The transcription factors NGN3 and NEUROD1 regulate the expression of INSM2 gene in pancreatic islet cells.
Abstract
The insulinoma-associated 2 (Insm2) gene is a member of the Snail/Gfi1/Insm1 transcriptional repressor superfamily. However, little is known about how the expression of human INSM2 or mouse Insm2 in neuroendocrine tissues is regulated. Here we report the expression of INSM2/Insm2 in human fetal pancreas and mouse embryos, as well as adult pancreatic islets, and its regulation by two major islet transcription factors. Mutagenesis and chromatin immunoprecipitation analysis demonstrated that the proximal E-boxes of the mouse Insm2 promoter are direct targets of neurogenin 3 and neurogenic differentiation 1 (NeuroD1). Furthermore, we found that endogenous Insm2 expression was activated in Ngn3/NeuroD1-transduced pancreatic epithelial duct cells. Our results suggest that Insm2 plays an important role in the differentiation cascade of Ngn3/NeuroD1 signaling in pancreatic islets.
The pancreatic islets regulate glucose metabolism through secretion of islet hormones such as insulin and glucagon. Factors that regulate islet cell differentiation are currently under intense investigation. In this study we show that the insulinoma-associated 2 gene, Insm2, also known as IA-6 (GenBank accession no. NM_032594) or Mlt1 (1), is closely associated with pancreatic endocrine cell differentiation.
The basic helix-loop-helix transcription factors, neurogenin 3 (Neurog3/Ngn3) and neurogenic differentiation 1 (NeuroD1), play central roles in the formation and function of the islets. Ngn3 is a proendocrine marker essential for islet cell development (2). NeuroD1, after being directly activated by Ngn3 (3), continues the endocrine differentiation process and participates in the maintenance of the differentiated phenotype of mature islet cells. Although loss-of-function analyses in mice have revealed important differences in the roles of these two factors during endocrine differentiation, the molecular base for the divergent aspects of Ngn3 and NeuroD1 functions in the islets is not completely understood. Therefore, we looked for novel targets in the downstream activation cascades elicited by Ngn3 and/or NeuroD1. Previously we isolated the Insm1 (IA-1) gene from an insulinoma library (4) and found that Insm1 is mainly expressed in neuroendocrine cells of approximately embryonic day (E) 10.5–13.5 mice embryos (5). Recent studies have demonstrated that Insm1 is a direct target of Ngn3 and NeuroD1 and a critical regulator of pancreatic β-cell development (6–9).
In the present study, we report that Insm2 is abundantly expressed in human fetal pancreatic islets and duodenal cells as well as mouse adult endocrine cells of pancreas and adrenals. Insm2 is expressed in various developmental stages with peak levels in mouse embryonic from E11.5 to E13.5 or in human fetal pancreas from 8 to 20 wk, an important period for the development of pancreatic endocrine cells. To search for the regulatory elements required for specific expression in pancreatic islets, we analyzed the 5′-upstream region of Insm2 using transient transfection experiments as well as chromatin immunoprecipitation (ChIP) analysis and demonstrated that Ngn3 and NeuroD1 are the upstream regulators for Insm2 gene activation.
Materials and Methods
Gene cloning, PCR, and mice
The human insulinoma-associated (INSM)-1 sequence (GenBank accession no. NM_002196) was used for blast searching of database expressed sequence tag (EST) sequences. A 2163-bp EST (GenBank accession no. AA046477) was obtained (Research Genetics, Huntsville, AL) and sequenced. To isolate the full-length cDNA, the primers (forward, 5′-CCATCCTAATACGACTCACTATAGGGC-3′; reverse, 5′-ACAGCTGAACACCTTGTCGCACTC-3′) were used to amplify the Marathon-Ready human brain cDNA library (BD CLONTECH, Palo Alto, CA). A 1.1-kb PCR product, overlapping the 5′ region of the EST, was confirmed by sequencing. A 3027-bp cDNA containing a full-length coding frame was identified and designated human INSM2. PCR analysis was performed using RNAs isolated from mouse tissues or cells (QIAGEN RNeasy minikit; Valencia, CA). First-strand synthesis was carried out using the RT-PCR kit (Stratagene, La Jolla, CA) with 10 μg of isolated RNAs that were treated with deoxyribonuclease I to remove the background genomic DNA. Semiquantitative RT-PCR for comparison of mRNAs in the above cells was conducted as previously described (10). Mice were housed under a 12-h light, 12-h dark schedule and maintained under specific-pathogen-free conditions. Wild-type C57BL mice used for the Insm2 project were approved by our Institutional Animal Care and Use Committee (approval no. 09-516).
Northern blot analysis
Northern blots (MTN) of adult mouse multiple tissues (BD CLONTECH) or mouse embryos or brains (Seegene, Inc., Seoul, Korea) were probed with the [32P]-labeled full-length mouse Insm2 cDNA as previously used (5). Northern blots (MTN) of adult human multiple tissues (BD CLONTECH) was probed with [32P]-labeled full-length human INSM2 cDNA. Approximately 2 μg of purified poly A+ RNA per lane on the MTN blots was adjusted for a consistent signal of a housekeeping gene across all lanes (BD CLONTECH).
Antisera, Western blots, and immunohistochemistry (IHC) analysis
INSM2 rabbit antibody against amino acids (a.a.) 5-SRQVLLLQMPLRPGC-3 (e.g. a.a. 553–566 of human INSM2 that share 100% identity with mouse INSM2) was generated and used at 1:1000 in Western blots and at 1:100 in IHC analysis. The antibody was purified with an AminoLink kit (Pierce, Rockford, IL) and stored as aliquots at −80 C. The specificity of the anti-INSM2 antibody was confirmed by blocking of the immunoreactive signal with specific peptide. Western blots were performed using anti-INSM2 antisera and GE Amersham ECL Plus Western blotting detection reagents (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). Antiglyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (clone 71.1; Sigma, St. Louis, MO) was used at 50 ng/ml in Western blots for protein loading controls. In IHC studies, insulin and glucagon mice antibodies (Sigma) were used at 1:500. Images were taken with a Carl Zeiss Microscopy and Axiovision software system (Thornwood, NY).
