Abstract
Molecular dynamics studies have been performed for 3.5 ns on the ETS domain of Ets-1 transcription factor bound to the 14-bp DNA, d(AGTGCCGGAAATGT), comprising the core sequence of high-affinity (GGAA), ETS–GGAA. In like manner, molecular dynamics simulations have been carried out for 3.9 ns on the mutant low-affinity core sequence, GGAG (ETS–GGAG). Analyses of the DNA backbone of ETS–GGAG show conformational interconversions from BI to BII substates. Also, crank shaft motions are noticed at the mutated nucleotide base pair step after 1,500 ps of dynamics. The corresponding nucleotide of ETS–GGAA is characteristic of a BI conformation and no crank shaft motions are observed. The single mutation of ETS–GGAA to ETS–GGAG also results in variations of helical parameters and solvent-accessible surface area around the major and minor grooves of the DNA. The presence of water contacts during the entire simulation proximal to the fourth base pair step of core DNA sequence is a characteristic feature of ETS–GGAA. Such waters are more mobile in ETS–GGAG at 100 ps and distant after 1,500 ps. Anticorrelated motions between certain amino acids of Ets-1 protein are predominant in ETS–GGAA but less so or absent in the mutant. These motions are reflected in the flexibility of amino acid residues of the protein backbone. We consider that these conformational features and water contacts are involved in stabilizing the hydrogen bond interactions between helix-3 residues of Ets-1 and DNA during the transcription process.
The Ets protein family of transcription factors includes species interacting with various genes that code for transcriptional activators and inhibitors involved in cell proliferation and differentiation (1, 2). The regulation of the initiation of gene transcription arises from the combined activity of different transcriptional regulators (2, 3). Ets family members found in species from invertebrates to humans share a conserved sequence of 85 amino acids, named the ETS domain. The ETS domain folds into a winged helix–turn–helix motif and binds to a consensus DNA sequence centered on the core GGAA motif, named the Ets-binding site. The sequences flanking this core motif (in the major groove) are variable and characterize the specificity of binding of the Ets transcription factor. Ets proteins have also been implicated in several types of cancer and other human diseases (4). Detailed conformational preferences that influence the sequence specificity of Ets proteins are essential for the design of anticancer drugs.
The high-affinity DNA contains the GGAA core sequence, ETS–GGAA (Fig. 1). The low-affinity DNA is the single-base-pair mutant, ETS–GGAG. Recently we reported molecular dynamics (MD) studies (5) dealing with the binding of the ETS domain of Ets-1 protein to the high- and low-affinity 14-bp DNA structures. We have observed that the most conserved residues Arg-391, Arg-394, along with Tyr-395 of Ets-1, jointly contribute to recognize the GGAA or GGAG core DNA sequences (5). The differential hydrogen bond interactions of Tyr-395 with the core DNA sequence are implicated in the strong affinity of GGAA as compared with GGAG helix. In this report, we have extended the MD analysis involving DNA conformations and solvent effects. Also, we investigated the collective motions of the Ets-1 residues. This approach was carried out to enhance our level of understanding of molecular recognition of a specific DNA base sequence in terms of the dynamic motions of the Ets-1 transcription factor.
Fig. 1.
Sequence and numbering of the 14-bp DNA of ETS–GGAA complex. The underlined A · · ·T base pair of the core binding sequence, G1G2A3A4, (red) is mutated to G · · ·C base pair in ETS–GGAG.
Methods
The starting structure of the Ets-1 ETS domain–DNA was obtained from x-ray studies (PDB ID code 1K79; ref. 6) and modeled with the charmm all-atom force field (7, 8). The ETS–DNA complex was immersed in an orthorhombic box (67.4. × 62.7 × 54.3 Å3) filled with TIP3P water molecules (9). The electrostatic interactions were evaluated by using the partial mesh Ewald method (10, 11). The structures with appropriate counterions and periodic boundary conditions were simulated with the charmm program (version c27b4; ref. 12) for a period of 3.5 and 3.9 ns for ETS–GGAA and ETS–GGAG, respectively. Details of the modeling and the adopted MD procedures have been presented elsewhere (5).
As the stability of the rms deviation values was observed ≈0.9 ns, reported earlier (5), the average structures of ETS–GGAA and ETS–GGAG were obtained for the period of 0.9–3.5 and 0.9–3.9 ns, respectively. The base step and base pair helical parameters of the DNA were evaluated by using the program freehelix98 (13). The solvent-accessible surface area (SASA) was estimated according to Lee and Richards (14) with a water probe of radius 1.4 Å. The dynamic crosscorrelation (DCC) (or normalized covariance) map of each protein–DNA complex was constructed by taking into account the fluctuations of two residues, averaged by residue over 103 Cα atoms of Ets-1 and the 26 phosphorous atoms of the DNA backbone. The order parameter S2, for π/ψ torsion angles of the protein backbone of the MD-averaged structures, was evaluated by using the model-free formalism (15). Details of SASA, DCC, and order parameter of the structures were given in Supporting Methods, which is published as supporting information on the PNAS web site.
