating sequence. The product of this reaction consists of a library of DNA oligomers, wherein the oligomer size range can be tuned by the primer concentration and number of thermal cycles (Fig. 2). As newly bound primers can generate products of the same or shorter length than the template strand, increasing the primer to template ratio in the reaction mixture generates more, albeit shorter products. In contrast, as the primer to template ratio is decreased, overlap extension of existing strands is favored since fewer primers are available to compete with their hybridization, resulting in a longer product at the expense of yield.
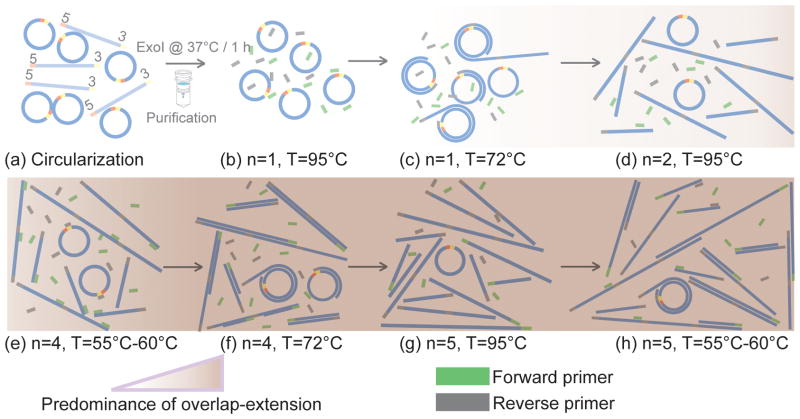
Figure 1.
Schematic of snapshots depicting the evolution of an OERCA reaction. (a) A linear oligonucleotide is first circularized yielding a population of circular DNA, which is enriched by removal of the remaining linear DNA before the start of the OERCA reaction (b–h). (b) The circularized oligonucleotide is added to a conventional PCR mixture containing forward and reverse primers. (c) After annealing of the reverse primer, extension in cycle 1 (n=1) occurs primarily in the form of rolling circle amplification. (d) Upon DNA denaturation in cycle 2, linear repeats of the original circularized sequence become available as extension templates. (e–f) As the reactions proceeds, primer annealing and overlap extension preferentially take place to amplify the linear DNA. (g–h) Upon DNA denaturation the repetitive single-stranded DNA is capable of priming/overlapping, which further promotes the extension of the original repeat unit.
Figure 2.
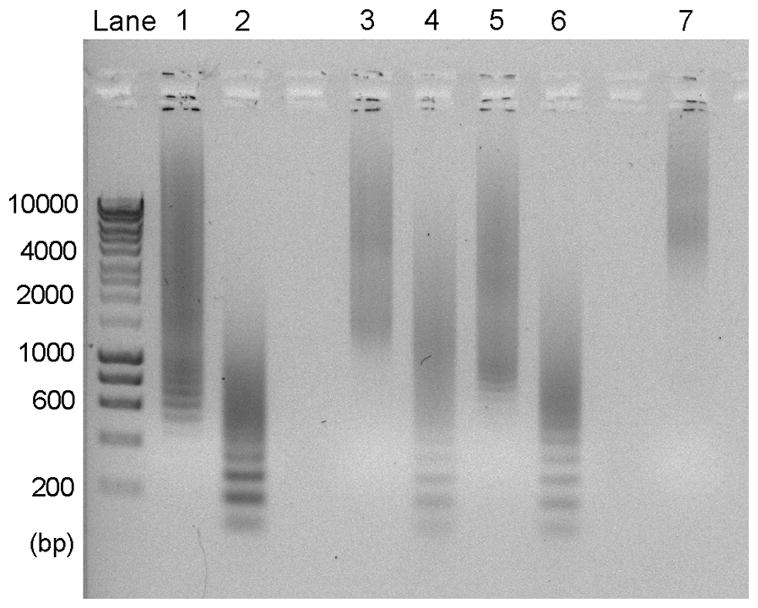
The effect of primer concentration and cycle number on the size range of the DNA product. The monomer gene for GLP-1 was used as a model. Lanes 1 and 2 are the products of 30 cycles with 10 and 40 pmol primer, respectively. All PCR reactions were column purified to remove excess primers and recover product. Products from lanes 1–2 were then subjected to 15 more cycles with no additional primer (lanes 3 and 5, respectively) or with 20 pmol primer (lanes 4 and 6). The size range of the DNA product can be further increased by 15 additional cycles without primer (lane 7 generated from lane 3).