In situ hybridization
The Allan Brain Atlas (http://www.brain-map.org) is a publicly accessible online gene expression digital library widely cited in numerous papers (11). In situ hybridization data for expression of Insm2 and NeuroD1 in adults were extracted from the Allan Brain Atlas. Ngn3 is not expressed in adult mouse brain. The Insm2 probe used for the hybridization is a 477-bp fragment from nt 1085–1561 (GenBank accession no. NM_020287.2); the NeuroD1 probe is 920 bp from nt 881-1800 (GenBank accession no. AK018781.1).
Human pancreatic tissue and immunofluorescent analysis
Human fetal pancreatic and duodenal tissue (8–21 wk fetal age) was collected according to protocols approved by the Health Sciences Research Ethics Board at the University of Western Ontario, in accordance with guidelines of the Canadian Council on Health Sciences Research Involving Human Subjects. Tissues were immediately processed for immunohistology with a minimum of three pancreases per age group (12). Pancreatic and duodenal tissues were fixed in 4% (vol/vol) paraformaldehyde. Sections (5 μm) were cut and stained with appropriate dilutions of primary antibodies: rat antihuman somatostatin (SST) (Abcam, Cambridge, MA), mouse antimouse NGN3 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), and mouse antihuman NEUROD1 (Santa Cruz Biotechnology, Santa Cruz, CA) were used 1:100. Fluorescein isothiocyanate- and Texas Red-labeled secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Images were recorded by a Leica DMIRE2 fluorescence microscope (Leica, Richmond Hill, Ontario, Canada) with Openlab image software (Improvision, Lexington, MA) (12).
Quantitative real-time PCR
RNAs were isolated from fetal (E13.5 and E15.5) and adult pancreas using the Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA was treated with deoxyribonuclease I (Promega, Madison, WI), extracted with phenol/chloroform, and precipitated using isopropanol. A total of 2 μg of the treated RNA was used for reverse transcription using a cDNA synthesis kit (Invitrogen) according to the manufacturer's instruction. For real-time PCR, 1 ml of the reverse-transcribed cDNA obtained from the above procedure was used in a 25-μl reaction mixture including SYBR Green PCR master mix (Bio-Rad, Hercules, CA) and 1 μl of 10 μm primers for the mouse Insm2 gene, Ngn3, NeuorD1, and Gapdh. The cycling conditions were held at 95 C for 5 min, followed by 30–45 cycles at 94 C for 30 sec, 60 C for 30 sec, and 72 C for 30 sec. The fluorescent signals were analyzed using an ICycler IQ multicolor real-time detection system (Bio-Rad). The PCR products were separated on 2% agarose gel. The PCR primers included Ngn3 (5′-AAGAGCGAGTTGGCACTGAGCAAG-3′; 5′-GCTGTGGTCCGCTATGCGCAG-3′), NeuroD1 (5′-CTTGGCCAAGAACTACATCTGG-3′; 5′-GGAGTAGGGATGCACCGGGAA-3′), Insm2 (5′-TCCCTCCTCTGACCTCCCGA-3′; 5′-ATCAGGAGTGGGAGCGGGTC-3′), and Gapdh (5′-GGTGAAGGTCGGTGTGAACG-3′; 5′-CTCGCTCCTGGAAGATGGTG-3′).
Promoter-reporter assays
A dual-luciferase reporter system was used for promoter activity measurement according to the manufacturer's instruction (Promega). A serial length of mouse Insm2 promoter fragment (−2500 to +49) was cloned in the pGL3 vector driving expression of the firefly luciferase reporter gene (8). Approximately 150-bp DNA fragments of Insm2 promoter, with or without E-box mutations, were synthesized (Genscript, Piscataway, NJ) and then subcloned into the pGL3 vector. Renilla luciferase was used as an internal control for transfection efficiency. Normalized luciferase data (firefly/renilla) was compared with the empty pGL3-Basic vector (t test). Full-length coding sequence of the mouse Ngn3 in pcDNA3.1 vector (8) or hamster NeuroD1 cDNA in pCR3.1-β2 vector (kindly provided by Dr. M. J. Tsai, Baylor College of Medicine, Houston, TX) was used in our experiments.
ChIP assay
The 2 × 106 MIN6 cells were used per immunoprecipitation assay (13) with modifications. Briefly, nuclei were collected from formaldehyde-fixed cells by homogenization. DNAs were sonicated into 200-1000 bp. Precleared chromatin was immunoprecipitated with 10 μg of NGN3 monoclonal antibody (Developmental Studies Hybridoma Bank) or 10 μg of NEUROD1 antibody G20 (Santa Cruz Biotechnology). Immunocomplexes were collected into 50 μl of protein A/G-Sepharose beads and purified with Qiaquick (QIAGEN) PCR purification columns. Immunoprecipitated DNAs were analyzed by PCR with primers encompassing the Insm2 E1-E2 boxes (TCCTGAGCTCCAGAGGTTTG; AACTGCAGGGCAGCTAAATC), the Insm2 E3-E4 boxes (GGTTCTTAACTACAACAACG; GTGAGAAATGCTGGTGGAATCG3), the Insm1 E3 box (TTTGGGGCCATTCTTCTCCTT; ACGTGCCTGGGCTTTGTGTCT), and the Brca1 (GAGTCCTAGCCCTTTCACCCATAC; GTGATGTTCCTGAGATGCCTTTG).
