Abstract
Genetic testing of cancer susceptibility genes is now widely applied in clinical practice to predict risk of developing cancer. In general, sequence-based testing of germline DNA is used to determine whether an individual carries a change that is clearly likely to disrupt normal gene function. Genetic testing may detect changes that are clearly pathogenic, clearly neutral or variants of unclear clinical significance. Such variants present a considerable challenge to the diagnostic laboratory and the receiving clinician in terms of interpretation and clear presentation of the implications of the result to the patient. There does not appear to be a consistent approach to interpreting and reporting the clinical significance of variants either among genes or among laboratories. The potential for confusion among clinicians and patients is considerable and misinterpretation may lead to inappropriate clinical consequences. In this article we review the current state of sequence-based genetic testing, describe other standardized reporting systems used in oncology and propose a standardized classification system for application to sequence based results for cancer predisposition genes. We suggest a system of five classes of variants based on the degree of likelihood of pathogenicity. Each class is associated with specific recommendations for clinical management of at-risk relatives that will depend on the syndrome. We propose that panels of experts on each cancer predisposition syndrome facilitate the classification scheme and designate appropriate surveillance and cancer management guidelines. The international adoption of a standardized reporting system should improve the clinical utility of sequence-based genetic tests to predict cancer risk.
Keywords: IARC, variants, cancer genetics, classification, recommendations
Genetic Testing for Cancer Susceptibility
Clinical testing is currently available for more than 1500 different genes or genetic conditions (www.genetests.org). The initial DNA-based tests introduced by clinically-certified genetic testing laboratories focused on identification of previously defined mutations in relatively rare genetic disorders. For example, molecular testing for cystic fibrosis and for multiple endocrine neoplasia type 2 (MEN2) was based on panels of known mutations in the CFTR (MIM# 602421) and RET (MIM# 164761) genes, respectively. Genetic tests based on sequencing of the entire coding region were not generally used clinically because of the expense involved in sequencing, the lack of sequence/polymorphism information, as well as the need to improve methodologies to efficiently interpret the sequence traces.
Full sequence analysis of the BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185) genes was introduced in 1996 by Myriad Genetic Laboratories, as one of the first sequence-based tests offered to identify increased risk for an otherwise common cancer (Collins, 1996). Sequence-based tests are now ubiquitous in genetic testing, due to advancement in high-throughput sequencing technologies and alternative approaches where mutation scanning methodologies are followed by sequencing of the targeted region. The vast majority of DNA sequence changes that result in an increased risk of developing cancer focus on identifying inactivating mutations that can occur throughout the length of the coding region of a tumor suppressor gene.
For sequence-based genetic tests, there are three possible results reported to physicians: (1) positive, in which a mutation that clearly disrupts gene function (and therefore is highly likely to cause clinical consequences) is detected, (2) negative, in which no variation in DNA sequence is detected, and (3) uncertain, in which a sequence “variant of uncertain/unclassified significance” (VUS) or “unclassified variant” (UV, UCV) is detected and it is not known whether the variant has any effect on gene function which might confer an increased cancer risk. One laboratory from the Netherlands reported that from one-third to one-half of the sequence changes reported for BRCA1 and BRCA2, respectively, were such variants (Gomez-Garcia et al., 2005). Similarly, analysis of the Myriad Genetics Laboratory data revealed that a physician who orders BRCA1 and BRCA2 testing has a similar likelihood of receiving a variant result (13%) as one with a pathogenic mutation (Frank et al., 2002). The likelihood of an unclassified variant result is even higher for individuals from understudied populations who undergo genetic testing due to insufficient information on the common polymorphisms in that population (Kean-Cowdin et al., 2005; John et al., 2007). In addition, less well studied or newly identified disease-associated genes present greater challenges for interpretation of sequence-based results.
Appropriate use of genetic testing in daily clinical practice requires clear reporting by laboratories, well informed interpretation by clinicians and clear communication of the consequences to patients. A classification system for variants with recommendations for action coupled to each class would help facilitate this goal. Clinical interpretation of sequence variants is not limited to testing for cancer susceptibility. Genetic testing is used for a variety of purposes from risk prediction, carrier testing and reproductive decision making which may impact how variants are classified. In this paper we focus on genetic testing for autosomal dominant cancer susceptibility with the expectation that success with this clinical application may lead to further expansion to other venues of genetic testing.
The question of how to report out complex data for prediction of cancer association is not unique to genetic testing. There are several clinical classifications systems in oncology that have been generally perceived to be effective in conveying the likelihood of a test result being associated with disease. We briefly review some of these systems, as they serve as a template for our recommendations with regard to genetic testing for cancer susceptibility.
Breast cancer triple assessment by clinical examination, imaging, and cytology
A consistent reporting framework using triple assessment is well established as a valuable tool in pre-operative breast cancer diagnosis (Boerner and Sneige, 1998; Roskell and Buley, 2004; Farshid and Downey, 2005). Following triple assessment of a breast lesion, the most suspicious category prevails in planning further management.
In many countries, the results of mammography examinations used as a screening tool for breast cancer are reported using the Breast Imaging Reporting and Data System (BIRADS) classification of the American College of Radiology (Kerlikowske et al., 1998). The BIRADS system includes 7 possible categories for the reporting of the results (Table 1). Of note, each category is paired with a clinical recommendation for further procedures, e.g. biopsy or follow up interval. This system has been in use in the United States since 1995 and is now accepted internationally for mammography reporting (Balleyguier et al., 2007).
Table 1.
BIRADS system for reporting mammography results
| Category | Diagnosis | Description; Recommended Action | Probability of Malignancy |
|---|---|---|---|
| 6 | Known Biopsy Proven Malignancy | Lesions known to be malignant that are being imaged prior to definitive treatment; assure that treatment is completed | N/A |
| 5 | Highly Suspicious of Malignancy | Lesion that has a high probability of being malignant; take appropriate action | ≥ 95% |
| 4 | Suspicious Abnormality | Not characteristic of breast cancer, but reasonable probability of being malignant; biopsy should be considered | 3 to 94% |
| 3 | Probably Benign | Findings that have a high probability of being benign six-month short interval follow-up | <2% |
| 2 | Benign | A definite benign finding; routine screening recommended | |
| 1 | Negative | There is nothing to comment on; routine screening recommended | |
| 0 | Incomplete | Your mammogram or ultrasound didn’t give the radiologist enough information to make a clear diagnosis; follow-up imaging is necessary |
Similarly, experienced cytopathologists analyze cells obtained from fine needle aspiration cytology of a palpable breast lesion. Cytological preparations cannot always be clearly interpreted (like sequence variants) and are also classified into five categories (malignant, suspicious, atypical, benign or inadequate). Clinicians use this data alongside evidence from radiological and clinical findings to reach a diagnosis.