Because this reaction is thermally cycled and uses primers that bind at the beginning and the end of each repeat unit, the vast majority of products have 5’- and 3’-ends that precisely match the target monomer sequence. Therefore, OERCA products are comprised of an integer number of repeats of the monomer gene and have blunt ends suitable for cloning directly into any blunt site. Hence, these products can be directly cloned into an expression vector to create an expression library. To illustrate this, Fig. 3a shows the size range of DNA oligomers generated by OERCA encoding for poly(AVPGVG), which were ligated to an expression vector linearized by a blunt restriction enzyme. We screened over 90% of ~200 total colonies and assessed insert orientation by directional cPCR, in which one primer binds to the vector and the other primer binds to a defined sequence in the insert, such that amplification is only achieved when the insert is in the correct orientation. Due to the repetitive nature of the insert, the internal primer can bind at multiple sites, so that a positive clone is identified by the presence of a smear of DNA on an agarose gel rather than a single band as typically observed for diagnostic PCR (Fig. 3b). We then selected ~50 positive clones based on directional cPCR analysis, and performed restriction analysis and direct DNA sequencing to confirm sequence accuracy and determine the distribution of oligomer sizes among these clones (Fig. 3c). Although we observed a higher efficiency of insertion for relatively short DNA fragments, medium-sized inserts occurred at high frequencies, and large genes of up to 1.5 kbs were also successfully isolated. Despite the well-known challenge of amplifying GC-rich repetitive templates by PCR 14,15, and the difficulty in sequencing such genes, we found that less than 0.05% of the > 26,000 sequenced bases were erroneous (mainly missing G or C nucleotides), and only 5 clones were internally truncated (likely due to incomplete overlap extension or secondary structure formation). Overall, this single “one-pot” reaction yielded a large number of sequence verified clones comprising a library of 11 different gene lengths and encoding for protein-polymers as small as 10 hexapeptides to remarkably long polypeptides over 80 hexapeptides in length, all in a single cloning reaction. Furthermore, we observed that the distribution of gene lengths in a library is readily tuned by adjusting the size range of the OERCA products prior to ligation (Supplementary Fig. S2).
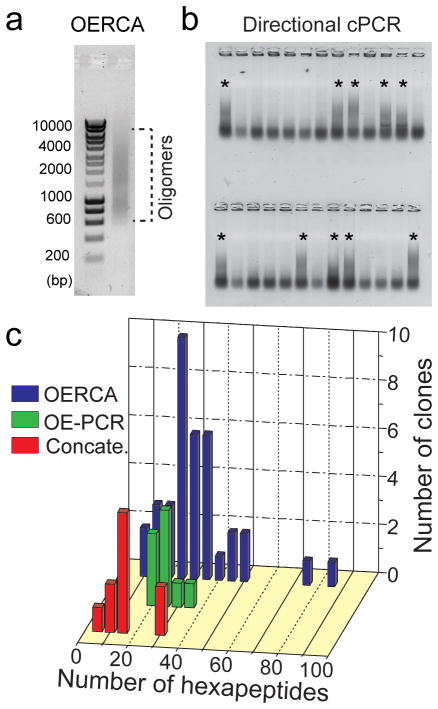
Figure 3.
Synthesis of polypeptide libraries by OERCA is simple and outperforms current synthesis methods. The product of an OERCA reaction (a) was ligated into a vector and positive colonies were screened by directional cPCR (b), wherein positive clones can be identified by the presence of a large DNA smear (*). Positive clones were subjected to restriction analysis and DNA sequencing to verify insert size and sequence fidelity (c). OERCA, unlike OE-PCR and concatemerization, enabled the synthesis of constructs with a wide size distribution (0.18 −1.5 Kbp). Insert sizes larger than 0.9 Kbp were estimated by restriction analysis.
To demonstrate the advantages of OERCA over standard methods for the synthesis of recombinant protein-polymer libraries, we compared the performance of OERCA, overlap elongation PCR (OE-PCR) and concatemerization in the synthesis of DNA oligomers encoding for poly(AVPGVG) (see supplementary information and Supplementary Fig. S3–S5). The number of colonies obtained from OE-PCR (~300) was similar to that of OERCA (~200), while concatemerization always yielded a lower number of colonies (~50–100 for AVPGVG and other repetitive genes shown in Fig. S5). Colonies obtained by the three methods were then screened by directional cPCR and positive clones (i.e., those with inserts in the correct orientation) were further subjected to restriction analysis and DNA sequencing to verify insert size and sequence fidelity. The size distribution of the resulting clones is shown in Figure 3c. The number of positive clones was far lower for OE-PCR as compared with OERCA, and DNA sequencing revealed a high incidence of internal truncation in clones generated by OE-PCR (data not shown), consistent with a previous report 16. Concatemerization, despite generating fewer clones, typically produced a large fraction of positive clones, likely due to the high proportion of small concatemers and the presence of 5’ overhangs. Libraries constructed by OEPCR and concatemerization were mostly comprised of small inserts and no inserts longer than ~0.6 Kbps were observed. In contrast, OERCA was the only method of the three to produce long constructs in the size range of 0.8 to 1.5 Kbp (~ 20 % of the clones) that encode for 45–85 hexapeptide repeats. Notably, around 50 % of all inserts obtained with OERCA were larger than 0.54 Kbp. Furthermore, libraries generated by OERCA and concatemerization for the GC-rich repetitive gene encoding for poly(VPGVA) demonstrated the inability of concatemerization to produce oligomers longer than 400 bp (Supplementary Fig. S5c), and affirmed the ability of OERCA to generate a large number of mutation-free constructs in the 400–825 bp range (upon screening an identical number of clones). Additionally, concatemerization resulted in an extremely high error rate for VPGVA (70 %) (Supplementary Fig. S5c), unlike OERCA (~30 %). These results, hence clearly demonstrate that OE-PCR and concatemerization are inferior to OERCA in ligation efficiency, size diversity of the library and sequence fidelity.
