Abstract
Single particle tracking (SPT) techniques were developed to explore bio-molecules dynamics in live cells at single molecule sensitivity and nanometer spatial resolution. Recent developments in quantum dots (Qdots) surface coating and bio-conjugation schemes have made them most suitable probes for live cell applications. Here we review recent advancements in using quantum dots as SPT probes for live cell experiments.
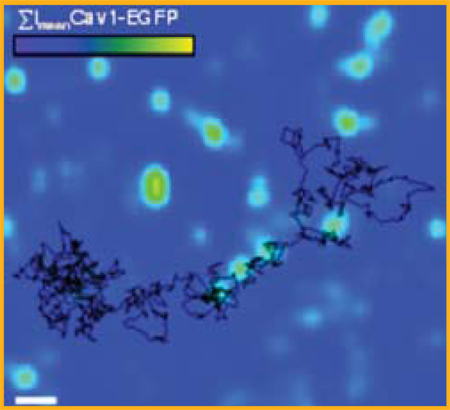
The trajectory of single quantum dot bound to avidin-GPI (in black) is overlaid with the mean intensity of caveolin-1-EGFP (in green) to allow colocalization studies of avidin-GPI with caveolae.
Keywords: quantum dots, single particle tracking, super-resolution, live cell tracking
I. Introduction
Observing real-time dynamics of bio-molecules in live cells can address many fundamental questions in cell biology: how molecules translocate, interact with partners, and respond to environmental cues. In the past two decades, various fluorescent microscopy methods have been developed, providing powerful tools to visualize molecular dynamics as macromolecules perform cellular functions.
Far field fluorescent optical microscopy has been the method of choice for studying biological systems, providing a non-invasive and sensitive approach for probing molecular dynamics in live cells. The major limitation of far field fluorescent microscopy, however, has been its limited resolution (~250 nm) due to the diffraction limit of light [1]. Because of this limitation, it was believed for over a century, that macromolecules inside the cell could not be imaged, or even localized, with the appropriate molecular resolution. In order to image biological macromolecules at work in the living cell, resolution on the order of nanometers is required. To achieve the nanometer resolution, several high resolution localization optical techniques were developed by localizing the centroids of the bright field or fluorescent images with high precision [2–5]. These localization techniques have been successfully applied to near-field microscopy [6], wide field microcopy [7], and confocal microscopy [8] at single molecule level.
Recently several super-resolution imaging techniques, that break or circumvent the diffraction limit of light, have been introduced: stimulated emission depletion microscopy (STED) [9], structured illumination microscopy [10], photoactivated localization microscopy (PALM) [11], fluorescence photoactivation localization microscopy (FPALM) [12], and stochastic optical reconstruction microscopy (STORM) [13]. The above mentioned super-resolution imaging methods rely on the presence, in the sample, of multiple fluorophores within the resolution limit, or the point-spread-function (PSF). These multiple fluorophores are randomly turned on and off, or their fluorescence is inhibited or reduced by structured illumination via stimulated emission and/or saturation. In the very low dilution limit, when on average less than one fluorophore occupies the PSF, super-resolution imaging is no longer attainable, but with recent advances in probes, optics and detectors, individual molecules can be detected and localized with super-resolution accuracy, even in the environment of live cells [14, 15]. For example, STED was recently being used to image and track vesicle movement within synaptic boutons at video rate with a lateral resolution of 62 nm, demonstrating the emerging ability of modern fluorescent microscopy to decipher cellular processes on the nanometer scale [16].
Single particle tracking (SPT) uses the same approach as some of the previously mentioned super resolution methods. For decades, researchers have used this approach in various ways to track single bio-molecules with high resolution [17, 18]. With the application of new imaging methods and the use of brighter and more stable probes, such as quantum dots, SPT has the capability to enter into a new era of high resolution and long timescale imaging. SPT techniques allow scientists to follow single molecules in real time and visualize the actual molecular dynamics in their habitant environment. Upon observing many molecules, a histogram of individual trajectories can be constructed. Such histogram depicts the stochastic dynamic distribution of the system, which is particularly useful to explore heterogeneous molecular behavior in complex environment. Such dynamical observations can lead to discovery of rare but important biological processes, which are often masked in ensemble measurements using traditional biochemical methods. Moreover, these un-averaged dynamic details provide valuable information about molecular mechanisms of biological interactions beyond what can be learnt through static snapshots of the cell.
This review intends to briefly survey recent advancements and achievements within the single particle tracking field with a primary focus on the rapid adaptation of semiconductor quantum dots (Qdots) for SPT in live cells. The organization of this article is as follows: section II summarizes techniques and probes used in SPT experiments; section III briefly discusses several examples of the use of organic fluorophores in SPT experiments; section IV introduces quantum dots (Qdots) and their functionalization methods for applications in live cells; and lastly, section V summarizes recent progress in SPT experiments using Qdots. Outlook for the use of Qdots for SPT is discussed in the concluding section.
II. Single particle tracking
Early SPT techniques utilized a similar approach to many of the currently developed super resolution methods (for review, see [18, 19]). To accomplish this, light-emitting or light-scattering particles were followed frame by frame with a camera and the particles fluorescence or scattered image is fit to a 2-dimensional Gaussian by least squares minimization. The center of this 2-dimensional fit corresponds to the x, y position of the particle, thus allowing the particle to be localized with nanometer precision. The localization precision of this approach depends on the number of detected photon per PSF image and therefore, it has no fundamental limit [20, 21]. Under favorable experimental conditions the localization precision is ~two orders of magnitude better than the diffraction limit itself. Using this approach, earlier experiments utilized latex or fluorescent microspheres (~20–500 nm), and colloidal gold particles (40 nm) [22, 23] to track single macromolecules in live cells. These labels were sufficiently bright to provide enough photons to track single molecules with fast sampling rates for a long time. Further improvements in SPT now allow ultrafast imaging and tracking of various biological molecules including lipids, membrane associated proteins, and cytosolic motor proteins. Temporal resolution as high as 25 micro-seconds has been achieved by Kusumi and coworkers with 40 nm immuno-gold nano-particles [24]. With this ultrafast SPT technique, they claimed to observe the partitioning of lipids and proteins into plasma membrane sub-domains formed by underlying cytoskeletal actin networks. This observation allowed the authors to suggest a “picket-fence model” which is a revised view of the “fluid mosaic” model for the plasma membrane proposed by Singer and Nicholson 30 years ago [23, 25]. Moreover, SPT analysis provides detailed description of the compartment sizes of micro-domains and the residence time of individual macromolecules in these compartments. Unfortunately, the size of the SPT probes may induce steric hindrance effects on the macromolecules they are attached to, thereby blocking normal molecular interactions or access to spatially constrained environments [26]. Using smaller probes can minimize steric effects. As an example, Lasne and others tracked 5 nm gold particles at video-rate by Laser Induced Scattering around a Nanoabsorber [27].
