Abstract
Clinical cancer genetic susceptibility analysis typically proceeds sequentially beginning with the most likely causative gene. The process is time consuming and the yield is low particularly for families with unusual patterns of cancer. We determined the results of in parallel mutation analysis of a large cancer-associated gene panel. We performed deletion analysis and sequenced the coding regions of 45 genes (8 oncogenes and 37 tumor suppressor or DNA repair genes) in 48 childhood cancer patients who also (1) were diagnosed with a second malignancy under age 30, (2) have a sibling diagnosed with cancer under age 30 and/or (3) have a major congenital anomaly or developmental delay. Deleterious mutations were identified in 6 of 48 (13%) families, 4 of which met the sibling criteria. Mutations were identified in genes previously implicated in both dominant and recessive childhood syndromes including SMARCB1, PMS2, and TP53. No pathogenic deletions were identified. This approach has provided efficient identification of childhood cancer susceptibility mutations and will have greater utility as additional cancer susceptibility genes are identified. Integrating parallel analysis of large gene panels into clinical testing will speed results and increase diagnostic yield. The failure to detect mutations in 87% of families highlights that a number of childhood cancer susceptibility genes remain to be discovered.
Keywords: Cancer susceptibility, tumor suppressor genes, oncogenes, mutation analysis, rhabdoid tumors
Introduction
Identifying mutations which result in susceptibility to childhood cancer has multiple clinical benefits including potential modification of cancer treatment, increased surveillance for subsequent malignancies and identification of at-risk relatives [1]. Selection of the appropriate gene to initiate testing often relies on recognition of specific patterns of cancer, such as familial clustering of sarcomas with other cancers in Li Fraumeni Syndrome or cancer occurring in a child with specific congenital anomalies (aniridia and Wilms tumor resulting from chromosome 11q13 microdeletion). However, relying on known patterns of family history or anomalies may underestimate the prevalence of constitutional/germline mutations in pediatric cancers. De novo mutations in either tumor suppressor genes (TSGs) or oncogenes can result in a child with cancer without a family history, e.g. mutations in SMARCB1/INI11 are found in approximately 35% of children with atypical teratoid/rhabdoid tumors (AT/RT) [2–4]. Unexpected patterns of childhood cancer can result from inheritance of biallelic mutations in TSGs previously associated with adult onset malignancies, e.g. Fanconi anemia resulting from biallelic BRCA2 mutations [5]. Unusual patterns of cancer can also result from inheriting mutations in more than one cancer-associated gene, termed oligogenic inheritance [6].
Physicians typically initiate clinical genetic testing starting with the gene most likely to carry a mutation and then moving down the list as negative results are received. This practice extends the time to obtain a positive result and testing is often halted after a small number of genes due to costs and inconvenience to the family. If testing is halted when the first pathogenic mutation is identified oligogenic mutations will be overlooked. For these reasons it is likely that we are under-diagnosing susceptibility mutations in children with cancer.
As sequence costs decline the ability to sequence larger numbers of genes should increase the yield of informative tests. We report here the results of in parallel sequencing of a panel of 45 cancer-associated genes for 48 childhood cancer patients with histories highly suggestive of genetic susceptibility. Our successful identification of clinically relevant mutations serves as a model for implementing larger scale sequencing platforms in clinical settings.
Materials and Methods
Accrual
Subjects were enrolled in longstanding institutional review board approved cancer genetics protocols with informed consent at three institutions: Baylor College of Medicine/Texas Children’s Hospital, University of Texas Southwestern and University of Texas MD Anderson Cancer Center. The database for each protocol was reviewed to identify families that met the inclusion criteria.
DNA Isolation
DNA was isolated from peripheral blood (majority of cases), fibroblast or lymphoblastoid cell lines (LCLs) using commercially available kits. In only one case where proband DNA was not available, DNA from both parents was used. For sequencing nine of the genes in the panel whole genome amplified DNA was produced using the Illustra Genome Amplification procedure (GE Healthcare-Amersham).
DNA Sequencing
Sequencing was focused on coding regions using primers based on tumor sequencing projects [7]. We avoided polymorphic residues within 5 bases of the 3’ end of the primer and performed in silico PCR analyses. Amplicons were tested on four control DNAs and redesigned as needed. Sequencing of both strands using Sanger sequencing chemistry on a robotic pipeline was performed in the Human Genome Sequencing Center (HGSC).
Data analysis
Sequence chromatograms underwent automated analysis using SNPdetector v3 [8] for single base changes and insertions/deletions and results stored in an Oracle database with password protected SNP Online Annotation Reports (SOAR) including predicted coding change for all mRNAs that map to that position, call confidence and presence in Entrez SNP. All deleterious mutations sequence traces were inspected manually and confirmed by PCR and sequencing in a second laboratory.