Results and Discussion
DNA Backbone Conformational Transitions. We have reported on the dynamics of deoxyribose interconversions from C2′-endo to C3′-endo conformations of certain DNA nucleotides of ETS–GGAA and ETS–GGAG structures (5). The dynamical interconversion of the DNA phosphate linkage from BI/BII substates (ref. 16 and Fig. 2) during complex formation has been suggested to play a role in the sequence recognition (17, 18). BI is characterized by torsions ε (C4′–C3′–O3′–P) and ζ (C3′–O3′–P–O5′) values between 120° and 210° (trans) and 235° and 295° (gauche–), respectively. For BII the ε lies between 210° and 300° (gauche–) and the ζ between 150° and 210° (trans). Analysis of ε and ζ torsions of the C–1G1G2A3A(G)4A5 region during dynamics indicates BI and BII transitions are observed at nucleotide base step, C–1/G1, G1/G2, A3/A4, T4′/T3′ and C1′/G-1′ for ETS–GGAA and at the C–1/G1, G2/A3, A3/G4, T5′/T4′, T3′/C2′, and G–1′/G–2′ for ETS–GGAG. The BI/BII conformational pattern of the phosphate linkage between T5′ and T(C)4′ of ETS–GGAA and ETS–GGAG are different. This fact is shown in the time variations plots of ε and ζ torsions (Fig. 3 a and b). In ETS–GGAG, the BII conformations are observed during the period of 1,750–2,050 ps and also in the latter part of the dynamics (red line of Fig. 3). This result opposes the report that the BII conformation does not occur at the pyrimidine/pyrimidine base step (19, 20).
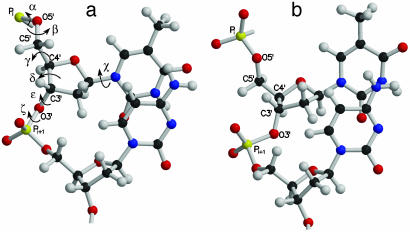
Fig. 2.
Molecular plot of the dinucleotide repeat with BI (a) and BII (b) conformations. The arrows indicate the backbone torsions around the central bond of the nucleotide. The torsions C4′–C3′–O3′–P(ε) and C3′–O3′–P–°O5′ (ζ) favor 210° (trans) and 261° (gauche–), respectively, in a. In b, the values of ε and ζ are 261° (gauche–) and 213° (trans), respectively. Notice the greater stacking of bases in a than in b.
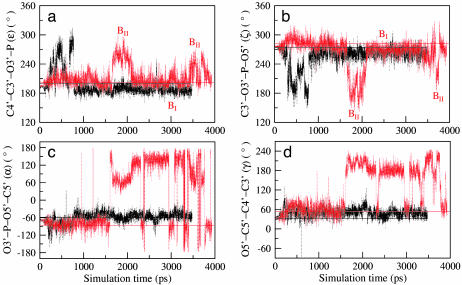
Fig. 3.
Time variation plot of the DNA backbone torsions at the nucleotide base step, T5′/T(C)4′, of the MD structures: ε (a), ζ (b), α (c), and γ (d) of ETS–GGAA (black) and ETS–GGAG (red). The regions of BI and BII conformations are shown in a and b. The corresponding crystal structure values of ETS–GGAA and ETS–GGAG are black and red straight lines, respectively.
The BII conformation is formed to overcome the energy barrier due to the destacking of adjacent bases. Consequently the phosphodiester governed by torsions ζ and α (O3′–P–O5′–C5′) prefer trans and gauche–values, respectively, in the extended conformation. However, in ETS–GGAA, after 900 ps, the conformations are in the BI state (black line). This finding results in a compact phosphodiester conformation (ζ, α: gauche–, gauche–), which is characterized by good stacking of bases. As sufficient equilibration is required for the stability of trajectories, the conformations before 900 ps are ignored.
The sugar pucker of the nucleotides T5′ and T(C)4′ fluctuates between C2′-endo and O4′-endo conformation in both complexes, except, in a few times, T5′ assumes C3′-endo conformation in ETS–GGAA (data not shown). A noticeable feature is that, in ETS–GGAG, there is a steep transition from C2′-endo to O4′-endo at 100 ps. The glycosyl torsions, χ (O4′–C1′–N1–C2) of T5′ and T4′ in ETS–GGAA fluctuate from high anti (280°) to anti (180°) values (data not shown). In ETS–GGAG, the χ of C4′ exhibits similar variations, but with steep transition to anti region at 100 ps. Also, at 2,100 ps, the χ of T5′ decreases from ≈280° and prefers values ≈230°.