Cell culture, adenoviral transduction, and DNA transfection
Cell lines (mPANC, MIN6, PC12, and HeLa) were cultured in RPMI 1640 or DMEM supplemented with 10% fetal calf serum as described previously (14–16). For adenoviral infection, cells in a six-well plate were incubated with adeno-Ngn3 or adeno-NeuroD1 at 40 multiplicity of infection for 2 h and then cultured for 48 h after the medium was replaced (17). Cells were transfected with appropriate plasmids such as the CMV-Ngn3 in pcDNA and the CMV-NeuroD1 in PCR3.1 using Lipofectamine 2000 (Invitrogen), which reached approximately 50% of average efficiencies at 48 h. The total amount of plasmid was kept constant between transfections. Cells were lysed 48 h after transfection and processed for analysis.
Results
INSM2 gene analysis
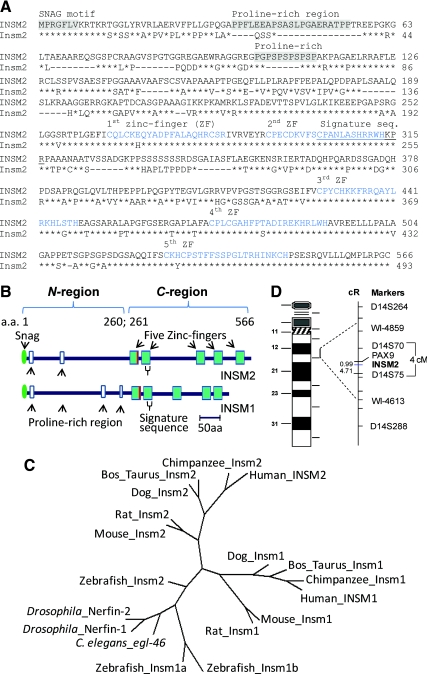
The full-length human INSM2 cDNA contains 3024 bp with an open reading frame of 1701 bp, encoding 566 a.a. with a molecular mass of 60 kDa. The N′-INSM2 (a.a. 1–260) contains a Snail/Gfi-1 motif and two proline-rich regions. The C'-INSM2 (a.a. 261–566) contains five C2H2 zinc-finger motifs. INSM2 shows 51% identity to the INSM1 (Fig. 1, A and B). BLAST analysis identified a signature sequence, e.g. the 15 residues CPANLASHRR(K)WHKPR(K) are exclusively present in all members of the Insm1/Insm2 family. Furthermore, phylogenetic analysis (Fig. 1C) revealed that Insm2 and Insm1 existed in all species we examined from Caenorhabditis elegans to human. The INSM2 gene was mapped to the chromosome 14q13.2 (Fig. 1D).
Fig. 1.
INSM2 analysis. A, Comparison of the human (upper lines) and mouse INSM2 (lower lines) shows their similarities in Snail/Gfi-1 (SNAG) motif (a.a. 1–7), two proline-rich regions (a.a. 35–55 and 102–115), and five zinc-finger motifs (a.a. 261–566). The signature sequence (a.a. 302–316) of the family is underlined. B, Schematic comparison of INSM2 structure with INSM1. The last residue of histidine of the first zinc-finger is replaced by arginine (in red). C, The phylogenetic tree was generated from multiple amino acid alignments (Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org). D, The integrated chromosome map shows the INSM2 location between D14S70 and D14S75.
Insm2 transcripts in embryo and postnatal tissues
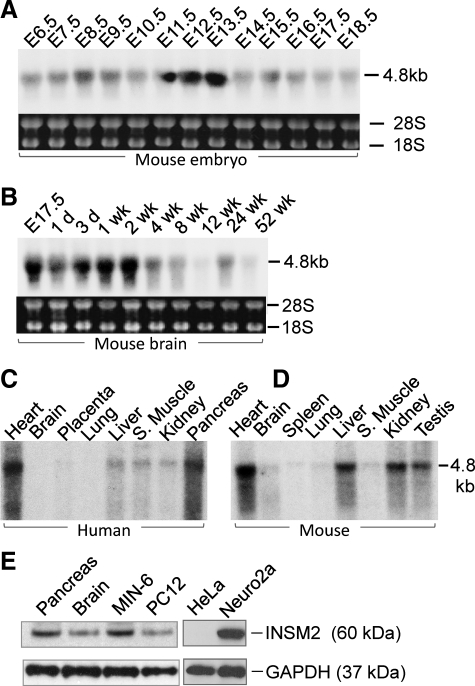
Northern blots with mouse Insm2 cDNA detected a 4.8-kb transcript across all E6.5-E18.5 embryos. Notably, the Insm2 levels were transiently enhanced up to 4- to 5-fold from E11.5 to E13.5 compared with the modest levels at other time points (Fig. 2A). In the developing mouse brain, Insm2 mRNA peaked 2 wk postnatally and gradually decreased thereafter but was still detectable at 52 wk (Fig. 2B).
Fig. 2.
Insm2 in developmental and adult tissues. A, Insm2 mRNA is shown in mouse embryonic development from E6.5 to E18.5. B, The Insm2 mRNA level in the mouse brain is gradually decreased after 4 wk postnatally. mRNAs are equally loaded as indicated by 18S and 28S. C, INSM2 transcript is shown in human adult tissues. D, Insm2 transcript is shown in adult mouse tissues. E, Western blots with INSM2 antibody detect a single band at 60 kDa in mouse pancreas and brain tissues as well as in mouse insulinoma MIN6 and rat pheochromocytoma PC12 cell lines. INSM2 is strongly expressed in mouse Neuro2a (N2a) cells, but not in the HeLa cells. GAPDH served as a loading control across all the lanes.