Copy number analysis
To detect copy number variants (CNVs) of the gene panel (except PMS2), we created a custom 4X44K oligonucleotide array (Agilent Technologies, Santa Clara, CA) including probes from 50 kbp upstream to downstream of each gene. For each sample, 1 µg of genomic DNA was restriction digested, Cy-5 dUTP-labeled and combined with gender matched Cy-3-dUTP-labeled reference DNA (G1521 and G1471, Promega, Madison, WI)[9]. Hybridization, washing, and scanning were performed according to Agilent specifications (Agilent Technologies, Santa Clara, CA).
CNVs were called based on a minimum of 3 probes with a minimum absolute average log ratio of 0.25 using the DNA Analytics 4.0 software (Agilent Technologies). The ADM-2 aberration algorithm was used at a 5.0 Threshold with deselected Fuzzy Zero and Centralization settings. Cytoreports were generated of all calls in which the log2 ratios were - 0.8 ≥ × ≤ 0.8. CNVs within exons were identified by uploading the data onto Genoboree (genboree.org) and intersecting with the UCSC RefSeq track. Calls were further analyzed manually to remove artifacts.
Given multiple PMS2 pseudogenes, detection of CNVs in PMS2 was performed using the MLPA kit P008 (MLPA-Holland, Amsterdam, The Netherlands). The labeled fragments were mixed with POP-7 polymers, LIZ500 internal size standards, and HiDi formamide and run on the ABI3130XL capillary sequencer (Life Technologies, Carlsbad CA).
Reverse transcriptase – polymerase chain reaction (RT-PCR) analysis of SMARCB1
LCLs were maintained in RPMI 1640 media (Invitrogen, Carlsbad, CA) - 15% fetal bovine serum (FBS). HEK 293T cells were grown in DMEM (Invitrogen) - 10% FBS. Total LCL cellular RNA was isolated using RNeasy mini kit (Qiagen, Chatsworth, CA) after incubation with or without 100 µg/ml emetine (Sigma, St. Louis, MO) for 7 hours. RT-PCR was performed in two steps: random hexamer primed Superscript 1st strand synthesis system (Invitrogen) using 5ug of total RNA followed by PCR. RT-PCR products were TOPO cloned (Invitrogen) and sequenced.
Immunoprecipitation assays
HEK 293T cells underwent Fugene 6-mediated transfection (Roche, Indianapolis, IN) with pFLAG-CMV2-MLH1 and pCS2-MT-PMS2 wildtype or mutant versions generated using the Quick-Change mutagenesis kit (Stratagene, La Jolla, CA). Cells were lysed after 16 hours with radioimmunoprecipitation (RIPA) buffer[10] and 750 µg of lysate was rotated overnight with 25 µl of anti-FLAG M2 affinity gel (Sigma). Pellets were washed and analyzed on 7% sodium dodecyl sulfate polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membrane (Millipore, Bedford, MA). Western blotting was performed with primary mouse monoclonal antibodies (anti-FLAG (Sigma) and anti-myc (Ab-1) (Calbiochem, San Diego, CA)) followed by IR800 dye-labeled goat anti-mouse IgG (Rockland Immunochemicals, Gilbertsville, PA) and visualized using an Odyssey infrared imager (LiCor, Lincoln, NE).
Results
Study design
The eligibility criteria were designed to focus on patients/families highly likely to carry cancer susceptibility mutation(s) and included 48 probands diagnosed with cancer in childhood (< age 18y) and who met at least one of three criteria (1) a second malignancy diagnosed before age 30 (n=20), (2) a sibling with cancer diagnosed prior to age 30 (n=24) or (3) diagnosis of developmental delay and/or major congenital anomalies (n=10). Several of the 48 families met more then one criterion and at the time of entry studies performed by DNA diagnostic laboratories had been uninformative. The families were ethnically diverse with the probands being 4% Asian, 14% African-American, 23% Hispanic-Caucasian and 59% non-Hispanic Caucasian subjects. We excluded families with well-defined dysmorphology syndromes, e.g. Beckwith-Wiedemann syndrome. For criteria 1, we attempted to exclude patients where the second primary malignancy was likely a therapy-related tumor, e.g. AML after etoposide treatment. The pattern of cancer in the families varied from extensive pedigrees to isolated cases.
