Abstract
Glycoproteins gB and gH/gL are required for entry of Epstein-Barr virus (EBV) into cells, but the role of each glycoprotein and how they function together to mediate fusion is unclear. Analysis of the functional homology of gB from the closely related primate gammaherpesvirus, rhesus lymphocryptovirus (Rh-LCV), showed that EBV gB could not complement Rh gB due to a species-specific dependence between gB and gL. To map domains of gB required for this interaction, we constructed a panel of EBV/Rh gB chimeric proteins. Analysis showed that insertion of Rh gB from residues 456-807 restored fusion function of EBV gB with Rh gH/gL, suggesting this region of gB is important for interaction with gH/gL. Split YFP bimolecular complementation (BiFC) provided evidence of an interaction between EBV gB and gH/gL. Together, our results suggest the importance of a gB-gH/gL interaction in EBV-mediated fusion with B cells requiring the region of EBV gB from 456-807.
Keywords: Epstein-Barr virus, Glycoprotein gB, Glycoprotein gH, Glycoprotein gL, Lymphocryptovirus, Rhesus, Fusion
Introduction
Epstein-Barr Virus (EBV) is a gammaherpesvirus that has a high prevalence in humans, with more than 90% of the population latently infected with the virus (Rickinson 2007). Primary infection during childhood is typically asymptomatic, but infection in adolescents can result in the development of infectious mononucleosis. Virions are transmitted through the saliva into epithelial cells of the oral pharynx, and subsequently to resting B cells. Following this initial infection, EBV persists in a latent state in memory B lymphocytes where it can remain indefinitely (Babcock, Decker et al. 1998; Thorley-Lawson and Babcock 1999). The expansion and proliferation of these cells can lead to the development of a number of EBV-related malignancies including Hodgkin's disease, Burkitt's lymphoma, some T-cell lymphomas, and tumors associated with epithelial tissues such as nasopharyngeal carcinoma and gastric carcinoma (Rickinson 2007). EBV is also associated with the lymphoproliferative disorders in immunocompromised patients, such as oral hairy leukoplakia and post-transplant lymphoproliferative disorders (Rickinson 2007).
Entry of herpesviruses requires the concerted efforts of multiple glycoproteins in a multi-step process that results in binding of the virus to the cell surface, interaction with cellular entry receptors, membrane-virion fusion, and internalization of the virion (Spear and Longnecker 2003). For Epstein-Barr virus (EBV), the viral glycoproteins required for fusion are glycoprotein B (gB), the complex of gH and gL (gH/gL), and gp42 (Spear and Longnecker 2003). Infection of B cells is initiated by an interaction between glycoprotein gp350/220 and the CD21/CR2 cellular receptor (Fingeroth, Weis et al. 1984; Nemerow, Wolfert et al. 1985; Tanner, Weis et al. 1987; Speck, Haan et al. 2000). Following initial binding, viral glycoprotein gp42 binds to human leukocyte antigen (HLA) class II molecules and triggers fusion, which is mediated by glycoproteins gB, gH, and gL (Li, Spriggs et al. 1997; Speck, Haan et al. 2000). The mechanism of infection of epithelial cells by EBV is less clear, though it has recently been found that fusion is triggered upon binding of gH/gL with cellular integrins αvβ6 or αvβ8 (Chesnokova, Nishimura et al. 2009). Glycoprotein gB and the heterodimer gH/gL constitute the conserved fusion machinery that is required for fusion of the virion and cell membrane across all members of the herpesviridae (Spear and Longnecker 2003; McShane and Longnecker 2004). However, the mechanism of how these glycoproteins function to mediate fusion is still unclear and the subject of much investigation.
Glycoprotein B is highly conserved throughout the herpesvirus family, and has been shown to be an essential component of virus-cell fusion (Spear and Longnecker 2003). The crystal structure of EBV gB in a presumed postfusion trimer conformation was recently reported and has provided valuable information on the structural domains important for mediating fusion (Backovic, Longnecker et al. 2009). Based on the distinct structural properties of the glycoprotein, EBV gB was added to the class III group of viral fusion proteins that previously included glycoprotein G of Vesicular Stomatatis virus (VSV) and glycoprotein B of Herpes Simplex virus 1 (HSV-1) (Heldwein, Lou et al. 2006; Roche, Bressanelli et al. 2006; Backovic and Jardetzky 2009). The fusion domain of gB contains the two putative fusion loops that are thought to insert into the target membrane through a conformational change that brings about fusion between the two membranes. Mutagenic analysis of the hydrophobic residues within the putative fusion loops of EBV and HSV-1 gB significantly reduced fusion function, demonstrating that the putative fusion loops are essential for the ability of gB to mediate fusion (Backovic, Jardetzky et al. 2007; Hannah, Heldwein et al. 2007; Hannah, Cairns et al. 2009). These studies, as well as others, have demonstrated that gB is likely the major fusogen that mediates fusion of the virion and cell membrane. However, a distinct deviation from other class III fusion proteins is that EBV and HSV-1 gB require the gH/gL complex for virus entry.
A central focus in understanding the entry of herpesviruses is deciphering the interactions that occur between viral glycoproteins and how these associations drive fusion. Much of our present knowledge on this matter has come from studies of HSV-1. In addition to interactions between the HSV-1 viral glycoproteins gD and gH/gL and gD and gB, a direct interaction between glycoproteins gB and gH/gL was detected using split fluorescence bimolecular fluorescence complementation (BiFC) assays (Atanasiu, Whitbeck et al. 2007; Avitabile, Forghieri et al. 2007). For EBV, evidence for an interaction between gB and gH/gL first came from a study of gL from the closely related rhesus lymphocryptovirus (Rh-LCV) that suggested the role of gL in fusion involves the activation or recruitment of gB with the gH/gL complex (Omerovic and Longnecker 2007; Plate, Smajlovic et al. 2009). The Rh-LCV glycoproteins share a high sequence similarity with EBV but Rh gL could not mediate fusion with B cells when paired with EBV gB. However, a complete restoration of the fusion function of Rh gL was observed when paired with Rh gB, demonstrating a species-specific reliance between these two glycoproteins that is essential for fusion. The essential dependence between gB and gL was used to map the regions and specific amino acid residues of gL that are necessary for mediating fusion function with gB. Subsequent studies found that EBV gB was unable to mediate B cell fusion when expressed with the Rh-LCV glycoproteins necessary for fusion (gp42, gH, and gL) while expression of all Rh-LCV glycoproteins together mediated fusion with B cells very efficiently. Given that the sequence similarity between EBV and Rh gB is near 86%, we hypothesized that regions of gB essential for mediating fusion function with gH/gL could be identified using chimeric EBV/Rh gB proteins.
The objective of this study was to further analyze the interaction between gB and gH/gL in EBV and Rh-LCV and map regions of gB that are involved in the interaction with gH/gL. Analysis of the EBV/Rh gB chimeras in the cell-cell fusion assay identified the region from residues 456-807 as being important for a functional interaction with gH/gL, with evidence that sites from both regions 456-628 and 628-807 are required. Further analysis of chimeras using BiFC confirmed that insertion of these regions of Rh gB into EBV gB could modestly enhance the ability of EBV gB to interact with Rh gH/gL. These results provide further evidence of an interaction between gB and gH/gL and highlights that there are likely multiple domains of gB required for an interaction with gH/gL.
Results
Rh-LCV gB does not complement EBV gB for fusion function
The glycoproteins of the closely related EBV and Rh-LCV share a high degree of sequence homology and have proven to be a useful tool in studying the required domains for glycoprotein fusion function. We previously found that Rh gL can only mediate fusion with EBV glycoproteins when expressed together with Rh gB, suggesting that there is a species-specific dependence between gL and gB that is necessary for these two glycoproteins to mediate fusion together (Omerovic and Longnecker 2007; Plate, Smajlovic et al. 2009).
