Abstract
It has been shown that the invasive trypomastigote forms of Trypanosoma cruzi use and modulate components of the extracellular matrix (ECM) during the initial process of infection. Infective trypomastigotes up-regulate the expression of laminin γ-1 (LAMC1) and thrombospondin (THBS1) to facilitate the recruitment of trypomastigotes to enhance cellular infection. Silencing the expression of LAMC1 and THBS1 by stable RNAi dramatically reduces trypanosome infection. T. cruzi gp83, a ligand that mediates the attachment of trypanosomes to cells to initiate infection, up-regulates LAMC1 expression to enhance cellular infection. Infective trypomastigotes interact with LAMC1 through galectin-3 (LGALS3), a human lectin, to enhance cellular infection. Silencing the expression of LGALS3 also reduces cellular infection. Some trypanosome surface molecules also interact with the ECM to facilitate infection. Despite the role of the ECM in T. cruzi infection, almost nothing is known about the ECM interactome networks operating in the process of T. cruzi infection. In this mini review, we critically analyze and discuss the regulation of the ECM by T. cruzi and its gp83 ligand, and present the first elucidation of the human ECM interactome network, regulated by T. cruzi and its gp83 ligand, to facilitate cellular infection. The elucidation of the human ECM interactome regulated by T. cruzi is critically important to the understanding of the molecular pathogenesis of T. cruzi infection and developing novel approaches of intervention in Chagas’ disease.
Keywords: Extracellular matrix (ECM), Trypanosoma cruzi, cellular infection, ECM interactome
Regulation of the ECM by T. cruzi and its gp83 ligand
The importance of the extracellular matrix (ECM) in T. cruzi infection has been highlighted [1, 2]. Our group and others have employed gene transcription microarray technology and have shown that certain host genes, including ECM genes, are significantly modulated by the parasite to facilitate the process of infection [3–8]. An integrative analysis of the observed gene expression profile at the transcriptome level, as well as protein-protein interaction (PPI) networks, can provide insights into the molecular mechanisms that contribute to the pathogenesis of Chagas’ disease. PPI network analysis has thus far not been exploited to elucidate the complex biological interactions occurring during the initial phases of T. cruzi infection. Our group studied the gene expression profile of human coronary artery smooth muscle cells (HCASM) modulated by T. cruzi trypomastigotes and the T. cruzi gp83 ligand [9–11] and here we report the interactome network operating in HCASM cells during the early process of T. cruzi invasion. The interactome data has shown that specific interactions between some ECM proteins are regulated by T. cruzi during early infection. THBS1, LAMC1, LGALS3 and fibronectin are part of the ECM interactome that T. cruzi regulates and uses in order to facilitate host cell trypanosome interactions leading to cellular infection.
We have demonstrated that T. cruzi up-regulates the levels of THBS1 expression in host cells during early infection. Stable RNA interference (RNAi) of host cell THBS1 knocks down THBS1 transcripts and protein expression causing inhibition of T. cruzi infection [4]. Exogenous THBS1 restored the level of infection of these THBS1 knock-down cells. Thus, host THBS1, regulated by the parasite, plays a crucial role in early infection. T. cruzi gp83, a ligand that T. cruzi uses to attach to host cells [9], increases the level of LAMC1 transcript and its expression in mammalian cells, leading to an increase in cellular infection. Stable RNA interference (RNAi) of host cell LAMC1 knocks down LAMC1 transcript levels and protein expression in mammalian cells, causing a dramatic reduction in cellular infection by T. cruzi [3]. Furthermore, human LGALS3 binds to a trypomastigote surface mucin [12, 13] and to HCASM cells in a lectin-like manner [14] to significantly increase the cellular adhesion of trypomastigotes. Silencing LGALS3 expression in mammalian cells by antisense approach significantly reduces trypomastigote adhesion to cells.
T. cruzi infection causes extensive fibrosis and severe heart cardiomyopathology, which includes vasculopathy, leading to cardiac arrest that is frequently followed by death [15]. The fact that T. cruzi trypomastigotes modulate the expression of ECM proteins and subsequent PPI networks suggest that the altered ECM may contribute to the pathology manifested in Chagas’ disease. Here we present the first elucidation of the host cell ECM interactome network induced by T. cruzi and its gp83, a critical ligand used by the trypanosome to bind host cells, to facilitate cellular infection. This is consistent with the first gene network analysis induced by T. cruzi and its gp83 in human cells [16].