Insm2 is widely expressed in adult tissues
Northern blot with the human INSM2 showed a 4.8-kb transcript abundantly expressed in heart, liver, skeletal muscle, kidney, and pancreas (Fig. 2C). Northern blot with mouse Insm2 showed a stronger expression in liver and kidney in addition to a strong expression in testis (Fig. 2D). The band size of Insm2 or INSM2 transcript in Northern blots are about 1.8 kb larger than the cloned cDNA, suggesting additional unidentified RNAs located at the 5′- and/or 3′-untranslated regions. INSM2 or Insm2 is expressed to a lesser extent in brain, lung, and spleen tissues. Western blots with the INSM2 antibody detected a single approximately 60-kDa band in mouse pancreas, brain, mouse islet MIN6 cells, rat adrenal pheochromocytoma PC12 cells, and mouse neuroblastoma N2a cells but not in the human epithelial cervical cancer HeLa cells (Fig. 2E).
INSM2 in adult islets and neuroendocrine tissues
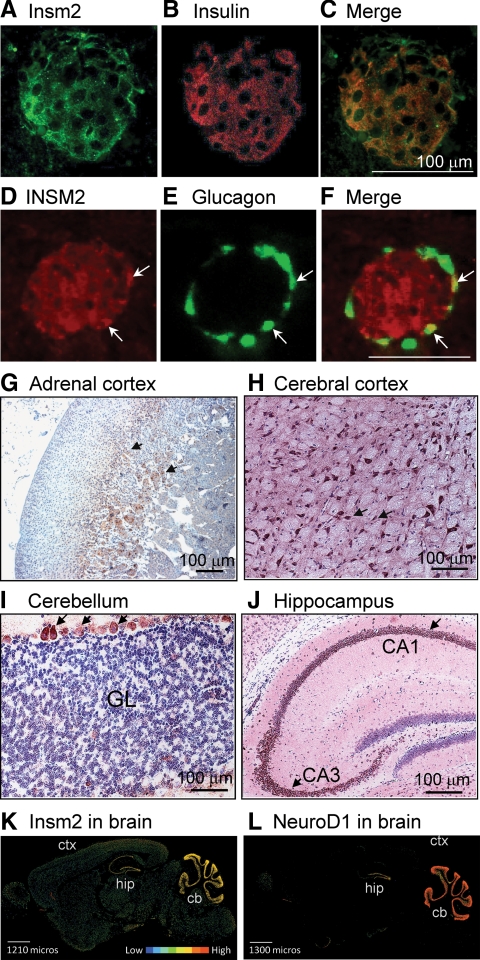
By immunofluorescent stainings with INSM2 antibody, we found that INSM2 was expressed in mouse and human pancreatic islet cells but not in acinar cells. INSM2 was colocalized with insulin in β-cells (Fig. 3, A–C) and glucagon in α-cells (Fig. 3, D–F). INSM2 also was detected in adrenal glands, particularly in the deeper layer of the cortex (Fig. 3G). Furthermore, Insm2 was found in the neuronal cells of the cerebral cortex (Fig. 3H), the Purkinje cells of the cerebellum (Fig. 3I), and the hippocampal region including CA1 and CA3 (Fig. 3J). It appears that Insm2 is present in the cytoplasmic region and, to a less extent, in the nuclei. Insm1 in medaka, and other transcription factors, also was found in the cytoplasm (18). The distribution of INSM2 protein in brain (Fig. 3K) was consistent with its mRNA as shown in the in situ hybridization study of the Allan Brain Atlas, a publicly accessible online gene expression digital library. Comparative analysis of Insm2 to other images in the Allen Brain Atlas revealed that the expression pattern of Insm2 is nearly identical to that of NeuroD1 (Fig. 3L). Ngn3 expression was not found in adult mouse brain tissues (image not shown).
Fig. 3.
INSM2 in neuroendocrine tissues. A–C, INSM2 was expressed in mouse pancreatic islet β-cells and colocalized with insulin. D–F, INSM2 also was detected in human islet α-cells because it was colocalized with glucagon (arrows). G, By histochemistry stainings, INSM2 was found in the deeper layers of the adrenal glands (arrows). INSM2 also was expressed in neuronal cells of the cerebral cortex (H), Purkinje cells of the cerebellum [granule layer (GL)], (I), and the hippocampus (J). K and L, Representative images of in situ hybridization in the adult mouse brain (sagittal). mRNAs of Insm2 (K) and NeuroD1 (L) are detected with a similar pattern in cerebellum (cb), hippocampal region (hip), and to a much lesser extent in cerebral cortex (ctx). Expression intensities are shown in color bars.