The 45 genes in the sequencing pipeline (Table 1) were selected to keep sequencing costs at the time under $3000/subject using a robotic platform and based on the following rationale; (1) eight oncogenes commonly mutated in solid tumors including the MYC and RAS families, ERBB2, SRC and RET given that rare, inherited oncogene mutations can result in childhood cancer, e.g. de novo mutations in HRAS in Costello syndrome;[11] (2) TSGs where heterozygous mutations result in genetic susceptibility to either childhood or adult-onset cancers, (3) a sampling of DNA damage response or repair genes, e.g. FANCG, many of which are associated with childhood cancer. Genes like RB1 for which a distinct early childhood cancer phenotype has already been established were not included. Also, very large, extensively studied genes, e.g. ATM, were excluded to limit sequencing costs.
Table 1.
Genes in sequencing pipeline
| Oncogenes | Tumor Suppressor and DNA Repair Genes | ||
|---|---|---|---|
| ERBB2 | APC | FANCG | PMS1 |
| HRAS | ATR | FANCM | PMS2 |
| KRAS | BARD1 | FH | PTCH |
| NRAS | BRCA1 | FLCN | PTEN |
| MYCC | BRCA2 | LIG4 | REST |
| MYCN | BRIP1 | MLH1 | SMAD4 |
| SRC | CDH1 | MLH3 | SMARCB1 |
| RET | CHEK2 | MSH2 | STK11 |
| CTNNB1 | MSH3 | SUFU | |
| EXO1 | MSH6 | TP53 | |
| FANCD2 | MUTYH | VHL | |
| FANCE | PALB2 | XRCC5 | |
| XRCC6 | |||
Sequencing results
The sequencing pipeline consisted of 670 amplicons totaling 133 kilobases of coding region for each proband. There were high quality reads in over 94% and 85% of the amplicons in one or both directions, respectively. For all genes there were two or fewer amplicon failures except PMS2 which had nearly 50% amplicon failures even after using published conditions [12].
There were over 800 variant calls defined as bases that differed from the reference genome. Despite focusing on coding regions, over 60% of the variant calls were intronic, highlighting the variability of non-coding sequences. There were 75 novel missense variants, the majority of which will require further analyses to determine their association with disease. For all missense changes identified in oncogenes we performed literature searches and used protein prediction algorithms but identified no previously reported activating mutations or missense changes in protein domains with other cancer-associated mutations. Thus, no clearly pathogenic mutations in the eight oncogenes included in this study were found.
The sequencing data were then analyzed for mutations likely to be deleterious (nonsense, frameshift and splice site) and rare or disease-associated missense variants in TSGs or DNA repair genes. We identified known single nucleotide polymorphisms (SNPs) for which previous data suggested a small impact on cancer risk, e.g. MLH1 I219V and PTCH P1315L [13;14]. However, given the frequency of these SNPs in the general population it is unclear if their presence in this small cohort is significant. Mutations were identified in genes previously implicated in both dominant and recessive cancer syndromes including nonsense mutations in PMS2 and SMARCB1, frameshift in TP53, splice site mutation in SMARCB1 and previously reported missense mutations in TP53 and FANCD2 in subject ALS034 (Table 2). Overall, disease-associated mutations were identified in six of the 48 families for a yield of 13%.
Table 2.
Putative deleterious mutations identified in 6 of 48 families
| Subject# | Gene | Alteration | Entrez SNP | Type | Group | Proband; family history |
|---|---|---|---|---|---|---|
| 5 | PMS2 | c.2404C>T R802X |
- | nonsense | 2 | Gliosarcoma brain tumor @ 9y; sibling with similar |
| PMS2 | c.1866G>T M622V |
- | missense | cancer in adolescence | ||
| 9 | SMARCB1 | c.233-1G>C | - | splice-site | 2 | Medulloblastoma @ 2y, atypical encapsulated neuroma of tongue @ 12y; sibling with astrocytoma @ 3y and ionizing radiation sensitivity |
| 23 | SMARCB1 | c.859G>T Q287X |
- | nonsense | 2 | Soft tissue rhabdoid @ 4m; sibling ATRT @ 1y; positive family history |
| 34 | TP53 | c.96+50del G |
- | frameshift | 1 | Osteosarcoma @ 14y, breast cancer @ 25y; no |
| FANCD2 | c.1367T>G L456R |
rs35782247 | missense homozygo us |
family history | ||
| 46 | TP53 | c.614A>G Y205C |
- | missense | 2 | Osteosarcoma @ 15y; sibling embryonal sarcoma liver @ 13y |
| 48 | TP53 | c.659A>G Y220C |
- | missense | 1 | Brainstem GBM @ 4y, choroid plexus papilloma @ 4y |
All mutations were heterozygous unless noted. Group = eligibility group.