We next decided to test the robustness and high-throughput potential of OERCA by synthesizing libraries of structural variants of ELPs. ELPs are soluble in aqueous solution below their inverse transition temperature (Tt), but when the temperature is raised above their Tt, they undergo a sharp (~2°C range) phase transition, leading to the subsequent formation of an ELP-rich coacervate phase. We chose ELPs for two reasons: first they present a synthetic challenge, as they are comprised of highly repetitive, GC-rich sequences 14,15. Second, the sequence requirements and constraints that govern their stimulus responsive behavior remain somewhat of a mystery, despite four decades of investigation. To date, it is unclear to what extent the generalized VPGXG motif (where X is any amino acid except Proline) can be altered without losing thermally responsive coacervation behavior, partly because methods to synthesize genes for ELPs that span a range of compositions and MWs are tedious and difficult to implement in parallel. Motivated by the large gap in our understanding of the phase behavior of ELPs and the increasing interest in these polymers for a wide range of applications 5,17, we used OERCA to demonstrate the ease of performing a systematic study of the sequence constraints that govern the phase transition behavior of these protein-polymers.
We generated libraries of genes that encode diverse chain lengths of 9 different Alanine (A) insertion and substitution mutants of poly(VPGVG), as well as the parent motif (Supplementary Table S1). We refer to these polypeptides as aELPs for brevity. We screened 96 colonies (of ~200) for each aELP motif, which typically yielded ~5 different gene lengths encoding protein-polymers with 10–55 repeats of the pentapeptide or hexapeptide motifs. We then characterized the thermally responsive behavior of this family of aELPs, with the exception of polypeptides with the repeat unit VAGVG, as they expressed poorly.
These experiments resulted in the exciting finding that the entire set of aELPs that were characterized display thermally responsive behavior, despite the various mutations introduced to the VPGVG motif (Fig. 4a). This finding is significant because it hints at the existence of a large and diverse set of motifs – far larger than the canonical VPGXG motif – that are capable of exhibiting stimulus responsive phase behavior. In previous work by Urry and others 18 , non-canonical ELP motifs often failed to display fully reversible phase transition behavior with negligible thermal hysteresis, as observed for ELPs 19,20. In contrast, we demonstrate, for the first time, the existence of more complex hexapeptide motifs AVPGVG, VPAGVG, VPGAVG and VPGVAG capable of displaying fully reversible thermally triggered phase transition behavior (Fig. 4b) and environmental sensitivity to both solution temperature and solute concentration (Fig. 4 and Fig. 5).
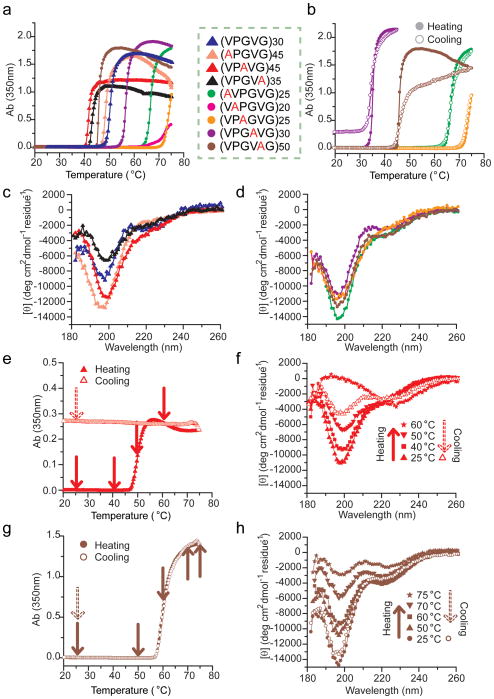
Figure 4.
Thermally responsive behavior of aELPs constructed by OERCA. (a) The turbidity profiles for all aELPs exhibit a sharp transition with temperature, characteristic of the inverse phase transition behavior displayed by canonical ELPs. We discovered 4 new hexapeptide motifs (b) that display reversible phase transition behavior. All polypeptides in (a–b) were prepared at a concentration of 50 μM in PBS (i.e., 0.14 M NaCl), except VAPGVG (0.64 M NaCl) and APGVG (2.14 M NaCl). Circular dichroism spectra at 25 °C revealed highly disordered conformations predominant in aELPs with both pentapeptide (c) and hexapeptide (d) motifs, similar to that of the canonical ELP. (e–h) The coacervation of aELPs with two distinct phase transition behaviors was studied by CD at various stages (arrows in e and g) of the coacervation process. The CD spectra and associated turbidity profiles were acquired in water at a polypeptide concentration of 5 μM, and θ indicates the mean residue ellipticity. The pentapeptide motif VPAVG (e–f) underwent quasi-irreversible phase separation (e) and lost its highly disordered conformers (i.e., negative peak at 197 nm) in the mature coacervate and upon cooling to 25 °C (f). In contrast, the hexapeptide motif VPGVAG (g–h) exhibited fully reversible phase transition behavior (g), highly disordered conformers were preserved in the mature coacervate, and the secondary structure of the polypeptide was completely recovered upon cooling (h). We also confirmed the reversible behavior of VPAVG upon cooling to 4 °C (Supplementary Fig. S7).
Figure 5.