INSM2 is coexpressed with NGN3 and NEUROD1 in the developing pancreas
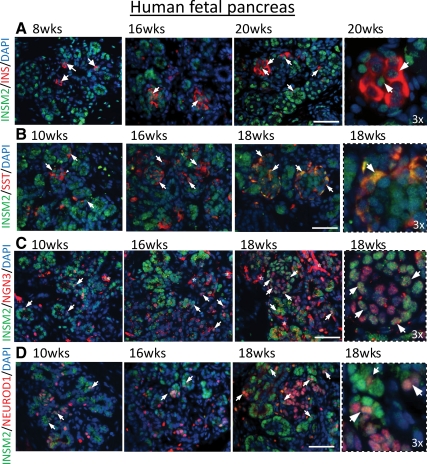
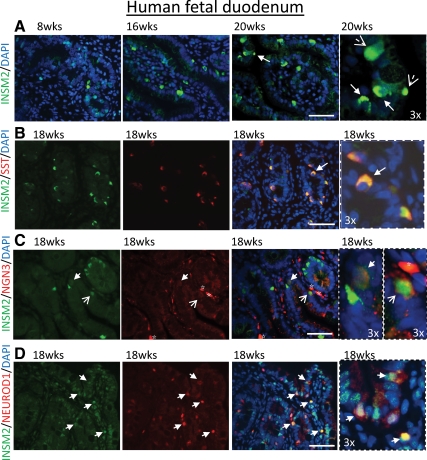
To see whether INSM2 was expressed in the endocrine cells and coexpressed with the same cells with NGN3 and NEUROD1, we examined human fetal pancreas and duodenum tissues using anti-INSM2 and endocrine cell-specific antibodies such as insulin and SST markers. We observed that INSM2 was specifically expressed in islet-like cells of pancreas (Fig. 4, A and B, in green) and epithelia cells of duodenum (Fig. 5A, in green) from 8 to 20 wk of fetal age, colocalizing with SST-positive D cells in pancreas (Fig. 4B, in yellow) and duodenum (Fig. 5B, in yellow) as well as coexpressing with insulin in islet β-cells (Fig. 4A, indicated by arrows). Furthermore, double immunostaining for INSM2 with NGN3 (Figs. 4C and 5C) or NEROD1 (Figs. 4D and 5D) showed that INSM2 expression is correlated with the expression of NGN3 and NEUROD1 in the developing human pancreas and duodenum.
Fig. 4.
INSM2 in human fetal pancreas. Expression of INSM2 (green) is present in the nuclei of most INS+ islet β-cells (red) (A). However, INSM2 is strongly colocalized with SST (red) in the cytoplasm of all D cells (yellow in B) of developing human pancreas. INSM2 is coexpressed with NGN3 (C) or NEUROD1 (D). Arrows indicate the INSM2+ costained cells. Images on fourth column are the amplification (×3) of the regions as indicated by arrows on the third column. Nuclei were stained by 4′,6′-diamino-2-phenylindole (DAPI; blue). Scale bar, 50 μm.
Fig. 5.
INSM2 in human fetal duodenum. A, Expression of INSM2 (green) in developing human duodenum (8–20 wk). B, INSM2 is colocalized with SST (red)-positive cells. INSM2 is coexpressed with NGN3 (C) or NEUROD1 (D) as indicated by arrows. Arrows (B–D) indicate INSM2+-costained cells. Images on fourth column are the amplification (×3) of the regions as indicated by the arrows on the third column. Nuclei were stained by 4′,6′-diamino-2-phenylindole (DAPI; blue). Scale bar: 50 μm.
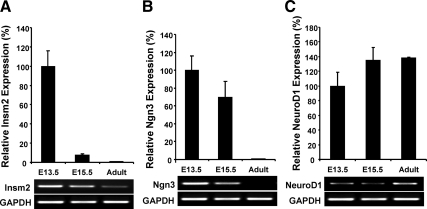
Quantitative real-time PCR showed that Insm2, Ngn3, and NeuroD1 transcripts also were expressed at the same time (E13.5) in mouse pancreas. By E15.5, both Insm2 and Ngn3 were markedly decreased, but Insm2 was still detectable at adult pancreas (Fig. 6, A and B). On the other hand, NeuroD1 expression was increased at E15.5 (∼35%, P < 0.05), and remained at the level in adult pancreas (Fig. 6C).
Fig. 6.
Quantitative analysis of Ngn3, NeuroD1, and Insm2 transcripts in fetal and adult mouse pancreas by real-time PCR. Gapdh was used as an internal control. The expression level at E13.5 was defined as 100%, and the level of each gene at E15.5 and in adult pancreas was calculated as the percentage of E13.5. A representative PCR (A, Insm2; B, Ngn3; C, NeuroD1) is shown below each graph.
Ngn3 and NeuroD1 regulate Insm2 promoter activity
To search for regulatory elements required for specific expression of Insm2 in the islets, we analyzed the sequence of the 5′-flanking region of Insm2. Ten putative E-boxes (CANNTG) were identified in the 2.5-kb region of the mouse or human Insm2 promoter (Fig. 7A). For consistence in our future studies in determining the biological function of Insm2 in an animal model, we chose the mouse Insm2 promoter in this study. Based on our earlier studies with INSM1 (6, 8), we then examined the effect of Ngn3 and NeuroD1 on the Insm2 promoter by using a series of deletion constructs with luciferase reporter (Fig. 7A).
Fig. 7.