Near-neighboring bond correlations between the torsions α and γ (O5′–C5′–C4′–C3′) (21) are observed at the nucleotide base step T5′/T(C)4′. This occurrence is referred to as crank shaft motion (22) of the nucleotide noted in relation to the stacked arrangement of bases with the interconversion of α from gauche– to trans, and γ from gauche+ to trans values. These motions are distinctly observed after 1,500 ps in ETS–GGAG (red line of Fig. 3 c and d). No such motions are observed in ETS–GGAA. Mutation of a single DNA base pair at A(G)4 thus likely exerts an influence on the backbone DNA conformations by allowing BI → BII interconversions and crank shaft motions between the nucleotides T5′ and T(C)4′ of ETS–GGAG.
DNA Helical Parameters. Investigation of the DNA helical parameters of specific base sequences provides the conformational features associated with differential deformability of the helices, which is essential to understand the recognition process. The nucleotide base-step parameters rotation (roll, tilt, and twist) and translation (slide and rise) for the region of core sequence G1G2A3A(G)4 determined from the MD structures of ETS–GGAA and ETS–GGAG are given in Table 2, which is published as supporting information on the PNAS web site. The corresponding values of the crystal structures (6) (PDB ID codes 1K79 and 1K7A for ETS–GGAA and ETS–GGAG, respectively) are in italic. The tilt is more negative at the A(G)4/A5 nucleotide base step in ETS–GGAA compared with ETS–GGAG. With the exception of the A5/T6 base step, the roll angles are positive (4.6° to 7.8°) in the MD structures (Table 2), indicating that the base pairs are bent toward the major groove. In ETS–GGAA and ETS–GGAG, the relative displacement between two adjacent base pair steps, slide, is negative and in the range –0.99 to –0.17 Å and –0.92 to 0.03 Å, respectively. The twist of the MD structures vary with high values at the A(G)4/A5 and A5/T6. Significant differences between the MD and x-ray structures are noticed for twist at the A5/T6 base step. The pattern of rise alternates between low and high values in the MD and x-ray structures.
The nucleotide base pair parameters rotation (tip, inclination, propeller twist, and buckle) and translation (X-displacement and Y-displacement) of C-1G1G2A3A(G)4A5 of the MD and x-ray structures are given in Table 1. In both of the MD structures, the inclination (angle between each base pair with respect to a chosen axis) is positive and is characteristic of A-type helix. The range of inclination suggests that the values are slightly higher in ETS–GGAG (7.2° to 12.6°) than in ETS–GGAA (7.8° to 10.1°). In MD structures, the average propeller twist of the base pairs is negative except for C–1 · · · G–1′. This finding is because the value for C–1 · · · G–1′ undergoes variations during the period 900–2,000 ps. Noticeable is the large negative values of the propeller twist for the A · · · T base pair when compared with the G · · · C base pair. This comparison is to avoid intrastrand steric clashes between the thymine methyl group and the 5′ neighboring sugar (23). A positive buckle is observed for the base-pairs C–1 · · · G–1′ and A5 · · · T5′ in the MD and x-ray structures. Base pairs are displaced from the helix axis (X-displacement) into the major groove by –2.67 to –1.96 Å in ETS–GGAA, and to a greater extent (–3.0 to –1.41 Å) in ETS–GGAG. This is a result of conformational changes brought about by the mobility of the DNA bound to helix-3 of Ets-1.
Table 1. Base-pair helical parameters of C-1G1G2A3A(G)4A5 sequence region of the DNA and their SD (in parentheses) of the MD-averaged structures.
| ETS—GGAA (900-3,480 ps)
|
