Abstract
Observations that β-sheet proteins form amyloid fibrils under at least partially denaturing conditions has raised questions as to whether these fibrils assemble by docking of preformed β-structure or by association of unfolded polypeptide segments. By using α-helical protein apomyoglobin, we show that the ease of fibril assembly correlates with the extent of denaturation. By contrast, monomeric β-sheet intermediates could not be observed under the conditions of fibril formation. These data suggest that amyloid fibril formation from apomyoglobin depends on disordered polypeptide segments and conditions that are selectively unfavorable to folding. However, it is inevitable that such conditions often stabilize protein folding intermediates.
Amyloid fibrils are polypeptide aggregates in which the polypeptide backbone is arranged in a specific type of β-sheet conformation, termed “cross-β” structure (1, 2). Within this structure, the β-strands extend transversely to the main fibril axis. Amyloid structures do not depend for their existence on the presence of distinctive sequence patterns or specific intramolecular side chain interactions (3). Hence, they differ in this property from globular protein structures that are always encoded in the amino acid sequence. In addition, it has been suggested that conserved main chain properties enable many, if not all, polypeptide chains to form amyloid (4–6) and that the extent and rate of this reaction is modulated by the side chains (3, 7). The latter properties enable protein structures to be stable under physiological conditions. Consistent with this view, only a small fraction of all natural polypeptide chains has been observed to form amyloid structures under pathological conditions, such as Alzheimer's disease, type II diabetes, or transmissible prion disorders (8–10).
Considerable debate exists as to the mechanism of fibril assembly. One issue of particular interest concerns the question of whether or not this process involves the docking of monomeric precursors that contain specific preformed elements of amyloid structure (2, 4). Evidence that might support such a proposal comes from observations that some proteins form amyloid fibrils in vitro under conditions where partially folded states are present (11–14). Nevertheless, the generic nature of the ability of polypeptide chains, including small peptides, to form amyloid structures suggests that this might not be necessary (3, 4) and, even for large proteins, it is generally unclear whether it is the folded or the unfolded regions of these partially denatured states that facilitates amyloid formation. In the case of acylphosphatase, at least, folding and aggregation are controlled by different sets of residues (7). Moreover, the proteins examined in these previous studies possess, in their native states, significant amounts of β-sheet structure and can therefore be expected to form β-rich intermediates.
We have used apomyoglobin (AMB) to explore the mechanism and principles of amyloid fibril formation in further detail. This protein is readily able to form amyloid fibrils in vitro, although its native state is predominantly α-helical and lacks elements of β-sheet structure (6, 15). These properties suggested to us that it might be possible, in this case, to distinguish whether the structured parts of a given partially folded state are relevant to native folding or whether they could possibly act as precursors of amyloid cross-β structure.
Materials and Methods
Preparation and Fibril Formation of Peptide and Protein Samples. Myoglobin (horse skeletal muscle) and all chemicals were obtained from Sigma, and the G-helix peptide was obtained from JERINI AG (Berlin). AMB was prepared by using a phase extraction procedure (16). Holomyoglobin (HMB) was dissolved in 0.01 M HCl and mixed thoroughly in equal proportion with 2-butanone on ice. After phase separation, the organic phase was removed and the extraction procedure was repeated until completion the solutions was clear. Residual 2-butanone was removed by dialysis in water, before lyophilization. The buffer at pH 9.0 was 50 mM sodium borate. Myoglobin concentrations were determined by quantitative amino acid analysis (A. Willis, Medical Research Council Immunochemistry Unit and Oxford Centre for Medical Sciences Sequencing Facility, personal communication) and UV absorption, by using a calculated molar extinction coefficient of 13,940 M–1·cm–1 at 280 nm in 6 M guanidine hydrochloride/20 mM sodium phosphate, pH 6.5. Centrifugation was carried out by using a Beckman (Unterschleissheim, Germany) Airfuge and A-95 or A-110 rotors at 1.8 to 2.0 × 105 × g for 30 min.
Assessment of the Amyloid Properties by Microscopy and X-Ray Diffraction. EM samples were typically diluted 1:10 in buffer or in water before specimen preparation by using the floating carbon method as described (3). Samples were analyzed by using an EM 400 T Philips (Eindhoven, The Netherlands) transmission electron microscope, operated at an acceleration voltage of 80 kV and by using magnifications of ×20,000 to ×60,000. The test for apple green birefringence was performed by drying 6-μl aliquots of a fibril suspension (10 mg/ml in H2O at pH 5.0), each placed between a glass slide and a coverslip, and left to dry over night. The coverslip was then removed, and the fibrils were stained by using a freshly filtered Congo red solution (0.1% in H2O). Excessive and nonspecific Congo red stain was removed by three washing steps of 1 min each using 20% ethanol. Samples were analyzed by using a Leica DM/DR ×450 polarization microscope. For x-ray diffraction analysis, alignment was performed by drying off aliquots of the peptide solution (10 mg/ml in H2O, pH 5.0) placed onto a glass slide as described (3).