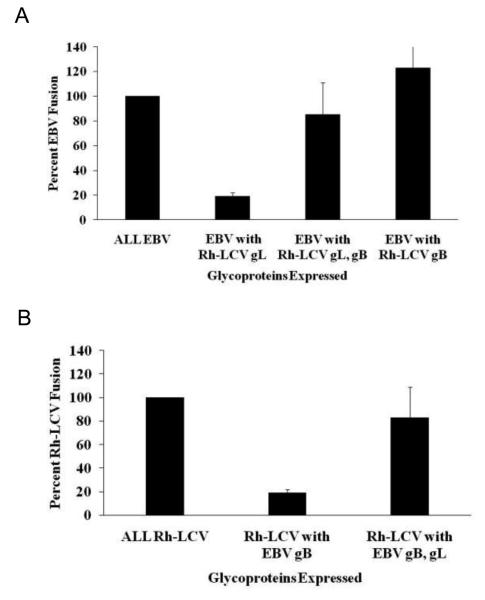
To examine the dependence between gB and gL in mediating fusion with B cells further, we examined the functional homology of gBs from EBV and the closely related Rh-LCV. Similar to what we previously found (Omerovic and Longnecker 2007; Plate, Smajlovic et al. 2009) but somewhat different from others (Wu and Hutt-Fletcher 2007), fusion with EBV gp42, gB, and gH and Rh gL was near background levels while fusion with EBV gp42, gH, and Rh gL and gB was restored to wild-type EBV fusion activity (Fig 1A). Rh-LCV has been shown to infect human B cells in vitro (Moghaddam, Koch et al. 1998) so it was not surprising that expression of Rh gp42, gB, gH, and gL can also mediate fusion with B cells in our cell-based fusion assay (Fig. 1B, column 1). However, the ability of the Rh-LCV glycoproteins to mediate fusion was significantly reduced when EBV gB was expressed in place of Rh gB (Fig. 1B, column 2). This observation is in agreement with our previous conclusion that there is a species-specific dependence between gB and gL, and the inability to mediate fusion is specifically due to the expression of EBV gB with Rh gL. This is further supported by the finding that fusion of the Rh-LCV glycoproteins with EBV gB is enhanced when EBV gL is expressed with EBV gB (Fig. 1B, column 3).
Figure 1. Fusion is blocked by EBV gB and Rh gL co-expression.
Fusion function of EBV and Rh-LCV glycoproteins with B cells. (A) CHO-K1 cells were transiently transfected with all of the EBV glycoproteins necessary for fusion (gp42, gB, gH, and gL) or the indicated Rh-LCV glycoprotein. Effector CHO-K1 cells were overlaid with target Daudi B cells and luciferase activity was measured. Luciferase activity was normalized to wild-type EBV levels, which was set to 100%. (B) CHO-K1 cells were transiently transfected with all of the Rh-LCV glycoproteins necessary for fusion (gp42, gB, gH, and gL) or the indicated EBV glycoprotein. Effector CHO-K1 cells were overlaid with target Daudi B cells and luciferase activity was measured. Luciferase activity was normalized to wild-type Rh-LCV levels, which was set to 100%.
Interestingly, we found that Rh gB is functional in mediating fusion with EBV glycoproteins gp42, gH, and gL (Fig. 1A, column 4). This finding would suggest that Rh gB can interact with EBV gH/gL while the converse, an interaction between EBV gB with Rh gH/gL, does not occur.
Generation of EBV/Rhesus chimeric gB glycoproteins
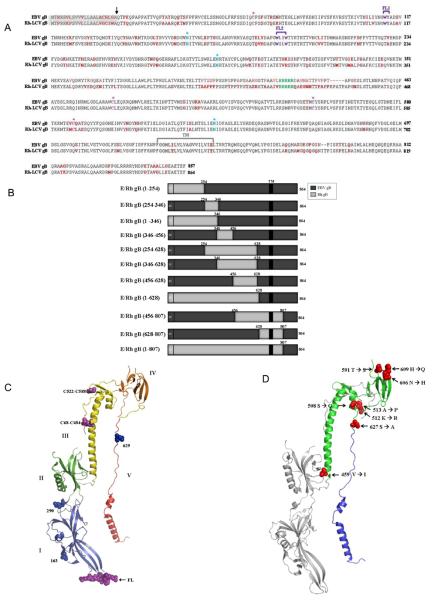
It is intriguing that EBV gB cannot substitute for Rh gB in mediating fusion with the Rh-LCV glycoproteins since these two proteins are extremely homologous, sharing about 86% sequence similarity. The amino acid sequence alignment of EBV and Rh gB is shown (Fig. 2A) and highlights that many features of gB are conserved between these two closely related glycoproteins. The position of the signal sequence, sites of predicted glycosylation, disulfide bonding, furin cleavage and predicted structural domains are shown. The three asparagine residues 163, 290, and 629, that are likely N-glycosylated in EBV gB (shown Fig. 2A and 2C, blue asterisks) are conserved in Rh gB and are also likely N-glycosylated to yield the mature, functional glycoprotein. In EBV gB, 4 conserved cysteines are predicted to form 2 disulfide bonds, between Cys68-Cys484 and Cys522-588 (Backovic, Longnecker et al. 2009). In Rh gB, these bonds are likely conserved and predicted to form between Cys68-489 and Cys556-Cys592 (Fig. 2A and 2C, pink asterisks). A stretch of five arginine residues (428-432) that form a cleavage site important for EBV fusion are also conserved between EBV and Rh gB (Fig. 2A) (Sorem and Longnecker 2009). The six hydrophobic residues in EBV gB that form the putative fusion loops (FL) that facilitate interaction with target membranes, WY112-113 and WLIW193-196, are also highly conserved in Rh gB. The residues corresponding to EBV FL1 and FL2 in Rh gB are WY112-113 and WILW193-196 (Fig. 2A and 2C, purple brackets). Due to the high degree of sequence similarity, as well as the conservation of predicted features, we predicted that a majority of the EBV/Rh gB chimeras would be properly processed and could assist in the identification of regions important for fusion function of gB in concert with gH/gL.
Figure 2. EBV and Rh-LCV gB amino acid sequence alignment, schematic diagram of EBV/Rh-LCV gB chimeras, and location of nonconserved residues implicated in the gB-gH/gL interaction on the EBV gB crystal structure.
(A) The 857 amino acid EBV gB sequence (accession number YP_401713) and 864 amino acid Rh-LCV gB sequence (accession number YP_068009) were aligned using the MultiAlin program. Nonconserved residues between EBV and Rh-LCV are highlighted in red. The 22 amino acid signal sequence is highlighted in grey and the transmembrane (TM) domain is depicted by the gray bracket. Residues modified by N-linked glycans are highlighted in blue with asterisks and the five arganine residues that form the putative furin cleavage site are highlighted in green. The conserved cysteine residues that were shown to form disulfide bonds in the crystal structure of EBV gB are highlighted in magenta with asterisks. The known fusion loops residues (WY, WLIW) of EBV gB and the predicted fusion loop residues (WY, WILW) of Rh gB are highlighted with purple brackets to show both fusion loop 1 (FL1) and 2 (FL2). (B) Chimeric EBV/Rh gB proteins were generated using regions from Rh gB (light grey) and EBV gB (dark grey). The amino acid residues of Rh gB insertions are indicated above each chimera. The location of the signal sequence (ss) and transmembrane (TM) domains are shown. Nonconserved amino acid residues between EBV gB and Rh gB were mapped onto the EBV gB crystal structure using PyMol. The numbers indicate the Rh-LCV amino acid substitution within the EBV gB amino acid sequence. (C) The structure of the EBV ectodomain is colored from blue to red, with the five domains labeled by roman numerals I through V. Numbers indicate the location of the Asn residues that are modified by N-linked glycans, which are shown in blue. Cysteine residues that form disulfide bonds are shown in magenta and fusion loops (FL) in purple. (D) Rh gB amino acid residues 456-628 correspond to EBV gB residues 451- 623 and are colored green on the cartoon structure of EBV gB. Nonconserved residues in this region are indicated in red based on the residue location in the EBV gB amino acid sequence (459, 508, 512, 513, 591, 606, and 609). Residues located in regions of the structure that exhibited discontinuous electron density of lower quality and were omitted from the gB structure are not shown (549, 587, 589). Rh gB amino acid residues 628-807 correspond to EBV gB residues 623-800 are colored blue on the cartoon structure of EBV gB. Most of the nonconserved residues between EBV and Rh gB in this region are located outside of the crystallized gB ectodomain and only residue 627 is shown.