ETS—GGAG (900-3,930 ps)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Base pair | Tip, ° | Inclination, ° | Propeller twist, ° | Buckle, ° | X-Disp., Å | Y-Disp., Å | Tip, ° | Inclination, ° | Propeller twist, ° | Buckle, ° | X-Disp., Å | Y-Disp., Å |
| C-1/G-1 | 1.8 (± 4.4) | 7.8 (± 3.8) | 1.2 (± 7.7) | 8.8 (± 10.5) | -2.62 (± 0.63) | -0.26 (± 0.63) | 2.5 (± 4.4) | 10.6 (± 3.9) | -0.4 (± 10.7) | 12.9 (± 11.4) | -2.64 (± 0.52) | -2.64 (± 0.52) |
| 5.3 | 12.01 | 2.38 | 15.54 | -2.46 | -0.66 | -4.7 | 13.31 | 0.6 | 6.4 | -4.95 | -1.21 | |
| G1/C1′ | 2.7 (± 3.9) | 10.1 (± 3.8) | -3.1 (± 8.1) | -4.9 (± 15.4) | -1.96 (± 0.50) | 0.11 (± 0.55) | 2.5 (± 4.4) | 12.6 (± 4.1) | -5.2 (± 8.5) | -4.1 (± 12.6) | -2.87 (± 0.70) | -0.11 (± 0.66) |
| 2.4 | 14.93 | -1.88 | -6.21 | -1.67 | 0.57 | -5.1 | 13.1 | -0.7 | -12.6 | -4.50 | 1.48 | |
| G2′/C2 | 2.8 (± 4.6) | 8.4 (± 3.6) | -5.8 (± 7.3) | -2.1 (± 10.9) | -2.67 (± 0.39) | -0.10 (± 0.49) | 1.1 (± 4.9) | 12.1 (± 3.8) | -8.2 (± 7.7) | 2.7 (± 10.4) | -3.0 (± 0.67) | -0.19 (± 0.43) |
| -0.42 | 10.22 | -16.00 | -7.37 | -2.44 | 0.73 | -5.4 | 8.7 | -9.8 | -3.8 | -4.20 | 3.12 | |
| A3′/T3′ | 4.2 (± 3.9) | 9.3 (± 3.0) | -12.4 (± 6.6) | -9.7 (± 9.4) | -2.40 (± 0.42) | 0.33 (± 0.50) | 0.8 (± 4.9) | 12.6 (± 3.6) | -10.5 (± 8.7) | -7.7 (± 9.8) | -2.57 (± 0.57) | 0.90 (± 0.55) |
| 5.5 | 7.86 | -15.54 | -6.03 | -2.29 | 0.94 | 1.7 | 6.3 | -9.7 | -8.5 | -2.60 | 4.01 | |
| A4′/T4′ | 5.2 (± 3.8) | 8.9 (± 3.0) | -16.7 (± 5.9) | -3.0 (± 9.4) | -2.29 (± 0.49) | 0.35 (± 0.56) | 0.3 (± 4.6) | 10.0 (± 4.0) | -13.5 (± 7.2) | -4.1 (± 10.7) | -2.13 (± 0.55) | 0.99 (± 0.65) |
| (G4/C4′) | 3.62 | 9.52 | -21.04 | -6.88 | -1.73 | 1.05 | 5.0 | 4.1 | -9.8 | -5.1 | -0.32 | 4.83 |
| A5′/T5′ | 4.3 (± 4.1) | 8.5 (± 3.1) | -17.3 (± 7.1) | 8.7 (± 8.8) | -2.19 (± 0.62) | 0.46 (± 0.63) | -1.6 (± 4.7) | 10.4 (± 3.1) | -14.2 (± 8.7) | 3.4 (± 9.5) | -1.41 (± 0.74) | 0.76 (± 0.79) |
| 0.23 | 7.22 | -22.71 | 8.39 | -0.77 | 1.18 | 4.8 | 3.7 | -15.8 | 6.5 | 2.75 | 2.92 | |
The corresponding values of the crystal structures are given in italics. Disp., displacement.
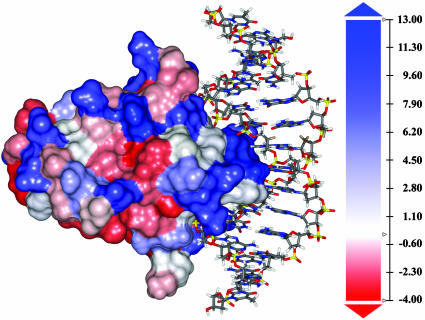
SASA of ETS–DNA Complexes. Fig. 4 indicates the hydrophobic nature of the portion of Ets-1 protein bound to the DNA duplex. It can be seen that 70% of the amino acid residues (blue) in contact with the major groove of the helix are hydrophobic. The difference in SASA is used to estimate the fit of the contacts between the protein and DNA. During dynamics, this value fluctuates in both the structures (Fig. 10, which is published as supporting information on the PNAS web site). With ETS–GGAG the values are higher (by 120–150 Å2) than with ETS–GGAA and exhibit a sharp rise and fall at ≈2,200 ps.
Fig. 4.
Molecular plot of the hydrophobic surface of the Ets-1 protein bound to 14-bp DNA. The scale of hydrophobicity is indicated with the maximum and minimum values in blue and red, respectively.
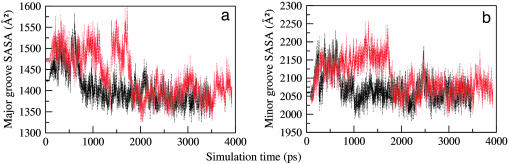
Conformations and transitions are influenced by solvation of the phosphates and nucleobases in the major and minor grooves. The variation of SASA around the DNA phosphodiester linkages shows similar pattern in both structures (data not shown) with an average value of 4,606 Å2. As can be seen in Fig. 5, the magnitude of SASA around the major groove indicates the groove to be relatively less accessible than the minor groove due to protein binding in the major groove. The accessibility of the grooves decreases steeply by a magnitude of 100–200 Å2 in both structures. In ETS–GGAA, the value of SASA falls at 750 ps (Fig. 5a), and, in ETS–GGAG, it decreases at about one-half of the dynamics period, 1750 ps (Fig. 5b). The loss in DNA solvation can be related to a tighter binding to protein.
Fig. 5.
Time variation plot of the SASA of the MD structures around the major (a) and minor (b) groove of the DNA of ETS–GGAA (black) and ETS–GGAG (red).