Fourier Transform Infrared (FTIR) Spectroscopy. Labile hydrogens in AMB were replaced by deuterium through repeated cycles of lyophilization and dissolution in D2O. Deuterated AMB (1 mg/ml) was dissolved in 50 mM sodium borate in D2O, pD 9.02 (measurements not corrected for isotope effects), and heated at 65°C for 4 days. The fibrils were concentrated by centrifugation before analysis. For the peptide corresponding to the G-helix, the lyophilized material was dissolved in D2O to reach a final concentration of 20 mg/ml. After recording a spectrum of the soluble peptide, the fibrils were formed in situ within the FTIR spectrometer by heating the samples at 75°C for 2 h. IR spectra were recorded at 20°C with a Jasco (Great Dunmow, U.K.) 615 Fourier transform spectrometer. Samples were inserted between CaF2 windows (Spectra-Tech, Waltham, MA) by using a 100-μm spacer. The spectra, the average of 1,000 scans, were recorded with a resolution of 4 cm–1. For each spectrum, signal corresponding to the water vapor was subtracted and the baseline was corrected. The decomposition of the spectra was carried out by using the curve-fitting software provided by Jasco.
CD and Fluorescence Spectroscopy. All CD spectra were recorded on a Jasco J-720 spectropolarimeter using thermostated quartz cells of 0.1 or 1 mm path length. Thermal denaturation studies were performed by exposing the protein to different temperatures within the range of 25–95°C. Spectra were acquired after equilibration for 5 min. The temperatures of the sample were measured precisely by using a thermocouple. Tryptophan fluorescence was excited at 279 nm and recorded at 340 nm. Thioflavin-T (ThT) fluorescence was recorded at 482 nm, whereas exciting was recorded at 450 nm. To measure the ThT fluorescence, protein samples were mixed with buffer and ThT stock solutions, effectively reducing the protein concentration by a factor of two. The final concentration of dye was always 20 μM. ANS (1-anilino-naphthalene-8-sulfonate) fluorescence was recorded by using 20 μM dye and a protein concentration of 2 μM (excitation at 350 nm). The spectra shown here represent averages of two scans. Unless stated otherwise, all fluorescence measurements were carried out at room temperature and in 50 mM sodium borate, pH 9.0. Sample temperatures were determined with a calibrated mercury thermometer. Fluorescence experiments were performed by using a Perkin–Elmer LS50B spectrofluorimeter equipped with a thermally adjusted sample holder or a Shimadzu RF-5301PC fluorimeter (Jena, Germany).
Results
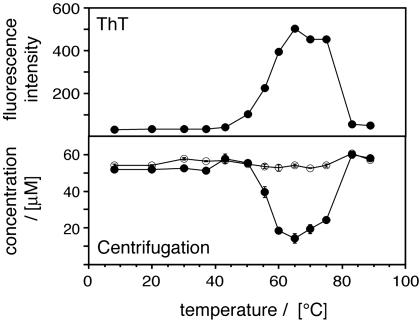
Myoglobin Readily Forms Amyloid Fibrils Only Under Specific Thermal Conditions. It has been well established that amyloid formation requires an appropriate physicochemical environment (4–6). In the case of AMB dissolved at pH 9.0, incubation at 65°C for 24 h was found to be sufficient to induce fibril formation (6). Hence, we wondered whether the dependence of amyloid formation on solution conditions might provide information concerning the underlying mechanism of fibril formation. Initially we focused on temperature, for variation of this physical property avoids many complications associated with changes in the solution composition. AMB samples were incubated at temperatures ranging from 8 to 90°C and aggregation was quantified by electron microscopy (EM), ultracentrifugation and the fluorescence of an aggregate-specific dye, ThT (Fig. 1). All three techniques were consistent in showing that fibrils formed most readily at temperatures from 55 to 75°C. Nevertheless, when we used EM, we could detect small amounts of nonfibrillar aggregates in solutions exposed to higher temperatures; such species are commonly observed to form from polypeptides under high temperature conditions. The temperature range of fibril formation and the position of its maximum (65°C) were not affected significantly by variations in the incubation time between 1 and 5 days (data not shown). We conclude that amyloid formation from AMB becomes progressively suppressed when the temperature is raised or decreased from 65°C.
Fig. 1.
Temperature dependence of amyloid fibril formation by AMB. Shown is temperature dependence of amyloid formation monitored by ThT-fluorescence (Upper) and centrifugation (Lower; open circles, sample before centrifugation; filled circles, supernatant after centrifugation). AMB solutions (53 μM, pH 9.0 buffer) were incubated at the various temperatures for 5 days before analysis. Error bars (close to symbol size) represent the variation between two measurements.