III. Single particle tracking using organic fluorophores
While small-sized gold nano-particles have proven to be good labeling reagents for SPT experiments, they are not well suited for multiplexed detection and simultaneous tracking of different macromolecules. Fluorescent microscopy, which allows one to simultaneously image multiple emission channels, offers the best multiplexing platform for studying dynamic cellular machinery. Thanks to the development of a large library of different-color fluorescent proteins (FPs), the development of genetic tags that allow site-specific in vivo targeting and labeling of permeable organic fluorophores, and the advancement of fast imaging technologies [28], simultaneous observation of multiple bio-molecules in different hues was possible.
These genetic encoded fluorophores and organic dye molecules are significantly smaller than synthetic beads and gold particles, and, as a consequence, are less likely to induce undesirable size-dependent artifacts in SPT experiments. However because of the relative weak fluorescent signal against strong cellular autofluorescence background and their fast photobleaching rate, tracking single molecules labeled with either FPs or organic dyes remains a challenging task. Practically, the excitation level needs to be carefully adjusted to balance between photobleaching rate and fluorescence emission intensity in order to achieve decent signal-to-background (S/B). Under the best conditions, these fluorescent probes could be traced for about 10 seconds, far less than the time scale achieved by SPT experiments using synthetic nano-particles. This article does not intend to review SPT using organic fluorophores or in vivo fluorophore labeling methodology, and thus only selected examples were discussed in the following section.
Despite these difficulties of tracking single FPs/organic fluorophores in vivo, several studies have traced organelles or vesicles loaded with multiple copies of FPs. For example, fluorescence imaging with one nanometer accuracy (FIONA) was used to track the movement of individual green fluorescent protein (GFP) tagged peroxysomes transported by conventional kinesins and cytosolic dyneins [29]. The large number of photons emitted from individual peroxysome containing multiple copies of GFPs allowed Kural et al to push the temporal imaging resolution down to 1.1 milliseconds, thus enabling the determination of the moving velocity and stepping characteristics of these microtubules associated molecular motors in great details. While a step size of ~8 nm is consistent with single molecule in vitro studies using optical trap [30] for both kinesin and dynein, the moving velocity appeared a lot faster than what was found in in vitro assays. Based on this observation, Kural et al. proposed a coordinated mechanism for peroxysome transport [29]. Alternatively, Cai et al. developed a three-tandom monomeric Citrine tag and used it to directly track single molecules of tagged kinesin-1 in live cells with ~20 nm resolution at video rates. This much better defined molecular system provides strong evidence that individual kinesin-1 molecules behave similarly in vivo and in vitro, thereby ruling-out the hypothesis that kinesins move at different speed in a cellular environment [31].
In addition to the genetic tagging of FPs, organic fluorophores with high quantum yield and slow photobleaching rate were also used in SPT experiments. These small organic dyes could provide much more versatility in single molecule tracking experiments, especially in applications where FP fusions perturb the biological system [32, 33]. Seisenberger et al. described the adeno-associated viruss infection pathway in great detail by tracking single viruses labeled with single Cy5 dye [34]. For most biological systems, however, tracking specific molecules among many others in live cells requires the development of efficient in vivo targeting strategies. A variety of enzymatic strategies combined with specific molecular tags have been developed over the years to allow the covalent attachment of small organic dyes to tags that were genetically fused to specified proteins. For example, modifying the target protein with a 15 amino-acid peptide sequence tag allows the protein to be specifically labeled with biotin by recombinant biotin ligase BirA [26]. Tetracysteine-biarsenical system utilizes a 12-residue peptide sequence that includes four cysteines capable of binding membrane-permeable biarsenical FIAsH and ReAsH dyes [35, 36], thus enabling labeling of cytosolic protein targets. Other systems include acyl carrier protein (ACP) tag for specific labeling by phosphopantetheine transferase [37], or Q-tag for the use of transglutaminase [38].
When combining the power of FPs and fluorophores, multi-color single molecule tracking and imaging can provide valuable description of molecular distribution, activation, and trafficking in a dynamic fashion. For example Jacquier et al. used double labeling of both a cytosolic GFP tag and an extracellular ACP tag. This extracellular tag permits specific pulse labeling of the membrane integrated subpopulation of the odorant receptor, therefore allowing the monitoring of individual odorant receptors from early, dynamic processes in the receptors signaling to ligand-induced endocytosis and recycling [39]. Alternatively, another labeling scheme that was utilized to study membrane receptors was the conjugation of fluorophores to the functional ligands themselves. For example, Yanagida and coworkers used functional Cy3-labeled EGF to study the dynamics of EGF Receptor, indicating the existence of preformed EGFR dimers and revealing structural fluctuations between the dimers [40].
IV. Quantum dots as cellular probes
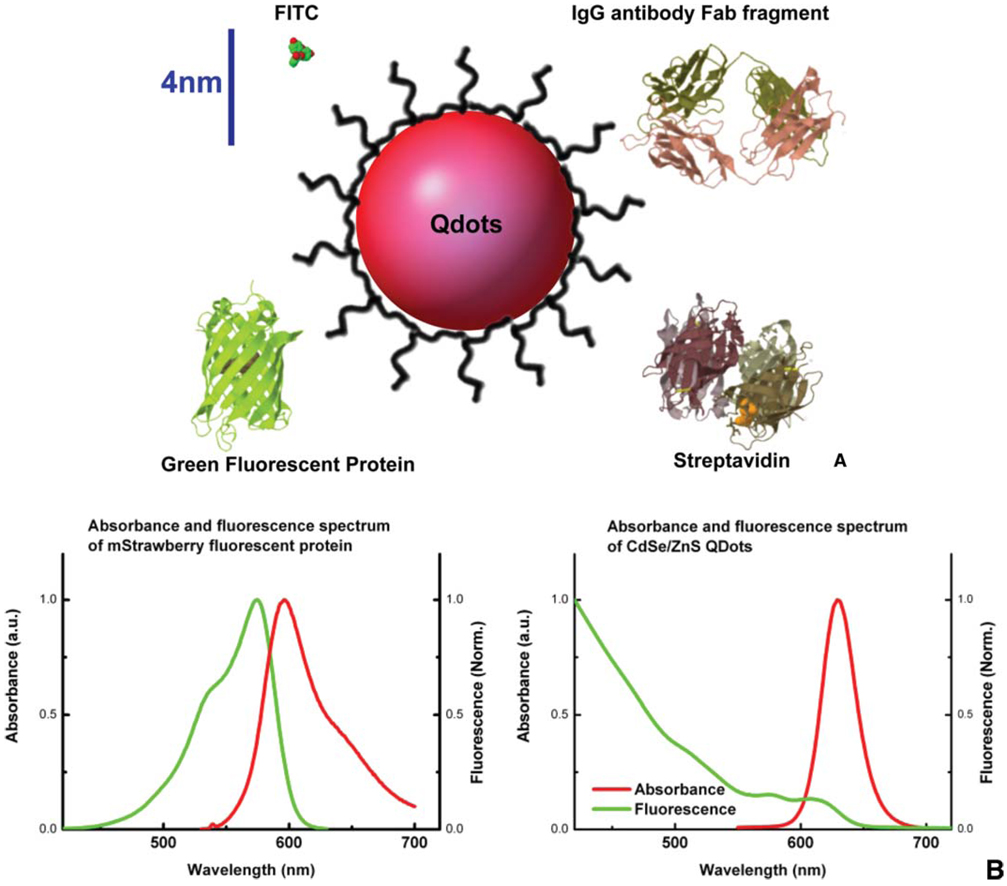
Quantum dots (Qdots) are semiconductor nanocrystals that have unique fluorescent properties (For review, see [41–43]). Their photophysical properties are highly dependent on the chemical composition, shape, and size, all of which can be controlled during synthesis. The unique wide absorption spectra and narrow emission band that can be tuned from UV to infrared wavelengths distinguish them from conventional organic fluorophores and provide many advantages for multi-color fluorescent imaging (see Figure 1). In addition, Qdots are orders of magnitude more photostable than organic dyes and FPs, making them very attractive candidates for long term SPT experiments in live cells, obtaining long tracking trajectories.