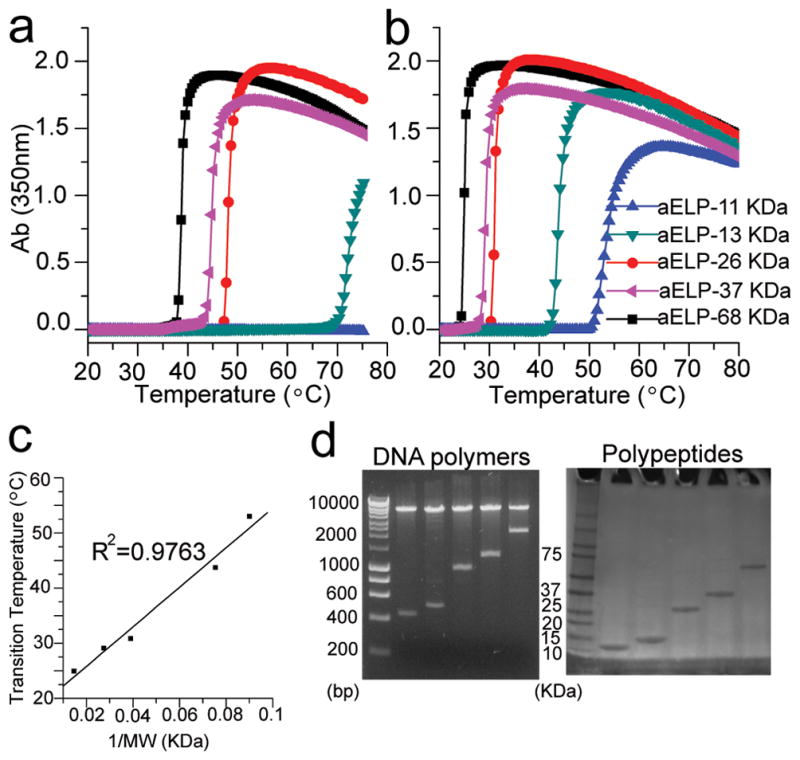
Gene synthesis, expression and characterization of a library of aELPs with the repeating sequence AVPGVG. Turbidity profiles for 5 constructs in this library in PBS (a) and PBS supplemented with NaCl to 1.14 M (b). The transition temperatures calculated from (b) varied linearly as the reciprocal of molecular weight of the aELP as expected for canonical ELP sequences (c). The wide distribution of molecular weights in this library synthesized by OERCA is illustrated at both the DNA and polypeptide level, which demonstrates the ability of OERCA to readily generate both low and large molecular weight protein-polymers (d).
The thermally-responsive behavior of canonical ELPs is easily and quantitatively tuned by controlling the polypeptide molecular weight 21. Therefore, we were interested in studying the molecular weight dependence of the thermally-responsive behavior of these novel aELP motifs. We harnessed the ability of OERCA to tune gene length by synthesizing a library encoding for aELPs with the hexapeptide motif AVPGVG. This library was assembled from the transformation reactions of two OERCA products, in which the reaction conditions had been modified to yield two different molecular weight distributions (Supplementary Fig. S2). This enabled the construction of a library spanning insert sizes from ~270 to ~2500 bp (Fig. 5). We then expressed, purified and characterized the phase transition behavior of 5 constructs spanning the entire range of molecular weights in this library. The transition temperature of these polypeptides decreased in a linear fashion as a function of the reciprocal of the molecular weight, as previously described for canonical ELPs (Fig. 5c–d) 21. Hence, the thermo-responsive behavior of this aELP is easily tunable by controlling polypeptide molecular weight. The functionality – thermal responsiveness – of this library of poly(AVPGVG) spanning polypeptides from ~10 KDa to ~70 KDa also serves to demonstrate that OERCA can be used to readily synthesize repetitive protein-polymers over an unprecedented wide range of sizes without the need for recursive cloning steps.
The behavior of aELPs also provided some insight into the role of hydrophobic interactions on the phase transition of elastin and ELPs. Despite having the same amino acid composition (and hence hydrophobicity) and molecular weight, the Tt of polypeptides composed of repeats of AVPGVG and VPAGVG that only differ in amino acid arrangement, are substantially different (Fig. 4a). This suggests that the overall hydrophobicity of these polypeptides fails to explain their propensity for coacervation (i.e., whether the Tt occurs at high or low temperatures), and in turn suggests that other factors such as their secondary structure are likely to contribute to the differences in their phase transition behavior. This motivated a study of the secondary structure of aELPs by circular dichroism (CD), which revealed that the aELPS are characterized by an ensemble of highly disordered structures populated by random coil, β-turns and distorted β-sheet conformations (Fig. 4c–d), reminiscent of the secondary structure of tropoelastin and ELPs 19,20. Interestingly, these studies suggest that different aELPs have different secondary structure propensities (Fig. 4c–d and Supplementary Fig. S6), as seen differences in their degree of disorder (i.e., the magnitude of the negative peak at ~197 nm) and the fraction of β-turns and/or distorted β-sheets (i.e., negative shoulder around 210 nm) 20. These differences may eventually lead to a better understanding of coacervation propensity, but we note that this “conformational” promiscuity is observed even among polypeptides that display ideal fully reversible phase transition behavior.