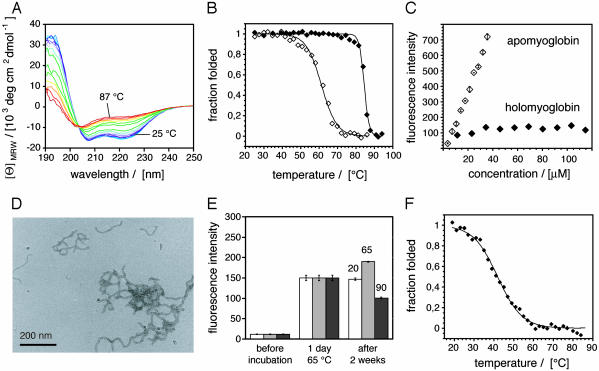
Amyloidogenic Environments Are Selectively Unfavorable to Protein Folding. Starting with the low temperature transition, we examined a number of possibilities to explain the suppression of amyloid formation under such conditions. Denaturing gel electrophoresis (in laurylsulfate), mass spectrometry, or C18 reverse phase chromatography reveal that the fibrils did not contain significant levels of covalently modified polypeptides (data not shown). Consistent with this notion, fibril formation declines sharply under the conditions that are most favorable to such modifications, i.e., at temperatures >75°C. Hence, we focused on the possibility that temperature might affect the noncovalent interactions within AMB. At 20°C, AMB is known to be present in a globular protein form. This structure is progressively lost, however, when the temperature is increased above 40°C (Fig. 2A). In addition, an unfolded ensemble of species prevails at temperatures above 80°C. The thermal unfolding transition fits to a two state mode, and the far-UV CD spectra obtained upon heating possess an isodichroic point (203.5 nm) indicating that temperature shifts the equilibrium between two major states, the folded and the unfolded form. The midpoint of this transition (Tm) was at 61.1 ± 1.0°C (Fig. 2B), consistent with previous measurements of the stability of native AMB at pH 7.5 (17). From these data it can be inferred that aggregation is unfavorable below 65°C because protein folding competes with and suppresses amyloid formation.
Fig. 2.
Thermal stability of AMB. (A) Thermal denaturation of native AMB (6.5 μM) from 25°Cto87°C, monitored by CD. (B) Thermal denaturation of AMB (open diamonds) and HMB (filled diamonds) monitored by CD at 222 nm and fitted to a two-state model (40). (C) ThT signal of AMB and HMB solutions after incubation at pH 9.0 and 65°C for 5 days. Error bars indicate the variation of two separate measurements. (D) Electron micrograph of AMB fibrils. (E) Effect of temperature on AMB fibrils monitored by ThT fluorescence. As a first step, fibrils were formed in all samples by incubation at 65°C for 1 day and fibril quantities were determined by ThT. Then, these fibril samples were incubated at 20, 65, and 90°C for the remaining period and the ThT fluorescence was determined again. Error bars indicate standard deviations from three measurements. (F) Thermal denaturation of AMB monitored by tryptophan fluorescence (filled diamonds) fitted to a two-state model (40).
Additional support for this conclusion was provided by the observation that fibril formation was abolished upon binding of a heme group (the natural cofactor of myoglobin) to the apoprotein (Fig. 2C). HMB, i.e., the protein with its heme group incorporated in its structure, is significantly more thermostable than AMB and possesses a Tm value of 84.5 ± 1.0°C (Fig. 2B). Similar observations concerning the effect of ligands have been reported previously for several proteins including transthyretin and acylphosphatase (12, 18). However, if destabilization of the native fold is the key to the differences between fibril formation at 65°C and lower temperatures, why does amyloid formation become unfavorable at higher temperatures, i.e., under conditions in which protein folding is highly destabilized? Two possible explanations were considered: (i) fibrils do not form at high temperatures because they are unstable under such conditions or (ii) fibrils do not form because amyloid formation depends on defined elements of residual native-like structure that become unfolded above 75°C.
To test hypothesis i, preformed fibrils of the type shown in Fig. 2D were exposed to temperatures of 20°C, 65°C, and 90°C for 2 weeks, and aggregate levels were assessed by using the ThT assay (Fig. 2E). Although incubation at 20°C was found to prevent the further formation of fibrils, it did not result in the dissolution of preformed fibrils. By contrast, prolonged incubation at 90°C led to a substantial decrease in the quantity of fibrils present in solution, indicating that very high temperatures are unfavorable to both the native protein and the amyloid form of AMB. Consistent with this view, other conditions are known to exist that also interfere strongly with noncovalent interactions between polypeptide groups and permit neither folding nor amyloid formation; such conditions include, for example, incubation in 1.4 M guanidine hydrochloride or in 10 mM acetic acid/HCl solution at pH 2.3. Taken together, we conclude that AMB forms amyloid fibrils under conditions that are selectively unfavorable to the folding of AMB, i.e., environments that prevent the formation of a globular protein structure but allow the formation of amyloid structures. These conclusions are consistent with hypothesis i described above.