To map regions of gB critical for fusion function with gH/gL, chimeras were generated that contain regions of both Rh and EBV gB. Chimeras were constructed using EBV gB as the backbone, with varying regions of Rh gB replacing the corresponding region in EBV gB (Fig. 2B). The regions of EBV gB were removed using two preexisting restriction sites. Regions of Rh gB from residues 1-628, 1-807, and 628-807 were isolated using restriction sites that were common for both EBV and Rh gB. Regions of Rh gB from residues 1-254, 254-346, 1-346, 346-456, 254-628, 346-628, and 456-807 were amplified using oligonucleotides containing the unique restriction sites for EBV gB. The generated chimeras cover a majority of the ectodomain of gB in overlapping fragments that span regions of high sequence similarity as well as less conserved regions.
Expression and cellular localization of EBV/Rhesus gB chimeras
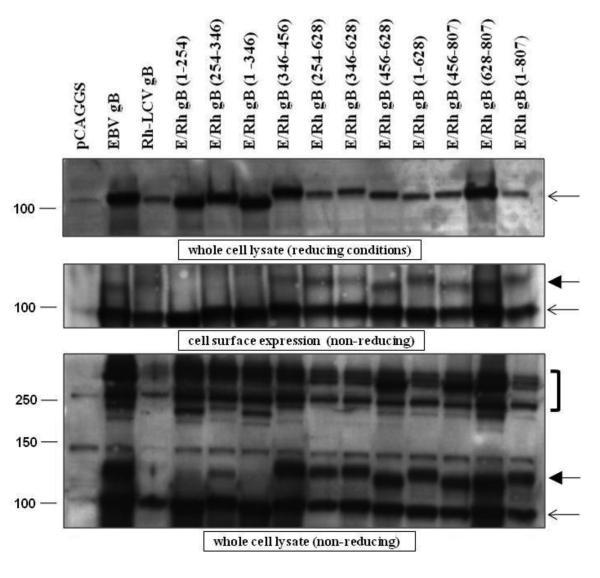
The protein expression of the EBV/Rh gB chimeras was examined by transient expression of the constructs in CHO-K1 cells. Analysis of expression by western blot showed that all chimeras were expressed, although Rh gB and some of the chimeras were expressed at slightly lower levels (Fig. 3, top panel). Whether this is due to a reduction in protein expression or a slight impairment of the EBV gB rabbit polyclonal antibody to recognize Rh gB is not clear.
Figure 3. Expression levels of most of the EBV/Rh gB chimeras are similar to that of EBV gB.
CHO-K1 cells were transfected with EBV gB, Rh-LCV gB, and the EBV/Rh gB chimeras as described in Materials and Methods. (A) Western blot analysis of EBV/Rh gB chimera expression in whole cell lysates was determined under reducing conditions. Molecular weight marker in kDa is indicated and the open arrow indicates the position of full length EBV gB. (B) The cell surface expression of EBV/Rh gB chimeras was determined by biotinylation of cell membranes followed by immunoprecipitation with a monoclonal gB antibody. Western blot analysis of EBV/Rh gB chimeras was done under non-reducing conditions with a polyclonal gB antibody. Molecular weight marker in kDa is indicated and the open arrow indicates the position of full length EBV gB. (C) Oliogomer formation of EBV/Rh gB chimeras was examined in whole cell lysates under nonreducing conditions. Oligomers are identified by brackets to the right of the blot, monomeric gB is indicated by an open arrow, and the fully glycosylated mature form of gB indicated by a closed arrow.
EBV gB predominantly localizes to the perinuclear region of cells (Gong, Ooka et al. 1987; Gong and Kieff 1990), but small amounts of gB can be detected at the cell surface using biotinylation of cell surface proteins, followed by western blot analysis (Backovic, Jardetzky et al. 2007; Reimer, Backovic et al. 2009). Presence of EBV and Rh gB and the EBV/Rh gB chimeras on the cell surface was analyzed using sulfosuccinimidyl-1-6-(biotinamido)hexanoate as previously described (Reimer, Backovic et al. 2009). All EBV/Rh gB chimeras and wild-type Rh gB were expressed on the cell surface at levels similar to wild-type EBV gB (Fig. 3 middle panel). This result indicates the variability of gB reactivity in the whole cell lysates in the top panel of Fig. 3 is likely not significant.
Herpes simplex virus (HSV) gB has been shown to form oligomers in vivo (Claesson-Welsh and Spear 1986; Grunewald, Desai et al. 2003) and the crystallized ectodomains of EBV and HSV gB formed trimers (Backovic, Longnecker et al. 2009; Heldwein, Lou et al. 2006), suggesting that the formation gB oligomers is a common feature in herpesviruses. A previous study of EBV gB found that mutants that could not oligomerize did not mediate fusion with epithelial or B cells (Reimer, Backovic et al. 2009). Therefore, the formation of oligomers was analyzed in whole cell lysates for EBV gB, Rh gB, and the EBV/Rh gB chimeras using SDS-PAGE under non-reducing conditions. While detection of oligomers was reduced for Rh gB, all EBV/Rh gB chimeras were able to form higher molecular weight oligomers similar to EBV gB (Fig. 3, bottom panel bracket).
The apparent molecular weight of EBV gB has been shown to vary, depending on the amount of glycosylation the protein undergoes during maturation and processing (Emini, Luka et al. 1987; Gong, Ooka et al. 1987; Papworth, Van Dijk et al. 1997; Lee 1999). While EBV gB is typically reported as a 110-kDa protein, the presence of a higher molecular size gB variant that migrates just above monomeric gB was reported and shown to be functionally important for fusion (Reimer, Backovic et al. 2009). This N-glycosylated modified form of monomeric gB likely represents the fully mature form of EBV gB. EBV/Rh gB chimeras that contain insertions of Rh gB in the amino terminus, ERh gB (1-254) and ERh gB (1-346), did not exhibit this higher molecular size band above monomeric gB (Fig. 3, closed arrows). The EBV/Rh gB chimera that contains the small portion of Rh gB from residues 254-346 was variable in the expression of the higher molecular size form of gB, and when detected migrated at a smaller molecular size than that for EBV gB and the other chimeras (compare bands indicated by closed arrows in middle and bottom panel). This chimera, as well as the other two chimeras that lack the variant band of gB, were unable to mediate fusion with either the EBV or Rh-LCV glycoproteins. As the function of these three EBV/Rh gB chimeras is likely hampered by the improper processing and maturation of gB, we did not examine their functional properties further.