GBM = glioblastoma multiforme.
Reference sequences utilized for mutation notation - PMS2 cDNA (NM_000535), protein (NP_000526); SMARCB1 cDNA (NM_003073), protein (NP_003064); TP53 cDNA (NM_000546), protein (NP_000537); FANCD2 cDNA (NM_033084), protein (NP_149075)
Deletion analysis
Deleterious mutations can also result from deletions or rearrangements which are not detected by Sanger sequencing. Therefore we analyzed the same DNA samples for copy number changes on a custom oligonucleotide array which densely covered all sequenced genes except PMS2 under study. Analysis of the array hybridization data revealed a large number of deletions and duplications buried within introns. However, there were no deletions confirmed by independent analysis of genomic DNA which encompassed coding exons or promoter regions in any TSG or DNA repair gene. MLPA analysis of PMS2 in 44 of the 48 samples also revealed no deletions or rearrangements.
SMARCB1 Mutation Analysis
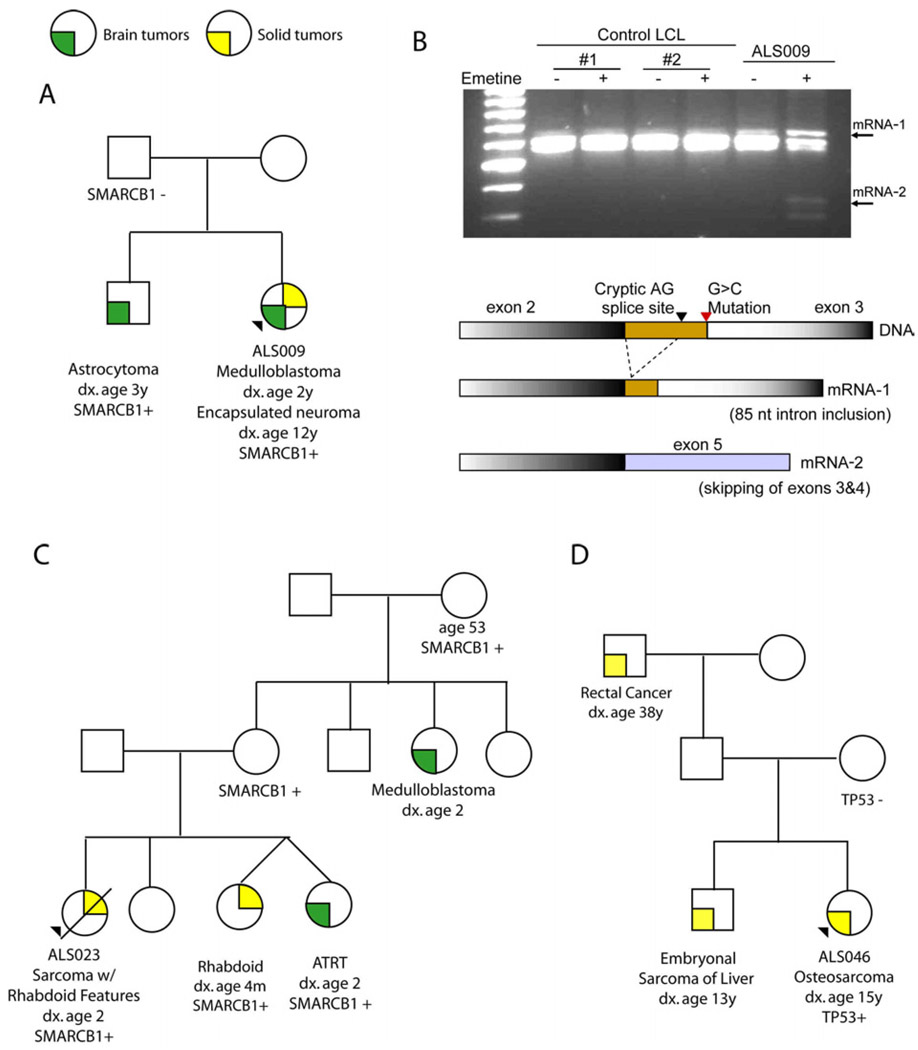
SMARCB1/SNF5/INI1 encodes a chromatin remodeling protein that functions as a tumor suppressor gene in patients with AT/RT or other rhabdoid tumors [3]. Proband ALS009 and the affected sibling are both heterozygous for a canonical splice site mutation in intron 2 (Figure 1A). RT-PCR of RNA from wild type and ALS009 LCLs demonstrated the expected splice product from exons 2 and 3 (Figure 1B). However, pretreatment with emetine to block nonsense mediated mRNA decay generated abnormal splice products only in ALS009 resulting from either activation of a cryptic splice site in intron 2 or exon skipping. Both abnormal messages contain a downstream nonsense allele resulting in truncation. This confirms that the splice site mutation found in subject ALS009 is deleterious.