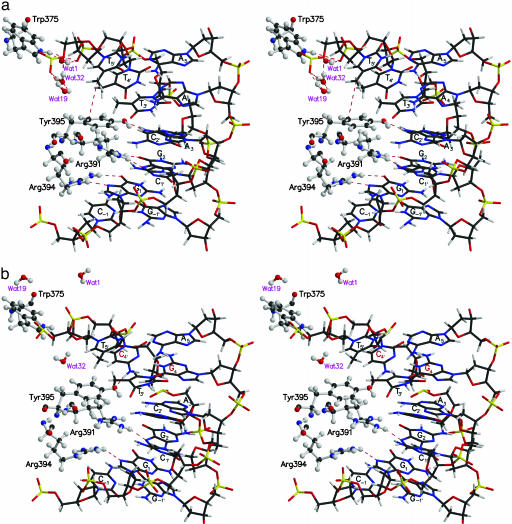
The Role of Waters in Structural Stabilization. The motion of 45 crystal waters in ETS–GGAA and ETS–GGAG was investigated. Of interest are the interactions of Wat1, Wat16, and Wat32 that exhibit different features in the high- and low-affinity complexes. Time variation plots related to the separations of these waters either from protein or DNA phosphate oxygens are given in Fig. 11, which is published as supporting information on the PNAS web site. In the x-ray structure of ETS–GGAA (6), Wat1 is at a distance 3.5 Å from the indole nitrogen NE1 of Trp-375. In the simulations of ETS–GGAA, wide fluctuations are observed in the distance between Wat1 and the NE1 of Trp-375, although a hydrogen bond is seen for extended period (Fig. 6a). Wat1 is also in close relationship to the phosphate oxygens at the nucleotide base step T5′/T4′. Wat16 and Wat32 are hydrogen-bonded to phosphate oxygens at the T5′/A6′. These water hydrogen bonds likely stabilize the position of the DNA helix. Consequently, the interactions between the conserved residues of Ets-1, Arg-391, Arg-394, and Tyr-395 with the nucleobases are retained in ETS–GGAA.
Fig. 6.
Stereoplot of the interactions of crystal waters in the MD-averaged structures of ETS–GGAA (a) and ETS–GGAG (b). The amino acids of Ets-1 are represented as a ball-and-stick model. The P atoms of the nucleotides are yellow. The legends of the mutated base pair and the non-bonded interactions are red. Notice the presence of interactions of waters with Ets-1 and DNA phosphate oxygens in a.
The mutation of a single base pair to provide ETS–GGAG renders the crystal waters more mobile. Due to the DNA-conformational variations (related to sugar pucker and glycosyl torsion) at the nucleotide base-step, T5′/C4′, waters are 4.5–6.5 Å away at 100 ps (red line of Fig. 11). Subsequently, these waters move farther during the period of 1,500–1,800 ps. This finding is a characteristic feature of ETS–GGAG, because the previously mentioned water contacts of ETS–GGAA are absent nearly after one-half period of the dynamics (Fig. 6b). This result can be attributed to the DNA-conformational variations at the T5′/C4′ that can no longer accommodate water molecules. This feature is consistent with the studies on EcoRI endonuclease, which indicates the BII conformation to be associated with the decrease of water content (20). The subsequent displacement of nucleotides in ETS–GGAG also results in disruption of hydrogen bonding of Arg-394 to G1 nucleotide after 2,000 ps of dynamics (5). The presence of water contacts thus appears to be pivotal for the stable interactions of ETS–GGAA.
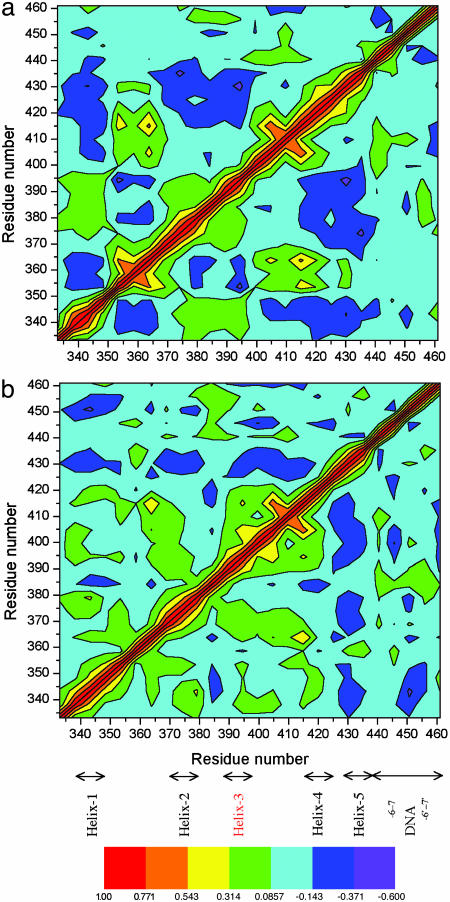
DCCs. The overall motion and dynamical structure of the protein and the DNA can be characterized by the analysis of the correlated motions between the Cα atoms of protein residues and the phosphorous (P) atoms of DNA. The extent of correlated motion is indicated by the magnitude of the corresponding correlation coefficient, Cij, displayed as DCC maps, Fig. 7. In ETS–GGAA, an extended region of anticorrelated motions (blue and violet regions) are observed between various protein residues (Fig. 7a). The region comprising strand-1 and strand-2 (353–366) anticorrelates with helix-1 (334–348), turn (between helix-2 and helix-3, 378–385) and helix-3 (392–397). The helix-4 and helix-5 (405–440) anticorrelates with regions of helix-1 (333–349), and helix-2–turn–helix-3 (370–400). The protein–DNA anticorrelations are noticed between 435–442, the P atoms of DNA (–6 to 1) and helix-1 (340–346) and helix-2 (370–380) of Ets-1. Also, limited anticorrelated motions occur between the other strand of DNA (P atoms, –6′ to –1′) and helix-1 (337–346), and helix-3 (380–390) regions of Ets-1. The characteristic anticorrelations of DNA helix are seen between P atoms of –3′ to 6′ and P atoms of 1 to 6. The positive correlations, except for the movements (of the residue with itself) along the diagonal (red, orange, and yellow), are limited. These are observed mostly between β-strand regions of Ets-1 (in yellow): strand-4 (412–420) with strand-2 (358–365), and strand-1 (352–355).