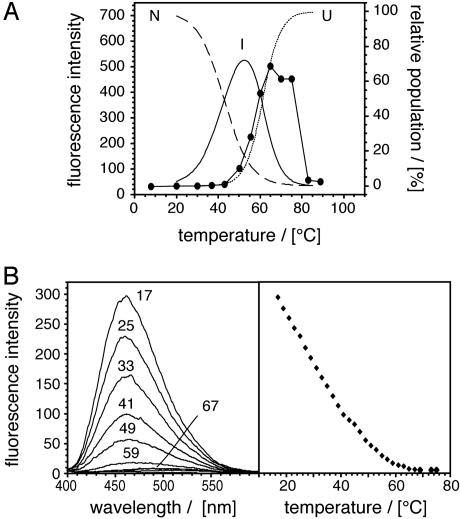
Amyloid Fibril Formation Does Not Correlate with the Presence of Partially Folded Species. We then tested hypothesis ii, i.e., whether specific partially folded states could be detected in AMB solutions exposed to amyloidogenic conditions. These species are most clearly detected by deviations from two-state unfolding, i.e., by a pronounced shoulder in the equilibrium denaturation curve. In the case of AMB, such a transition occurs in the acidic pH range (19). By contrast, when the thermal unfolding of AMB was monitored by using either far-UV CD (to assess the secondary structure) or tryptophan fluorescence (which is more sensitive toward tertiary structural changes), we obtained two sigmoidal transitions corresponding to a two-state unfolding reaction (Fig. 2 B and F). Nevertheless, the transition midpoint was much lower when unfolding was monitored by fluorescence (41.8 ± 0.9°C) than by far-UV CD (61.1 ± 1.0°C). The difference in the two transition midpoints implies that the proteins forms, before full thermal unfolding, an intermediate state that contains the same (α-helical) secondary structural composition as the native protein (Fig. 3A). It is of note that all tryptophane residues are located on the A-helix. The observed changes may therefore reflect only a local structural rearrangement, such as an increase in the dynamics of the A-helix.
Fig. 3.
Amyloid formation does not correlate with the population of partially folded species. (A) Overlay of the experimental data from Fig. 1 (filled symbols) with a fit of the data displayed in Fig. 2 B and F to define the population of the various monomeric AMB states: native (N, broken line), intermediate (I, continuous line), and unfolded AMB (U, dotted line). (B) Temperature dependence of the ANS fluorescence induced by AMB. (Right) Fluorescence reading at 460 nm.
Taken together, of the three states observed in pH 9.0 solutions (native, intermediate, and unfolded), only the unfolded ensemble differs substantially in its secondary structural composition from native AMB, whereas the presence of an aberrant (nonnative) β-sheet structure was only found in conjunction with intermolecular aggregates. Moreover, the formation of amyloid structures correlates with the formation of the most unfolded species, but not with the population of the intermediate or the native form (Fig. 3A). As a further test for the possible presence of partially folded states, we examined AMB by using the dye ANS. This dye has long been utilized to probe for exposure of hydrophobic regions present in partially folded states (20), and a recent study on the formation of amyloid fibrils from acidic fibroblast growth factor demonstrates that this spectroscopic probe can be used under high-temperature conditions (21). As expected, HMB does not bind ANS at 65°C or room temperature, consistent with the absence of stable of stable hydrophobic clusters (data not shown). By contrast, AMB possesses a significant capacity to bind ANS at room temperature, presumably because the dye binds in the heme-binding pocket (22). However, shifting the temperature to a value where amyloid fibrils form readily (65°C) resulted in a marked decay of the fluorescence signal (Fig. 3B). Based on the dye ANS, we could not obtain any evidence, such as a shoulder in the fluorescence transition, that would have indicated the presence of partially folded structure under the conditions of amyloid fibril formation. Finally, we used FTIR to test for the presence partially folded structure within the monomeric amyloid fibril precursors. Albeit the Bruker CONFOCHECK FTIR system (Ettlingen, Germany) equipped with a BIO-ATR II cell allowed very fast acquisitions of FTIR spectra, i.e., within 33 sec, which includes heating the sample from 25 to 65°C, aggregation was found to be extremely fast under the very high AMB concentrations required for these measurements, and the β-sheet structure detected under such conditions could be attributed to the rapid formation of aggregates (Fig. 7, which is published as supporting information on the PNAS web site). We conclude that unfolded chain segments are the precursors of amyloid cross-β structure. These ideas imply, however, that the initial formation of partially folded structures may not be necessary for the formation of amyloid fibrils from AMB.