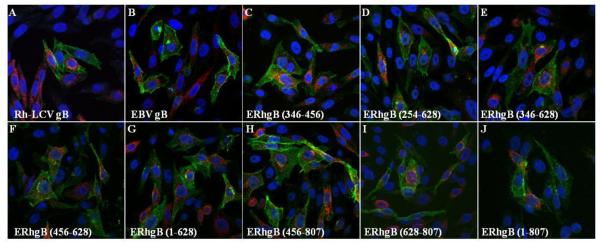
While EBV gB is primarily localized to the perinuclear membrane and the endoplasmic reticulum, the EBV gH/gL complex is largely detected at the cell surface (Gong, Ooka et al. 1987; Gong and Kieff 1990; Hutchinson, Browne et al. 1992; Li, Turk et al. 1995; Lee 1999; Neuhierl, Feederle et al. 2002). Expression of the glycoproteins together in cells does not alter the localization of either gB or the gH/gL complex. Immunofluorescence analysis of the Rh-LCV glycoproteins confirmed that Rh gB and gH/gL have the same cellular localization as EBV gB and gH/gL (Fig. 4, A and B). We then examined the intracellular expression of Rh gB, EBV gB, and the EBV/Rh gB chimeras to determine if localization was disrupted for the chimeras. The EBV/Rh gB chimeras localized predominantly to the perinuclear membrane and endoplasmic reticulum, similar to what was observed for both EBV and Rh gB (Fig. 4). The localization of Rh gH/gL was not altered upon expression of the EBV/Rh chimeras. In summary, analysis of expression and localization showed that eight of the EBV/Rh gB chimeras were expressed intracellularly and at the cell membrane, processed to yield a fully glycosylated mature form of gB, formed higher molecular weight oligomers and were properly localized within transfected cells.
Figure 4. Cellular localization of EBV/Rh gB chimeras and Rh gH/gL is similar to wild-type gB.
CHO-K1 cells were transfected with Rh-LCV gH/gL and either EBV gB (A), Rh gB (B), or the indicated EBV/Rh gB chimera (C - J). Cells were fixed at 24 hours after transfection and immunostained using the monoclonal E1D1 gH/gL specific antibody (green channel) and the polyclonal gB antibody (red channel) with DAPI nuclear staining.
Evaluation of the ability of EBV/Rh gB chimeras to mediate fusion with B cells
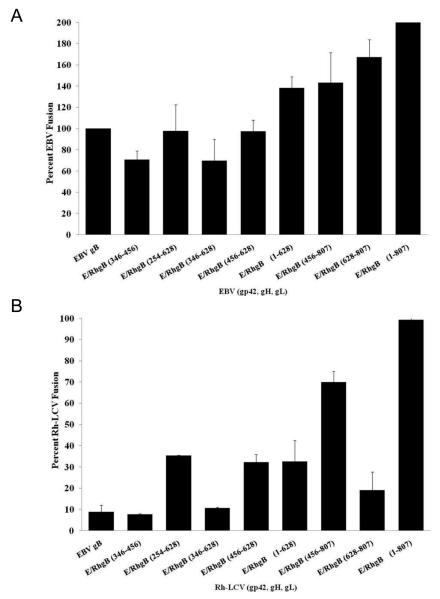
In the current study, we found that Rh gB is able to mediate fusion when expressed with the EBV glycoproteins, demonstrating that Rh gB does not have the same species specificity as EBV gB. This observation provides a means of testing the general fusion function of the chimeras. Since both EBV gB and Rh gB mediate fusion with the EBV glycoproteins, we would expect the EBV/Rh gB chimeras to also function in fusion if they are properly processed. Fusion of the effector and target cells led to the expression of luciferase, which was quantified and normalized to the EBV wild-type level, set at 100% fusion activity. As expected, all of the EBV/Rh gB chimeras were able to mediate fusion with EBV glycoproteins at levels near or above EBV gB (Fig. 5A). As we have previously observed, Rh gB mediates fusion with B cells at levels above wild-type EBV gB and therefore fusion activity of EBV/Rh gB chimeras containing larger portions of Rh gB also elicited fusion at levels above wild-type EBV gB. While these results do not elucidate regions of gB that are essential for mediating fusion in complex with gH/gL, we verified that the EBV/Rh gB chimeras that are properly processed to yield the fully glycosylated mature form of gB are also likely in the correct structural conformation to allow gB to execute fusion.
Figure 5. Fusion function of EBV/Rh gB chimeras with EBV and Rh-LCV glycoproteins.
(A) CHO-K1 cells were transiently transfected with all of the EBV glycoproteins necessary for fusion (gp42, gB, gH, and gL) or the indicated EBV/Rh gB chimera. Effector CHO-K1 cells were overlaid with target Daudi B cells and luciferase activity was measured. Luciferase activity was normalized to wild-type EBV levels, which was set to 100%. (B) (A) CHO-K1 cells were transiently transfected with all of the Rh-LCV glycoproteins necessary for fusion (gp42, gB, gH, and gL) or the indicated EBV/Rh gB chimera. Effector CHO-K1 cells were overlaid with target Daudi B cells and luciferase activity was measured. Luciferase activity was normalized to wild-type Rh-LCV levels, which was set to 100%.
To identify regions of gB that are essential for fusion function with gH/gL, the EBV/Rh gB chimeras were tested in the cell-cell fusion assay with the Rh-LCV glycoproteins. As we predict that certain gB residues are critical for fusion function with gH/gL, chimeras that contain essential Rh gB domains are predicted to be active for fusion with Rh gH/gL. As previously observed, EBV gB cannot mediate B cell fusion when expressed with Rh gp42, gH, and gL (Fig. 5B, first column). The level of fusion with EBV gB is roughly 10% of the level of wild-type Rh gB fusion and similar to vector only control. Insertion of a majority of the Rh gB sequence from residues 1-807 into EBV gB fully restored fusion function of EBV gB to 99% of wild-type Rh gB. This region includes the transmembrane domain and the first 69 amino acids of the cytoplasmic tail. Insertion of the Rh gB sequence from residues 1-628 only restored fusion function of EBV gB to 33% of wild-type Rh gB, suggesting that the region from 628-807 is essential for the fusion function observed with the chimera containing Rh gB residues 1-807. Insertion of Rh gB residues 456-807 significantly restored fusion function, mediating fusion at 70% of the level of wild-type Rh gB. Interestingly, insertion of Rh gB residues 456-628 or 628-807 resulted in a restoration of fusion function to 30% or 20%, respectively. This observation indicates that sites from both regions are important for fusion function with gH/gL, since one region alone is insufficient to significantly restore fusion. The region between residues 456-807 is highly conserved between EBV and Rh gB, with almost 93% sequence similarity. The remaining chimeras that contain Rh gB residues 346-456, 254-628, and 346-628 did not significantly restore fusion function of EBV gB with B cells, indicating that these regions are likely not important for gB fusion function with gH/gL.
Location of non-conserved residues important for gB and gH/gL fusion function
There are 25 nonconserved amino acid residues between EBV and Rh gB between residues 456 and 807; with 10 nonconserved residues present in the region from 456-628, and the remaining 15 in the region from residues 628-807. To analyze the location of residues that may be important for fusion function with gH/gL, the crystal structure of the EBV gB ectodomain from residues 23 to 685 was used to illustrate nonconserved residues between EBV and Rh gB. The 10 nonconserved residues between EBV and Rh gB from 456-628 are all located in domains III and IV of EBV gB (Fig. 2D).
The most prominent element of domain III is the long central alpha-helix that spans 42 residues, which is highly conserved between Rh and EBV gB. There is a single amino acid variation at the base of this αC-helix at valine residue 459 of EBV gB that is an isoleucine at residue 464 of Rh gB. The serine at residue 508 of EBV gB, located in the αD-helix, is a glycine at that residue in Rh gB. The two remaining amino acid variations in domain III of EBV gB, a lysine at residue 512 and alanine at 513, are both located in a turn that connects the αD-helix and the β24 strand.
Domain IV of EBV gB is composed entirely of β-sheets, and appears more disordered than the same domain in HSV-1 gB. The threonine at residue 591 of EBV gB is a serine residue in Rh gB and is located just beyond the disordered region between the β30 and β31 strands. The asparagine at residue 606 and the histidine at residue 609 of EBV gB are located at the end of the β32 and beginning of the β33 strands, respectively. Interestingly, the DY residues that are located in the turn between nonconserved residues 606 and 609 are highly conserved across many herpesviruses, including HSV-1 and 2, CMV, and HHV-6 and 8 (Backovic, Longnecker et al. 2009). The three remaining nonconserved residues at 549, 587, and 589 are located in areas of EBV gB that were omitted from the final crystal structure.