Fig. 7.
DCC map of the residue–residue fluctuations of the Cα atoms of Ets-1 protein and P atoms of DNA of the MD structures: ETS–GGAA (averaged 900–3,480 ps) (a) and ETS–GGAG (averaged 900–3,930 ps) (b). The range of correlations is shown by the various colors. The regions related to the DNA strands and α-helices of the protein are indicated.
In ETS–GGAG, a small region of positive correlated motions are observed (Fig. 7b) between protein residues of strand-4 (412–417) and strand-2 (360–365). The negative correlated motions are seen in the following: helix-3 (384–387) with helix-1 and coil regions (340–352) and helix-4 (412–424) with turn region (376–383). Also, helix-5 (427–434) anticorrelates with helix-1 (333–350), helix-2 (365–380), and helix-3 and strand-3 regions (385–415). Extended anticorrelated motions occur between the P atoms (–6′ to 3′) of the second strand of DNA and amino acids of helix-1 (333–354) in ETS–GGAG. A comparison of DCC maps of ETS–GGAA and ETS–GGAG complexes (Fig. 7) indicate that anticorrelations between protein residues are significantly extended in ETS–GGAA. Some of these motions are distinctly absent in ETS–GGAG. This occur in the following regions: strand-1 and strand-2 (353–366) with helix-1 (334–348); strand-1 (352–365) with turn (378–385) and helix-3 (392–397) regions. Besides, some of the anticorrelations related to the protein–DNA and DNA–DNA interactions are absent in ETS–GGAG. However, the motions between the helix-1 of Ets-1 and the P atoms of DNA (–6′ to 3′) are more pronounced in ETS–GGAG than in ETS–GGAA.
The correlated and anticorrelated motions of the protein can be appreciated in terms of corresponding secondary structure elements as depicted in the stereoplots of ETS–GGAA and ETS–GGAG complexes (Fig. 12, which is published as supporting information on the PNAS web site). Except for the anticorrelated motions of helix-1 with strand-1 and strand-2, similar, but less extended anticorrelated motions, are observed in ETS–GGAG (Fig. 12b). Interestingly, some of the anticorrelated motions are associated with core-binding region of Ets-1, helix-3 (385–396) that is in motion either with strand-1 and strand-2 or helix-4 and helix-5. Thus, in ETS–GGAA, the extended protein–protein anticorrelations and other anticorrelated motions related to the protein–DNA and DNA–DNA would propagate information and promote the recognition of GGAA sequence by Ets-1 transcription factor.
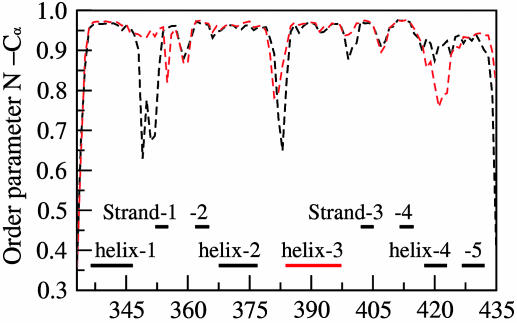
Dihedral Order Parameters of Protein. The order parameter, S2, for N–Cα and Cα–C vectors (related to π and ψ torsions, respectively), provides additional insights about correlated dynamics between different residues of protein bound to the DNA (24, 25). As observed in Fig. 8, and Fig. 13, which is published as supporting information on the PNAS web site, the protein backbone is rigid, with the great majority of the π/ψ dihedral angles visiting a relatively small conformational space in both the structures. Based on the observed low values of S2 (of highly mobile region) in ETS–GGAA and ETS–GGAG, the conformations of a few residue regions of Ets-1 protein are probed in detail with the aid of Ramachandran plots (Fig. 9). The plots in Fig. 9 indicate that the Ets-1 residues of MD structures sample conformations close to the crystal structure (6) during dynamics.