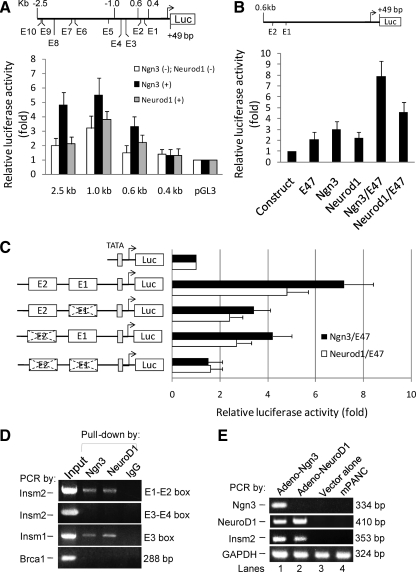
Activation of Insm2 promoter by Ngn3 and NeuroD1. A, Schematic representation of the E-boxes within the proximal −2.5-kb region of Insm2 promoter-driven luciferase (Luc) reporter. Luciferase reporter-gene assays were performed with various lengths of the Insm2 promoter-Luc constructs. HeLa cells were transiently cotransfected with 0.5 μg of each plasmid DNA in the absence or presence of CMV-Ngn3 or CMV-NeuroD1 for 48 h. B, Schematic diagram of two proximal E-boxes in the 0.6 kb of Insm2 promoter-Luc reporter. To test the effects of Ngn3/E47- or NeuroD1/E47-mediated activation on the reporter, 0.5 μg of the plasmid DNA was transfected alone (first lane) or cotransfected with E47, Ngn3, NeuroD1, Ngn3/E47, and NeuroD1/E47 (lane 2–6, respectively) into HeLa cells for 48 h. C, Constructs containing the two proximal E-boxes, either wild type or mutated (×), of the Insm2 promoter and the TATA-Luc reporter were established as shown. Then 0.5 μg of each reporter plasmid DNA was cotransfected into HeLa cells with CMV-Ngn3/E47 or CMV-NeuroD1/E47 reagents. The effects are shown as fold activation over basal values of vectors or constructs (means ± se). Transfection was carried out in triplicates and repeated three times. D, Ngn3 and NeuroD1 bind the Insm2 promoter. ChIP with Ngn3 or NeuroD1 antibodies or IgG was performed on chromatin isolated from MIN6 cells. Input DNA was serially diluted and used as a positive control for PCR analysis. The Insm2 promoter is coimmunoprecipitated using the Ngn3 or the NeuroD1 antibodies but not by the preimmune IgG. The positive control Insm1, but not the negative control Brca1, was specifically coprecipitated by the Ngn3 or NeuroD1 antibodies. E, Ngn3 and NeuroD1 induced endogenous Insm2 expression. Expression of Insm2 was induced in adeno-Ngn3- or adeno-NeuroD1-transduced mPANC cells for 48 h (lanes 1–2), but not detectable from the adenovector-infected cells (lane 3) or parental mPANC cells (lane 4) as determined by PCR. NeuroD1 was also induced in adeno-Ngn3-transduced mPANC cells (second panel, lane 1). GAPDH was the template control (lanes 1–4). Representative images from three separate experiments are shown.
The CMV-Ngn3 or CMV-NeuroD1 and Insm2 promoter reporter constructs were cotransfected in HeLa cells, which did not show endogenous Insm2 expression. We consistently found that, among the five constructs tested, the 1.0-kb promoter reporter (containing four E-boxes: E1, −477 to −482; E2, −550 to −555; E3, −836 to −841; E4, −925 to −930) had the highest level of activation by Ngn3 (means ± se, 5.5 ± 1.2, P < 0.01) or NeuroD1 (3.8 ± 0.6, P < 0.01) in HeLa cells; the 0.6-kb promoter reporter (containing the most proximal E1-E2 boxes) could still be stimulated by the Ngn3 (3.3 ± 0.7, P < 0.05) or NeuroD1 (2.2 ± 0.5, P < 0.05) to a modest level (Fig. 7A). However, the 0.4-kb promoter reporter, which eliminated the E1-E2 boxes, resulted in the loss of Ngn3- or NeuroD1-mediated activation (Fig. 7A). We thus identified a small region less than 200 bp (−400 to −600) in the Insm2 promoter that was active for the Ngn3 or NeuroD1 activity.
To verify whether the cofactor E47 has a synergetic role with Ngn3 or NeuroD1 for the proximal Insm2 promoter, we cotransfected HeLa cells with the 0.6-kb Insm2 luciferase reporter and the CMV-Ngn3/E47 or the CMV-NeuroD1/E47 (Fig. 7B). We found that the luciferase activity increased 7.9-fold in the coexpression of Ngn3/E47 (7.9 ± 1.4, P < 0.01) or 4.6-fold in the coexpression of NeuroD1/E47 (4.6 ± 0.9, P < 0.01) as a heterodimer compared with a 2.1-fold (2.1 ± 0.6, P < 0.05) and a 3.0-fold (3.0 ± 0.7, P < 0.05) or a 2.2-fold (2.2 ± 0.5, P < 0.05) increase in the E47, Ngn3, or NeuroD1 alone, respectively (Fig. 7B). Thus, NeuroD1/E47 was found capable of increasing luciferase activity but to a lesser extent than Ngn3/E47.
To further evaluate whether the E1 and/or E2 boxes were required for the activity of Ngn3 or NeuroD1, we synthesized and cloned the 150-bp promoter region (−438 to −587) covering E1-E2 boxes in either a wild-type (CANNTG) or a mutated (TANNCT) form into a TATA-luciferase reporter (Fig. 7C). Wild-type and mutated constructs were cotransfected into HeLa cells with Ngn3/E47 or NeuroD1/E47 expression vectors. In the wild-type constructs, the luciferase activities of the Ngn3/E47- or the NeuroD1/E47-treated cells are 7.2-fold (7.2 ± 1.2, P < 0.01) or 4.8-fold (4.8 ± 0.9, P < 0.01), respectively, compared with that of the vector controls (Fig. 7C). Mutation of the E1 or E2 box was reduced by 53% (3.4 ± 0.7 vs. 7.2 ± 1.2, P < 0.01) and 42% (4.2 ± 0.8 vs. 7.2 ± 1.2, P < 0.01), respectively, in the Ngn3/E47-mediated activity and by 50% (2.4 ± 0.5 vs. 4.8 ± 0.9, P < 0.01) and 43% (2.7 ± 0.6 vs. 4.8 ± 0.9, P < 0.01), respectively, in the NeuroD1/E47-mediated activity compared with the reductions in their wild-type controls (Fig. 7C). Mutations of both E1 and E2 boxes decreased Ngn3/E47-mediated or NeuroD1/E47-mediated activation to a level close to that of the control vector (TATA-luciferase) (Fig. 7C).