The 15 nonconserved residues between EBV and Rh gB from 628-807 are located outside of the crystallized gB ectodomain, with the exception of the serine at residue 627 of EBV gB. This residue is located in domain V, before the αE and αF C-terminal helices, and is an alanine at the corresponding residue in Rh gB (Fig. 2D).
Generation and characterization of EBV and Rh-LCV glycoproteins in bimolecular fluorescence complementation (BiFC) assay
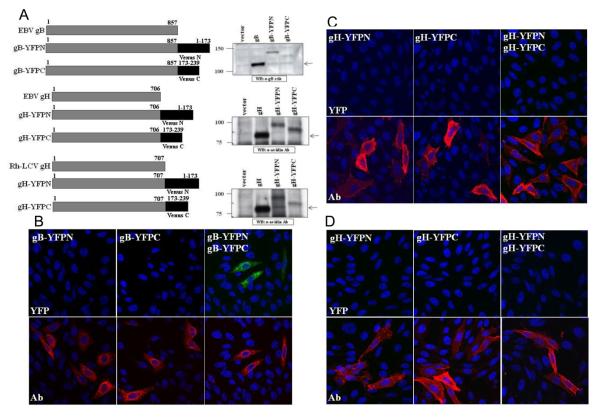
To specifically address the interaction of EBV gB and gH/gL, we utilized the BiFC assay that was previously used to demonstrate the interaction between HSV-1 gB and gH/gL (Atanasiu, Whitbeck et al. 2007; Avitabile, Forghieri et al. 2007). BiFC utilizes a fluorescent protein that has been split into the amino and carboxyl terminal halves and thus no longer produces fluorescence. By fusing each of the two non-fluorescent fragments to putative interacting partners, restoration of fluorescence provides visual evidence of a protein-protein interaction (Kerppola 2006; Kerppola 2006; Kerppola 2006). The amino and carboxy terminal fragments of the YFP-variant Venus fluorescent protein (YFPN or YFPC) were fused in frame to the carboxy terminus of EBV glycoproteins gB and gH and Rh gH (Fig. 6A) (Nagai, Ibata et al. 2002). Western blot analysis of cells transfected with the YFP-tagged glycoproteins showed that the electrophoretic mobility of the constructs was increased relative to wild-type, as expected (Fig. 6A). Depending on the construct examined, variable levels of fusion activity were observed when the YFP-tagged glycoproteins were examined in the cell-cell fusion assay. A significant reduction in fusion function was observed when the split YFP-tagged gBs were analyzed in the cell-cell fusion assay, with levels close to background (data not shown). In contrast, the fusion function of EBV gH and Rh gH YFP-tagged constructs were less impaired, with fusion levels near 50% or 100%, respectively. Previous studies have indicated that the EBV gB cytoplasmic tail is an important determinant of fusion function and the YFP tag may be interfering with this function. However, our analysis shows that the YFP-tagged gB constructs form oligomers, demonstrating that protein-protein interactions are not impaired and the structure of gB is maintained. Due to the impairment of fusion activity, the BiFC analysis we performed was not in the context of EBV-mediated fusion and was only utilized to examine glycoprotein interactions in transiently transfected cells.
Figure 6. Generation and characterization of EBV and Rh-LCV BiFC constructs.
(A) Schematic diagram of EBV gB, EBV gH, and Rh gH with the N- and C-terminal fragments of Venus YFP and western blot analysis. CHO-K1 cells were transfected with EBV gB, EBV gH/gL, Rh gH/gL and the YFP-tagged constructs. Cell lysates were analyzed using the polyclonal gB antibody for EBV gB and gB-YFP constructs. CHO-K1 cells expressing EBV gH/gL, Rh gH/gL, and gH-YFP constructs were biotinylated as previously described and cell lysates analyzed using the HRP conjugated Avidin antibody. (B-D) Homologous glycoprotein interactions. (B) CHO-K1 cells were transfected with EBV gB-YFPN, EBV gB-YFPC, or both constructs together. Cells were fixed, permeabilized and stained with the polyclonal gB antibody and analyzed for gB expression (red channel) and YFP fluorescence (green channel) with DAPI staining of nuclei (blue channel). (C) CHO-K1 cells were transfected with EBV gH-YFPN, EBV gH-YFPC, or both constructs together with EBV gL. Cells were fixed, permeabilized and stained with the monoclonal E1D1 gH/gL antibody and analyzed for gH/gL expression (red channel) and YFP fluorescence (green channel). All confocal images were taken at 40X magnification. (D) CHO-K1 cells were transfected with Rh gHYFPN, Rh gH-YFPC, or both constructs together with Rh gL. Cells were fixed, permeabilized and stained with the monoclonal E1D1 gH/gL antibody and analyzed for gH/gL expression (red channel) and YFP fluorescence (green channel) with DAPI staining of nuclei (blue channel). All confocal images were taken at 40X magnification.
To analyze homologous glycoprotein interactions, CHO-K1 cells were transfected with each of the glycoprotein constructs alone or with their corresponding YFP pair. As expression of gH and gL together is necessary for the proper processing and transport of the gH/gL complex, wild-type EBV or Rh gL was transfected in all samples that contained the corresponding YFP-tagged gH construct. Following a 20-hour transfection, the cells were fixed and stained with either the polyclonal gB antibody or monoclonal E1D1 gH/gL antibody. In cells that expressed single glycoprotein constructs, expression of all the gB and gH constructs was detected by antibody staining (Fig. 6B-D, bottom panels). No fluorescence in the YFP channel was detected in any cell expressing a single construct (Fig. 6B-D, top panels). Since previous analysis has shown that EBV gB forms oligomers, we predicted that an interaction between gB constructs would allow the two YFP fragments to reform. A strong YFP signal was detected in cells transfected with both EBV gB-YFPN and EBV gB-YFPC constructs, demonstrating that the interaction between gB constructs was sufficient to permit BiFC (Fig. 6B, top panel). Interestingly, coexpression of the EBV gH-YFP constructs together with EBV gL did not result in YFP fluorescence, suggesting that the EBV gH/gL complex does not form oligomers (Fig. 6C, top panel). A similar result was found when the Rh gH-YFP constructs were expressed together with Rh gL (Fig. 6D, top panel). These results are in contrast to what is observed for HSV-1 gH/gL oligomer formation using BiFC (Atanasiu, Whitbeck et al. 2007), which suggests that the formation of gH/gL oligomers may be unique to specific herpesvirus subgroups.
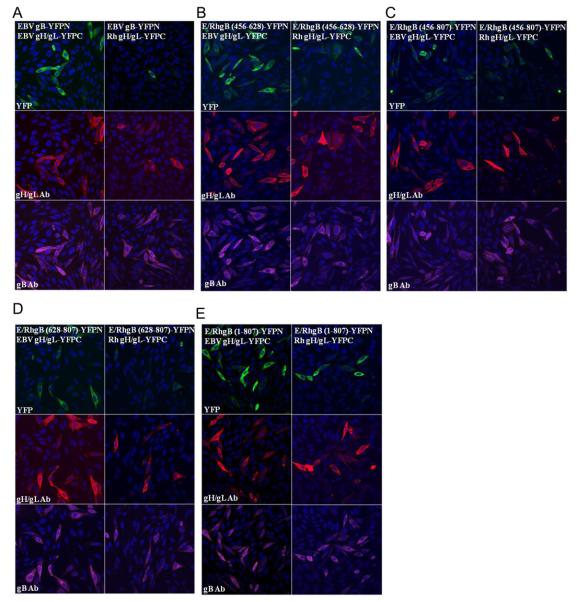
To analyze the interaction between gB and the gH/gL complex, CHO-K1 cells were transfected with the EBV gB-YFPN and EBV gH-YFPC constructs together with EBV gL. Following the 20 hour transfection, the cells were fixed and stained with both the polyclonal gB antibody and monoclonal E1D1 gH/gL antibody. Bright YFP fluorescence was observed in cells that expressed both gB and gH/gL constructs, indicating that an interaction between the two glycoproteins was mediating BiFC (Fig. 7A, left panel). To determine if the reduction of fusion function when EBV gB is expressed with the Rh gH/gL complex is due to an impairment in the ability of these glycoproteins to interact, the EBV gB-YFPN construct was transfected with the Rh gHYFPC construct together with Rh gL. There was a noticeable decrease in the YFP fluorescence, both in the number of cells expressing YFP and the intensity of YFP fluorescence (Fig. 7A, right panel). To quantify the BiFC results, multiple fields of cells from two separate experiments were counted to establish the number of cells expressing gB, gH/gL, and YFP fluorescence. The percentage of YFP cells was then calculated based on the total number of cells stained with both the gB or gH/gL antibody. The data is summarized in Table 1. Based on the quantitative analysis of the gB-gH/gL interaction, there was a reduction in the percentage of YFP cells for EBV gBYFPN with Rh gH/gL-YFPC by 15% (83% - 68%).