As reported previously, T. cruzi trypomastigotes are able to increase the levels of THBS1 expression in host cells during early infection. Thrombospondins constitute a set of unique multidomain proteins that are synthesized, secreted and incorporated into the ECM by many cell types [17, 18]. Thrombospondins are unique members of the ECM; they have been described as ‘matricellular’ proteins. Most thrombospondin isoforms are expressed in myocytes, smooth muscle cells, fibroblasts and endothelial cells, all of which T. cruzi infects.
Little is known about the role that host THBS1 isoforms play in the process of microbial infections. It was suggested that T. cruzi might have receptors for thrombospondin [19]. Since nearly all cells that T. cruzi infects are surrounded by basement membranes (of which THBS1 is a key component), the ability of the parasite to effectively regulate and interact with THBS1 is also critically important for its passage through the host cell membrane.
The kinetics of change in gene transcript profiles of THBS1, THBS2, THBS3 and THBS4 was evaluated by real time PCR during the early process of infection of HCASM cells by T. cruzi. It was shown that THBS1 transcript levels increase approximately 6-fold at 60 min, followed by an increase of 2.7-fold at 120 min during infection of HCASM cells by T. cruzi. The transcript levels of THBS2, THBS3 and THBS4 do not change significantly during the same period of time of T. cruzi infection of HCASM cells [4].
TGF β-1 induced over-expression of THBS1 in human epithelial cells, causing a significant increase in the number of trypanosomes bound to cells and parasite load as compared to cells transfected with vector alone or scrambled THBS1 antisense. Thus, there is a direct correlation between over expression of THBS1 in human cells with an increase of infection of these cells. T. cruzi infection of mammalian cells is inhibited by stable RNAi of host cell THBS1 [4]. Addition of THBS1 to these cells restores infection. Thus, host THBS1, regulated by the parasite, plays a crucial role in early infection. RNAi silencing of THBS1 does not alter the expression of other ECM proteins such as fibronectin and LAMC1 and/or LGALS3, nor their abilities to adhere to substrate, supporting the specificity of these studies. Therefore, these results indicate that THBS1 is required for the process of T. cruzi infection.
Since T. cruzi modulates the ECM by up-regulating THBS1 expression, the parasite is able to exploit THBS1 in addition to LAMC1 to navigate through the ECM and facilitate infection. THBS1 is required for the infection process of T. cruzi as evidenced by RNAi of that specific isoform. The T. cruzi up-regulation of host THBS1 expression to facilitate infection of human cells represents a new mechanism that may contribute to elucidating the pathogenesis of T. cruzi infection.
Similarly to THBS1, LAMC1 is also up regulated by T. cruzi, specifically by its gp83 surface glycoprotein. An additional T. cruzi trypomastigote surface glycoprotein, Tc-85 that binds to laminin, has also been implicated in infection [20]. LAMC1 is the most abundant laminin isoform in humans [17] and stable RNAi of host cell LAMC1 knocks down both LAMC1 mRNA transcript and protein levels in mammalian cells. As a result of silencing LAMC1, there is a dramatic reduction in cellular infection by T. cruzi [3]. Thus, host LAMC1, which is regulated by the parasite, also plays a crucial role in the early process of infection.
The fact that a T. cruzi trypomastigote ligand, gp83, increases LAMC1 transcript levels in HCASM cells correlates with the finding that laminin is deposited in the hearts of patients infected with Chagas’ disease [21]. This suggests that the regulation of LAMC1 in cardiomyocytes by gp83 may partially explain the cause of Chagas’ disease pathology.
Our findings indicating that T. cruzi gp83 ligand remodels the ECM by up-regulating the expression of LAMC1, together with the report that T. cruzi presents laminin receptors on its surface [20], suggests that the parasite exploits LAMC1 to navigate through the ECM and facilitate infection. Thus, the T. cruzi gp83 ligand is a virulence factor that modifies LAMC1 expression in the ECM to contribute to the pathogenesis of T. cruzi infection in human cardiomyocytes.