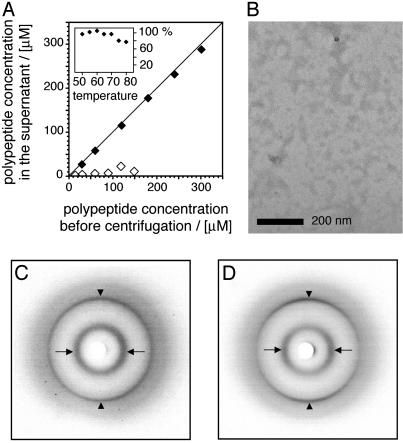
The Amyloid β-Sheet Structure Involves Sequence Segments That Are α-Helical in Globular AMB. The best-characterized partially folded form of AMB can be obtained when the protein is incubated in 10 mM sodium acetate, pH 4.1, at 50°C (19). Although this state contains regions of α-helical structure (which cannot serve as a direct structural precursor of amyloid cross-β structure), aggregation of sperm whale AMB has been reported to occur under such conditions (19). We confirmed these observations by using equine AMB, and incubation times of >4 days led to the formation of flake-like aggregates that settled at the bottom of the sample tube, whereas shorter incubation times did not produce such effects, even at concentrations of 300 μM (Fig. 4A). The structural analysis of these aggregates shows that they possess an internal structure that is highly similar to amyloid fibrils. We used FTIR spectroscopy to obtain an amide I′ maximum at 1,617 cm–1 and x-ray diffraction revealed main chain and side chain Bragg spacings at 4.63 and 10.03 Å (Fig. 4 C and D). Nevertheless, these aggregated species do not possess the fibrillar overall structure of amyloid (Fig. 4B) and the diffraction pattern we obtained lacked anisotropy. These properties imply that the β-sheets in nonfibrillar aggregates do not have the well defined orientations that are characteristic of amyloid fibrils.
Fig. 4.
Partially folded AMB forms nonfibrillar aggregates. (A) Quantity of polypeptide remaining in solution after incubation for 88 h, examined by centrifugation. Open diamonds, AMB, pH 9.0 buffer, 65°C. Filled diamonds, AMB, 10 mM Na acetate, pH 4.1 buffer, 50°C. (Inset) Percentage of polypeptide remaining in solution after incubation at various temperatures for 4 days, pH 4.1. (B) Electron micrograph of AMB in pH 4.1 buffer after 4 days at 50°C. (C and D) X-ray diffraction pattern of nonfibrillar aggregates (C) and AMB fibrils (D). Arrows (along the equatorial direction), side chain spacing; arrowheads (along the meridional direction), main chain spacing. Spacings in C, 4.65 and 10.05 Å; spacings in D, 4.63 and 10.03 Å.
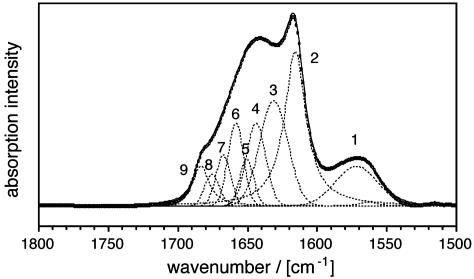
A decomposition analysis of the amide I′ region of AMB amyloid fibrils (Fig. 5) gave the following distribution of the different types of conformation: β-sheet structure (35%), α-helical conformation (11%), turns or 310-helices (16%), and random coil structure (38%). Similar values were obtained previously by quantitative far-UV CD measurements (23). Because native myoglobin possesses 78% α-helical structure (15) and because amyloid fibrils are always associated with β-sheet structure, >30% of the fibril-specific β-sheets is constructed from residues that form α-helices in the folded protein. To enable fibril formation, these structural elements must be unfolded. Hence, fibril formation is likely to be inhibited if folding is initiated within these critical regions. Consistent with this view, we found that the α-helical partially folded state from pH 4.1 solutions formed only nonfibrillar aggregates.
Fig. 5.
Internal structure of AMB amyloid. FTIR amide I′ region of myoglobin fibrils. The spectrum was decomposed by fitting with Gaussian or Lorentzian curves (dotted lines). The sum of the fitted curves is displayed as a continuous line, closely overlapping the experimental data points (open symbols). Assignments: 1, side chain carboxyl groups; 2, β; 3, β-turns; 4, random coil structure; 5, α; 6, 7, 8, turns or 310 helices; 9, β.