Figure 1.
(online colour at: www.biophotonics-journal.org) (A) Comparison of fluorophores and commonly used targeting proteins shown to scale. While Qdots as synthesized core/shell structure have diameters ranging from 2–10 nm, the solubilization interface and biological functionality coating significantly increase the size of the final products. Here a CdSe/ZnS quantum dot coated with peptides (Qdot in red, and peptide coating in black) is shown to compare with FITC fluorophore, green fluorescent protein, IgG Fab fragment, and streptavidin. The hydrodynamic diameter of peptide-coated red-emitting CdSe/ZnS quantum dots is ~12 nm [45]. Streptavidin coated Qdots usually end up with diameters ~15–20 nm, and antibody Fab fragments coupled Qdots can be as large as 25 nm in diameter. Scale bar, 4 nm. (B) Comparison of absorption and fluorescence spectra of FPs to Qdots. Normalized absorbance is in green, and normalized fluorescence emission is in red. The spectra of one red emitting fluorescent protein mStrawberry [71] is shown on the left, and the spectra of red emitting CdSe/ZnS Qdot is shown on the right. The absorption band of typical organic fluorophores is narrow, and the emission peak is broadened on the red shoulder. Qdots, on the contrary, have a broadband absorption spectrum and symmetric, narrow emission peak.
Much of the challenge of using Qdots in live cells comes from surface solubilization and functionalization. Since Qdots are initially synthesized in non-polar solvents and are hydrophobic, their surface needs to be decorated with ampiphilic coatings, rendering them hydrophilic for biological applications. The surface chemistry needs to be stable enough to survive in biological fluids. Over the years, scientists have developed a wide array of surface chemistries for Qdots biological interfacing (For review, see [42]). These surface coatings not only have to retain the advantageous photophysical and size properties of the nanocrystal, but also provide additional reactive groups for subsequent conjugation of biomolecular recognition molecules.
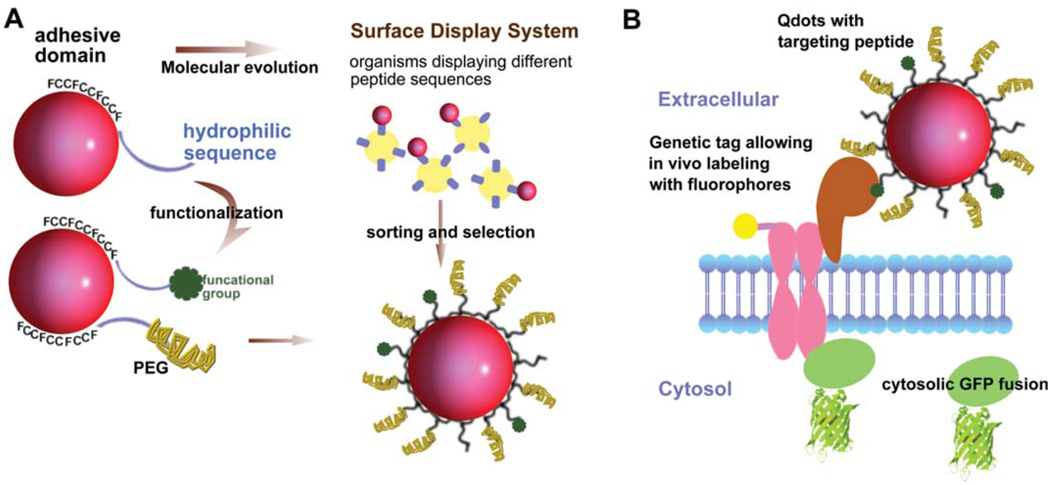
In an effort to achieve small and bio-compatible Qdots, our laboratory has developed Qdots coated with phytochelatin-related peptides, where carefully selected amino-acid sequences solubilize Qdots in aqueous solution, thus providing a biological interface necessary for live cell imaging [44]. These small peptide-coated Qdots have been proven to maintain excellent colloidal and photophysical properties [45, 46]. Furthermore, the coating peptide sequence can be easily customized to acquire different functionalities. For example adding an N-terminal cysteine to the peptide sequence can react with maleimide-biocytin after reduction with dithiothreitol [42, 44]. To further reduce non-specific adhesion between the Qdots and cellular surface, polyethylene glycol (PEG) modified peptides can also be introduced on the Qdot surface as shown in Figure 2 [47].
Figure 2.
(online colour at: www.biophotonics-journal.org) (A) Design and evolution of peptide-coated Qdots. The natural peptide used for Qdots solubilization has a hydrophobic adhesive domain and a hydrophilic sequence [44, 48]. The hydrophilic tail can be functionalized by adding reactive groups or functional groups such as biotin at the N-terminus. Polyethylene glycol (PEG)-conjugated peptide can be introduced simultaneously to reduce non-specific binding to cell surfaces. The adhesive domain sequence can be selected via peptide display libraries such as phage or yeast surface display system. This method can be used to select for peptides that specifically bind to Qdots, or even peptides that improve the photophysical properties such as reduced blinking or enhanced brightness. (B) Schematic representation of multiplexing fluorescent labeling methods to study protein recruitment, trafficking, and activation. Qdots (in red) with specific targeting peptides, genetic tags allowing organic fluorophores to be covalently attached (fluorophore in yellow), and FP tags on cytosolic proteins (in green) can be used in parallel to study complex, dynamic system such as signal transduction pathway.