We then investigated the relationship between the phase behavior and secondary structure of two aELPs with distinct thermally responsive behavior by concurrently measuring their turbidity profiles and their CD spectra at various stages of the thermally triggered coacervation process (CD spectra were recorded at temperatures indicated by arrows in Fig. 4e, g). The aELP consisting of repeats of the pentapeptide motif VPAVG exhibited quasi-irreversible phase separation with large hysteresis (Fig. 4e), and complete solubility was only recovered upon cooling the coacervate down to 4 °C (Supplementary Fig. S7a). In contrast, the aELP consisting of repeats of the hexapeptide motif VPGVAG exhibited fully reversible phase transition behavior as a function of solution temperature (Fig. 4g). CD spectroscopy illuminated the structural origins of these differences in their thermally triggered coacervation. Whereas both polypeptides showed significant loss of structural disorder as they were heated, as seen by a decrease in intensity of the negative peak at ~197 nm, their CD spectra differed significantly in the mature coacervate stage (i.e., at temperatures corresponding to the maximum absorbance in Fig. 4e-, g) and upon subsequent cooling below the Tt. The quasi-irreversible phase behavior of poly(VPAVG) was accompanied by a dramatic loss of structural disorder (i.e., a nearly positive peak at 197 nm) that persisted to a large extent upon cooling well below the Tt (Fig. 4f), and the original degree of disorder was only recovered upon cooling to 4 °C, in accordance with the reversal of the coacervation process as assessed by turbidity data (Supplementary Fig. S7b). In contrast, poly(VPGVAG) exhibited significant residual disorder in the mature coacervate (i.e., negative peak at 197 nm) and showed complete reversal of its secondary structure upon cooling to 25 °C (Fig. 4h) . These results clearly suggest that residual disorder in the mature coacervate and recovery of conformational disorder in aELPs are the key to their thermal reversibility.
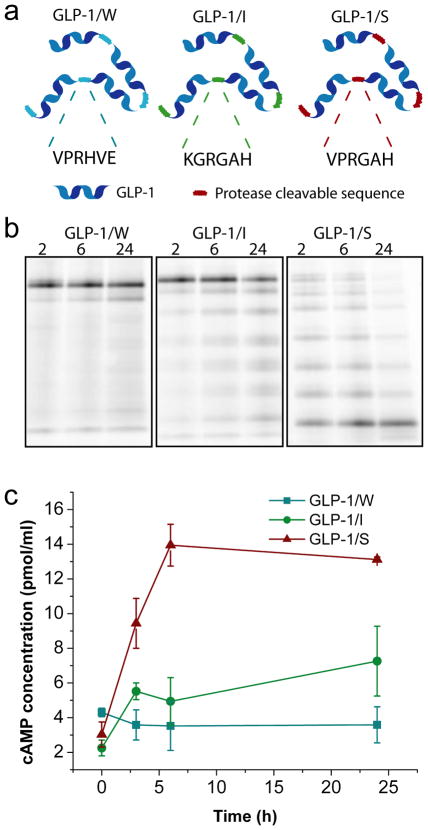
To further illustrate the broad applicability of OERCA, we next synthesized protein-polymers comprised of long (e.g., 30 amino acids) peptides, as opposed to the short 5–6 amino acids long aELP motifs. An emerging strategy for temporally sustained peptide delivery involves peptide polymerization with intervening protease-cleavable linkers 11. However, as existing designs employed protease sites that are rapidly cleaved, these constructs still required repeated daily injections. Others have demonstrated release of serum albumin-bound peptides by thrombin-cleavable linkers, enabling once-a-day injection regimens 22. To further prolong the sustained release of peptide drugs, we sought to generate protease-responsive protein-polymers of a model peptide drug, GLP-1, with intervening thrombin cleavable sequences of variable strengths to optimize monomer release. We used OERCA to create expression libraries of synthetic genes that encode protein-polymers of modified GLP-1 monomer sequences (Supplementary Fig. S8) with intervening thrombin cleavage sequences designed to be weak (GLP/W), intermediate (GLP/I) and strong (GLP/S) (Fig. 6a and Supplementary Table S1). We then chose a clone from each library that contained 6 repeats of each GLP-1 monomer, and demonstrated their sequence dependent protease-responsive behavior by cleavage with thrombin (Fig. 6b). Whereas the most thrombin-sensitive protein-polymer (GLP-1/S) was rapidly cleaved within 2 h (top band in Fig. 6b), the intermediate construct GLP-1/I was cleaved less efficiently, and the weak GLP-1/W only produced visible fragments after a 24 h incubation. We further demonstrated that the sequence dependent efficiency of thrombin cleavage is biologically relevant, as GLP-1 polymers incubated in mouse plasma were capable of activating the GLP-1 receptor overexpressed by Baby Hamster Kidney (BHK) cells. Moreover, Fig. 6c shows that the magnitude of receptor activation was proportional to the thrombin responsiveness of the GLP-1 protein-polymers, with GLP-1/S showing the highest and most sustained receptor activation, followed by GLP-1/I and GLP-1/W, respectively.
Figure 6.
Characterization of protease mediated cleavage of GLP-1 protein-polymers with variable thrombin recognition sequences. (a) Schematic illustration of GLP-1 polymers with variable protease cleavable sequences. Thrombin cleaves C-terminal to the Arginine (R), and in vivo the enzyme DPPIV can then cleave the “GA” dipeptide to leave a free N-terminal Histidine (H), the first amino acid in active GLP-1. (b) SDS-PAGE analysis of Alexa-488 labeled constructs incubated with 1 U thrombin for 2, 6 and 24 hours. (c) Differential activation of GLP-1 protein-polymers by incubation with mouse plasma demonstrated by cAMP production following GLP-1 binding to the GLP-1R in Baby Hamster Kidney cells.