IARC Carcinogen Classification
Another well-established classification system for cancer association is the determination of whether compounds are carcinogenic to humans. The IARC Monographs program was established in 1971. Its objective is to prepare, with the help of international Working Groups of experts, and to publish in the form of Monographs, critical reviews and evaluations of evidence on the carcinogenicity of a wide range of human exposures (see http://monographs.iarc.fr/ENG/Preamble/CurrentPreamble.pdf).
The first step is evaluating the strength of the evidence for carcinogenicity arising from human and experimental animals, using standard terms. Evidence relevant to carcinogenicity from human and animal studies is classified into one of four categories: Sufficient evidence, limited evidence, inadequate evidence or evidence suggesting lack of carcinogenicity. Mechanistic data are evaluated for the strength of evidence that an observed carcinogenic effect is due to a particular mechanism, using terms such as ‘weak’, ‘moderate’ or ‘strong’. The body of evidence for carcinogenicity derived from human, experimental animal, and mechanistic studies are combined into an overall evaluation of the carcinogenicity of the agent to humans into one of five categories (Figure 1 and Table 2). A working group made up of experts in the exposure under consideration may assign the agent to a higher or lower category than a strict interpretation of human and animal data would indicate.
Figure 1.

The algorithm used for data combination during evaluation of possible carcinogens
Table 2.
IARC (International Agency for Cancer Research) Classification of Carcinogens
| Group | Description |
|---|---|
| 1 | The agent is carcinogenic to humans |
| 2A | The agent is probably carcinogenic to humans |
| 2B | The agent is possibly carcinogenic to humans |
| 3 | The agent is not classifiable as to its carcinogenicity to humans |
| 4 | The agent is probably not carcinogenic to humans |
Other classification systems for sequence variants
In 2007 practice guidelines were proposed for classification of variants as a corporate action of the Dutch and British societies for clinical molecular genetics (as presented on their website: http://cmgsweb.shared.hosting.zen.co.uk/BPGs/Best_Practice_Guidelines.htm). These guidelines set quality standards and lines of evidence from a clinical molecular genetic point of view that should be reviewed in assessing whether a variant might be pathogenic. This guideline concludes that it is essential to report all sequence variants. It proposed reporting uncertain variants in three classes: a) certainly not pathogenic, b) unlikely to be pathogenic and c) likely to be pathogenic. However, the levels of pathogenicity to place a variant into any given category, and clinical recommendations associated with each category, were not included. Similarly, recently published recommendations by the ACMG for standards for interpretation and reporting of sequence variations into six categories focus on the provision of quality clinical laboratory genetic services. However, they do not recommend specific quantitative information to categorize variants. There is a broad category 3 - “sequence variation previously unreported and is of the type which may or may not be causative of the disorder” (Richards et al., 2008).
Proposed Classification System
We propose a classification system that more clearly conveys information about the relevance of a variant to clinical practice including cancer risk assessment and the need for future studies. This system will encompass variants that are definitely pathogenic and definitely neutral as well as those currently of unknown clinical significance. We recommend dropping the term “unclassified” in describing variants, unless it applies to variants identified in the laboratory prior to being designated a specific class. Similar to the systems described above, we propose that clinical and research purposes will best be served by five classes (see Table 3).
Table 3.
Proposed Classification System for Sequence Variants Identified by Genetic Testing
| Class | Description | Probability of being Pathogenic |
|---|---|---|
| 5 | Definitely Pathogenic | >0.99 |
| 4 | Likely Pathogenic | 0.95–0.99 |
| 3 | Uncertain | 0.05–0.949 |
| 2 | Likely Not Pathogenic or of Little Clinical Significance | 0.001–0.049 |
| 1 | Not Pathogenic or of No Clinical Significance | <0.001 |
Class 1 corresponds to the qualitative classification “Not Pathogenic” or “No Clinical Significance”. Assigning a variant to Class 1 rules out a major clinically significant effect of that variant on cancer risk. Some apparently neutral variants might actually confer low increases (or decreases) in cancer risk, of the order of magnitude associated with common polymorphisms reported in genome wide association studies (GWAS) (Easton et al., 2007b) (Hunter et al., 2007). However, the proposed reporting and classification system described here pertains only to clinical testing for mutations of likely high penetrance.
The major advance of the proposed classification system is the creation of Class 2 and Class 4, “Likely Not Pathogenic/Little Clinical Significance (LCS)” and “Likely Pathogenic” with a consistent definition of the likelihood of pathogenicity of 5% and 95%, respectively. The working group felt that it was important to differentiate these two groups of variants from Class 3 (“Uncertain”) in order to differentiate variants for which there is really too little information to make any recommendation (Class 3) from those for which there is significant, but not irrefutable, evidence against (Class 2) or for (Class 4) pathogenicity. The initial test report should convey that the classification category may be altered if more evidence becomes available in the future, eg. segregation studies, tumour histopathology, in vitro assays, etc. The classification category can therefore be continually refined as data accumulate.
The ideal system would enable a quantitative assessment for each variant. In some cases, a probability of pathogenicity can be assigned based on likelihood ratios or odds ratios that are determined empirically from multiple lines of evidence (Goldgar et al., 2004) (Easton et al., 2007a) as described below. Although at the present time accurate quantification is only possible for a small number of variants in a few genes, this number will increase. Laboratory and computational methods for classifying variants are improving, and as more families are identified with variants, more definitive results will come from segregation analysis and statistical genetic methods with comprehensive and widely accessible gene/disease specific databases curating evidence on variants as this becomes available (see companion papers in this special issue: Couch et al, 2008; Easton et al, 2008; Goldgar et al, 2008; Tavtigian et al, 2008; Greenblatt et al, 2008).
However, even when some types of information cannot be quantified, we recommend that all available data be used to assign each variant to one of these five classes. This classification may incorporate information including the strength of multiple lines of evidence, similar to the IARC system for classifying carcinogens (see http://monographs.iarc.fr/ENG/Preamble/CurrentPreamble.pdf). For example, an arbitrary point-based system has been suggested for (MMR) repair gene variants, but is not currently in general use (Barnetson et al., 2008). We recognize that for genes with limited information this may result in the majority of variants being assigned to Class 3.