Interestingly because the photophysical properties of Qdots are somewhat influenced by the peptide adhesive domain [46], it is possible to generate Qdots with higher emission rates, increased stability and reduced blinking by varying the binding sequence via molecular evolution [42]. Pilot experiments showed that hydrophobic unnatural amino acid 3-cyclohexylalanines of our original peptide sequence design could be replaced by other natural hydrophobic amino acids such as phenylalanine [48]. Such natural sequences can be easily produced in large quantities in E. coli. High throughput screening of new candidate sequences to optimize Qdots properties and binding is currently underway using yeast surface display (see Figure 2).
Except for few applications using non-functionalized Qdots [49], most experiments require specific binding of Qdots to the bio-molecule under study. Among many binding moieties and labeling strategies, the ultra-high affinity streptavidin-biotin is the most commonly used binding pair, utilizing commercially available streptavidin-Qdots (SA-Qdots) [50]. For example Tings group tracked AMPA receptors tagged with biotin acceptor sequence using SA-Qdots [26, 51].
Otherwise, extra-cellular parts of most membrane proteins can be readily targeted using a three-layer strategy: primary antibody against the target molecule, followed by a biotinylated secondary antibody, followed by a streptavidin-coated Qdot. However since both primary and secondary antibodies are divalent, this three-layer strategy could result in cross-linking of target proteins and form undesired high molecular weight aggregates. A similar effect occurs when multiple binding moieties present on the surface of the Qdots. Such cross-linking could induce oligomerization and possibly even unwanted activation of signaling cascades. Controlling the number of ligands on the Qdots surface is therefore sometimes necessary.
Membrane proteins were also targeted via Qdots conjugated to biotinylated primary high affinity antibodies [52]. Yet in another approach, several membrane receptors were studied using biotinylated ligands. For example, SA-Qdots decorated with biotinylated epidermal growth factor (EGF) were used to study EGF transport by erbB (HER2) receptors [53]. The sizes of these immuno-targeted Qdots complexes were quite large (~50 nm), sometimes blocking access to narrow or crowded locations on the cells surface [26], or inhibiting molecular interactions due to steric hindrance. Qdots decorated with other reactive groups such as carboxyl or amine are also commercially available, allowing a wide variety of coupling chemistries for conjugating recognition molecules of interest to the nanoparticles. For example, Vu et al. coupled Qdots to nerve growth factor (NGF) and demonstrated that it maintained its biological activity after coupling [54]. Each conjugation scheme requires optimization of reaction conditions and stoichiometry for achieving best labeling results.
In summary, Qdots provide unique properties as cellular imaging probes. Because of their relative large size (with respect to the target), Qdots are best utilized when tagging sparsely distributed target molecules, and when long time observation is desired. In particular, single molecule tracking experiments benefit from the high brightness and high photostability of Qdots. As discussed below, Qdots have been used in numerous live cell studies, proving their utility for single particle tracking experiments.
V. Qdots as probes in single molecule tracking experiments
V.1 Qdots as probes to study membrane receptors
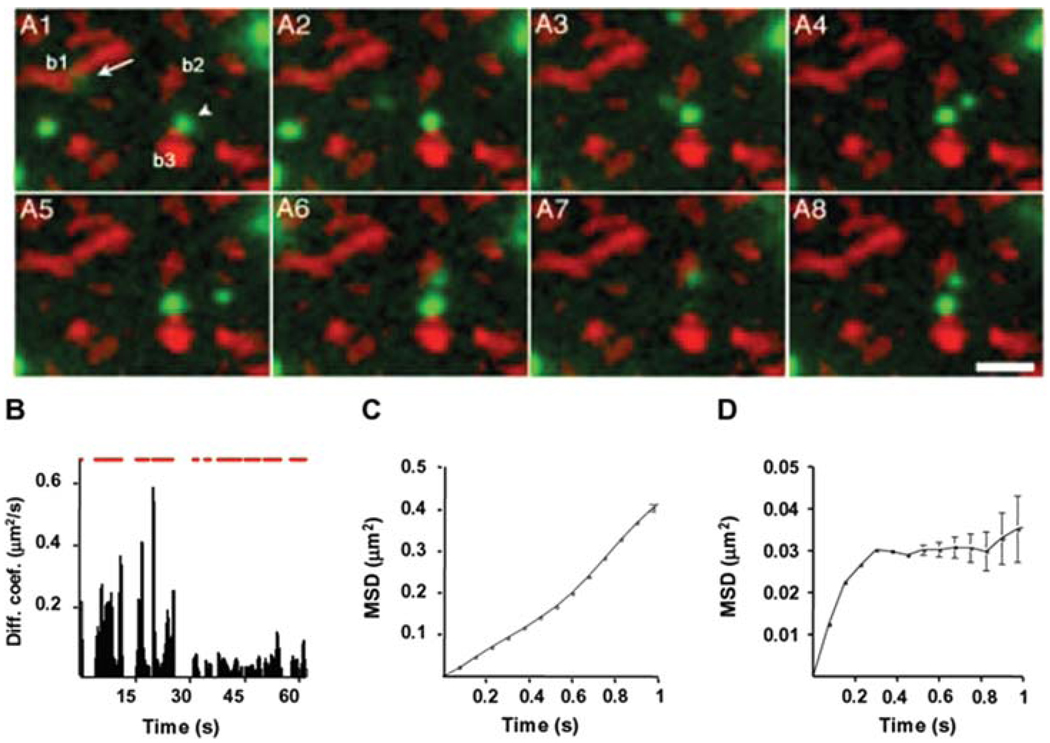
Membrane receptors or membrane associated proteins are intuitive targets for Qdots imaging because they do not require intracellular delivery through the impermeable plasma membrane. Dahan et al. were first to track the lateral diffusion of glycine receptors in living neurons for up to 20 minutes whereas CY3 labeled receptors could only be tracked for 5 seconds before photobleaching [55]. As shown in Figure 3, this work nicely characterized multiple diffusion behaviors during a single trajectory and correlated the receptors behavior to their synaptic localization. Although the size of these Qdots labeled glycine receptors were on the same order as the size of the synaptic cleft, Qdots could be seen entering and exiting this confined environment.
Figure 3.
(online colour at: www.biophotonics-journal.org) Diffusion dynamics of Qdots bound glycine receptors. (A) Selected images from a sequence of 850 frames. The acquisition time for each frame is 75 ms. Qdots fluorescence is in green and FM4–64 labeled synaptic boutons are in red. One green Qdot (arrow) in A1 moves from bouton b1 to b2 by diffusion. (B) Instantaneous diffusion coefficient over time. The upper red bar represents the on time of the tracking Qdot. This blinking behavior indicates that one single Qdot was being followed. For the first 30 seconds, the diffusion coefficient is ~0.1 µm 2/sec and the mean square displacement (MSD) showed a linear relationship with time as showed in (C). For 30–63 seconds of the trajectory, the diffusion coefficient decreased to ~0.02 µm 2/sec and MSD curve in (D) indicated confined diffusion. During this period, the Qdot was located at the vicinity of bouton b2 (A6). Reprinted by permission from Science [55], copyright 2003.