LAMC1 also interacts with LGALS3, a human lectin which binds β-galactoside. This interaction is thought to play an important role in the early steps of T. cruzi infection of human cells [14]. LGALS3 is known to bind to the surface of HCASM cells, as well as to the surface of invasive T. cruzi trypomastigotes [14]. The binding of LGALS3 to the surface of both T. cruzi trypomastigotes and HCASM cells is almost completely inhibited by lactose (but not by sucrose), indicating that LGALS3 binds to the surface of both trypomastigotes and HCASM cells in a lectin-like manner [14]. LGALS3 increases by five-fold the adhesion of trypomastigotes to HCASM cells; this effect is specific since it is inhibited by lactose in a concentration-dependent and saturable manner. On cells which LGALS3 has been knocked down by antisense approach there was a significant reduction of bound trypanosomes compared to control cells. This effect of LGALS3 knock-down is reversed by exogenous LGALS3, indicating that cellular expression of LGALS3 is required for T. cruzi adhesion to human cells.
The fact that exogenous human LGALS3 specifically binds to the surface of HCASM cells and to T. cruzi suggests that LGALS3 bridges trypanosomes and cells, resulting in an enhancement of T. cruzi binding to human cells. The proposed mechanism by which this interaction occurs is through the association of LGALS3 molecules with T. cruzi 45 (mucin), 32 and 30 kDa surface proteins and with LAMC1 via its carbohydrate recognition domains [13,22,23]. The 45 kDa mucin found on the surface of invasive T. cruzi trypomastigotes specifically interacts with LGALS3 and plays an important role in trypomastigote adhesion [12, 13]. These studies suggest that this is a trypanosome trapping mechanism which enables the organisms to accumulate in the basement membrane prior to invasion, making LGALS3 a candidate molecule which enhances the pathogenesis of T. cruzi.
LGALS3 is also expressed in B cells from T. cruzi-infected mice [24] and is up-regulated by T. cruzi infection of mice [25]. The concentrations of LGALS3 that increase trypanosome adhesion to HCASM cells in vitro are similar to the concentrations of LGALS3 present in vivo [26]. Furthermore, LGALS3 levels increase approximately 300-fold during microbial infection in vivo [27]. These observations suggest that the parasite modulates the host and takes advantage of a host inflammatory molecule, LGALS3, to bind to host cells. These findings may contribute to determining a cause of the pathology manifested in Chagas’ disease.
Fibronectin is an adhesion ECM glycoprotein which has been shown to be involved in target cell invasion by trypomastigotes via the RGDS peptide (fibronectin cell attachment site) binding to the trypomastigote surface leading to inhibition of trypomastigote internalization [28]. The trypomastigote ligand for fibronectin was purified by affinity chromatography, and identified as an 85 kDa protein which interacts with cells bearing fibronectin molecules, such as human monocytes, neutrophils and 3T3 fibroblasts [28]. Fibronectin also mediates the uptake of T. cruzi by human monocytes via RGDS [29]. The involvement of fibronectin in T. cruzi cardiomyocyte invasion has been observed during T. cruzi infection both in vivo and in vitro. Pre-treatment of trypomastigotes with fibronectin before cardiomyocyte interaction reduced T. cruzi infection significantly. Further, enhancement of ECM components was detected in the myocardium during late acute infection with T. cruzi [30]. However, molecular genetic approaches have not been used to validate the role of fibronectin in the process of infection.