Utilization of the Classification System
Use of this classification system will facilitate the processing and transmission of appropriate information to patients and improve clinical utilization of the information in the following ways: 1) Genetics professionals, and clinicians who regularly deal with high risk patients, will be able to transmit to their patients the classification, the general rationale, and the clinical recommendations based on each class of variant. 2) Health professionals without genetics training who care for patients who have undergone genetic testing will be better able to understand the information if a clear and consistent classification system is used and if that system includes instructions for appropriate clinical actions for each category as exemplified by the BIRADS classification. The risk of misinterpretation and misinformation reaching patients is reduced.
Understanding what evidence was considered in classifying each variant, and how the data were integrated will be beyond the expertise of most clinicians. Experts will have to develop standards for each class of information, evaluate the data available to set those standards for each variant, and transmit the conclusions and a summary of the data to the clinical and molecular testing communities. Expert panels already exist through international consortia on cancer susceptibility including the Breast Cancer Information Core (BIC) Steering Committee which interprets sequence variants in the BRCA1 and BRCA2 genes and posts conclusions on the BIC web site http://research.nhgri.nih.gov/bic/. The International Society of Gastrointestinal Hereditary Tumors (InSiGHT; http://www.insight-group.org/) has established a committee to classify variants in MMR genes, in partnership with the Human Variome Project (HVP; http://www.humanvariomeproject.org/) to achieve the goal of classifying MMR gene variants (Cotton 2007). Its additional goals are to 1) establish standards for each type of evidence and for integrating them, 2) validate each type of evidence, and quantify if possible with predictive values, and 3) promote transparency in the process of evaluating data and displaying conclusions. The goal is that the BIC, InSiGHT and HVP will first establish standards for the two most common inherited cancer syndromes with extension to other genes later.
Currently, many groups are cautious in classifying variants as pathogenic unless there is overwhelming epidemiologic evidence of association with the cancer. For example, the BIC database currently uses a three way classification for “clinically important,” either Yes, No or Unknown (Couch and Weber, 1996). All variants between 0.1–99% pathogenicity are listed as “unknown”. Many diagnostic laboratories avoid using the category “Likely Pathogenic” because of the potential liability if the classification proves to be incorrect based on new data. Furthermore clinicians may be inclined to interpret this comment as meaning pathogenic but no formal proof yet. . These variants are commonly simply reported as “VUS” or “unclassified”. Some laboratories will cite supporting evidence without reporting a conclusion (for example, if the variant is annotated in a database as pathogenic, or the variant occurs at a conserved residue) and some will not. The outcome of this inconsistency for clinicians and patients in such cases is uninformative; unhelpful at best and, at worst, open to misinterpretation.
This working group noted that in contrast to this conservative approach, health care professionals are accustomed to making important clinical decisions based on predictive values of 80–85% in many other aspects of oncology. For example, transrectal ultrasound (with an estimated accuracy of 70–90% (Giovannini and Ardizzone, 2006) is the recommended procedure (or “test”) to use to determine decisions about surgery for rectal cancer. Clinical genetics professionals should be able to discuss rational strategies for variants with 95–99% (or conversely 0.1–5%) likelihood of cancer association. Currently, with no accepted standard for a category of “Likely Pathogenic” or “Likely Not Pathogenic”, individuals carrying both of these classes of variants are treated based on family history although vast differences in cancer risks are clearly recognizable (based on expert consensus). This can lead to increased risk for individuals. For example, in hereditary non-polyposis colon cancer (HNPCC), in the absence of a genetic diagnosis all first degree relatives of affected individuals are recommended to have frequent (often annual) colonoscopies which carry a risk for major complications of 1:1000 colonoscopies, and for death is 1-5:10,000 (Gatto et al., 2003; Becker et al., 2007). Classifying some variants as Class 4 and testing at-risk relatives for the Class 4 variant can reduce the number of unnecessary colonoscopies in non-carriers.
Degree of Risk Associated with Each Category
We believe that a Bayesian system to generate a posterior probability should ultimately be the standard for all variants. Probabilistic information can be transmitted to patients in a variety of ways to increase understanding (Trevena et al., 2006). The thresholds of 99% and 0.1% for Class 5 and Class 1 were chosen as sufficiently likely to be pathogenic or neutral/little clinical significance, respectively, that further data are not likely to change the classification. For Classes 2–4 (posterior probability of pathogenicity 0.1% to 99%), additional supporting data accumulated over time would help reclassify the variant into a category with increased confidence.
Carriers of a Class 4 variant (95–99% likelihood of pathogenicity) can be advised to undergo surveillance and cancer prevention treatments advised for class 5 variant carriers even if there is a small risk that they will be over treated. Class 2 variants (0.1–5% likelihood of pathogenicity) are classified as “Likely Not Pathogenic”. They could change to Class 3 (uncertain) if more data favoring pathogenicity are found. However, they are unlikely to convert to Class 4 or 5. Starting with a prior probability of 0.05, new data would need to have a likelihood ratio of >1900:1 in order to convert a variant from Class 2 to Class 4 or 5 (posterior probability of ≥ 0.95). Therefore, given the unlikeliness of Class 2 converting to a pathogenic class, Class 2 mutations can be considered to have negligible clinical relevance, although further data should be sought to definitively classify them as Class 1 and not Class 3.
From the clinical perspective, the most important distinction will thus be between Class 3 and Class 4. When considered quantitatively, a posterior probability of 0.95 (Class 4) corresponds to posterior odds of 19:1 (95%/5%). One reason for choosing this cut-off is that a small change in probability below this level results in a larger change in posterior odds (e.g., 90% probability, posterior odds = 90/10 = 9:1; for 85% probability, posterior odds = 85/15 = 5.7:1). This degree of uncertainty could be significantly altered by new data. Thus, our Class 3 variants carry sufficient uncertainty such that no clinical predictive testing should be done on other relatives and further data are needed before any action is undertaken based on the test result. It is also possible that Class 3 variants with these uncertain odds ratios represent variants with intermediate clinical effects or low penetrance alleles. Further data are needed to verify this hypothesis. Overall, given the uncertainty, clinical advice for carriers of a Class 3 variant (and their family members) should depend on the pattern of cancer in the family, tumor histology, etc. rather than the presence or absence of the variant.