Following this original work, several other neurotransmitter receptors have been followed by either antibody or ligand functionalized Qdots at the single molecule level. For example, Bouzigues et al. followed single GABA receptors as they redistribution on nerve growth cones in response to external ligand gradient in a microtubule dependent manner [56]. Croquet and coworkers compared the diffusion behaviors of single dye or single Qdots tagged AMDA glutamate receptors and found out that all antibody-based labeling strategies yielded similar diffusion coefficients [57]. All these receptors showed periods of fast diffusion (Diffusion coefficient D = 10−1 − 10−2um2/sec) and slow diffusion (10−5 − 10−2 um2/sec), where slow diffusion corresponded to entry into synapses. The diffusion coefficients from the Qdots tracking experiments were comparable with experiments using single dye labeling, indicating that Qdots are faithful reporters of the lateral diffusion of membrane receptors. Cui et al. and Echarte et al. both tracked NGF axonal transport by biotin-NGF and SA-Qdots conjugates, reporting that internalized NGF-Qdots containing endosomes underwent active retrograde transportation in a stop-and-go fashion [58], providing a much more detailed description of the dynamic behavior than previous tracking experiments using fluorophores cy3 did [59].
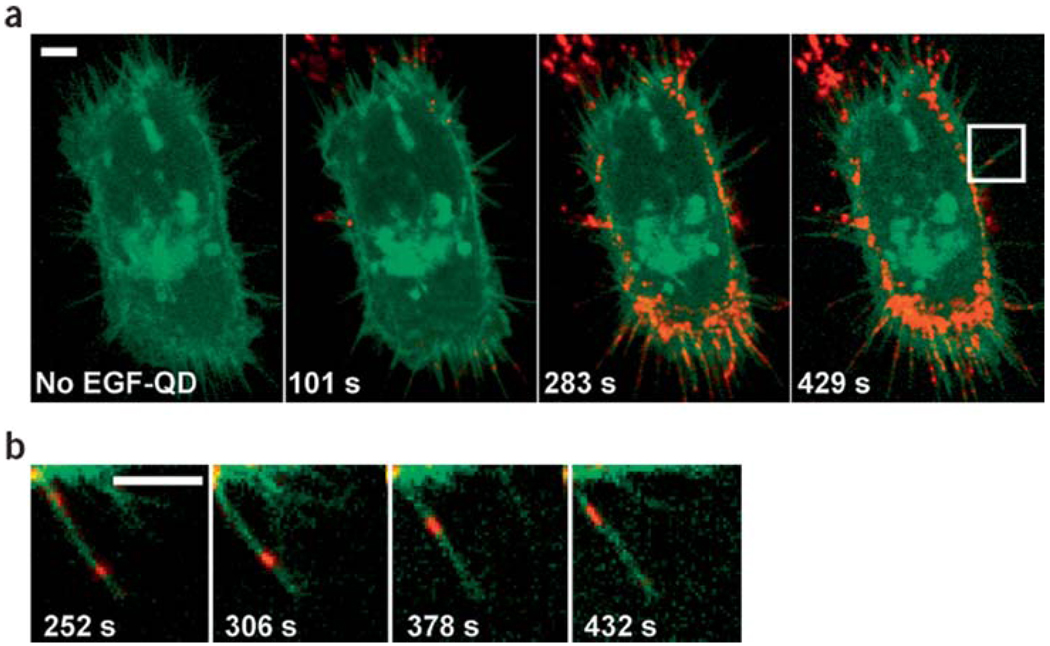
Qdots were also used to study other membrane receptor mediated signal transduction mechanisms in non-neuronal cell systems. Lidke et al. first used biotinylated epidermal growth factor (EGF) conjugated Qdots to study the endocytosis and trafficking of erbB receptors, and were first to report retrograde transport along filopodia towards the cell body as shown in Figure 4 [53]. Haggie et al. tracked cystic fibrosis transmembrane conductance regulator (CFTR) receptors and found them to be immobile due to C-terminal PDZ interactions [60]. Taking together, the above mentioned examples prove Qdots to be a versatile tool for studying diffusion, recruitment, and activation of membrane bound receptors.
Figure 4.
(online colour at: www.biophotonics-journal.org) Retrograde transport of EGF conjugated Qdots along filopodia. (a) A431 cells expressing endogeneous erbB1 and erbB3-eGFP (in green) were labeled with Qdots (in red) conjugated to epidermal growth factor (EGF), which binds to the transmembrane receptor erbB1. Confocal images at different time points were shown in series, indicating the endocytosis of Qdots-ligand bound receptors. (b) A single filopodium of the cell indicated in (a) was magnified, demonstrating the migration of EGF-Qdots towards the cell body at a velocity ~10 nm/sec. This figure was reprinted by permission from Macmillan Publishers Ltd: Nature Biotechnology [53], copyright 2004.