Figure 7. Analysis of glycoprotein interactions by BiFC assay.
Interactions between YFP-tagged gB and gH/gL were detected using BiFC (green channel). CHO-K1 cells were transfected with the indicated YFP-tagged glycoprotein pairs for 20 hours, fixed, permeabilized and stained with the polyclonal gB antibody (purple channel) and monoclonal E1D1 gH/gL antibody (red channel) with nuclei stained with DAPI (blue channel). All confocal images were taken at 40X magnification. (A) Glycoprotein interactions between EBV gB-YFPN and EBV gH/gL-YFPC are exhibited by YFP fluorescence (left panel). A weaker interaction between EBV gB-YFPN and Rh gH/gL-YFPC is exhibited by minimal YFP fluorescence (right panel). (B-D) Glycoprotein interactions between EBV/Rh gB-YFPN chimeras and EBV gH/gL-YFPC (left panels) or Rh gH/gL-YFPC (right panels) is exhibited by YFP fluorescence.
Table 1. Quantification of glycoprotein interactions by BiFC assay.
Quantification of gB – gH/gL interactions. For each of the glycoprotein combinations, the number of cells expressing gB, gH/gL, and YFP were counted from two separate experiments. A ratio of the number of YFP cells to the number of total cells expressing gB and gH/gL was calculated and presented as a percentage.
| gB construct | gH/gL |
# YFP cells # gB and gH/gL stained |
|---|---|---|
| EBV gBN | EBV gHC EBV gL | 83% |
| EBV/Rh (456-628) gBN | EBV gHC EBV gL | 93% |
| EBV/Rh (456-807) gBN | EBV gHC EBV gL | 84% |
| EBV/Rh (628-807) gBN | EBV gHC EBV gL | 83% |
| EBV/Rh (1-807) gBN | EBV gHC EBV gL | 93% |
| EBV gBN | Rh-LCV gHC Rh-LCV gL | 68% |
| EBV/Rh (456-628) gBN | Rh-LCV gHC Rh-LCV gL | 72% |
| EBV/Rh (456-807) gBN | Rh-LCV gHC Rh-LCV gL | 79% |
| EBV/Rh (628-807) gBN | Rh-LCV gHC Rh-LCV gL | 59% |
| EBV/Rh (1-807) gBN | Rh-LCV gHC Rh-LCV gL | 91% |
Generation and characterization of EBV/Rh gB chimeras in BiFC assay
Analysis of EBV/Rh gB chimeras in the fusion assay identified the region of Rh gB from residues 456-807 as essential for mediating fusion with the Rh gH/gL complex. To further characterize the region of gB necessary for fusion function with gH/gL, we generated EBV/Rh gB chimeric constructs containing the N-terminal fragment of YFP for analysis in the BiFC assay. For this analysis, we used EBV gB YFPN constructs that contained the regions of Rh gB from 1-807, 456-807, 456-628, and 628-807.
The ability of the EBV/Rh gB-YFPN chimeras to mediate BiFC was first examined in association with the EBV gH/gL-YFPC complex. All of the EBV/Rh gB chimeras can mediate fusion with the EBV gH/gL complex so we would expect the YFPN-tagged version would also be capable of interacting with the YFPC-tagged gH/gL complex. In cells expressing each of the EBV/Rh gB-YFPN constructs and EBV gH/gLYFPC, bright YFP fluorescence was observed in a large percentage of transfected cells (Fig. 7C-F, left panels). Upon quantifying the number of YFP expressing cells, we found the percentage of YFP stained for gB and gH/gL was near or above the levels observed for EBV gB-YFPN with EBV gH/gL-YFPC (Fig. 7B). All of the EBV/Rh gB-YFPN chimeras were also able to mediate BiFC with EBV gB-YFPC, verifying that these constructs are able to form gB oligomers (data not shown).
We then examined the ability of the EBV/Rh gB-YFPN chimeras to mediate BiFC in association with Rh gH/gL-YFPC. While all of the EBV/Rh gB-YFPN chimeras were able to mediate BiFC at moderate levels, there were subtle variations between chimeric gBs. We observed the highest level of YFP fluorescence when the EBV/Rh (1-807) gBYFPN chimera was expressed with Rh gH/gL-YFPC, near levels detected for the EBV gB - gH/gL interaction (Fig. 7C, right panel). The lowest level of YFP fluorescence was observed for the EBV/Rh (628-807) gB-YFPN chimera, with only 59% of dual labeled cells expressing YFP (Table 1). Insertion of the region from 456-807 of Rh gB resulted in a modest enhancement in the ability to mediate BiFC with the Rh gH/gL complex. The percentage of YFP cells observed for the interaction of EBV/Rh (456-807) gBYFPN with Rh gH/gL-YFPC increased by 11% (68% to 79%) over what was observed for the EBV gB – Rh gH/gL interaction.
Discussion
We used functional analysis of chimeric proteins and BiFC to provide further evidence of an interaction between gB and gH/gL. EBV gB cannot functionally complement Rh gB and as a result does not mediate fusion with Rh gH/gL. However, a chimeric form of EBV gB that contained domains of Rh gB restored fusion with Rh gH/gL. Specifically, the region from residues 456 to 807 of Rh gB was critical in mediating fusion with Rh gH/gL and is likely important for an interaction between gB and gH/gL.
Of the lymphocryptoviruses within the gammaherpesvirus subgroup, the viruses that infect humans (EBV) and primates (Rh-LCV) are the most closely related, sharing a high sequence similarity, similar genome organization, biological properties, epidemiology, and pathogenesis (Moghaddam, Koch et al. 1998; Cho, Gordadze et al. 1999; Wang, Rivailler et al. 2001; Rivailler, Cho et al. 2002; Rivailler, Carville et al. 2004). Rh-LCV has been extremely useful in complementation studies with EBV to identify functional domains of the glycoproteins necessary for EBV-mediated fusion (Omerovic and Longnecker 2007; Wu and Hutt-Fletcher 2007; Plate, Smajlovic et al. 2009). The glycoproteins that make up the core fusion complex for herpesviruses; gB, gH, and gL, share a sequence similarity with Rh-LCV of 86%, 85%, and 82%, respectively (Rivailler, Jiang et al. 2002). In the current study, we found that EBV gB could not substitute for Rh gB to mediate fusion with Rh gH/gL. However, by replacing a portion of EBV gB with Rh gB, fusion function was restored. Together, these studies demonstrated that amino acid changes in gB and gL can alter their ability to mediate fusion together, providing evidence of a functional interaction between gB and the gH/gL complex.