Fig. 8.
Order parameter, S2, for the π torsion of Ets-1 residues of the MD-averaged structures: ETS–GGAA (black) and ETS–GGAG (red). The secondary structural elements of Ets-1 are indicated.
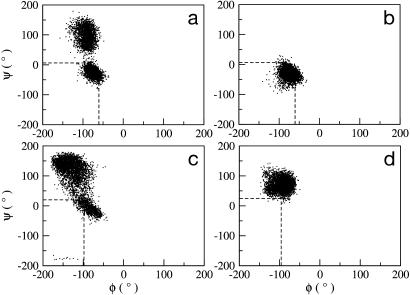
Fig. 9.
Ramachandran plot representing the protein π and ψ conformations visited by residues Ser-349 (a and b) and Lys-383 (c and d) of the MD structures of ETS–GGAA and ETS–GGAG, respectively. The dashed lines indicate the corresponding crystal structure values.
In ETS–GGAA Ser-349 (between helix-1 and strand-1), is one of the highly flexible residues and visits two different conformations of ψ (Fig. 9a). During the simulation time of 0–1,700 ps, the values of ψ are in the range of –50° to 0° and, beyond 1,700 ps, there is a steep increase with the preferred values from 50° to 150° (data not shown). Consequent to the observed variations in ψ, low S2 (0.70) of Ser-349 are seen. This feature is consistent with high values of the Cα atom fluctuations of Ser-349 evaluated from crystal structure B factors (6) and MD simulations (5). The flexibility of Ser-349 influences the conformations of neighboring residues. The Lys-348 and Cys-350 prefer two different values of ψ similar to that noticed in Ser-349. Also, the torsion π of Gln-351 is affected with fluctuations from –150° to 170° (beyond 1,700 ps; data not shown). In ETS–GGAG, the Ser-349 favors ψ values in a small range between–50° to 0° (Fig. 9b), indicating less flexibility (S2 is 0.92). This observation reflects the absence of anticorrelations between residues 353–366 with 334–350 in ETS–GGAG, in contrast to ETS–GGAA (Fig. 7).
The other residues that display high flexibility (Fig. 8) are Asn-380 and Lys-383, which are located in the turn region between helix-2 and helix-3 (characteristic of helix–turn–helix motif) and Gly-423 (between helix-4 and helix-5). The conformational differences of Asn-380 and Gly-423 residues between ETS–GGAA and ETS–GGAG are given in Fig. 14, which is published as supporting information on the PNAS web site. The S2 values of π and ψ for Lys-383 in ETS–GGAA are low (0.65 and 0.75 respectively,) due to the wide fluctuations (Fig. 9c), whereas the protein backbone is rigid in ETS–GGAG (Fig. 9d). These conformational variations appear to be associated with the differential anticorrelations shown in the DCC map in the neighborhood of these residues (Fig. 7). It may appear that the large-amplitude motions involving a few residues of Ets-1 also contribute to preferential DNA sequence binding.
Conclusions
The results of our past (5) and present study provide a detailed description of dynamical structural variations of the Ets-1–DNA complexes. The replacement of a single base pair A · · · TbyG · · · C in ETS–GGAG results in significant DNA backbone conformational interconversions. These BI to BII transitions and crank shaft motions lead to the loss of water contacts at the region of mutation after half the period of dynamics and renders the DNA helix of ETS–GGAG to be more mobile. Consequently, the nucleotides of the core sequence undergo displacement and some of the essential hydrogen bond interactions between the helix-3 region of the Ets-1 protein and the DNA bases are disrupted. In ETS–GGAA, the specific DNA backbone conformations include congenially stacked bases. This finding is in agreement with the crystal structure studies, which indicated the adenine bases (similar to stretch A3A4A5 of ETS–GGAA) are capable of stacking without any discontinuity, whereas guanine bases result in dislocation in the stack, affecting the backbone of the helix (26). Also, in ETS–GGAA, the presence of water interactions during the entire simulation stabilizes the DNA and the vital interactions between Ets-1 and nucleobases.
Tyr-395 contacts with the fourth base pair (A or G) and the flanking fifth base pair of DNA and is suggested to transmit information about the sequence to the contacts made with the first two base pairs of the core sequence (5). Direct readout of DNA by Ets-1 is possible, in view of the differential hydrogen bond interactions of the hydroxyl group of Tyr-395 observed during dynamics either with the N6 nitrogen of A3 or the O4(N4) of T(C)4′ nucleobases in ETS–GGAA and ETS–GGAG. In our prior investigation (5), we considered the unlikely possibility of indirect readout of DNA, where protein recognizes the sequence specific inherent DNA conformations before binding or the induced conformations of DNA after binding. This result is based on similar helix-bending pattern and ETS-domain–DNA phosphate interactions in ETS–GGAA and ETS–GGAG. The current study shows that the mutation of a single base pair influences the fine structure of the double helix that renders the GGAG sequence to be of low affinity than GGAA. The results, then, also support the indirect readout mechanism for the recognition of the GGAA sequence by Ets-1, as suggested for ETS family of transcription factors (27). Hence, direct and indirect readout mechanisms of DNA recognition both play a role, as reported in other protein–DNA structures (28–30).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant 5R37DK0917136.