V.2 Qdots as probes to study membrane organization
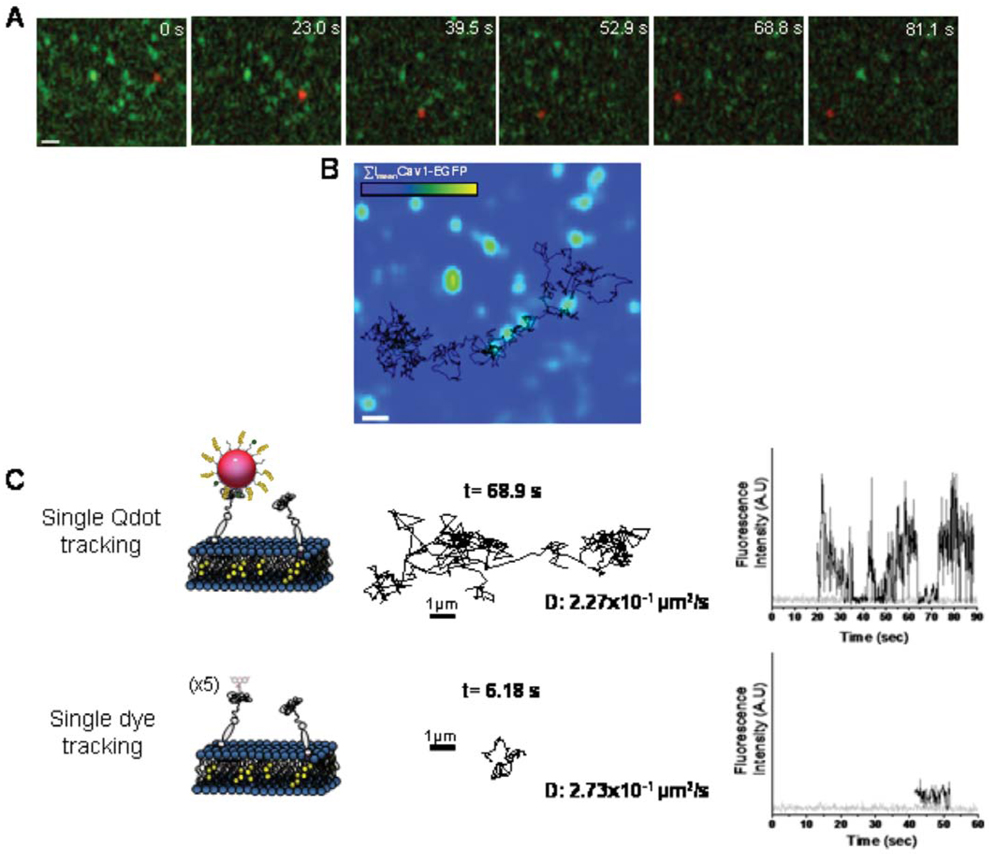
Our lab has used biotinylated peptide-coated Qdots to track single GPI-anchor avidin protein (Av-GPI) expressed in HeLa cells, serving as a model system to study plasma membrane organization and structure (Pinaud et al., submitted). By combining single particle tracking and simultaneous fluorescent imaging in a dual-color total internal reflection microscopy setup, we were able to identify different diffusion behaviors of Av-GPI and to correlate these behaviors with the location of plasma membrane components such as glycosphingolipids GM1 domains and caveolae. Comparative single Qdot tracking and single dye tracking of Av-GPI confirmed that the high photostability of Qdots is advantageous in studying complex membrane organization since it allows the acquisition of diffusion trajectories much longer and much more informative than those obtained with conventional and rapidly photobleached fluorophores (see Figure 5).
Figure 5.
(online colour at: www.biophotonics-journal.org) (A) Time series images from a dual color TIRF microscopy movie of an HeLa cell co-expressing GPI-anchored avidin proteins (Av-GPI) labeled with Qdots (red) and EGFP fused to caveolin-1, a marker of caveolae (green). Scale bar: 1 Dm. (B) The trajectory of the single Av-GPI labeled in A is overlaid with the mean intensity projection image of all frames (EImean) for the caveolin-1-EGFP green channel. This approach allows colocalization studies of Av-GPI with caveolae for the entire movie despite the photobleaching EGFP. (C) Single molecule tracking with Qdots or organic dyes. Schematic representation of plasma membrane anchored proteins targeted with 620 nm emitting Qdots or alexa 488 dyes. Tracking was performed by Gaussian fitting, frame by frame, the PSF of a single Qdot (integration: 100 ms/frame) or a single alexa 488 labeled GPI-av protein (integration: 60 ms/frame) diffusing in the membrane of a HeLa cell. Trajectories for Qdots and dye tracking are plotted together with their duration length, the diffusion coefficient determined from the measurement and the probe fluorescent intensity along the diffusion trajectory (bottom, black) compared to fluorescent background signal from the cell (grey). Notice that for a diffusion coefficient similar to that of the dye, the Qdot trajectory is much longer, thus allowing access to more complex dynamic information than would be inferred from the single dye trajectory. Notice also the typical blinking behavior of the single Qdots and the rapid disappearance of the single dye fluorescence by photobleaching.
V.3 Qdots as probes to study intracellular motor proteins
While Qdots have been used to target membrane receptors and GPI-anchored proteins, it is still challenging to deliver Qdots across the plasma membrane and direct them to cytosolic targets. When cells are presented with Qdots, they internalize them via endocytosis [61], and often trap Qdots in membrane-bound organelles. Nan et al. used this property to study the movement of Qdots-containing endosomes, presumably driven by microtubule associated kinesin and dynein motors [49]. Although the fluorescent signal from each Qdots aggregates was strong, such vesicle-tracking experiments do not give information about the number or the type of motor proteins on each endosome, complicating data interpretation. To avoid such drawbacks, Courty et al. conjugated kinesin to Qdots in vitro and introduced the conjugates into live cells with a cell-loading technique based on the osmotic lysis of pinocytic vesicles. By tracking single Qdots attached to single kinesin motors, they reported velocity and processivity comparable to those measured in in vitro single molecule experiments using purified components [62]. However, due to the relative weak signal from single Qdots, the spatio-temporal resolutions of their experiment did not allow the detection of the 8 nm steps that were measured in in vitro single molecule assays.
Other attempts to transport Qdots across membrane have been made using membrane translocation peptides or transfection agents [63–65]. Unfortunately all these methods tend to result in inhomogeneous distribution of Qdots in the cytoplasm or trapping inside endosomes. As of today, single cell microinjection, albeit very low throughput and time consuming, remains the most reliable way to deliver Qdots and target them to cytosolic proteins. This method was successfully used to target Qdots carrying specific organelle targeting sequence to transport into the nucleus [65] or the mitochondria [63]. Development of new translocating methods for Qdots will greatly accelerate our ability to follow bio-molecules in the cytosol, where most biological processes take place.
V.4 Tracking Qdots in three-dimensions
While two-dimensional single molecule tracking has been applied to many live cell systems, it is often-times limited to biological events that take place on the plasma membrane or a cytoskeleton track. Since many biological processes occur in the cytosolic space, the development of a three-dimensional (3D) single molecule tracking method is highly desirable. Such a 3D tracking capability will provide valuable information about macromolecular processes such as trafficking, translocation and self-assembly. Because of their photostability and brightness, Qdots are well suited for this task.
Several groups have developed different optical techniques to solve this challenging problem: Defocused imaging was used to track a fluorescent bead in a porous polymer network [66]; multi-plane imaging have been used to follow the recycling pathway that leads to exocytosis of the GFP-tagged MHC Class I-related receptor, FcRn [67]; Bifocal imaging was used to track phagocytosed fluorescent beads and organelles [68]; Orbiting a beam around single particle was used to track GFP-labeled chromatin movement in interphase cells in a two-photon excitation microscopy setup [69, 70]; A cylindrical lens was placed in the detection path of a microscope in order to introduce a slight astigmatism, which in turn provided full 3D information.
This last method was applied to tracking endocytosed Qdots and identified several transport modes including active transport and passive 3D diffusion within the trajectories [63]. Holtzer et al. achieved position accuracy of 43 nm in lateral and 130 nm in axial directions at a frame rate of 167 Hz, thereby demonstrating the acquisition of detailed three-dimensional molecular trajectories within the crowded environment of the cell.