Regulation of the ECM Interactome by T. cruzi and its gp83 ligand
The genetic architecture in the early T. cruzi infection process of human cells is unknown. To understand this aspect of infection, we conducted gene transcription microarray analysis followed by gene network construction of the host cell response in primary HCASM cells infected with T. cruzi or exposed to T. cruzi gp83. Using THBS1, LAMC1, and LGALS3 as the seed nodes for biological network construction, we built an interactome network of the early T. cruzi infection process which centered on the ECM. After seeding the initial three nodes, the network was expanded to one degree of direct biological interaction, resulting in base interaction networks with the seed node as a center point in each. In order to populate and build our interaction network, the Michigan Molecular Interactions (MiMI) Cytoscape plugin (version 3.2) was used. The MiMI interface uses a database which is itself constructed from merged data of well-known protein interaction databases, including BIND, DIP, HPRD, RefSeq, SwissProt, IPI and CCSB-HI1 [31]. The MiMI query engine was used to seed the THBS1, LAMC1, and LGAL3 initial nodes and their respective nearest neighbors to one degree of biological interactions. The networks were then merged for interconnections and the global interactome was visualized in Cytoscape (version 2.6.3) [32], thus, yielding a snapshot of the ECM interactome involved in early T. cruzi infection. This ECM-focused interactome contains 104 nodes representing protein coding genes connected by 218 edges representing biological interactions between nodes (Fig. 1).
Fig. 1. ECM interactome of early Trypanosoma cruzi infection.
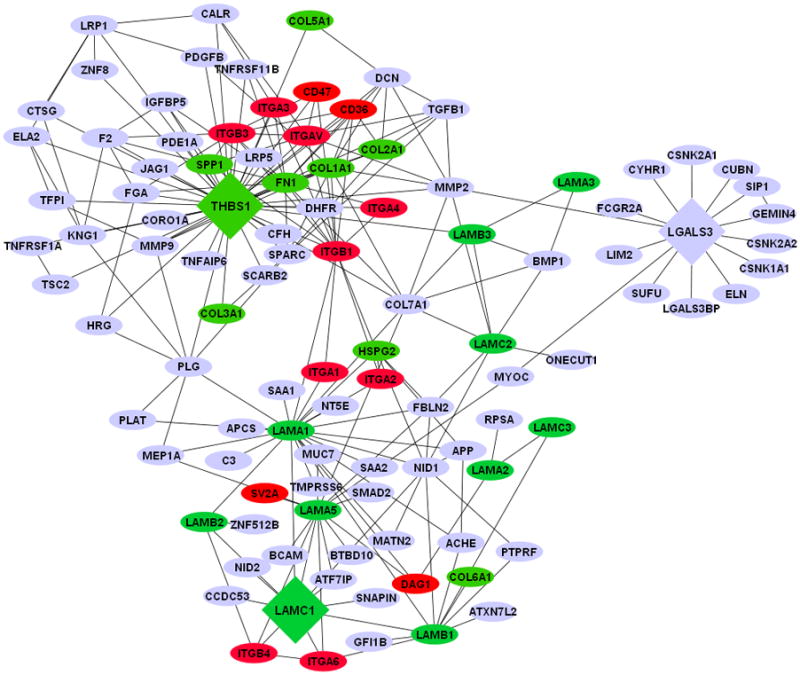
The network was connected by using Laminin gamma-1 (LAMC1), Thrombospondin-1 (THBS1), and Galectin-3 (LGALS3) as initial seed nodes (large DIAMOND nodes). Node expansion was done to one degree of biological protein-protein interaction partners of the seed nodes using the Michigan Molecular Interaction Database. Proteins mapping to the canonical “ECM- Receptor Interaction Pathway” of KEGG (hsa04512) are highlighted as following: GREEN nodes are ECM ligands: (laminins, collagens, fibronectin, heperan sulfate, and thrombospondin-1). RED nodes are ECM receptors: (integrins, cd35, cd47, and SV2A). BLUE nodes are not part of the KEGG ECM-ECM Receptor Pathway, but have established biological interactions with ECM-Receptor interaction nodes.
Increased THBS1 expression significantly modulates the interactome cross talk between cells. This change in network topology potentially favors parasite invasion and infection of host cells. As such, THBS1 interacts with several proteins ranging from adhesion receptors (CD36 and CD47) to structural proteins (COL7A1) and zymogens (PLG) as shown in Fig. 1. CD47 is a receptor for the C-terminal domain of THBS1 and this interaction may be important in membrane transport and signal transduction. In addition, CD47 is involved in the intracellular calcium increase which occurs when the cell adheres to the ECM. An increase in cytosolic Ca++ in T. cruzi trypomastigotes was detected at the single cell level after association of the parasites with myoblasts. Ca++ mobilization in the host cells was also detected upon contact with trypomastigotes. Confirmatively, pretreatment of the parasites with the Ca++ chelators decreased the trypomastigotes' association to myoblasts indicating that calcium mobilization is required for cell invasion [33].