Generality of this Classification
Theoretically, any cancer susceptibility gene can be classified by this system. The challenge will be to develop mechanisms to quantify the posterior probability based on the various methods used to classify variants. At this time, BRCA1 and BRCA2 are the most commonly analyzed human genes in genetic diagnostic laboratories, and a large amount of information is available on their sequence variation. Many BRCA variants can be classified based on posterior probabilities from statistical genetic studies (Easton 2007 2008). For genes involved in rarer cancer susceptibility syndromes, the problem cannot easily be addressed statistically by comparing cases and controls. For genes that are frequent targets of somatic changes, such as TP53 (MIM# 191170), CDKN2A (MIM# 600160) and RET, somatic mutations that are recurrent in tumor samples can provide quantifiable evidence in favor of pathogenicity (Greenblatt 2003). However, limited data on somatic mutation spectra are available for most genes involved in cancer syndromes. Evolutionary conservation data, while unlikely to be definitive alone, have also been quantified and can provide odds ratios that can be used to help determine probability of pathogenicity (reviewed in Tavtigian et al, 2008). In vitro assays that measure the impact of the variant on function of the encoded protein are promising classifiers for some cancer susceptibility genes. Although their contribution cannot yet be quantified, results from some assays (e.g., MMR) can contribute important qualitative information (see Couch et al. 2008). The same can be said for tumor pathology features (Hofstra et al, 2008). Convening expert panels, such as the InSiGHT committee described previously for MMR genes, to review the types of data available for each specific gene and condition will encourage the development of numeric scores for all these different types of data. Over time, we expect that these efforts will expand to include researchers and clinicians that focus on less common cancer susceptibility conditions.
Standardization of clinical and/or research testing recommendations for family members
Clearly, sequence variants will continue to represent a challenge for the scientific and medical community. The ultimate aim is to reclassify all Class 2–4 variants, and if possible to obtain conclusive assignment to either class 1 or 5. This will be aided by additional testing of family members in order to obtain information about whether the variant segregates with cancer in the family (referred to as “research testing” in Table 4) (Goldgar et al., 2004). It is also important that the results of genetic testing performed in the research setting be made available to the scientific community through peer-reviewed publications and/or locus-specific databases (Greenblatt et al., 2008), in order to allow more rapid progress in our understanding of their functional and clinical consequences.
Table 4.
Testing Recommendations Associated with Each Class of Variant
| Class | Clinical Testing | Surveillance Recommendations if At-Risk Relative is Positive | Research Testing of Family Members |
|---|---|---|---|
| 5 | Test at-risk relatives for variant | Full high-risk surveillance guidelines | Not indicated |
| 4 | Test at-risk relatives for variant* | Full high-risk surveillance guidelines | May be helpful to further classify variant |
| 3 | Do not use for predictive testing in at-risk relatives* | Based on family history (and other risk factors) | May be helpful to further classify variant |
| 2 | Do not use for predictive testing in at-risk relatives* | Treat as “no mutation detected” for this disorder | May be helpful to further classify variant |
| 1 | Do not use for predictive testing in at-risk relatives* | Treat as “no mutation detected” for this disorder | Not indicated |
Recommend continuing to test proband for any additional testing modalities available for the disorder in question, e.g. rearrangement testing.
Currently, presymptomatic testing for clinical use (Table 4 – “clinical testing”) is generally not recommended for any variant that is classified as uncertain (Vink et al., 2005). Members of families where such a variant has been detected are given the same clinical recommendations provided to families where no sequence alteration was detected based on family history and on the presence of other risk factors. In the circumstances where the proband carries a Class 4 “Likely Pathogenic” variant, the 50% of relatives who have not inherited the variant are highly likely to be at average risk for cancer, yet they are still receiving intensive screening and in some cases prophylactic surgery or other therapeutic measures. Conversely, clinicians unfamiliar with genetic terminology may interpret any sequence variant as a deleterious result. If clinicians and molecular pathology laboratories are confident in the standards and recommendations for Classes 2 – 4, then more rational decisions can be made for these individuals.
We anticipate that use of a five-category classification system will lead to improved interpretation and targeting of clinical protocols for cancer prevention and surveillance. In Table 4, we provide recommendations for clinical testing of at-risk individuals in the family if testing of the affected proband leads to identification of a sequence variant. Compared to previous standards, the clinical approach is unchanged for probands with variants in Classes 1– 3. Predictive testing of their relatives for Class 2–3 variants should not be offered, since knowledge of the genotype does not affect clinical recommendations. However, we recommend predictive testing available not only to family members of individuals with a clear-cut pathogenic sequence variant (Class 5), but also to those in Class 4, based on strong evidence that the variant is likely pathogenic (Table 4). Hence, recommendations for cancer surveillance for individuals carrying a Class 4 variant reflect those given for Class 5 carriers. In selected cases, where the disease gene locus is certain, a sequence variant may be used as a linked marker to predict carrier status and cancer risk within a given family. Before proceeding with linkage testing, two conditions must be met. First, a sufficient number of affected family members must be available for segregation analysis. Second, the variant is found to be co-transmitted with the phenotype in the family under study. Clinical use of a variant in this context is subject to the caveats of indirect testing, namely incomplete sensitivity and specificity due to potential recombination, and is typically carried out by a trained genetics professional aware of these limits. It is rarely appropriate in hereditary breast and ovarian cancer or the Lynch syndrome because of locus heterogeneity.
The biggest potential problem is advising those at-risk individuals in the family who test negative for a Class 4 variant that they are likely not to be at increased cancer risk. Such a discussion should include the 1–5% residual likelihood that the variant of interest may not be pathogenic. This explanation is similar to the discussion if no mutation or variant is detected in the proband. This 1–5% chance can be ascribed to one of three possible explanations and depends somewhat on the type of family history. First the technique used for mutation searching is not 100% sensitive (the methods used for mutation testing must be reviewed and it must be certain that a comprehensive mutation search has been conducted in the proband). Secondly the proband tested may not be a mutation carrier, the variant is co-incidental to disease and a pathogenic mutation is present in another affected family member; depending on family structure the appropriate strategy when no mutation has been found may be to comprehensively test another affected family member. However in the presence of a variant highly likely to be pathogenic (Class 4), one would test any other affected person in the family for the variant to check that it segregated as the first option rather than comprehensively testing a second family member for other mutations. Note that in this respect, although families segregating two separate mutations have been reported it is not routine if a class 5 pathogenic mutation has been detected to conduct a comprehensive full mutation screen on a second family member. Finally (and the most likely explanation in the majority of families tested) is that the disease is polygenic and not due to a single high risk gene segregating in the family. We have used the latter as it is the most likely explanation for most families where no BRCA1/2 mutation is found. The residual risk due to the family history arising as a result of polygenic factors (familial non BRCA1/BRCA2 or familial colorectal cancer not due to an MMR gene defect) can be calculated for each condition. This residual risk may be small in comparison to the residual population risk for many of the common cancers being assessed, e.g. breast or colon cancer. Examples of the potential implications for BRCA1/2 and MMR gene testing are given below.