VI. Conclusions
Direct observation of single molecules in live cells offers biologists an attractive tool to elucidate the spatio-temporal distribution of bio-molecules in their native environment when all cellular circuits are wired-up. Over the last several years, improvements in Qdots synthesis, surface coating, and bio-conjugation have resulted in very bright probes that are well suited for fluorescence imaging and single particle tracking. Qdots still suffer from drawbacks, owing mainly to size (~5–25 nm). This elicits steric effects that may exclude them from applications in crowded environments, non-specific binding, imprecise stoichiometry of functional groups, and yet not-optimized cytosolic delivery methods. Future improvements to Qdots will arise from synthesizing smaller (but still bright) particles, developing more robust surface coatings, increasing photostability, and devising more efficient cellular delivery methods. Advancements in three-dimensional tracking techniques and development of faster imaging detectors [64, 65] will also greatly enhance our ability to follow single molecules in vivo beyond video rate and the diffraction limit of light, thereby providing insights into fast biomolecular processes in live cells. Together with the development of fluorescent proteins and in vivo targeting strategies, multi-color Qdot fluorescence imaging with nanometer resolution will offer exciting possibilities for deciphering fundamental dynamic cellular processes.
Acknowledgement
We thank Dr. Maxime Dahan for providing us the original figure from [47]. We thank Dr. Xavier Michalet and Dr. Gopal Iyer for critical discussion of the manuscript. This work was supported by NIH-BRP (1R01EB006353-01A1), NSF-CBST (002865-UCLA2), and NIH-CCNE (#U54 CA119367)
Biographies

Yun-Pei Chang received her Ph.D. degree in Biophysics from the University of California at Berkeley, USA. Her research interests include single molecule manipulation, single molecule fluorescent imaging and tracking in live cells.

Fabien Pinaud received his Ph.D. degree in Chemistry and Biochemistry from the University of California at Los Angeles, USA. His research focused on functionalizing quantum dots for biological applications, and probing cell membrane organization by tracking single quantum dots in cell.
Joshua Antelman is a current graduate student in Department of Chemistry and Biochemistry in University of California at Los Angeles, USA. His research focuses on fluorescent imaging at nanometer scale.

Shimon Weiss is a professor in the Department of Chemistry and Biochemistry, University of California at Los Angeles, USA. His research focuses on single molecule fluorescent spectroscopy and microscopy. He also pioneered on quantum dots development for biological application.
References
- 1.Abbe E. Schultze Arch. Mikrosc. Anat. 1873;9:413–468. [Google Scholar]
- 2.Gelles J, Schnapp BJ, Sheetz MP. Tracking Kinesin-Driven Movements with Nanometre-Scale Precision. Nature. 1988;331(6155):450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 3.Betzig E. Proposed Method for Molecular Optical Imaging. Optics Letters. 1995;20(3):237–239. doi: 10.1364/ol.20.000237. [DOI] [PubMed] [Google Scholar]
- 4.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophysical Journal. 2002;82(5):2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yildiz A, Selvin PR. Fluorescence imaging with one manometer accuracy: Application to molecular motors. Accounts of Chemical Research. 2005;38(7):574–582. doi: 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- 6.Ha T, et al. Dual-molecule spectroscopy: Molecular rulers for the study of biological macromolecules. Ieee Journal of Selected Topics in Quantum Electronics. 1996;2(4):1115–1128. [Google Scholar]
- 7.Schmidt T, et al. Imaging of single molecule diffusion. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(7):2926–2929. doi: 10.1073/pnas.93.7.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacoste TD, et al. Ultrahigh-resolution multicolor colocalization of single fluorescent probes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(17):9461–9466. doi: 10.1073/pnas.170286097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hell SW, Wichmann J. Breaking the Diffraction Resolution Limit by Stimulated-Emission – Stimulated-Emission-Depletion Fluorescence Microscopy. Optics Letters. 1994;19(11):780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 10.Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. Journal of Microscopy-Oxford. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 11.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 12.Hess ST, Girirajan TPK, Mason MD. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys. J. 2006;91(11):4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bates M, et al. Multicolor Super-Resolution Imaging with Photo-Switchable Fluorescent Probes. Science. 2007;317(5845):1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nature Methods. 2008;5(2):155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 15.Hess ST, et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proceedings of the National Academy of Sciences. 2007;104(44):17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westphal V, et al. Video-Rate Far-Field Optical Nanoscopy Dissects Synaptic Vesicle Movement. Science. 2008:1154228. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 17.Cherry RJ. Keeping track of cell surface receptors. Trends in Cell Biology. 1992;2(8):242–244. doi: 10.1016/0962-8924(92)90312-b. [DOI] [PubMed] [Google Scholar]
- 18.Saxton MJ, Jacobson K. Single-particle tracking: Applications to membrane dynamics. Annual Review of Biophysics and Biomolecular Structure. 1997;263:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 19.Levi V, Gratton E. Exploring dynamics in living cells by tracking single particles. Cell Biochemistry and Biophysics. 2007;48(1):1–15. doi: 10.1007/s12013-007-0010-0. [DOI] [PubMed] [Google Scholar]
- 20.Ram S, Ward ES, Ober RJ. Beyond Rayleighs criterion: A resolution measure with application to single-molecule microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(12):4457–4462. doi: 10.1073/pnas.0508047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michalet X, Weiss S. Using photon statistics to boost microscopy resolution. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(13):4797–4798. doi: 10.1073/pnas.0600808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki KGN, et al. GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. Journal of Cell Biology. 2007;177(4):717–730. doi: 10.1083/jcb.200609174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annual Review of Biophysics and Biomolecular Structure. 2005;34:U351–U354. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara T, et al. Phospholipids undergo hop diffusion in compartmentalized cell membrane. Journal of Cell Biology. 2002;157(6):1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer SJ, Nicolson GL. Fluid Mosaic Model of Structure of Cell-Membranes. Science. 1972;175(4023):720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 26.Howarth M, et al. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(21):7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasne D, et al. Single Nanoparticle Photothermal Tracking (SNaPT) of 5-nm Gold Beads in Live Cells. Biophys. J. 2006;91(12):4598–4604. doi: 10.1529/biophysj.106.089771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giepmans BNG, et al. Review – The fluorescent toolbox for assessing protein location and function. Science. 2006;312(5771):217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 29.Kural C, et al. Kinesin and Dynein Move a Peroxisome in Vivo: A Tug-of-War or Coordinated Movement? Science. 2005;308(5727):1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 30.Vale RD, Milligan RA. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288(5463):88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 31.Cai DW, Verhey KJ, Meyhofer E. Tracking single kinesin molecules in the cytoplasm of mammalian cells. Biophysical Journal. 2007;92(12):4137–4144. doi: 10.1529/biophysj.106.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann C, et al. A FlAsH-based FRETapproach to determine G protein – coupled receptor activation in living cells. Nature Methods. 2005;2(3):171–176. doi: 10.1038/nmeth742. [DOI] [PubMed] [Google Scholar]