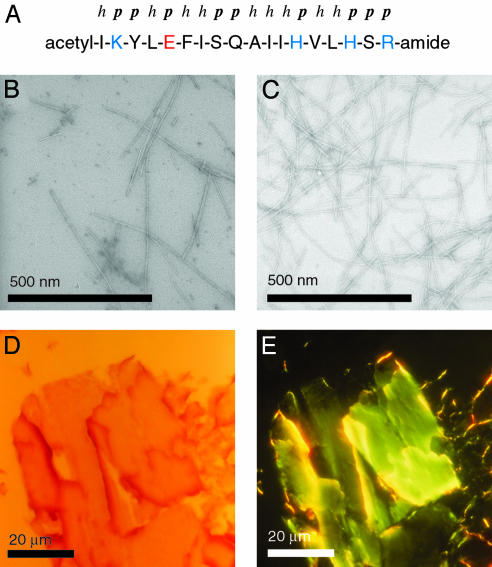
It is not known which helices might be involved in the transition from the partially structured soluble state of AMB to the β-sheet-rich amyloid fibril. However, peptide fragment corresponding to the G-helix (Fig. 6A) or to the N terminus of myoglobin have been shown previously to form species with extensive β-structure (24, 25). The G-helix is well defined within partially folded states of AMB and forms early during folding (19). We found that this peptide readily formed amyloid fibrils (Fig. 6 B and C) that possess a width of 9.7 ± 1.4 nm. These give rise to Congo red green birefringence when viewed in a polarizing microscope (Fig. 6 D and E), and x-ray fiber diffraction shows Bragg spacings at 4.73 Å and 10.9 Å. The maximum of their amide I′ region was centered at 1,617 cm–1 and the β-sheet content was estimated to be 50–55%. The potential of this peptide to form amyloid structures is interesting, because the G-helix lacks a clear polar–hydrophobic sequence pattern and represents a very stable element of secondary structure of the globular protein. Nevertheless, the evidence described here suggest that the G-helix might form part of the sequence segment that is relevant for the α to β transition of AMB from the globular protein to amyloid fibrils.
Fig. 6.
Amyloid fibrils formed by the G-helix peptide of myoglobin. (A) Sequence of the G-helix peptide and classification of the residues as polar (p) and hydrophobic (h), acidic (red) and basic (blue). (B and C) Electron micrographs of 1 mg/ml peptide incubated for 4 days at 60°C in 50 mM sodium phosphate pH 3.0 (B) or 50 mM sodium acetate pH 5.0 (C). (D and E) Fibrils viewed in a polarization microscope; bright field (with only one polarizer, D) or dark field with crossed polarizers (E).
Discussion
Myoglobin represents the case of a protein where amyloid fibril formation correlates with conditions in which the polypeptide chain is unfolded, rather than with environments that unfold the protein only partially. Moreover, all intermediate states detected in the course of our analysis were α-helical within their structured regions, and therefore, they represent species that may be relevant to native folding, albeit they cannot be relevant to aggregation. These data argue that the structural precursors of the amyloid cross-β structure are polypeptide chain segments that lack stable (hydrogen-bonded) elements of secondary structure. It is of note that even unfolded polypeptide chains, such as urea-denatured acidic AMB, can encompass significant amounts of nonrandom conformation (26) and, from the far-UV CD spectrum of heat-unfolded AMB, it is evident that it contains regions that are more structured than the “classical” random coil structure of poly(l-lysine) (27). The present observations differ, therefore, from previous analyses of fibril forming proteins that can form well defined partially folded states with β-sheet structure (11, 12, 14, 21, 28). On the basis of the present data, we propose that the core of AMB amyloid fibrils does not need to be preformed in terms of hydrogen-bonded elements within a monomeric precursor and that partial folding does not represent a general step in amyloid formation, although it may occur in specific cases, such as some β-sheet proteins. As for AMB, however, the characteristic main chain structure of amyloid fibrils was only found to be consolidated in conjunction with the formation of intermolecular hydrogen-bonds.