Endogenous Ngn3 and NeuroD1 bind Insm2 gene promoter (ChIP)
To establish whether Ngn3 and NeuroD1 directly bound to the Insm2 promoter in vivo, a ChIP assay was conducted in the insulinoma MIN6 cells that express Ngn3, NeuroD1, Insm1, and Insm2. After immunoprecipitation with either Ngn3 or NeuroD1 antibodies, precipitated DNAs were amplified by specific primers for the E1-E2 or E3-E4 box-containing region of Insm2, the proximal E box of Insm1, and the Brca1 promoter. Only the E1-E2 box-containing region in the proximal Insm2 promoter was efficiently precipitated and amplified by either Ngn3 or NeuroD1 antibody (Fig. 7D) or the positive control E3 box of Insm1 but not the E3-E4 box-containing region or the negative control Brca1. Taken together, these results show that Insm2 is a novel direct target of Ngn3 and NeuroD1.
Ngn3 and NeuroD1 activate endogenous Insm2 expression
Because both Ngn3 and NeuroD1 transcription factors are capable of binding to the Insm2 E-boxes, we further investigated whether Ngn3 or NeuroD1 expression is capable of inducing endogenous Insm2 in vivo. Adeno-Ngn3 or adeno-NeuroD1 was used to infect mouse pancreatic epithelial duct cells (mPANCs) for 48 h. mRNAs were then extracted from the infected cells for semiquantitative RT-PCR analysis with Ngn3-, NeuroD1-, and Insm2-specific primers (Fig. 7E). Transcripts of Ngn3 (Fig. 7E, lane 1) or NeuroD1 (Fig. 7E, lane 2) were successfully detected. Furthermore, we found that expression of adeno-Ngn3 or adeno-NeuroD1, but not the adeno-vector in mPANC cells, led to the induction of Insm2 mRNA (Fig. 7E, lanes 1–3). NeuroD1 also was activated in the infected mPANC cells by adeno-Ngn3 expression (Fig. 7E, lane 1). The parental mPANC cells did not show any detectable endogenous expression of Insm2 (Fig. 7E, lane 4). GAPDH was used as a positive control (Fig. 7E, lanes 1–4). These results suggest that Ngn3 and NeuroD1 regulate the Insm2 gene expression.
Discussion
The Snail/Gfi1/Insm1 transcriptional repressor superfamily plays an important role in the development and various diseases (19). The superfamily consists of three subfamilies of the C2H2 zinc-finger transcription factors: the Snail, the Gfi1, and the Insm1. Insm2 shares high similarities in its amino acid sequence with Insm1. However, Insm2 displays several different properties from Insm1. First, in contrast to Insm1, which has restricted expression patterns in embryonic neuroendocrine tissues, Insm2 has a much broader spatial-temporal expression pattern, suggesting that Insm2 could be regulated by additional transcriptional factors to fulfill physiological requirements in other tissues. Further studies using Insm2-deficient animals are under way to determine the physiological functions. Second, human INSM1 is a biomarker for neuroendocrine differentiation because it is strongly expressed in all small cells of lung cancers (20) but not in normal lung tissues, whereas the INSM2 homolog mlt-1 expression is silenced due to its promoter methylation during liver tumorigenesis initiated in Simian virus 40 T/t antigen transgenic mice (1). Third, Insm2, but not Insm1, is expressed in many adult tissues, suggesting different biological targets from that of Insm1. The fact that the Insm1−/− mouse is embryonic lethal (7) argues that Insm2 is not sufficient to compensate for the loss of Insm1.
Insm1 is a crucial transcription factor for islet development and insulin secretion (5–7). In the pancreas of Insm1−/− mice, endocrine precursor cells are formed, but they do not express any pancreatic islet hormones (7). In developing embryonic tissues, Insm2 expression is transiently enhanced in E11.5-E13.5, which overlaps the expression of Ngn3 (E11.5-E15.5). Similarly, immunostaining showed that INSM2, NGN3, and NEUROD1 are expressed in the same cells of human developing pancreas and duodenum at the E10- to E18-wk period. By quantitative RT-PCR analysis of fetal and adult mouse pancreas, we have confirmed that the peak level of Insm2 expression is overlapped with that of Ngn3 as well as NeuroD1. Moreover, the RNA Abundance Database revealed that deletion of Ngn3 in mice resulted in a 6.3-fold decrease of Insm2 mRNA in E18.5 embryonic islets compared with that in wild type (https://www.cbil.upenn.edu/RADQuerier/php/displayStudy.php?study_id=1330) (21). These observations point to a potential role of Insm2 in the development of mouse embryonic pancreas.
Insm2 promoter contains multiple E-boxes. The proximal E-boxes, as shown in our mutation and ChIP analysis, were regulated by the two basic helix-loop-helix transcription factors, Ngn3 and NeuroD1, which also were identified as regulators for Insm1 (8). NeuroD1 is known to be an important factor in neuronal and endocrine cell terminal differentiation, survival, and mature β-cell function (3). Mutations of the human NEUROD1 gene are associated with the development of type 2 diabetes (22) and cause maturity-onset diabetes of the young type 6 (23). Therefore, the identification of the target genes of NeuroD1 helps to elucidate the causes of diabetes. Because both Ngn3 and NeuroD1 are capable of activating Insm1 and Insm2 during early pancreas development or adult islets, we conclude that Insm1 and Insm2 are potential contributors to the transcriptional network of islet cells.
Acknowledgments
We thank Drs. B. Swaim, P. Yu, and C. Wohlenberg for technical help, Drs. M. German (University of California, San Francisco, San Francisco, CA) for adeno-NGN3 reagent and D. Hanahan (University of California, Los Angeles, Los Angeles, CA) for the mPANC cell line.
This work was supported in part by the Intramural Research Program of the National Institute of Dental and Craniofacial Research/National Institutes of Health. The human fetal pancreas and duodenum study has received funding from the Canadian Diabetes Association, and Dr. R. Wang has received a New Investigator Award from the Canadian Institute of Health Research.