Abbreviations: MD, molecular dynamics; SASA, solvent-accessible surface area; DCC, dynamic crosscorrelation.
References
- 1.Sementchenko, V. I. & Watson, D. K. (2000) Oncogene 19, 6533–6548. [DOI] [PubMed] [Google Scholar]
- 2.Lelievre, E., Lionneton, F., Soncin, F. & Vandenbunder, B. (2001) Int. J. Biochem. Cell Biol. 33, 391–407. [DOI] [PubMed] [Google Scholar]
- 3.Ogata, K., Sato, K. & Tahirov, T. H. (2003) Curr. Opin. Struct. Biol. 13, 40–48. [DOI] [PubMed] [Google Scholar]
- 4.Dittmer, J. & Nordheim, A. (1998) Biochim. Biophys. Acta 1377, F1–F11. [DOI] [PubMed] [Google Scholar]
- 5.Obika, S., Reddy, S. Y. & Bruice, T. C. (2003) J. Mol. Biol. 331, 345–359. [DOI] [PubMed] [Google Scholar]
- 6.Garvie, C. W., Hagman, J. & Wolberger, C. (2001) Mol. Cell 8, 1267–1276. [DOI] [PubMed] [Google Scholar]
- 7.MacKerell, A. D., Jr., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., et al. (1998) J. Phys. Chem. B 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- 8.Mackerell, A. D. & Banavali, N. (2000) J. Comput. Chem. 21, 105–120. [Google Scholar]
- 9.Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. (1983) J. Chem. Phys. 79, 926–935. [Google Scholar]
- 10.Darden, T., York, D. & Pedersen, L. (1993) J. Chem. Phys. 98, 10089–10092. [Google Scholar]
- 11.Petersen, H. G. (1995) J. Chem. Phys. 103, 3668–3679. [Google Scholar]
- 12.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S. & Karplus, M. (1983) J. Comput. Chem 4, 187–217. [Google Scholar]
- 13.Dickerson, R. E. (1998) Nucleic Acids Res. 26, 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, B. & Richards, F. M. (1971) J. Mol. Biol. 55, 379–400. [DOI] [PubMed] [Google Scholar]
- 15.Lipari, G. & Szabo, A. (1982) J. Am. Chem. Soc. 104, 4546–4559. [Google Scholar]
- 16.Gupta, G., Bansal, M. & Sasisekharan, V. (1980) Proc. Natl. Acad. Sci. USA 77, 6486–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szyperski, T., Fernandez, C., Ono, A., Kainosho, M. & Wuthrich, K. (1998) J. Am. Chem. Soc. 120, 821–822. [Google Scholar]
- 18.Chang, K. Y. & Varani, G. (1997) Nat. Struct. Biol. 4, Suppl., 854–858. [PubMed] [Google Scholar]
- 19.Grzeskowiak, K., Yanagi, K., Prive, G. G. & Dickerson, R. E. (1991) J. Biol. Chem. 266, 8861–8883. [DOI] [PubMed] [Google Scholar]
- 20.Winger, R. H., Liedl, K. R., Rudisser, S., Pichler, A., Hallbrucker, A. & Mayer, E. (1998) J. Phys. Chem. 102, 8934–8940. [Google Scholar]
- 21.Yathindra, N. & Sundaralingam, M. (1976) Nucleic Acids Res. 3, 729–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson, W. K. (1982) Nucleic Acids Res. 10, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter, C. A. (1993) J. Mol. Biol. 230, 1025–1054. [DOI] [PubMed] [Google Scholar]
- 24.Van der Spoel, D. & Berendsen, H. J. C. (1997) Biophys. J. 72, 2032–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chillemi, G., Fiorani, P., Benedetti, P. & Desideri, A. (2003) Nucleic Acids Res. 31, 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickerson, R. E., Goodsell, D. & Kopka, M. L. (1996) J. Mol. Biol. 256, 108–125. [DOI] [PubMed] [Google Scholar]
- 27.Szymczyna, B. R. & Arrowsmith, C. H. (2000) J. Biol. Chem. 275, 28363–28370. [DOI] [PubMed] [Google Scholar]
- 28.von Hippel, P. H. (1994) Science 263, 769–770. [DOI] [PubMed] [Google Scholar]
- 29.Bareket-Samish, A., Cohen, I. & Haran, T. E. (1998) J. Mol. Biol. 277, 1071–1080. [DOI] [PubMed] [Google Scholar]
- 30.Chen, S. F., Gunasekera, A., Zhang, X. P., Kunkel, T. A., Ebright, R. H. & Berman, H. M. (2001) J. Mol. Biol. 314, 75–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.