Figure 1.
Family History and RNA Analysis for a Subset of Probands in the study (A) Pedigree for proband ALS009 and mutation status of the SMARCB1 splice site mutation (c.233-1G>C). (B) Reverse transcription – PCR (RT-PCR) analysis of RNA from lymphoblastoid cells from wild-type controls and ALS009 after growth in culture with and without treatment with emetine. RT-PCR performed with SMARCB1-F (5’-GACGACGGCGAGTTCTACAT-3’) in exon 2 and SMARCB1-R (5’-GGCGTCATCAACTTCTCATTCATG-3’) overlying junction of exons 5 and 6. Small arrowheads mark abnormal RNA species detected after treatment. Schematic diagram of sequence data obtained from the different mRNA species is provided. (C) Pedigree and SMARCB1 Q287X mutation status for the family of proband ALSO23. (D) Pedigree and TP53 Y205C mutation status for the family of proband ALSO46.
The pedigree for family ALS023 demonstrates an extensive history of tumor histopathologies typically associated with SMARCB1 mutations (Figure 1C). Previous sequence efforts had not detected the Q287X nonsense mutation identified here. However, we did confirm in a second laboratory that multiple affected members of this pedigree carry the Q287X mutation. Analysis of a rhabdoid tumor from the sibling of the proband revealed a “second hit” as frameshift mutation c.847-848delAT. The inheritance pattern is consistent with autosomal dominant with incomplete penetrance demonstrated by several unaffected adult mutation carriers. This pattern is consistent with the concept of an early childhood window of susceptibility to rhabdoid tumors [15].
PMS2 mutations
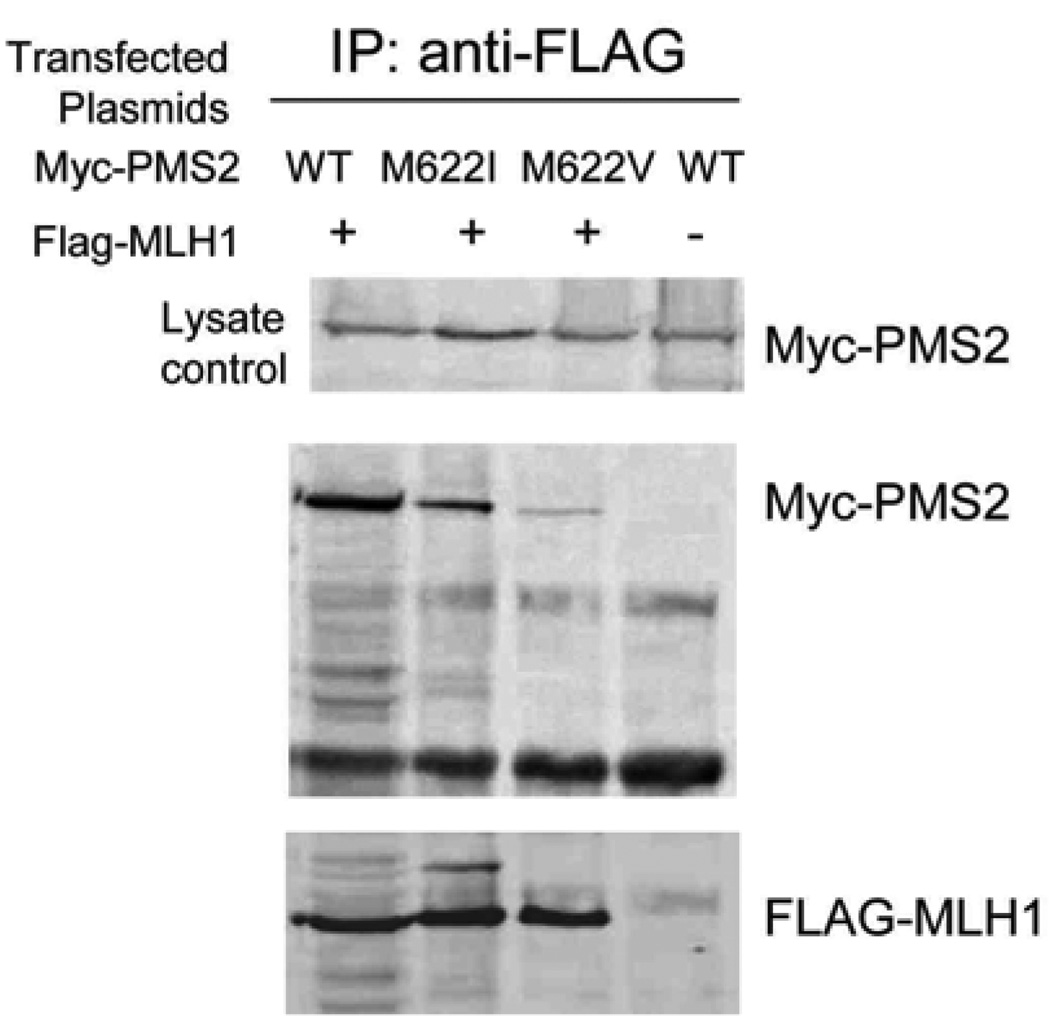
Subject ALS005 carries a nonsense mutation in PMS2 that has been described in other families with biallelic mutations, termed mismatch repair deficiency syndrome (MMR-D) [12]. The ALS005 clinical history, siblings with gliosarcomas, is also consistent with MMR-D. ALS005 also carries a novel M622V missense change. M622I is a rare polymorphism reported to diminish PMS2 protein function [16]. We tested the impact of M622V by co-expressing wild type, M622I or M622V alleles with its binding partner MLH1 in mammalian cells (Figure 2). The mutant and wild type PMS2 proteins are equally stable. Among replicate experiments, MLH1 binding to M622I protein was variably diminished. In contrast, binding of PMS2-M622V to MLH1 was consistently decreased. Thus, subject ALS005 carries one nonsense allele and a second PMS2 missense change likely to impact function.
Figure 2.
Analysis of the Impact of M622V PMS2 mutation on binding to MLH1. The results of co-transfection, immunoprecipitation and western blotting of HEK293 cells after transfection with flag epitope tagged - MLH1 with either wild-type or mutant myc epitope tagged - PMS2 constructs.
TP53 Mutations