From a number of previous observations it can be inferred that the mechanism proposed here may not be limited to AMB: (i) Amyloid fibrils can be formed by polypeptide chains that have no capacity to fold or to form partially folded species, such as various polyamino acids (3, 29) and very short peptides (2, 30). (ii) Solid-state NMR spectroscopy revealed that the amyloid fibrils formed from Aβ (1–40) and a peptide fragment of transthyretin lack intramolecular main chain hydrogen bonds (31, 32). Unfolded polypeptide chains represent, therefore, plausible candidates for the precursor structures of these amyloid fibrils. (iii) Increasing experimental and theoretical evidence (even for all β proteins or domains) suggests that the amyloid fibril precursors are more expanded than well defined and compact partially folded states (33–35). (iv) “Unfolded” conformations (including acidic urea-denatured AMB) can have discernible structure, in particular they seem to possess a bias toward extended main chain conformations (26, 36–38). This finding implies that unfolded ensembles are therefore rich in (non-hydrogen bonded) transient structures that are sufficiently extended to align into amyloid fibrils. (ν) The present results, along with previous data (5, 39), imply that the environments which enable amyloid fibril formation are highly selective as they must balance two opposing effects. On the one hand they must allow, in principle, the formation of noncovalent polypeptide interactions. On the other hand, they must be specifically unfavorable to protein folding. Hence, the formation of highly ordered and fibrillar aggregates would be inhibited by extensive levels of residual native-like structure. These characteristics are inevitably reminiscent of those involved in the partial folding of otherwise denatured states, and therefore the possibility cannot be excluded that the population of partially folded intermediates coincides, under some circumstances, with amyloid formation, although the two processes may not be directly related. Indeed, mutational studies have indicated that there is kinetic partitioning between folding and aggregation, with the two processes being nucleated by highly distinct regions of the denatured polypeptide chain (7). Amyloid fibrils represent, therefore, a generic conformational state of a polypeptide chain which reproduces the π- and φ-angle distribution that is most favorable to the main chain more closely than do folded structures. An improved understanding of the critical steps in amyloid formation may allow the identification of more successful strategies to interfere with the pathogenic processes of amyloidoses.
Supplementary Material
Acknowledgments
We thank M. A. Fletcher, G. Gellermann, and T. R. Appel for assistance in recording the CD and light microscopy data, P. E. Wright and S. Schwarzinger for helpful discussions, and M. Boese and M. Luft (Bruker) for access to their infrared spectrometers. M.F. is funded by a BioFuture grant (BMBF) and by a grant from the Deutsche Forschungsgemeinschaft. The research of C.M.D. is supported in part by a Program Grant from the Wellcome Trust.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AMB, apomyoglobin; HMB, holomyoglobin; FTIR, Fourier transform infrared; ThT, thioflavin-T.
References
- 1.Serpell, L. C., Fraser, P. E. & Sunde, M. (1999) Methods Enzymol. 309, 526–536. [DOI] [PubMed] [Google Scholar]
- 2.Sunde, M. & Blake, C. C. F. (1998) Q. Rev. Biophys. 31, 1–39. [DOI] [PubMed] [Google Scholar]
- 3.Fändrich, M. & Dobson, C. M. (2002) EMBO J. 21, 5682–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobson, C. M. (2001) Philos. Trans. R. Soc. London Ser. B. 356, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiti, F., Webster, P., Taddei, N., Clark, A., Stefani, M., Ramponi, G. & Dobson, C. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fändrich, M., Fletcher, M. A. & Dobson, C. M. (2001) Nature 410, 165–166. [DOI] [PubMed] [Google Scholar]
- 7.Chiti, F., Taddei, N., Baroni, F., Capanni, C., Stefani, M., Ramponi, G. & Dobson, C. M. (2002) Nat. Struct. Biol. 9, 137–143. [DOI] [PubMed] [Google Scholar]
- 8.Westermark, P., Benson, M. D., Buxbaum, J. N., Cohen, A. S., Frangione, B., Ikeda, S., Masters, C. L., Merlini, G., Saraiva, M. J. & Sipe, J. D. (2002) Amyloid 9, 197–200. [DOI] [PubMed] [Google Scholar]
- 9.Baskakov, I. V., Legname, G., Prusiner, S. B. & Cohen, F. E. (2001) J. Biol. Chem. 276, 19687–19690. [DOI] [PubMed] [Google Scholar]
- 10.Lindquist, S., Krobitsch, S., Li, J. & Sondheimer, N. (2001) Philos. Trans. R. Soc. London Ser. B 356, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booth, D. R., Sunde, M., Bellotti, V., Robinson, C. V., Hutchinson, W. L., Fraser, P. E., Hawkins, P. N., Dobson, C. M., Radford, S. E., Blake, C. C. & Pepys, M. B. (1997) Nature 385, 787–793. [DOI] [PubMed] [Google Scholar]
- 12.Kelly, J. W., Colon, W., Lai, Z., Lashuel, H. A., McCulloch, J., McCutchen, S. L., Miroy, G. J. & Peterson, S. A. (1997) Adv. Prot. Chem. 50, 161–181. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez-Alvarado, M., Merkel, J. S. & Regan, L. (2000) Proc. Natl. Acad. Sci. USA 97, 8979–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McParland, V. J., Kalverda, A. P., Homans, S. W. & Radford, S. E. (2002) Nat. Struct. Biol. 9, 326–331. [DOI] [PubMed] [Google Scholar]
- 15.Evans, S. V. & Brayer, G. D. (1988) J. Biol. Chem. 263, 4263–4268. [PubMed] [Google Scholar]
- 16.Teale, F. W. J. (1959) Biochim. Biophys. Acta 35, 543. [DOI] [PubMed] [Google Scholar]
- 17.Fink, A. L., Oberg, K. A. & Seshadri, S. (1997) Fold. Des. 3, 19–25. [DOI] [PubMed] [Google Scholar]
- 18.Chiti, F., Taddei, N., Stefani, M., Dobson, C. M. & Ramponi, G. (2001) Protein Sci. 10, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright, P. E. & Baldwin, R. L. (2000) in Mechanism of Protein Folding, ed. Pain, R. S. (Oxford Univ. Press, Oxford), 2nd Ed.