To provide further evidence of an interaction between the gB and gH/gL, we utilized the BiFC assay that was previously used to demonstrate an interaction between HSV-1 gB and gH/gL. The formation of gB oligomers was demonstrated by bright YFP fluorescence in cells expressing the YFPN and YFPC tagged forms of gB, confirming that BiFC was a viable assay to demonstrate EBV glycoprotein interactions. An interaction between EBV gB and gH/gL was detected by BiFC, providing evidence that these glycoproteins form a heterologous complex in cells. While expression of EBV gBYFPN and EBV gH/gL-YFPC together resulted in abundant YFP, expression of EBV gBYFPN and Rh gH/gL-YFPC yielded modest fluorescence in a smaller number of cells. These results support the interpretation that EBV gB does not mediate fusion with Rh gH/gL because the glycoproteins interact poorly.
To identify regions of gB that are involved in the interaction with gH/gL, we generated a panel of eleven chimeras in which portions of EBV gB were replaced with the corresponding region of Rh gB. Following a thorough examination of the expression and localization patterns of the chimeras, we determined that eight of the eleven chimeras were properly processed and retained the structural integrity of EBV gB. Analysis of these EBV/Rh gB chimeras in the fusion assay found that insertion of Rh gB from 456-807 permitted EBV gB to mediate fusion with Rh gH/gL at levels significantly above EBV gB. Chimeras that contained smaller portions of Rh gB from 456-628 and 628-807 were defective in mediating fusion with Rh gH/gL, demonstrating that both regions contribute to fusion function. The ability of some of the EBV/Rh gB chimeras to interact with gH/gL was also analyzed using the BiFC assay. While this assay is not a quantitative method to measure the glycoprotein interactions, we did observe a slight increase in YFP fluorescence for some of the BiFC construct pairs. Although the difference in YFP fluorescence between the EBV/Rh gB-YFPN constructs with Rh gH/gL-YFPC was very modest, it is intriguing that the BiFC data correlated with the functional data from the fusion assay. EBV/Rh (1-807) gB-YFPN and EBV/Rh (456-807) gB-YFPN had slightly increased levels of YFP above the other chimeras and EBV gB-YFPN. This data would suggest that these regions are important for the interaction with Rh gH/gL. The EBV/Rh gB chimeras containing regions (1-807) and (456-807) also exhibited increased fusion function with Rh gH/gL when examined in the cell fusion assay. The data obtained in this study is summarized in Table 2.
Table 2.
Summary of Results.
| Region of Rh gB inserted in EBV gB |
Expression similar to WT gB |
Higher molecular size gB variant |
Fusion with EBV gH/gLa |
Fusion with Rh gH/gLb |
%YFP with EBV gH/gL |
% YFP with Rh gH/gL |
Domain of inserion in EBV gB |
|---|---|---|---|---|---|---|---|
| 1 - 254 | Yes | No | 8 | 2 | NTc | NT | I and II |
| 254 - 346 | Yes | Variant | 16 | 2 | NT | NT | I and II |
| 1 - 346 | Yes | No | 8 | 2 | NT | NT | I and II |
| 346 - 456 | Yes | Yes | 71 | 8 | NT | NT | II |
| 254 - 628 | Yes | Yes | 98 | 35 | NT | NT | I, II, III and IV |
| 346 - 628 | Yes | Yes | 70 | 11 | NT | NT | II, III and IV |
| 456 - 628 | Yes | Yes | 97 | 32 | 93 | 72 | III and IV |
| 1 - 628 | Yes | Yes | 138 | 33 | NT | NT | I, II, III and IV |
| 456 - 807 | Yes | Yes | 143 | 70 | 84 | 79 | III, IV and V |
| 628 - 807 | Yes | Yes | 167 | 19 | 83 | 59 | V |
| 1 - 807 | Yes | Yes | 200 | 99 | 93 | 91 | I, II, III, IV and V |
Fusion activity presented as a percentage of wild-type EBV fusion
Fusion activity presented as a percentage of wild-type Rh-LCV fusion
NT, not tested.
The BiFC assay used in this study has been a valuable technique to study glycoprotein interactions in cells and has provides evidence of an interaction between EBV gB and gH/gL. However, one of the major limitations of this assay that has been frequently observed is the formation of non-specific interactions (Kerppola 2006; Kerppola 2006). We observed YFP fluorescence between a vast majority of the glycoprotein pairs that were tested, including gB constructs and chimeras that mediate fusion at low levels. The fact that the split YFP complementation assay can stabilize weak or transient interactions may explain why this technique was successful while other methods, like immunoprecipitation, have been unable to detect the gB-gH/gL interaction. The interaction observed in this study between EBV gB and gH/gL will therefore need to be further examined to confirm the biological significance of the interaction seen using BiFC. To control for non-specific interactions in our BiFC studies, we used the lowest concentration of our tagged constructs that would allow for the detection and quantification of YFP levels between samples. Our BiFC experiments were completed using transiently transfected cells that lack the EBV receptors, suggesting that the interaction we observe between EBV gB and gH/gL occurs prior to fusion initiation. The simultaneous analysis of the gB-gH/gL interaction using complementation studies of Rh-LCV and functional analysis of EBV/Rh gB chimeras provided additional evidence of a functional interaction between gB and gH/gL beyond BiFC analysis.
An interaction between gB and gH/gL was first demonstrated by BiFC analysis of HSV-1 glycoproteins in previous studies (Atanasiu, Whitbeck et al. 2007; Avitabile, Forghieri et al. 2007). These studies also demonstrated that a number of homologous and heterologous glycoprotein interactions occur between gD, gB, and gH/gL that are likely essential in the execution of fusion. Subsequent studies have characterized regions of HSV-1 gB important for its interaction with gH/gL, as well as recent analysis of possible gB binding sites on the structure of the HSV-2 gH/gL complex. Analysis of HSV-1 gB chimeras, in which portions of the glycoprotein were replaced with the corresponding region from human herpesvirus 8 (HHV-8) gB, found that while all chimeras could interact with gH/gL, some exhibited a reduced ability to interact with the complex (Avitabile, Forghieri et al. 2009). A separate study analyzed regions of HSV-1 gB that are involved in the interaction between gB and gH/gL using monoclonal gB antibodies that were able to block cell-cell fusion, gB-gH/gL BiFC, or both. Neutralizing antibodies that map to structural domains I, II and V of HSV-1 gB blocked the interaction between gB and gH/gL, as demonstrated by reduced BiFC, and fusion (Atanasiu, Whitbeck et al. 2010). Neutralizing antibodies that map to structural domain IV of HSV-1 gB were effective in blocking cell-cell fusion but had no effect on the interaction between gB and gH/gL, thus distinguishing the role of gB in mediating fusion and the interaction with gH/gL that precedes fusion. Taken together, data from these studies provide strong evidence that multiple sites on gB are required for the interaction with gH/gL. The findings from the current study are consistent with the results from these studies of HSV-1 gB, although the specific regions of gB required for the interaction with gH/gL may be different. We found that sites from two regions of gB (456-628 and 628-807) are both important for a functional interaction with gH/gL, similar to the observation that multiple sites on HSV-1 gB are likely required for the interaction with gH/gL. Our analysis of EBV gB identified sites important for the interaction with gH/gL in domains III, IV, and V, while HSV-1 gB domains I, II, and V were found to be important for the interaction with gH/gL (Avitabile, Forghieri et al. 2009; Atanasiu, Whitbeck et al. 2010). The gB binding site on the newly identified structure of HSV-2 gH/gL was mapped to a region proximal to the gH/gL interface (Chowdary, Cairns et al. 2010). Interestingly, the crystal structure of the HSV-2 gH/gL complex does not resemble any previously characterized viral fusion proteins, providing further evidence that gB acts as the sole fusogen (Chowdary, Cairns et al. 2010). Instead, it has been proposed that HSV gH/gL activates gB for fusion, likely through a direct interaction between these glycoproteins, similar to what we have proposed for the activation of EBV gB by the gH/gL complex.