Only one of three germline TP53 mutations identified here, encoding Y220C, has been reported as a germline mutation in the p53/Li Fraumeni syndrome database (http://www-p53.iarc.fr/Germline.html). Y220C was transmitted through an unaffected parent (Figure 1D). The c.96+50delG frameshift was de novo in the proband and although not previously described is expected to encode a truncated protein. Prior sequencing of this subject’s DNA had failed to identify this mutation. The Y205C mutation has been reported as a somatic mutation in a variety of cancer types and impacts an amino acid in the DNA binding domain.
Discussion
In this study, we analyzed germline DNA for mutations in a panel of cancer associated genes from families with patterns of childhood cancer suggestive of genetic susceptibility. The use of robotics and automated sequence calling resulted in a substantial decrease in sequencing costs compared to standard clinical laboratory methods. Truncating or pathogenic missense mutations were identified in six of 48 (13%) families which is consistent with the mutation yield for other clinical genetic tests and could be increased further by improved analysis of amplicons which failed in the robotic pipeline [17]. The in parallel approach (where all genes are sequenced for all subjects) was effective in rapidly identifying causative mutations even for patients that do not meet diagnostic criteria for a known cancer susceptibility syndrome.
Among the families with mutations there were varying pattern of inheritance including autosomal dominant inheritance with non-penetrant parents (SMARCB1, TP53), apparently autosomal recessive (biallelic PMS2 mutations) and de novo mutations (TP53). The mutations identified have clinical implications with regard to risk of second malignancies in the probands as well as cancer risk in family members.
All the identified mutations were in tumor suppressor or DNA repair genes. Although no activating mutations were found in this small set of oncogenes, expanding the analysis of oncogenes is needed as highlighted by activating mutations in the ALK gene underlying familial neuroblastoma [18].
SMARCB1
We were surprised to identify SMARCB1/INI1/SNF5 mutations in two families. SMARCB1 mutations were previously associated with renal and extra-renal rhabdoid tumors and central nervous system AT/RT [2;3]. There is an unexpected pattern of cancer in the family of ALS009. The mother may either be an unaffected carrier or one parent may demonstrate gonadal mosaicism for the mutation. The diagnosis of medulloblastoma in the proband might reflect an AT/RT given pathologic criteria at that time. Her brother was diagnosed with astrocytoma, a malignancy not associated with SMARCB1 mutations, however, this diagnosis was prior to the routine use of immunohistochemistry for SMARCB1 (utilizing the BAF47 antibody) to distinguish rhabdoid tumors from other brain tumors. Ten years after the medulloblastoma diagnosis the proband developed a benign encapsulated neuroma with pathologic features of a schwannoma. SMARCB1 mutations, including splice site mutations, result in familial schwannomatosis [19;20]. The novel splice site mutation identified in this family results in tumors overlapping both syndromes as reported in several other kindreds [4;21].