- 20.Semisotnov, G. V., Rodionova, N. A., Kutyshenko, V. P., Ebert, B., Blanck, J. & Ptitsyn, O. B. (1987) FEBS Lett. 224, 9–13. [DOI] [PubMed] [Google Scholar]
- 21.Srisailam, S., Wang, H.-M., Kumar, T. K. S., Rajalingam, D., Sivaraja, V, Sheu, H.-S., Chang, Y.-C. & Yu, C. (2002) J. Biol. Chem. 277, 19027–19036. [DOI] [PubMed] [Google Scholar]
- 22.Bismuto, E., Sirangelo, I., Irace, G. & Gratton, E. (1996) Biochemistry 35, 1173–1178. [DOI] [PubMed] [Google Scholar]
- 23.Fletcher, M. A. (2000) Part II thesis (Univ. of Oxford, Oxford).
- 24.Waltho, J. P., Feher, V. A., Merutka, G., Dyson, H. J. & Wright, P. E. (1993) Biochemistry 32, 6337–6347. [DOI] [PubMed] [Google Scholar]
- 25.Chow, C. C., Chow, C., Ragunathan, V., Huppert, T. J., Kimball, E. B. & Cavagnero, S. (2003) Biochemistry 42, 7090–7099. [DOI] [PubMed] [Google Scholar]
- 26.Schwarzinger, S., Wright, P. E. & Dyson, H. J. (2002) Biochemistry 41, 12681–12686. [DOI] [PubMed] [Google Scholar]
- 27.Greenfield, N. & Fasman, G. D. (1969) Biochemistry 8, 4108–4116. [DOI] [PubMed] [Google Scholar]
- 28.Jackson, G. S., Hosszu, L. L. P., Power, A., Hill, A. F., Kenny, J., Saibil, H., Craven, C. J., Waltho, J. P., Clarke, A. R. & Collinge, J. (1999) Science 283, 1935–1937. [DOI] [PubMed] [Google Scholar]
- 29.Perutz, M. F., Pope, B. J., Owen, D., Wanker, E. E. & Scherzinger, E. (2002) Proc. Natl. Acad. Sci. USA 99, 5596–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez De La Paz, M., Goldie, K., Zurdo, J., Lacroix, E., Dobson, C. M., Hoenger, A. & Serrano, L. (2002) Proc. Natl. Acad. Sci. USA 99, 16052–16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaroniec, C. P., MacPhee C. E., Astrof, N. S., Dobson, C. M. & Griffin, R. G. (2002) Proc. Natl. Acad. Sci. USA 99, 16748–16753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zurdo, J., Guijarro, J. I., Jimenez, J. L., Saibil, H. R. & Dobson, C. M. (2001) J. Mol. Biol. 311, 325–340. [DOI] [PubMed] [Google Scholar]
- 34.Canet, D., Last, A. M., Tito, P., Sunde, M., Spencer, A., Archer, D. B., Redfield, C., Robinson, C. V. & Dobson, C. M. (2002) Nat. Struct. Biol. 9, 308–315. [DOI] [PubMed] [Google Scholar]
- 35.Dima, R. I. & Thirumalai, D. (2002) Protein Sci. 11, 1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brant, D. A. & Flory, P. J. (1965) J. Am. Chem. Soc. 87, 663–664. [Google Scholar]
- 37.Smith, L. J., Bolin, K. A., Schwalbe, H., MacArthur, M. W., Thornton, J. M. & Dobson, C. M. (1996) J. Mol. Biol. 255, 494–506. [DOI] [PubMed] [Google Scholar]
- 38.Shi, Z., Olson, A., Rose, G. D., Baldwin, R. L. & Kallenbach, N. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9190–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villegas, V., Zurdo, J., Filimonov, V. V., Aviles, F. X., Dobson, C. M. & Serrano, L. (2000) Protein Sci. 9, 1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pace, C. N., Shirley, B. A. & Thomson, J. A. (1989) in Protein Structure: A Practical Approach, ed. Creighton, T. E. (Oxford Univ. Press, Oxford).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.