Disclosure Summary: The authors declare that they have no conflict of interest.
Footnotes
- a.a.
- Amino acids
- ChIP
- chromatin immunoprecipitation
- E
- embryonic day
- EST
- expressed sequence tag
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- IHC
- immunohistochemistry
- INSM
- insulinoma-associated
- mPANC
- mouse pancreatic epithelial duct cell
- NeuroD1
- neurogenic differentiation 1
- Ngn3
- neurogenin 3
- SST
- somatostatin.
References
- 1. Tateno M, Fukunishi Y, Komatsu S, Okazaki Y, Kawai J, Shibata K, Itoh M, Muramatsu M, Held WA, Hayashizaki Y. 2001. Identification of a novel member of the snail/Gfi-1 repressor family, mlt 1, which is methylated and silenced in liver tumors of SV40 T antigen transgenic mice. Cancer Res 61:1144–1153 [PubMed] [Google Scholar]
- 2. Gradwohl G, Dierich A, LeMeur M, Guillemot F. 2000. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci USA 97:1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. 1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev 11:2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goto Y, De Silva MG, Toscani A, Prabhakar BS, Notkins AL, Lan MS. 1992. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with “zinc-finger” DNA-binding motifs. J Biol Chem 267:15252–15257 [PubMed] [Google Scholar]
- 5. Xie J, Cai T, Zhang H, Lan MS, Notkins AL. 2002. The zinc-finger transcription factor INSM1 is expressed during embryo development and interacts with the Cbl-associated protein. Genomics 80:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mellitzer G, Bonné S, Luco RF, Van De Casteele M, Lenne-Samuel N, Collombat P, Mansouri A, Lee J, Lan M, Pipeleers D, Nielsen FC, Ferrer J, Gradwohl G, Heimberg H. 2006. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J 25:1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gierl MS, Karoulias N, Wende H, Strehle M, Birchmeier C. 2006. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic β cells and intestinal endocrine cells. Genes Dev 20:2465–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breslin MB, Wang HW, Pierce A, Aucoin R, Lan MS. 2007. Neurogenin 3 recruits CBP co-activator to facilitate histone H3/H4 acetylation in the target gene INSM1. FEBS Lett 581:949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breslin MB, Zhu M, Lan MS. 2003. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem 278:38991–38997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Serazin-Leroy V, Denis-Henriot D, Morot M, de Mazancourt P, Giudicelli Y. 1998. Semi-quantitative RT-PCR for comparison of mRNAs in cells with different amounts of housekeeping gene transcripts. Mol Cell Probes 12:283–291 [DOI] [PubMed] [Google Scholar]
- 11. Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176 [DOI] [PubMed] [Google Scholar]
- 12. Lyttle BM, Li J, Krishnamurthy M, Fellows F, Wheeler MB, Goodyer CG, Wang R. 2008. Transcription factor expression in the developing human fetal endocrine pancreas. Diabetologia 51:1169–1180 [DOI] [PubMed] [Google Scholar]
- 13. Párrizas M, Maestro MA, Boj SF, Paniagua A, Casamitjana R, Gomis R, Rivera F, Ferrer J. 2001. Hepatic nuclear factor 1-α directs nucleosomal hyperacetylation to its tissue-specific transcriptional targets. Mol Cell Biol 21:3234–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida T, Hanahan D. 1994. Murine pancreatic ductal adenocarcinoma produced by in vitro transduction of polyoma middle T oncogene into the islets of Langerhans. Am J Pathol 145:671–684 [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang G, Hirai H, Cai T, Miura J, Yu P, Huang H, Schiller MR, Swaim WD, Leapman RD, Notkins AL. 2007. RESP18, a homolog of the luminal domain IA-2, is found in dense core vesicles in pancreatic islet cells and is induced by high glucose. J Endocrinol 195:313–321 [DOI] [PubMed] [Google Scholar]
- 16. Gasa R, Mrejen C, Lynn FC, Skewes-Cox P, Sanchez L, Yang KY, Lin CH, Gomis R, German MS. 2008. Induction of pancreatic islet cell differentiation by the neurogenin-neuroD cascade. Differentiation 76:381–391 [DOI] [PubMed] [Google Scholar]
- 17. Gasa R, Mrejen C, Leachman N, Otten M, Barnes M, Wang J, Chakrabarti S, Mirmira R, German M. 2004. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci USA 101:13245–13250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Candal E, Alunni A, Thermes V, Jamen F, Joly JS, Bourrat F. 2007. Ol-insm1b, a SNAG family transcription factor involved in cell cycle arrest during medaka development. Dev Biol 309:1–17 [DOI] [PubMed] [Google Scholar]
- 19. Lan MS, Breslin MB. 2009. Structure, expression, and biological function of INSM1 transcription factor in neuroendocrine differentiation. FASEB J 23:2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lan MS, Russell EK, Lu J, Johnson BE, Notkins AL. 1993. IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res 53:4169–4171 [PubMed] [Google Scholar]
- 21. Juhl K, Sarkar SA, Wong R, Jensen J, Hutton JC. 2008. Mouse pancreatic endocrine cell transcriptome defined in the embryonic Ngn3-null mouse. Diabetes 57:2755–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malecki MT, Jhala US, Antonellis A, Fields L, Doria A, Orban T, Saad M, Warram JH, Montminy M, Krolewski AS. 1999. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet 23:323–328 [DOI] [PubMed] [Google Scholar]
- 23. Fajans SS, Bell GI, Polonsky KS. 2001. Molecular mechanisms and the clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med 345:971–980 [DOI] [PubMed] [Google Scholar]