Oligogenic Inheritance
One advantage of an in parallel approach is identification of patients who carry clinically significant mutations in more than one gene. For example, ALS034 is heterozygous for a frameshift TP53 mutation and homozygous for FANCD2 L456R missense change originally reported as disease associated [22]. However, genotyping reveals that L456R is a frequent polymorphism in the African-American population; ALS034 is African-American without evidence of a Fanconi phenotype. Thus, L456R is likely a benign polymorphism, highlighting the need to continually update mutation annotation in the medical literature as large-scale sequencing of control populations is completed [23].
In addition to the brain tumors diagnosed in ALS009 and sibling, these individuals have clinically significant sensitivity to ionizing radiation not related to ATM mutations [24]. Sensitivity to IR is not a feature of patients with SMARCB1 mutations and it is likely that these siblings inherited both a SMARCB1 mutation and mutation(s) in a DNA repair gene not included in this study.
Results based on eligibility criteria
Table 3 describes the results as a function of the eligibility criteria demonstrating that 10% of Group 1 patients with multiple malignancies carried TP53 missense mutations as previously reported in other cohorts [25]. The cancer history (sarcoma in childhood and breast cancer as young adult) is typical for TP53 mutation carriers. Although initial analysis of TP53 prior to more extensive sequence analysis is indicated, it should be noted that many Group 1 subjects without detectable mutations had very similar cancer histories including two subjects with three distinct cancer diagnoses.
Table 3.
Mutation Detection Stratified by Eligibility Criteria
| Eligibility Criteria | Probands | Deleterious Mutations Identifieda |
Comments |
|---|---|---|---|
| Group 1 - Proband with childhood malignancy and a second malignancy diagnosed <age 30 |
20 | 2 | TP53x2 |
| Group 2 - Proband with childhood malignancy and at least one sibling with a diagnosis of cancer diagnosed <age 30 |
24 | 4 |
TP53, SMARCB1x2, PMS2x2, FANCD2 |
| Group 3 - Proband with childhood malignancy and major congenital anomaly AND/OR developmental delay |
10 | 0 |
The second column does not total to 48 because a number of families met more than one entry criteria. 3 probands met both Group 1 and 2; 4 probands met Group 1 and 3.
Four of 24 probands with childhood cancer and a sibling with cancer (Group 2) had mutations detected from a diverse set of genes and genetic mechanisms. Extensive studies of families based on siblings with childhood (or young adult) cancer have not been performed. These families may particularly benefit from the in parallel analysis of a large gene set. As above, subjects without detectable mutations had similar histories to the positive subjects.
Childhood cancer patients with congenital anomalies and/or developmental delay (Group 3) did not have mutations detected by sequencing this gene set. This phenotype is more likely to result from larger scale deletions or duplications and use of genome-wide high resolution array hybridization techniques is indicated.
In summary, we demonstrate that analysis of genes in parallel allows efficient identification of clinically important cancer susceptibility mutations in families with unique patterns of childhood cancer. As sequencing costs decrease, this approach should decrease turnaround time and improve diagnostic yield in clinical laboratories. Given that the history of cancer was similar in those patients with and without detectable mutations there are clearly a number of novel childhood cancer susceptibility genes which may be identified through the use of massively parallel sequencing methodologies.