Utilization of the classification system by molecular geneticists and clinicians
We expect that the development of a well-delineated 5-class scheme is a significant step forward to standardizing both the classification of rare sequence variants and the clinical management of patients and their families at increased risk of cancer. However, several barriers will need to be overcome to implement this system internationally.
-
Who will assign/calculate the initial classification at the time of testing? Given that variant classification is based on information from multiple sources, it is unlikely that any individual would be sufficiently trained to comfortably derive and/or interpret data from all aspects used to classify. As already described, we expect that for most genes expert panels will need to determine the likelihood of pathogenicity for sequence variants to be accepted for use by the cancer genetics community. This is true even when expressed as quantitative scores based on a variety of data types. Coordination between relevant experts is required in order to provide classifications, and such coordination could take several forms.
Building on the approach originally developed by the BIC Steering Committee, and soon to be implemented by InSiGHT, classifications may be provided on well-curated databases by a panel which covers the range of expertise required. The information used and decision-making processes must be transparently displayed so that clinical and molecular laboratories can assess and interpret the conclusions. In the future, resources could be provided on well-curated databases to calculate probabilities for the variant from data provided by molecular geneticists and/or clinicians (e.g., Bayesian scores for segregation from pedigree information or tumour pathology) combined with other types of data (e.g., evolutionary conservation). This could be achieved by linking pedigree-based programs, such as BOADICEA (Antoniou et al., 2008) with databases such as BIC. Even in the absence of absolute probabilities, as is the case for MMR genes variant classification, a set of rules applied to a standard data set could be developed to formalize the “experience based” approach currently used by most clinical geneticists to assess the clinical significance of a particular variant (Lucci-Cordisco et al., 2006) (Barnetson et al., 2008). Similar systems are used for radiographic and other diagnostic tests.
Discussion is needed to assess where legal liability rests for those that decide on a classification and/or provide the resources for such classification (database, programming facilities). The overwhelming view of the IARC Unclassified Genetic Variants Working Group is that the current approach of non-standardized reporting creates a greater chance of inappropriate decision making and interferes with clinical care.
Time and resources are required to provide this classification. Any system which requires extensive input by molecular or clinical geneticists in individual laboratories/clinics would need to be recognized by the health care provider as a necessary function of these professionals, with provision of appropriate resources to enable improved classification and, consequently, clinical interpretation of these rare variants. Likewise, support would be required, preferably at an international level, to fund and standardize the development of databases to store additional information, or allow active programming for use by individual laboratories/clinics. Currently, the time provided by database curators and members of expert panels is essentially pro bono. Despite this limitation, curators of databases like the BIC and the MMR databases have remained committed over a decade or more because they have felt the need to fill a clear knowledge gap that is hampering the implementation of genetic tests into clinical genetics practice.
Presentation of consensus information on disease-specific databases using standard reporting criteria to facilitate and standardize classification. The task of reporting variants is too extensive to be taken on singly by either the molecular geneticist or the clinician dealing directly with the patient. Moreover, the sharing of information from different sources would facilitate the interpretation process by allowing accrual of larger data sets. It should be an international goal to strive for a single entry point for information pertaining to each disease gene, incorporating information from local, national and commercial databases to allow for compilation of information (see Greenblatt et al., 2008). The effort on the part of laboratories and national health services to integrate information has been launched by the Human Variome Project (Cotton 2007). If accomplished, it will improve classification, may allow for feedback to original submitting labs when classifications are updated, and ensure that the information provided to patients with the same variant is the same both within and between families. Because of the use of this information for clinical classification, it may require the formal approval of the relevant clinically recognized body for each country (e.g., American College of Medical Genetics, British Society for Human Genetics, etc).
Illustration of sequence variant classification and clinical application
Example 1: Breast Cancer – variant – BRCA2 D3095E
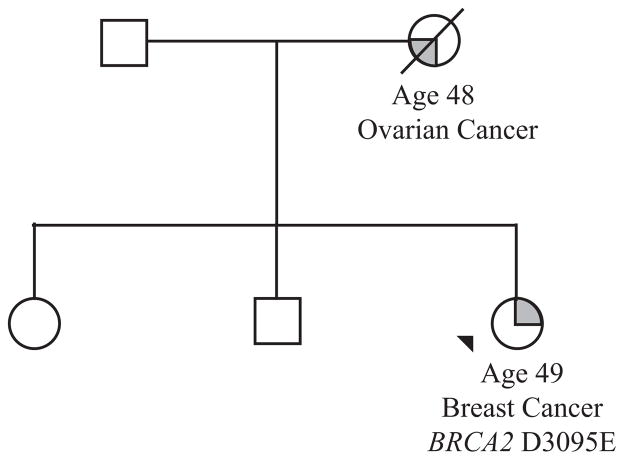
The variant encoding the putative missense change Asp3095Glu in the BRCA2 gene was identified in a 49 year old woman diagnosed with breast cancer as shown in Figure 2. At the time of identifying this variant the following evidence was available to assess pathogenicity:
Figure 2.
The index case (arrowed) was referred to the genetics service following the diagnosis of a grade 3 breast cancer. Her mother died of a serous papillary ovarian carcinoma aged 48 years. Genetic testing revealed variant D3095E in the BRCA2 gene.
Seen only once by the clinical diagnostic laboratory performing the test.
The family is small, with no other living affected relatives, so it is not possible to check co-segregation of cancer in the maternal lineage with the variant.
Both parents are deceased so we are not able to check if this mutation is de novo (de novo mutations are rarely reported in BRCA2).
Reported 12 times in the BIC database and listed as clinical significance “unknown”.
There are no reported observations in the literature of this mutation occurring in trans with deleterious mutation (if found this would argue against pathogenicity).