We developed OERCA in response to the limitations of current methods available for the synthesis of repetitive genes, and our own interest in these biopolymers for medical applications. Whereas step-wise oligomerization provides deterministic control over the number of repeats 13, this recursive synthetic strategy is slow and tedious as it requires multiple cycles of laborious cloning, and uses specific restriction enzymes that must be carefully chosen to avoid the introduction of extraneous nucleotides at the ligation junction 23. Concatemerization of repetitive genes, although simple and rapid, typically results in a low yield of inserts with more than 2–3 monomers and offers little control over molecular weight 23. Similarly, PCR-based methods, such as OE-PCR, have been unsuccessful in synthesizing high molecular weight constructs, most likely due to the high error rates associated with nonspecific self-priming of short overlapping sequences 24,25. Other PCR-based strategies also suffer from poor sequence fidelity and small insert sizes 24,26,27. Indeed, we confirmed the poor performance of OE-PCR and concatemerization for the synthesis of libraries of repetitive genes (Supplementary Fig S4 and Supplementary Fig S5). Thermally cycled RCA, in contrast, has not received significant attention as a method for high fidelity synthesis of repetitive polypeptides 28, although we note its previous use in bioanalytical applications 29, 30.
OERCA provides a useful new molecular biology methodology for the rapid and parallel synthesis of protein-polymers, and will likely accelerate the discovery and development of new protein-polymers as shown here in the case of protease-responsive drug polymers and stimulus responsive protein-polymers. Ongoing work in our laboratory that uses OERCA is focused on the design and in vivo evaluation of protease responsive GLP-1 polymers as novel drug delivery vehicles for the treatment of type II diabetes, and the combinatorial screening of short peptide motifs in an effort to identify design principles for engineering new stimulus responsive polymers. We also envision the use of OERCA for the synthesis of repetitive immunogenic epitopes, cell adhesion protein-polymers with repetitive peptide motifs to modulate cellular interactions 32 and new biomaterials with useful structural and functional properties 33.
Materials and Methods
The reader is encouraged to review the detailed methods in the supplementary information.
Circularization of ssDNA template
250 pmol of ssDNA template was synthesized (Supplementary Table 1) and circularized in a 50μL reaction containing 2.5 μL Circligase, 2.5 μL MnCl2 and 2.5 μL ATP. The reaction was incubated at 60 °C for 2 h, column purified and incubated with 20 U Exonuclease I at 37 °C for 1h, followed by column purification.
OERCA
The OERCA reaction consisted of 150 ng of circular ssDNA template, 10–40 pmol of sense and antisense DNA primers (Supplementary Table 1), 25 mM of a 70% G/C dNTP mixture and 1μl of Pfu polymerase in a volume of 50 μL. The reaction was incubated at 95 °C for 2 min, followed by 30–40 cycles at 95 °C for 20 s, 52–55°C for 20 s and 72 °C for 30 s, and finalized at 72 °C for 5 min. The product was purified and visualized on a 1% agarose gel. Where indicated, the PCR product was subjected to additional PCR cycles with varying primer concentration and/or temperature to optimize the size range of the DNA product. For the synthesis of repetitive genes encoding for aELPs, the OERCA products were typically further extended in the absence of primers at an annealing temperature of 60 °C.
Generation of gene libraries from OERCA products
Modified pET-25b+ expression vectors (supplementary information) were used to clone aELPs and GLP-1 oligomers, incorporating a hexahistidine-tag or an ELP tag 34 for protein purification, respectively. The vector for aELPs also encoded for the N-terminal leader sequence SKGP in order to maximize expression levels 35. Following verification by DNA sequencing, 2 μg of vector was digested with 2 μl of SmaI for 2 h at room temperature (followed by 1 h digestion with 2 μl of AleI at 37°C for GLP-1 vector), dephosphorylated with 1 μl CIP for 15 min - 1 h at 37°C, and column purified. 250 ng of OERCA product was ligated to the vector using 5 units of T4 DNA ligase, 2 μl PEG-4000 and ~250 ng of digested vector. The ligation mixture was incubated at 25 °C for 3 h, and BL21 cells were transformed with the mixture following the manufacturer’s instructions.
Screening of gene libraries
Colonies were added to 25 μl solutions containing 12.5 μl GoTaq green master mix, 10 pmol T7-promotor primer and 10 pmol insert-specific reverse primer. The PCR reaction conditions were: 95 °C for 2 min, followed by 30 cycles at 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 1 min. The results were visualized on a 1% agarose gel. Clones with inserts in the correct orientation were identified by the presence of large smears on the DNA gel. The selected clones were subjected to DNA restriction analysis and DNA sequencing.
Comparison of OERCA with OE-PCR and concatemerization
For OE-PCR, oligonucleotides were designed to have 50% or 100% overlapping regions. The OE-PCR reaction mixture consisted of 10–40 pmol of each ssDNA oligonucleotide, 10 nmol of a dNTP mixture, 0.8 μl of Pfu polymerase, 4μl of 10X Pfu buffer, and water for a final volume of 40 μl, in accordance with similar OE-PCR reactions reported in the literature 24 36. The final optimized concatemerization protocol consisted of two 1 hour concatemerization reactions of 1 μM gene in the presence of 400 U of T4 DNA ligase (New England Biolabs) and 0.1 μM of either the 5’ or 3’ adaptor followed by mixing the two reactions and allowing for further ligation for 2.5 h, all performed at room temperature.
Expression, purification and characterization of aELP and GLP-1 polymers
Protein-polymers were overexpressed by IPTG induction in 1 L cultures supplemented with 100 μg/mL ampicillin. The cells were harvested 24 h after inoculation and purified by ITC 37. To characterize the inverse transition temperature of aELPs, the optical density of aELP solutions (50 μM in PBS unless indicated) was monitored 34. The secondary structure of aELPs was studied by circular dichroism at multiple temperatures for 5 μM polypeptide solutions in water. In addition, the ability of anti GLP-1 antibodies to bind GLP-1 protein-polymers was assayed by western blot analysis, using a 1:3000 dilution of mouse anti-GLP-1 antibody.