The structural protein COL7A1, or collagen 7 alpha 1, is typically found in the basement membrane and also associates with THBS1. Once COL7A1 interacts with THBS1, it makes the ECM conducive to parasite motility and cellular invasion with the help of proteins from the laminin family of genes (Fig. 1). Plasminogen (PLG) links LAMC1 and THBS1. PLG (a zymogen), is cleaved into plasmin (a serine protease), and angiostatin (an angiogenesis inhibitor). Plasmin is known to cleave fibronectin, THBS1, and LAMC1, thus taking part in ECM modifications.
T. cruzi must navigate through the basal lamina, which contains LAMC1. The T. cruzi gp83 ligand modifies LAMC1 expression in the ECM and contributes to the pathogenesis of T. cruzi infection in human heart cells. The fact that LAMC1’s network is connected to THBS1 through PLG but also COL7A1 and MMP2 suggests that this network could facilitate parasite mobilization. Matrix metallo proteinase 2 (MMP2) is a type IV collogenase that participates in the rearrangement of the ECM, which could facilitate parasite mobilization. In the subnetwork of LAMC1, there is an indirect connection (through LAMA1) to Dystroglycan (DAG1). DAG1 is a dystrophin-associated glycol-protein responsible for transmembrane linkage between the ECM and host cell cytoskeleton. The extracellular form of DAG1 can bind to merosin alpha-2-laminin in the ECM [34]. If T. cruzi can alter the dystroglycan complex, it could consequently manipulate or weaken the host cell cytoskeleton without first gaining entry into the cell. This could yet be another mechanism by which T. cruzi invades host cells. In reference to the ECM network reviewed, LAMC1 also has a second-degree interaction (one node in between the genes) with LGALS3 through myocilin (MYOC). MYOC is a secreted protein believed to have a role in cytoskeletal function, specifically vesicular transport and extracellular matrix conformation [35]. Unlike other intracellular pathogens which avoid contact with host cell lysosomes, T. cruzi requires the low-pH environment of lysosomes to initiate egression from the vacuole and delivery to the host cell cytoplasm where replication takes place [36]. Therefore the control of vesicular trafficking by T. cruzi improves the rate of trypomastigote entry and amastigote replication in the host cell. The fact that LAMC1 is connected to LGALS3 through MYOC in the ECM interactome suggests the importance of LGALS3 in T. cruzi’s manipulation of host ECM.
Increased LGALS3 expression in the ECM promotes the adhesion of T. cruzi to host cells and subsequent infection [14]. In addition, LGALS3 has numerous ECM interacting partners [37] including collagen IV, hensin, laminins, fibronectin, vitronectin, tenascin and elastin. LGALS3 regulates adhesion of these ECM proteins to a variety of host cells. Matrix metalloproteases, which are more active in T. cruzi infected mice, regulate LGAL3 function [38]. When metalloproteases are activated in the ECM, they can cleave LGALS3 and negatively regulate its function. Consequently, increased activation of matrix metalloproteases 1 and 9 (MMP-1 and MMP-9) is associated with ECM destruction and myocarditis in T. cruzi infection.
Multiple types of collagen interact with the 3 central seed nodes, THBS1, LGALS3 and LAMC1. A glimpse of the importance of collagen in early T. cruzi infection was reported by Velge et al [39]. Their results suggest that a T. cruzi surface glycoprotein, made up of an 80–85 kDa subunit and a 58/68 kDa subunit, binds host cell fibronectin and collagen. In addition, this interaction may occur during the initial phase of parasite-cell recognition [39].
Thus, the elucidation of human ECM interactome regulated by T. cruzi is of paramount importance to the understanding of the molecular pathogenesis of T. cruzi infection and developing novel strategies of intervention in Chagas’ disease.