- 33.Andresen M, Schmitz-Salue R, Jakobs S. Short tetracysteine tags to beta-tubulin demonstrate the significance of small labels for live cell imaging. Molecular Biology of the Cell. 2004;15(12):5616–5622. doi: 10.1091/mbc.E04-06-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seisenberger G, et al. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 2001;294(5548):1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- 35.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281(5374):269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 36.Adams SR, et al. New biarsenical Ligands and tetracysteine motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. Journal of the American Chemical Society. 2002;124(21):6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 37.George N, et al. Specific labeling of cell surface proteins with chemically diverse compounds. Journal of the American Chemical Society. 2004;126(29):8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 38.Lin CW, Ting AY. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. Journal of the American Chemical Society. 2006;128(14):4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacquier V, et al. Visualizing odorant receptor trafficking in living cells down to the single-molecule level. Proceedings of the National Academy of Sciences. 2006;103(39):14325–14330. doi: 10.1073/pnas.0603942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell. Biol. 2000;2(3):168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 41.Alivisatos AP, Gu WW, Larabell C. Quantum dots as cellular probes. Annual Review of Biomedical Engineering. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 42.Pinaud F, et al. Advances in fluorescence imaging with quantum dot bio-probes. Biomaterials. 2006;27(9):1679–1687. doi: 10.1016/j.biomaterials.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michalet X, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307(5709):538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinaud F, et al. Bioactivation and cell targeting of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides. Journal of the American Chemical Society. 2004;126(19):6115–6123. doi: 10.1021/ja031691c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doose S, et al. Comparison of photophysical and colloidal properties of biocompatible semiconductor nanocrystals using fluorescence correlation spectroscopy. Analytical Chemistry. 2005;77(7):2235–2242. doi: 10.1021/ac050035n. [DOI] [PubMed] [Google Scholar]
- 46.Tsay JM, Doose S, Weiss S. Rotational and translational diffusion of peptide-coated CdSe/CdS/ZnS nanorods studied by fluorescence correlation spectroscopy. Journal of the American Chemical Society. 2006;128(5):1639–1647. doi: 10.1021/ja056162i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ballou B, et al. Noninvasive imaging of quantum dots in mice. Bioconjugate Chemistry. 2004;15(1):79–86. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- 48.Iyer G, et al. Solubilization of quantum dots with a recombinant peptide from Escherichia coli. Small. 2007;3(5):793–798. doi: 10.1002/smll.200600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nan X, et al. Observation of Individual Microtubule Motor Steps in Living Cells with Endocytosed Quantum Dots. J. Phys. Chem. B. 2005;109(51):24220–24224. doi: 10.1021/jp056360w. [DOI] [PubMed] [Google Scholar]
- 50.Quantum Dots. Invitrogen Corporation; Invitrogen. [Google Scholar]
- 51.Howarth M, et al. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nature Methods. 2008;5(5):397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe TM, Higuchi H. Stepwise movements in vesicle transport of HER2 by motor proteins in living cells. Biophysical Journal. 2007;92(11):4109–4120. doi: 10.1529/biophysj.106.094649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lidke DS, et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nature Biotechnology. 2004;22(2):198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 54.Vu TQ, et al. Peptide-conjugated quantum dots activate neuronal receptors and initiate downstream signaling of neurite growth. Nano Letters. 2005;5(4):603–607. doi: 10.1021/nl047977c. [DOI] [PubMed] [Google Scholar]
- 55.Dahan M, et al. Diffusion Dynamics of Glycine Receptors Revealed by Single-Quantum Dot Tracking. Science. 2003;302(5644):442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 56.Bouzigues C, et al. Asymmetric redistribution of GABA receptors during GABA gradient sensing by nerve growth cones analyzed by single quantum dot imaging. Proceedings of the National Academy of Sciences. 2007;104(27):11251–11256. doi: 10.1073/pnas.0702536104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groc L, et al. Surface trafficking of neurotransmitter receptor: Comparison between single-molecule/quantum dot strategies. Journal of Neuroscience. 2007;27(46):12433–12437. doi: 10.1523/JNEUROSCI.3349-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui BX, et al. One at a time, live tracking of NGF axonal transport using quantum dots. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(34):13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tani T, et al. Trafficking of a Ligand-Receptor Complex on the Growth Cones as an Essential Step for the Uptake of Nerve Growth Factor at the Distal End of the Axon: A Single-Molecule Analysis. J. Neurosci. 2005;25(9):2181–2191. doi: 10.1523/JNEUROSCI.4570-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haggie PM, et al. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Molecular Biology of the Cell. 2006;17(12):4937–4945. doi: 10.1091/mbc.E06-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jaiswal JK, et al. Use of quantum dots for live cell imaging. Nature Methods. 2004;1(1):73–78. doi: 10.1038/nmeth1004-73. [DOI] [PubMed] [Google Scholar]
- 62.Courty S, et al. Tracking individual kinesin motors in living cells using single quantum-dot imaging. Nano Letters. 2006;6(7):1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 63.Hoshino A, et al. Quantum dots targeted to the assigned organelle in living cells. Microbiology and Immunology. 2004;48(12):985–994. doi: 10.1111/j.1348-0421.2004.tb03621.x. [DOI] [PubMed] [Google Scholar]
- 64.Derfus AM, Chan WCW, Bhatia SN. Intracellular delivery of quantum dots for live cell labeling and organelle tracking. Advanced Materials. 2004;16(12):961–966. [Google Scholar]
- 65.Chen FQ, Gerion D. Fluorescent CdSe/ZnS nanocrystal-peptide conjugates for long-term, nontoxic imaging and nuclear targeting in living cells. Nano Letters. 2004;4(10):1827–1832. [Google Scholar]
- 66.Speidel M, Jonas A, Florin EL. Three-dimensional tracking of fluorescent nanoparticles with subnanometer precision by use of off-focus imaging. Optics Letters. 2003;28(2):69–71. doi: 10.1364/ol.28.000069. [DOI] [PubMed] [Google Scholar]
- 67.Prabhat P, et al. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(14):5889–5894. doi: 10.1073/pnas.0700337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toprak E, et al. Three-Dimensional Particle Tracking via Bifocal Imaging. Nano Lett. 2007;7(7):2043–2045. doi: 10.1021/nl0709120. [DOI] [PubMed] [Google Scholar]
- 69.Levi V, Ruan QQ, Gratton E. 3-D particle tracking in a two-photon microscope: Application to the study of molecular dynamics in cells. Biophysical Journal. 2005;88(4):2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levi V, et al. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophysical Journal. 2005;89(6):4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nature Biotechnology. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]