GLP-1 monomer release study
Purified GLP-1 oligomers were conjugated to Alexa-488 and digested with 1 U thrombin for 2, 6 and 24 h. The peptides were separated on a Tris-tricine SDS PAGE and imaged using a Typhoon 9410.
GLP-1 activity assay
GLP-1 activity assay was conducted using Baby Hamster Kidney (BHK) cells stably transfected with rat GLP-1 receptor (GLP-1R) (a gift from Prof. Drucker, University of Toronto). Cells were incubated with 100 μM 3-isobutyl-1-methylxanthine 38 followed by 10 min incubation with GLP-1. Intracellular cAMP concentrations triggered by GLP-1-R activation were measured by ELISA (Assay Designs).
Supplementary Material
Acknowledgments
A.C. acknowledges the financial support of NIH grants R21 EB009904 and R01 GM61232, M.A. acknowledges the support of a graduate fellowship from the Center for Biologically Inspired Materials and Material Systems, and F.G.Q. acknowledges the support of a fellowship from the Medtronic Foundation.
Footnotes
Author contributions. A.C. designed experiments, analyzed data and prepared the manuscript. M.A. and F.G.Q. designed and performed experiments, analyzed data and prepared the manuscript. D.C. designed experiments.
Competing financial interest. The authors declare that they have no competing financial interests.
References
- 1.Mackay JA, Chilkoti A. Temperature sensitive peptides: Engineering hyperthermia-directed therapeutics. International Journal of Hyperthermia. 2008;24:483–495. doi: 10.1080/02656730802149570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A. Peptide-based biopolymers in biomedicine and biotechnology. Materials Science & Engineering R-Reports. 2008;62:125–155. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nettles DL, et al. In situ crosslinking elastin-like polypeptide gels for application to articular cartilage repair in a goat osteochondral defect model. Tissue Engineering Part A. 2008;14:1133–1140. doi: 10.1089/ten.tea.2007.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreher MR, et al. Temperature triggered self-assembly of polypeptides into multivalent spherical micelles. Journal of the American Chemical Society. 2008;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simnick AJ, Lim DW, Chow D, Chilkoti A. Biomedical and biotechnological applications of elastin-like polypeptides. Polymer Reviews. 2007;47:121–154. [Google Scholar]
- 6.Rao XC, et al. Design and expression of peptide antibiotic hPAB-beta as tandem multimers in Escherichia coli. Peptides. 2005;26:721–729. doi: 10.1016/j.peptides.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Kim MS, Cho JH, Kim SC. Enhanced expression of tandem multimers of the antimicrobial peptide buforin II in Escherichia coli by the DEAD-box protein and trxB mutant. Applied Microbiology and Biotechnology. 2002;58:790–796. doi: 10.1007/s00253-002-0962-3. [DOI] [PubMed] [Google Scholar]
- 8.Wang YQ, Cai JY. High-level expression of acidic partner-mediated antimicrobial peptide from tandem genes in Escherichia coli. Applied Biochemistry and Biotechnology. 2007;141:203–213. doi: 10.1007/BF02729062. [DOI] [PubMed] [Google Scholar]
- 9.Hou JH, et al. High-level expression of fusion protein containing 10 tandem repeated GLP-1 analogs in yeast Pichia pastoris and its biological activity in a diabetic rat model. Bioscience Biotechnology and Biochemistry. 2007;71:1462–1469. doi: 10.1271/bbb.60694. [DOI] [PubMed] [Google Scholar]
- 10.Kempe T, et al. Multiple-Copy Genes - Production and Modification of Monomeric Peptides from Large Multimeric Fusion Proteins. Gene. 1985;39:239–245. doi: 10.1016/0378-1119(85)90318-x. [DOI] [PubMed] [Google Scholar]
- 11.Ma X, et al. Poly-GLP-1, a novel long-lasting glucagon-like peptide-1 polymer, ameliorates hyperglycaemia by improving insulin sensitivity and increasing pancreatic beta-cell proliferation. Diabetes Obes Metab. 2009;11:953–965. doi: 10.1111/j.1463-1326.2009.01070.x. [DOI] [PubMed] [Google Scholar]
- 12.Prasad S, Mathur A, Jaggi M, Mukherjee R. Delivering multiple anticancer peptides as a single prodrug using lysyl-lysine as a facile linker. J Pept Sci. 2007;13:458–467. doi: 10.1002/psc.867. [DOI] [PubMed] [Google Scholar]
- 13.Meyer DE, Chilkoti A. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: Examples from the elastin-like polypeptide system. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- 14.Clarke LA, Rebelo CS, Goncalves J, Boavida MG, Jordan P. PCR amplification introduces errors into mononucleotide and dinucleotide repeat sequences. J Clin Pathol-Mol Pa. 2001;54:351–353. doi: 10.1136/mp.54.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey UH, Bachmann HS, Peters J, Siffert W. PCR-amplification of GC-rich regions: ‘slowdown PCR’. Nat Protoc. 2008;3:1312–1317. doi: 10.1038/nprot.2008.112. [DOI] [PubMed] [Google Scholar]
- 16.White MJ, Fristensky BW, Thompson WF. Concatemer chain reaction: a Taq DNA polymerase-mediated mechanism for generating long tandemly repetitive DNA sequences. Anal Biochem. 1991;199:184–190. doi: 10.1016/0003-2697(91)90087-a. [DOI] [PubMed] [Google Scholar]