Acknowledgments
This work was supported by NIH grant AI080580 to FV. PNN, is supported by NIH grant AI083925. TCC and CAJ are fellows supported by NIH grant 2T32AI007281-21. MFL is supported by NIH grant 5R25GM059994-10. Experimental set-up, interpretation and data analysis were performed through the use of the Meharry Medical College Microarray, Proteomics and Bioinformatics Cores (http://www.mmc.edu/bioinformatics/) which are supported by NIH grants 5R25GM059994-S1, 5G12RR003032-25, and 1U54RR026140-01.
References
- 1.Villalta F, Madison MN, Kleshchenko YY, Nde PN, Lima MF. Molecular analysis of early host cell infection by Trypanosoma cruzi. Front Biosci. 2008;13:3714–3734. doi: 10.2741/2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villalta F, Scharfstein J, Ashton AW, Tyler KM, Guan F, Mukherjee S, Lima MF, Alvarez S, Weiss LM, Huang H, Machado FS, Tanowitz HB. Perspectives on the Trypanosoma cruzi-host cell receptor interactions. Parasitol Res. 2009;104:1251–1260. doi: 10.1007/s00436-009-1383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nde PN, Simmons KJ, Kleshchenko YY, Pratap S, Lima MF, Villalta F. Silencing of the laminin gamma-1 gene blocks Trypanosoma cruzi infection. Infect Immun. 2006;74:1643–1648. doi: 10.1128/IAI.74.3.1643-1648.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmons KJ, Nde PN, Kleshchenko YY, Lima MF, Villalta F. Stable RNA interference of host thrombospondin-1 blocks Trypanosoma cruzi infection. FEBS Lett. 2006;580:2365–2370. doi: 10.1016/j.febslet.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 5.Vaena de Avalos S, Blader IJ, Fisher M, Boothroyd JC, Burleigh BA. Immediate/early response to Trypanosoma cruzi infection involves minimal modulation of host cell transcription. J Biol Chem. 2002;277:639–644. doi: 10.1074/jbc.M109037200. [DOI] [PubMed] [Google Scholar]
- 6.Shigihara T, Hashimoto M, Shindo N, Aoki T. Transcriptome profile of Trypanosoma cruzi-infected cells: simultaneous up- and down-regulation of proliferation inhibitors and promoters. Parasitol Res. 2008;102:715–722. doi: 10.1007/s00436-007-0819-x. [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee S, Belbin TJ, Spray DC, Iacobas DA, Weiss LM, Kitsis RN, Wittner M, Jelicks LA, Scherer PE, Ding A, Tanowitz HB. Microarray analysis of changes in gene expression in a murine model of chronic chagasic cardiomyopathy. Parasitol Res. 2003;91:187–196. doi: 10.1007/s00436-003-0937-z. [DOI] [PubMed] [Google Scholar]
- 8.Imai K, Mimori T, Kawai M, Koga H. Microarray analysis of host gene-expression during intracellular nests formation of Trypanosoma cruzi amastigotes. Microbiol Immunol. 2005;49:623–631. doi: 10.1111/j.1348-0421.2005.tb03654.x. [DOI] [PubMed] [Google Scholar]
- 9.Villalta F, Smith CM, Ruiz-Ruano A, Lima MF. A ligand that Trypanosoma cruzi uses to bind to mammalian cells to initiate infection. FEBS Lett. 2001;505:383–388. doi: 10.1016/s0014-5793(01)02853-8. [DOI] [PubMed] [Google Scholar]
- 10.Villalta F, Zhang Y, Bibb KE, Burns JM, Jr, Lima MF. Signal transduction in human macrophages by gp83 ligand of Trypanosoma cruzi: trypomastigote gp83 ligand up-regulates trypanosome entry through the MAP kinase pathway. Biochem Biophys Res Commun. 1998;249:247–252. doi: 10.1006/bbrc.1998.9127. [DOI] [PubMed] [Google Scholar]