Acknowledgments
We thank Weidong Jin, Ping Yang and Stephanie Gutierrez for management of clinical samples. The authors gratefully acknowledge the efforts of the families described here in support of research on the causes of childhood cancer. The authors acknowledge grant support from Alex’s Lemonade Stand Foundation to SEP, The Ann Parsons Endowment for Pediatric Genetics to LCS, grants NIHP01 CA34936 to LCS, 5R01CA138836 to SEP and NIHCA46274 to JAB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Plon SE, Nathanson K. Inherited susceptibility for pediatric cancer. Cancer J. 2005;11:255–267. doi: 10.1097/00130404-200507000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 3.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germline and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 4.Eaton K, Tooke LS, Wainwright LM, Judkins AR, Biegel JA. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer. 2010 doi: 10.1002/pbc.22831. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch B, Shimamura A, Moreau L, Baldinger S, Hag-alshiekh M, Bostrom B, et al. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004;103:2554–2559. doi: 10.1182/blood-2003-06-1970. [DOI] [PubMed] [Google Scholar]
- 6.Plon SE, Pirics ML, Nuchtern J, Hicks J, Russell H, Agrawal S, et al. Multiple tumors in a child with germ-line mutations in TP53 and PTEN. N Engl J Med. 2008;359:537–539. doi: 10.1056/NEJMc0800627. [DOI] [PubMed] [Google Scholar]
- 7.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Wheeler DA, Yakub I, Wei S, Sood R, Rowe W, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong LJ, Dimmock D, Geraghty MT, Quan R, Lichter-Konecki U, Wang J, et al. Utility of oligonucleotide array-based comparative genomic hybridization for detection of target gene deletions. Clin Chem. 2008;54:1141–1148. doi: 10.1373/clinchem.2008.103721. [DOI] [PubMed] [Google Scholar]
- 10.Shimodaira H, Yoshioka-Yamashita A, Kolodner RD, Wang JY. Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc Natl Acad Sci U S A. 2003;100:2420–2425. doi: 10.1073/pnas.0438031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gripp KW, Lin AE, Stabley DL, Nicholson L, Scott CI, Jr, Doyle D, et al. HRAS mutation analysis in Costello syndrome: Genotype and phenotype correlation. Am J Med Genet A. 2006;140:1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- 12.De Vos M, Hayward BE, Picton S, Sheridan E, Bonthron DT. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74:954–964. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picelli S, Zajac P, Zhou XL, Edler D, Lenander C, Dalen J, et al. Common variants in human CRC genes as low-risk alleles. Eur J Cancer. 2010;46:1041–1048. doi: 10.1016/j.ejca.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Liboutet M, Portela M, Delestaing G, Vilmer C, Dupin N, Gorin I, et al. MC1R and PTCH gene polymorphism in French patients with basal cell carcinomas. J Invest Dermatol. 2006;126:1510–1517. doi: 10.1038/sj.jid.5700263. [DOI] [PubMed] [Google Scholar]
- 15.Janson K, Nedzi LA, David O, Schorin M, Walsh JW, Bhattacharjee M, et al. Predisposition to atypical teratoid/rhabdoid tumor due to an inherited INI1 mutation. Pediatr Blood Cancer. 2006;47:279–284. doi: 10.1002/pbc.20622. [DOI] [PubMed] [Google Scholar]
- 16.Yuan ZQ, Gottlieb B, Beitel LK, Wong N, Gordon PH, Wang Q, et al. Polymorphisms and HNPCC: PMS2-MLH1 protein interactions diminished by single nucleotide polymorphisms. Hum Mutat. 2002;19:108–113. doi: 10.1002/humu.10040. [DOI] [PubMed] [Google Scholar]
- 17.American Society of Clinical Oncology policy statement update: genetic testing for cancer susceptibility. J Clin Oncol. 2003;21:2397–2406. doi: 10.1200/JCO.2003.03.189. [DOI] [PubMed] [Google Scholar]
- 18.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007;80:805–810. doi: 10.1086/513207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyd C, Smith MJ, Kluwe L, Balogh A, Maccollin M, Plotkin SR. Alterations in the SMARCB1 (INI1) tumor suppressor gene in familial schwannomatosis. Clin Genet. 2008;74:358–366. doi: 10.1111/j.1399-0004.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 21.Swensen JJ, Keyser J, Coffin CM, Biegel JA, Viskochil DH, Williams MS. Familial occurrence of schwannomas and malignant rhabdoid tumour associated with a duplication in SMARCB1. J Med Genet. 2009;46:68–72. doi: 10.1136/jmg.2008.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalb R, Neveling K, Hoehn H, Schneider H, Linka Y, Batish SD, et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am J Hum Genet. 2007;80:895–910. doi: 10.1086/517616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuehn BM. 1000 Genomes Project promises closer look at variation in human genome. JAMA. 2008;300:2715. doi: 10.1001/jama.2008.823. [DOI] [PubMed] [Google Scholar]
- 24.Alsbeih G, Story MD, Maor MH, Geara FB, Brock WA. Chromosomal fragility syndrome and family history of radiosensitivity as indicators for radiotherapy dose modification. Radiother Oncol. 2003;66:341–344. doi: 10.1016/s0167-8140(02)00327-4. [DOI] [PubMed] [Google Scholar]
- 25.Malkin D, Jolly KW, Barbier N, Look AT, Friend SH, Gebhardt MC, et al. Germline mutations of the p53 tumor-suppressor gene in children and young adults with second malignant neoplasms. N Engl J Med. 1992;326:1309–1315. doi: 10.1056/NEJM199205143262002. [DOI] [PubMed] [Google Scholar]