- 17.MacEwan SR, Chilkoti A. Elastin-Like Polypeptides: Biomedical Applications of Tunable Biopolymers. Biopolymers. 2010;94:60–77. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 18.Urry DW, Urry KD, Szaflarski W, Nowicki M. Elastic-contractile model proteins: Physical chemistry, protein function and drug design and delivery. Adv Drug Deliv Rev. 2010 doi: 10.1016/j.addr.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Bochicchio B, Pepe A, Tamburro AM. Investigating by CD the molecular mechanism of elasticity of elastomeric proteins. Chirality. 2008;20:985–994. doi: 10.1002/chir.20541. [DOI] [PubMed] [Google Scholar]
- 20.Yamaoka T, et al. Mechanism for the phase transition of a genetically engineered elastin model peptide (VPGIG)(40) in aqueous solution. Biomacromolecules. 2003;4:1680–1685. doi: 10.1021/bm034120l. [DOI] [PubMed] [Google Scholar]
- 21.Meyer DE, Chilkoti A. Quantification of the effects of chain length and concentration on the thermal behavior of elastin-like polypeptides. Biomacromolecules. 2004;5:846–851. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 22.Li H, et al. A protease-based strategy for the controlled release of therapeutic peptides. Angew Chem Int Ed Engl. 2010;49:4930–4933. doi: 10.1002/anie.201000287. [DOI] [PubMed] [Google Scholar]
- 23.Mi LX. Molecular cloning of protein-based polymers. Biomacromolecules. 2006;7:2099–2107. doi: 10.1021/bm050158h. [DOI] [PubMed] [Google Scholar]
- 24.Kurihara H, Morita T, Shinkai M, Nagamune T. Recombinant extracellular matrix-like proteins with repetitive elastin or collagen-like functional motifs. Biotechnology Letters. 2005;27:665–670. doi: 10.1007/s10529-005-4477-8. [DOI] [PubMed] [Google Scholar]
- 25.Kurihara H, Nagamune T. DNA polymerase-catalyzed elongation of repetitive hexanucleotide sequences: Application to creation of repetitive DNA libraries. Biotechnology Progress. 2004;20:1855–1860. doi: 10.1021/bp049925z. [DOI] [PubMed] [Google Scholar]
- 26.Bang DH, Church GM. Gene synthesis by circular assembly amplification. Nature Methods. 2008;5:37–39. doi: 10.1038/nmeth1136. [DOI] [PubMed] [Google Scholar]
- 27.Lyons RE, et al. Design and facile production of recombinant resilin-like polypeptides: gene construction and a rapid protein purification method. Protein Engineering Design & Selection. 2007;20:25–32. doi: 10.1093/protein/gzl050. [DOI] [PubMed] [Google Scholar]
- 28.Fire A, Xu SQ. Rolling Replication of Short DNA Circles. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang DY, Brandwein M, Hsuih TCH, Li HB. Amplification of target-specific, ligation-dependent circular probe. Gene. 1998;211:277–285. doi: 10.1016/s0378-1119(98)00113-9. [DOI] [PubMed] [Google Scholar]
- 30.Zhang WD, et al. Detection of Chlamydia trachomatis by isothermal ramification amplification method: a feasibility study. J Clin Microbiol. 2002;40:128–132. doi: 10.1128/JCM.40.1.128-132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel JR, Mackay JA, Quiroz FG, Chilkoti A. Recursive directional ligation by plasmid reconstruction allows rapid and seamless cloning of oligomeric genes. Biomacromolecules. 2010;11:944–952. doi: 10.1021/bm901387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee BW, et al. Strongly Binding Cell-Adhesive Polypeptides of Programmable Valencies. Angew Chem Int Edit. 2010;49:1971–1975. doi: 10.1002/anie.200906482. [DOI] [PubMed] [Google Scholar]
- 33.Lv S, et al. Designed biomaterials to mimic the mechanical properties of muscles. Nature. 2010;465:69–73. doi: 10.1038/nature09024. [DOI] [PubMed] [Google Scholar]
- 34.Meyer DE, Chilkoti A. Quantification of the effects of chain length and concentration on the thermal behavior of elastin-like polypeptides. Biomacromolecules. 2004;5:846–851. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 35.McDaniel JR, MacKay JA, Quiroz FG, Chilkoti A. Recursive Directional Ligation by Plasmid Reconstruction Allows Rapid and Seamless Cloning of Oligomeric Genes. Biomacromolecules. 2010;11:944–952. doi: 10.1021/bm901387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurihara H, Morita T, Shinkai M, Nagamune T. Recombinant extracellular matrix-like proteins with repetitive elastin or collagen-like functional motifs. Biotechnol Lett. 2005;27:665–670. doi: 10.1007/s10529-005-4477-8. [DOI] [PubMed] [Google Scholar]
- 37.Christensen T, et al. Fusion order controls expression level and activity of elastin-like polypeptide fusion proteins. Protein Science. 2009;18:1377–1387. doi: 10.1002/pro.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baggio LL, Huang QL, Brown TJ, Drucker DJ. A recombinant human glucagon-like peptide (GLP)-1-albumin protein (Albugon) mimics peptidergic activation of GLP-1 receptor-dependent pathways coupled with satiety, gastrointestinal motility, and glucose homeostasis. Diabetes. 2004;53:2492–2500. doi: 10.2337/diabetes.53.9.2492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.