Splicing software predicts normal splicing and no RNA studies have been published.
Asp 3095 is located in the functionally important BRCA2 DNA binding domain and BRCA2 protein containing this variant demonstrates loss of transactivation in vitro (Farrugia et al., 2008).
Asp 3095 is invariant in an alignment of 12 BRCA2 protein sequences (11 vertebrates plus sea urchin, online at <http://agvgd.iarc.fr/alignments.php>). The prior probability that missense substitutions falling at invariant positions in this domain are pathogenic is 0.73 (Spurdle et al., 2008).
Using other algorithims based on sequence conservation demonstrated that Align-GVGD and SIFT predict that the asp to glu substitution is “probably deleterious” and PolyPhen predicts that the substitution is benign.
An integrated likelihood ratio based on family histories, co-occurrence, and cosegregation was available from the literature and shows odds of 23:1 in favour of causality (Easton et al., 2007a). However, this calculation did not consider sequence conservation, predicted effect on function, or functional assay results. Our current assessment of these additional factors yields a probability in favor of pathogenicity of 0.73, or an Odds Ratio of 2.7. Taking all the available evidence into account and combining this probability with the integrated likelihood ratio, the Posterior Probability of causality is 98.4%, a Class 4 variant – likely pathogenic.
BRCA2 mutations result in a high risk of both breast and ovarian cancer. We next considered the consequences of identifying this Class 4 variant in the BRCA2 gene in the affected proband and her family. Under our classification, testing of the unaffected sister is recommended. If she tests positive for the Class 4 variant, she has a less than 2% chance that she will be given a falsely higher level of risk than her family history and position in the family alone would predict. The lower limit of pathogenicity for calling a variant Class 4 is 95% (or a 5% chance that the information given will be based on a false premise of pathogenicity). At this level of posterior probability we should follow the same clinical recommendations as given to a carrier of a Class 5 pathogenic mutation.
To illustrate the robustness of these recommendations we have estimated the impact on risk for making these recommendations in a Class 4 variant at the lower limit of 95%.
-
The sister’s genetic test shows that she carries the variant
-
Interpretation - she has a high risk for developing BRCA2-associated cancers.
Cumulative lifetime breast cancer risk is 0.95probability × 0.8penetrance PLUS 0.05probability × 0.05 × 0.30[estimated risk based on family history] = 0.76 + 0.015 = 0.775
Cumulative lifetime ovarian cancer risk is 0.95probability ×0.2penetrance PLUS 0.05 × 0.015[estimated risk based on family history] = 0.19 + 0.00075 = 0.19.
-
Based on 78% breast cancer and 19% ovarian cancer risk, clinical advice in the United Kingdom will include
Annual mammogram and MRI from 30–35 years.
Ovarian surveillance research trials from 35 years (NB. no evidence of efficacy for ovarian screening at present)
Bilateral salpingo–oophorectomy available if requested from 35–40 years and advised from 40 years
Bilateral prophylactic mastectomy available if requested
-
-
The sister tests negative for the Class 4 variant with a 5% chance that the wrong test has been done and that the risk is due to other gene(s) or to an occult mutation.
-
Interpretation – if the family history is due to polygenic factors (familial non-BRCA1/BRCA2) then her cancer risk is 95% of the population risk PLUS 5% of the familial estimated risk.
Cumulative lifetime breast cancer risk 0.95[population risk] ×0.1 PLUS 0.05 × 0.3[familial risk] = 0.095 + 0.015= 0.11
Cumulative lifetime ovarian cancer risk is 0.95 × 0.01[population risk] PLUS 0.05 × 0.015[familial risk] = 0.0095 + 0.00075 = 0.01
-
The calculated risks of breast and ovarian cancer including the population risks give a maximum lifetime cumulative risk for breast cancer of approximately 11% and for ovarian cancer approximately 1% which is very similar to the population risk. It therefore seems reasonable to manage the patient in the same way as for someone in the general population.
Example 2: Variant in the mismatch repair gene (MMR) MLH1 P654L
Similarly, the new classification system would also have the most impact in families carrying Class 4 variants of MMR gene variants associated with HNPCC with a 5% chance risk that the variant is actually neutral. An example of such a variant is MLH1 (MIM# 120436) P654L, which falls in the PMS2 binding domain. The variant has been seen multiple times, and tumor pathology, computational studies, and in vitro assays for MMR and protein binding have been reported.
P654L has been reported in at least seven families, with 20 cases of HNPCC-related tumors. In some families, Amsterdam Criteria were present. Whether the variant segregates with cancer is unknown (Raevaara et al. 2005, Mangold et al. 2005).
Two tumors were reported to show microsatellite instability (MSI-H), consistent with MMR defects in the tumor cells.
Immunohistochemical staining for MLH1 protein was lost in 5/6 tumors tested, and focally positive in the other. MSH2 (MIM# 609309) staining was normal.
In vitro assays of P654L protein variant demonstrated reduced expression and reduced nuclear localization compared to wild type protein.
In vitro MMR activity of MLH1 P654L was similar to wild type MLH1.
The corresponding yeast variant P667Y showed deficient MMR activity in a yeast reversion assay (Wanat et al., 2007).
P654 is invariant in evolution. Several computational algorithms all predict that P654L would be deleterious (Positive Predictive Value of mutation at an invariant site 96.8% (Chan et al., 2007)).
The conclusion of the MMR Gene Missense Mutation Database and of the relevant publications is that the variant is likely pathogenic (Raevaara et al., 2005) (Chan et al., 2007) (Mangold et al. 2005) (www.mmrmissense.net). However, some uncertainty remains regarding pathogenicity, since some but not all assays showed functional deficiency, co-segregation has not been established, and the presence of P654 in relevant control populations has not been reported. Currently that uncertainty cannot be quantified by a Likelihood Ratio, but we approximate it as 95–99% likelihood of pathogenicity. In the new classification system this would be a Class 4 variant.