- 11.Villalta F, Lima MF, Ruiz-Ruano A, Zhou L. Attachment of Trypanosoma cruzi to host cells: a monoclonal antibody recognizes a trypomastigote stage-specific epitope on the gp 83 required for parasite attachment. Biochem Biophys Res Commun. 1992;182:6–13. doi: 10.1016/s0006-291x(05)80104-4. [DOI] [PubMed] [Google Scholar]
- 12.Turner CW, Lima MF, Villalta F. Trypanosoma cruzi uses a 45-kDa mucin for adhesion to mammalian cells. Biochem Biophys Res Commun. 2002;290:29–34. doi: 10.1006/bbrc.2001.6189. [DOI] [PubMed] [Google Scholar]
- 13.Moody TN, Ochieng J, Villalta F. Novel mechanism that Trypanosoma cruzi uses to adhere to the extracellular matrix mediated by human galectin-3. FEBS Lett. 2000;470:305–308. doi: 10.1016/s0014-5793(00)01347-8. [DOI] [PubMed] [Google Scholar]
- 14.Kleshchenko YY, Moody TN, Furtak VA, Ochieng J, Lima MF, Villalta F. Human galectin-3 promotes Trypanosoma cruzi adhesion to human coronary artery smooth muscle cells. Infect Immun. 2004;72:6717–6721. doi: 10.1128/IAI.72.11.6717-6721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanowitz HB, Kirchhoff LV, Simon D, Morris SA, Weiss LM, Wittner M. Chagas' disease. Clin Microbiol Rev. 1992;5:400–419. doi: 10.1128/cmr.5.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nde PN, Johnson CA, Pratap S, Cardenas TC, Kleshchenko YY, Furtak VA, Simmons KJ, Lima MF, Villalta F. Gene network analysis during early infection of human coronary artery smooth muscle cells by Trypanosoma cruzi and Its gp83 ligand. Chem Biodivers. 7:1051–1064. doi: 10.1002/cbdv.200900320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawler J. The functions of thrombospondin-1 and-2. Curr Opin Cell Biol. 2000;12:634–640. doi: 10.1016/s0955-0674(00)00143-5. [DOI] [PubMed] [Google Scholar]
- 18.Jurk K, Clemetson KJ, de Groot PG, Brodde MF, Steiner M, Savion N, Varon D, Sixma JJ, Van Aken H, Kehrel BE. Thrombospondin-1 mediates platelet adhesion at high shear via glycoprotein Ib (GPIb): an alternative/backup mechanism to von Willebrand factor. FASEB J. 2003;17:1490–1492. doi: 10.1096/fj.02-0830fje. [DOI] [PubMed] [Google Scholar]
- 19.Ulrich H, Magdesian MH, Alves MJ, Colli W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J Biol Chem. 2002;277:20756–20762. doi: 10.1074/jbc.M111859200. [DOI] [PubMed] [Google Scholar]
- 20.Giordano R, Fouts DL, Tewari D, Colli W, Manning JE, Alves MJ. Cloning of a surface membrane glycoprotein specific for the infective form of Trypanosoma cruzi having adhesive properties to laminin. J Biol Chem. 1999;274:3461–3468. doi: 10.1074/jbc.274.6.3461. [DOI] [PubMed] [Google Scholar]
- 21.Milei J, Sanchez J, Storino R, Yu ZX, Denduchis B, Ferrans VJ. Antibodies to laminin and immunohistochemical localization of laminin in chronic chagasic cardiomyopathy: a review. Mol Cell Biochem. 1993;129:161–170. doi: 10.1007/BF00926364. [DOI] [PubMed] [Google Scholar]
- 22.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 23.Kasai K, Hirabayashi J. Galectins: a family of animal lectins that decipher glycocodes. J Biochem. 1996;119:1–8. doi: 10.1093/oxfordjournals.jbchem.a021192. [DOI] [PubMed] [Google Scholar]
- 24.Acosta-Rodriguez EV, Montes CL, Motran CC, Zuniga EI, Liu FT, Rabinovich GA, Gruppi A. Galectin-3 mediates IL-4-induced survival and differentiation of B cells: functional cross-talk and implications during Trypanosoma cruzi infection. J Immunol. 2004;172:493–502. doi: 10.4049/jimmunol.172.1.493. [DOI] [PubMed] [Google Scholar]