Recommendations for individuals that carry deleterious MMR gene variants thought to cause HNPCC and who have a high lifetime risk (estimated on average at about 70% cumulative lifetime risk) of developing colorectal cancer have been published (Lindor et al., 2006). Intensive colonoscopic screening starting in the third decade of life is standard. For carriers of MMR gene variants that are judged to have a 95% probability of pathogenicity, full screening recommendations seem reasonable. Using a similar rationale as for the breast cancer example, lifetime risk for colorectal cancer for a family member testing positive for this mutation is:
(0.95prob × 0.7penetrance) + (0.05prob × 0.06gen pop risk) = 0.665 + 0.003 = 0.668
The cancer risk assessment of a non-carrier of P654L in this family must include the residual 5% probability that this Class 4 variant is neutral and that there is an undetected MMR mutation in the family that is causing the HNPCC, so it is slightly higher than the 6% risk of the general population. Assuming a 50% chance of carrying the occult variant, and using the lifetime risk of colorectal cancer in a MMR mutation carrier of 70%, the residual additional cancer risk is the chance of additional mutation × Penetrance × 0.5:
Lifetime risk = population risk + residual genetic risk
(0.95prob × 0.06 gen pop risk) + (0.05prob × 0.7pen × 0.5) = 0.057 + 0.018= 0.075
The relative colon cancer risk in HNPCC is higher at younger age. The median age of onset of cancer in most HNPCC reports has been in the mid-40s (Lindor et al., 2006). Risk of colon cancer before age 40 in an HNPCC carrier is approximately 30% (Plaschke 2004). Thus, the proportion of the residual risk prior to age 40 or 50 may be roughly estimated
Residual genetic risk to age 50 = 0.05 × 0.5 × 0.35 = 0.00875 (approximately 0.009)
Residual genetic risk to age 40 = 0.05 × 0.5 × 0.30 = 0.0075
Only about 5% of colorectal cancers in the general population are diagnosed before age 50; with the absolute risk of 0.003, see (http://info.cancerresearchuk.org/cancerstats/types/bowel/incidence/, ACS Cancer Facts and Figures 2008). The risk of colon cancer in the US population before age 40 is about 1 in 1350 (0.00074), about 1/10 that of a non-carrier of a Class 4 variant (ACS Cancer Facts and Figures 2008). Using this information, the cumulative risk for a non carrier to ages 40 and 50 works out at
Residual cumulative risk to age 50 for a non carrier = 0.003 + 0.009 = 0.012
Residual cumulative risk to age 40 for a non-carrier = 0.00074 + 0.0075 = 0.00824
A screening strategy for age of initiation and frequency should be developed by an expert panel and studied in this population, but it seems reasonable to discuss with individual patients whether they wish to undergo an invasive screening examination like colonoscopy in light of a 1% chance of developing colorectal cancer before age 50 years. A recommendation for screening in line with a modest increase in genetic risk for a patient choosing to be screened would be one option. These recommendations currently vary among centers and among countries. A strategy of intermediate intensity might include colonoscopies beginning at age 35–40 with a frequency between the extremes of 1–2 years (HNPCC) and 7–10 years (general population). The panel would also need to consider recommendations for surveillance of other HNPCC associated cancers including endometrial and ovarian.
Conclusion
We have presented in this paper a scheme for clinical reporting of genetic variants particularly orientated towards inherited mutations in tumour suppressor genes conferring a high risk of cancer. Both under- and over-interpretation of such a result are common in clinical practice. The purpose of this classification and reporting framework is to improve the clinical utilization of genetic testing results, to maximize the opportunity to learn more about variants for the benefit of other families and to minimize the risk of incorrect interpretation of variants in the clinical setting. If applied successfully, the principles of this classification could be expanded to other genes where inactivating mutations can occur throughout the gene and where analysis of the gene detects sequence variants for which a clear functional consequence cannot be immediately assigned.
Acknowledgments
The authors acknowledge: Kurt Straif for information on the IARC classification of carcinogens, Jelena Blagojevitch for data from her survey of the VUS results from a year of BRCA1 and BRCA2 testing in the Wessex Regional Genetics Laboratory and Claire Noll for review of literature. The authors acknowledge the following grant or financial support: SEP – NIH HG004064, MSG- NIH CA 96536, Lake Champlain Cancer Research Organization, ABS -Australian National Health Medical Research Council, MG - Ente Cassa di Risparmio Firenze, WDF - Canadian Breast Cancer Research Alliance, SVT - NIH CA116167, and DFE is a Cancer Research UK Principal Research Fellow.
Appendix
Members of the IARC Working Group on Unclassified Genetic Variants:
Fergus Couch, Mayo Clinic, USA; Niels de Wind, Leiden University, the Netherlands; Diana Eccles, University of Southampton, UK; Douglas Easton, Cambridge University, UK; William Foulkes, McGill University, Canada; Maurizio Genuardi, University of Florence, Italy David Goldgar, University of Utah, USA; Marc Greenblatt, University of Vermont, USA; Robert Hofstra, University Medical Center Groningen, the Netherlands; Frans Hogervorst, Netherlands Cancer Institute, the Netherlands; Nicoline Hoogerbrugge, University Medical Center Neimejen, the Netherlands; Sharon Plon, Baylor University, USA; Paolo Radice, Istituto Nazionale Tumori, Italy; Lene Rasmussen, Roskilde University, Denmark; Olga Sinilnikova, Hospices Civils de Lyon, France; Amanda Spurdle, Queensland Institute of Medical Research, Australia; Sean Tavtigian, IARC, France; Paolo Boffetta, IARC, France.
References
- Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA, Risch HA, Eyfjord JE, Hopper JL, Southey MC, Olsson H, Johannsson O, Borg A, Passini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, nton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tryggvadottir L, Syrjakoski K, Kallioniemi OP, Eerola H, Nevanlinna H, Pharoah PD, Easton DF. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008 doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleyguier C, Ayadi S, Van NK, Vanel D, Dromain C, Sigal R. BIRADS classification in mammography. Eur J Radiol. 2007;61:192–194. doi: 10.1016/j.ejrad.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Barnetson RA, Cartwright N, van VA, Haq N, Drew K, Farrington S, Williams N, Warner J, Campbell H, Porteous ME, Dunlop MG. Classification of ambiguous mutations in DNA mismatch repair genes identified in a population-based study of colorectal cancer. Hum Mutat. 2008;29:367–374. doi: 10.1002/humu.20635. [DOI] [PubMed] [Google Scholar]
- Becker F, Nusko G, Welke J, Hahn EG, Mansmann U. Benefit-risk analysis of different risk-related surveillance schedules following colorectal polypectomy. Hepatogastroenterology. 2007;54:2249–2258. [PubMed] [Google Scholar]
- Boerner S, Sneige N. Specimen adequacy and false-negative diagnosis rate in fine-needle aspirates of palpable breast masses. Cancer. 1998;84:344–348. [PubMed] [Google Scholar]
- Collins FS. BRCA1 -- Lots of mutations, lots of dilemmas. N Engl J Med. 1996;334:186–188. doi: 10.1056/NEJM199601183340311. [DOI] [PubMed] [Google Scholar]
- Couch FJ, Rasmussen L, Hofstra R, Monteiro AANM, Greenblatt MS, de Wind N. Assessment of Functional Effects of Unclassified Genetic Variants. Hum Mutat. 2008:29. doi: 10.1002/humu.20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ, Weber BL. Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Breast Cancer Information Core. Hum Mutat. 1996;8:8–18. doi: 10.1002/humu.1380080102. [DOI] [PubMed] [Google Scholar]
- Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, len-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, Goldgar DE. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007a;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le ML, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo KY, Noh DY, Ahn SH, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low YL, Bogdanova N, Schurmann P, Dork T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den OA, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BA. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007b;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, et al. Hum Mutat. 29 in preparation. [Google Scholar]
- Farrugia DJ, Agarwal MK, Pankratz VS, Deffenbaugh AM, Pruss D, Frye C, Wadum L, Johnson K, Mentlick J, Tavtigian SV, Goldgar DE, Couch FJ. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 2008;68:3523–3531. doi: 10.1158/0008-5472.CAN-07-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshid G, Downey P. Combined use of imaging and cytologic grading schemes for screen-detected breast abnormalities improves overall diagnostic accuracy. Cancer. 2005;105:282–288. doi: 10.1002/cncr.21280. [DOI] [PubMed] [Google Scholar]
- Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR, Critchfield GC. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- Gatto NM, Frucht H, Sundararajan V, Jacobson JS, Grann VR, Neugut AI. Risk of perforation after colonoscopy and sigmoidoscopy: a population-based study. J Natl Cancer Inst. 2003;95:230–236. doi: 10.1093/jnci/95.3.230. [DOI] [PubMed] [Google Scholar]
- Giovannini M, Ardizzone S. Anorectal ultrasound for neoplastic and inflammatory lesions. Best Pract Res Clin Gastroenterol. 2006;20:113–135. doi: 10.1016/j.bpg.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Deffenbaugh AM, Monteiro AN, Tavtigian SV, Couch FJ. Integrated Evaluation of DNA Sequence Variants of Unknown Clinical Significance: Application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75 doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Byrnes GB, Spurdle AB, Iversen ES, Greenblatt MS. Integration of various data sources for classifying uncertain variants into a single model. Hum Mutat. 2008;29 doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Garcia EB, Ambergen T, Blok MJ, van den WA. Patients with an unclassified genetic variant in the BRCA1 or BRCA2 genes show different clinical features from those with a mutation. J Clin Oncol. 2005;23:2185–2190. doi: 10.1200/JCO.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Greenblatt MS, Brody LC, Foulkes W, Genuardi M, Hofstra R, Plon S, Sijmons RH, Sinilnikova OM, Spurdle AB. Locus-Specific Databases (LSDBs) and the Classification of Variants in Cancer Susceptibility Genes. Hum Mutat. 2008;29 doi: 10.1002/humu.20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstra RW. Tumor characteristics as an analytic tool for classifying genetic variants of uncertain clinical significance. Hum Mutat. 2008;29 doi: 10.1002/humu.20894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John EM, Miron A, Gong G, Phipps AI, Felberg A, Li FP, West DW, Whittemore AS. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. JAMA. 2007;298:2869–2876. doi: 10.1001/jama.298.24.2869. [DOI] [PubMed] [Google Scholar]
- Kean-Cowdin R, Spencer FH, Xia LY, Pearce CL, Thomas DC, Stram DO, Henderson BE. BRCA1 variants in a family study of African-American and Latina women. Hum Genet. 2005;116:497–506. doi: 10.1007/s00439-004-1240-5. [DOI] [PubMed] [Google Scholar]
- Kerlikowske K, Grady D, Barclay J, Frankel SD, Ominsky SH, Sickles EA, Ernster V. Variability and accuracy in mammographic interpretation using the American College of Radiology Breast Imaging Reporting and Data System. J Natl Cancer Inst. 1998;90:1801–1809. doi: 10.1093/jnci/90.23.1801. [DOI] [PubMed] [Google Scholar]
- Lindor NM, Petersen GM, Hadley DW, Kinney AY, Miesfeldt S, Lu KH, Lynch P, Burke W, Press N. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. JAMA. 2006;296:1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- Lucci-Cordisco E, Boccuto L, Neri G, Genuardi M. The use of microsatellite instability, immunohistochemistry and other variables in determining the clinical significance of MLH1 and MSH2 unclassified variants in Lynch syndrome. Cancer Biomark. 2006;2:11–27. doi: 10.3233/cbm-2006-21-203. [DOI] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- Roskell DE, Buley ID. Fine needle aspiration cytology in cancer diagnosis. BMJ. 2004;329:244–245. doi: 10.1136/bmj.329.7460.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurdle AB, Lakhani SR, Healey S, Parry S, Da Silva LM, Brinkworth R, Hopper JL, Brown MA, Babikyan D, Chenevix-Trench G, Tavtigian SV, Goldgar DE. Clinical classification of BRCA1 and BRCA2 DNA sequence variants: the value of cytokeratin profiles and evolutionary analysis--a report from the kConFab Investigators. J Clin Oncol. 2008;26:1657–1663. doi: 10.1200/JCO.2007.13.2779. [DOI] [PubMed] [Google Scholar]
- Tavtigian SJ, Greenblatt MS, Lesueur F, Byrnes GB. In silico analysis of missense substitutions using sequence-alignment based methods. Hum Mutat. 2008;29 doi: 10.1002/humu.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevena LJ, Davey HM, Barratt A, Butow P, Caldwell P. A systematic review on communicating with patients about evidence. J Eval Clin Pract. 2006;12:13–23. doi: 10.1111/j.1365-2753.2005.00596.x. [DOI] [PubMed] [Google Scholar]
- Vink GR, van Asperen CJ, Devilee P, Breuning MH, Bakker E. Unclassified variants in disease-causing genes: nonuniformity of genetic testing and counselling, a proposal for guidelines. Eur J Hum Genet. 2005;13:525–527. doi: 10.1038/sj.ejhg.5201379. [DOI] [PubMed] [Google Scholar]