- 25.Vray B, Camby I, Vercruysse V, Mijatovic T, Bovin NV, Ricciardi-Castagnoli P, Kaltner H, Salmon I, Gabius HJ, Kiss R. Up-regulation of galectin-3 and its ligands by Trypanosoma cruzi infection with modulation of adhesion and migration of murine dendritic cells. Glycobiology. 2004;14:647–657. doi: 10.1093/glycob/cwh068. [DOI] [PubMed] [Google Scholar]
- 26.Sano H, Hsu DK, Yu L, Apgar JR, Kuwabara I, Yamanaka T, Hirashima M, Liu FT. Human galectin-3 is a novel chemoattractant for monocytes and macrophages. J Immunol. 2000;165:2156–2164. doi: 10.4049/jimmunol.165.4.2156. [DOI] [PubMed] [Google Scholar]
- 27.Sato S, Hughes RC. Regulation of secretion and surface expression of Mac-2, a galactoside-binding protein of macrophages. J Biol Chem. 1994;269:4424–4430. [PubMed] [Google Scholar]
- 28.Ouaissi MA, Cornette J, Capron A. Identification and isolation of Trypanosoma cruzi trypomastigote cell surface protein with properties expected of a fibronectin receptor. Mol Biochem Parasitol. 1986;19:201–211. doi: 10.1016/0166-6851(86)90002-2. [DOI] [PubMed] [Google Scholar]
- 29.Noisin EL, Villalta F. Fibronectin increases Trypanosoma cruzi amastigote binding to and uptake by murine macrophages and human monocytes. Infect Immun. 1989;57:1030–1034. doi: 10.1128/iai.57.4.1030-1034.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvet CM, Meuser M, Almeida D, Meirelles MN, Pereira MC. Trypanosoma cruzi-cardiomyocyte interaction: role of fibronectin in the recognition process and extracellular matrix expression in vitro and in vivo. Exp Parasitol. 2004;107:20–30. doi: 10.1016/j.exppara.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Gao J, Ade AS, Tarcea VG, Weymouth TE, Mirel BR, Jagadish HV, States DJ. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics. 2009;25:137–138. doi: 10.1093/bioinformatics/btn501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreno SN, Silva J, Vercesi AE, Docampo R. Cytosolic-free calcium elevation in Trypanosoma cruzi is required for cell invasion. J Exp Med. 1994;180:1535–1540. doi: 10.1084/jem.180.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada H, Shimizu T, Tanaka T, Campbell KP, Matsumura K. Dystroglycan is a binding protein of laminin and merosin in peripheral nerve. FEBS Lett. 1994;352:49–53. doi: 10.1016/0014-5793(94)00917-1. [DOI] [PubMed] [Google Scholar]
- 35.Caballero M, Rowlette LL, Borras T. Altered secretion of a TIGR/MYOC mutant lacking the olfactomedin domain. Biochim Biophys Acta. 2000;1502:447–460. doi: 10.1016/s0925-4439(00)00068-5. [DOI] [PubMed] [Google Scholar]
- 36.Burleigh BA. Host cell signaling and Trypanosoma cruzi invasion: do all roads lead to lysosomes? Sci STKE. 2005;2005:pe36. doi: 10.1126/stke.2932005pe36. [DOI] [PubMed] [Google Scholar]
- 37.Dumic J, Dabelic S, Flogel M. Galectin-3: an open-ended story. Biochim Biophys Acta. 2006;1760:616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez FR, Lalu MM, Mariano FS, Milanezi CM, Cena J, Gerlach RF, Santos JE, Torres-Duenas D, Cunha FQ, Schulz R, Silva JS. Increased activities of cardiac matrix metalloproteinases matrix metalloproteinase (MMP)-2 and MMP-9 are associated with mortality during the acute phase of experimental Trypanosoma cruzi infection. J Infect Dis. 2008;197:1468–1476. doi: 10.1086/587487. [DOI] [PubMed] [Google Scholar]
- 39.Velge P, Ouaissi MA, Cornette J, Afchain D, Capron A. Identification and isolation of Trypanosoma cruzi trypomastigote collagen-binding proteins: possible role in cell-parasite interaction. Parasitology. 1988;97:255–268. doi: 10.1017/s0031182000058467. [DOI] [PubMed] [Google Scholar]