Results from the current study demonstrate the importance of specific domains of gB that are required for fusion function with gH/gL and the ability of the virus to mediate fusion with target cells. Previous studies have shown that EBV glycoproteins gp42 and gH/gL form a tripartite complex in cells that is also essential for fusion (Li, Turk et al. 1995; Kirschner, Omerovic et al. 2006; Omerovic and Longnecker 2007). Furthermore, analysis of Rh gL suggested that the interaction between EBV gB and the gH/gL complex may be through a direct interaction between gB and gL (Plate, Smajlovic et al. 2009). Thus, it appears that multiple interactions between the glycoproteins necessary for fusion likely occur prior to and during EBV entry, as has also been recently suggested for HSV (Atanasiu, Whitbeck et al. 2010). The significance of each interaction and the sequence in which these events occur will need to be the subject of further investigation to elucidate the mechanism of EBV entry. However, based on our current knowledge of these interactions we would propose a model in which EBV-mediated fusion with B cells occurs through sequential binding of the glycoproteins upon binding of gp42 to HLA class II. Receptor binding induces a conformational change in the gp42/gH/gL tripartite complex that allows recruitment and/or activation of gB to the glycoprotein complex to mediate fusion through insertion of gB fusion loops into the target cell membrane. In conclusion, our studies, in line with studies of other herpesviruses, highlight the complexity of herpesvirus-induced membrane fusion and the requirement for multiple interactions of the virus-encoded glycoproteins for the overall process.
Materials and Methods
Cells
Chinese Hamster Ovary cells (CHO-K1) kindly provided by Nanette Susmarski were grown in Ham's F-12 medium (BioWhittaker) containing 10% FetalPlex animal serum complex (Gemini Bio-Products) and 1% penicillin-streptomycin (100 U penicillin/mL, 100 μg streptomycin/mL; BioWhittaker). Cells were grown in 75-cm2 cell culture flasks and detached using either trypsin-Versene (BioWhittaker) or Versene (phosphate-buffered saline [PBS]-1 mM EDTA. The Daudi 29 cell line stably expressing T7 RNA polymerase (Silva, Omerovic et al. 2004) were grown in RPMI 1640 medium (BioWhittaker) containing 10% Fetal Bovine Serum (HyClone) and 1% penicillin-streptomycin with 900 μg/mL G418 (Sigma).
Antibodies
The monoclonal gB antibody CL55 and the monoclonal antibody E1D1 that recognizes the gH/gL complex were generously provided by Lindsay Hutt-Fletcher (Louisiana State Health Sciences Center, Shreveport, LA) (Balachandran, Oba et al. 1987). The polyclonal gB antibody was generated by immunization of rabbits with EBV gB expression vectors (Aldevron) (McShane and Longnecker 2004).
Construction of EBV/Rhesus gB chimeric glycoproteins
Amino acid residues 1-254, 254-346, 1-346, 346-456, 254-628, 346-628, 456-628, and 456-807 of Rh-LCV gB were amplified by PCR using primers designed with flanking restriction enzyme digest sites unique to EBV gB in pSG5 (Stratagene). The amplified regions of Rh gB were then ligated into the predigested EBV gB using the restriction sites listed for each chimera: ERh gB (1-254) EcoRI/PmlI ; ERh gB (254-346) PmlI/SapI; ERh gB (1-346) EcoRI/SapI ; ERh gB (346-456) SapI/PasI; ERh gB (254-628) PmlI/PstI; ERh gB (346-628) SapI/PstI; ERh gB (456-628) PasI/PstI; and ERh gB (456-807) PasI/AfeI. Amino acid residues 1-628, 1-807, and 628-807 of Rh gB were isolated using restriction enzyme digestion using the unique EcoRI, PstI, and AfeI sites and ligated into the corresponding sites in EBV gB in pSG5. EBV/Rh-LCV chimeras (EBV/Rh) were named based on the amino acid sequence location of Rh gB inserted. All mutants were sequenced by the Northwestern Biotech core sequencing facility to confirm that the appropriate sequences were present and no mutations were introduced by PCR. DNA was isolated using Qiagen Maxi Kits (Qiagen).
Transfections
All transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen). For the fusion assay cells were seeded into six well plates twenty hours before transfection in order to reach 80% confluency and transiently transfected with 0.5 μg each of EBV or Rh gB, gH, and gL; 2 μg of gp42; and 0.8 μg of a luciferase reporter plasmid with a T7 promoter (Okuma, Nakamura et al. 1999; Haan, Lee et al. 2001). For Western blot experiments, CHO-K1 cells were plated in T-25 cm2 cell culture flasks to reach 70% confluency 24 hours later for transfection with 10 μg of gB. For all transfections, cells were washed 6 hours after transfection and serum-containing Ham's F-12 medium was added.
Fusion assay
The virus-free cell-based fusion assay was described previously (Haan, Lee et al. 2001; McShane and Longnecker 2004). Effector CHO-K1 cells were transfected with EBV or Rh-LCV glycoproteins or mutant proteins as stated above. Twelve hours post-tranfection cells were detached with Versene, counted with a Beckman Coulter Z1 particle counter and mixed 1:1 with target B cells (0.25 × 106 per sample) into a 24-well plate in 1 mL Ham's F-12 medium. The target cell, Daudi 29 B cells, stably express T7 RNA polymerase and were maintained in G418 selection media (Omerovic, Lev et al. 2005) . Twenty-four hours later, the cells were washed with PBS and lysed with 100 μL of passive lysis buffer (Promega). Luciferase was quantified in duplicate by transferring 20 μL of lysed cells to a 96-well plate and adding 100 μL of luciferase assay reagent (Promega) and luminescence was measured on a Perkin-Elmer Victor plate reader. Luciferase activity was normalized to wild-type levels and set to 100%.
Biotinylation and immunoprecipitation of biotinylated gB samples
Biotinylation of cell-surface expressed gB was performed as previously described (Reimer, Backovic et al. 2009). CHO-K1 cells were transfected with gB as described above, washed with PBS, and detached with 1 mM EDTA in PBS 24 hours after transfection. EBV, Rh, and chimeric gB expressing cells were washed twice with ice cold PBS and counted using a Z1 Coulter particle counter (Beckman Coulter). 2.5 × 107 cells were incubated in a final concentration of 2 mM EZ-Link Sulfo-NHS-LC-Biotin (Pierce) and rotated at 4°C for 30 minutes. Samples were washed three times with PBS containing 100 mM Glycine to stop the reaction before the cells were pelleted and resuspended in 1% Triton X-100 lysis buffer. Cell lysates were incubated at 4°C for 30 minutes before insoluble material was removed by centrifugation at 13,000 rpm at 4°C. 20 μL of protein G-Sepharose beads (GE Healthcare) were rotated with 1 μL CL55 gB antibody and lysates overnight at 4°C to precipitate gB. Beads were washed four times with 1 mL of Triton X-100 lysis buffer. Bound biotinylated protein was eluted in SDS buffer without β-mercaptoethanol and boiled at 70°C for 10 minutes.
SDS-PAGE and Western blot analysis of gB
CHO-K1 cells were transfected as described above and washed twice with PBS 24 hours after transfection. Cells were detached using 1mM EDTA in PBS, pelleted by centrifugation, and counted using a Z1 Coulter particle counter (Beckman Coulter). Cell pellets were lysed with 1% Triton X-100 lysis buffer (at 1 mL lysis buffer per 10 million cells) and insoluble material was removed by centrifugation at 13,000 rpm at 4°C. For intracellular protein expression samples were mixed with SDS buffer containing β-mercaptoethanol, boiled at 95°C for 10 minutes and pelleted by centrifugation. For analysis of oligomer formation, SDS buffer lacking β-mercaptoethanol was used. All gB sample supernatants were separated on 7.5% Criterion sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (Bio-Rad) and transferred to Immobilon membranes (Millipore). Membranes were blocked in Tris-buffered saline plus 1% Tween-20 with 5% milk for 30 minutes and probed overnight at 4°C with the polyclonal gB antibody (1:2000 dilution). Membranes were washed in Tris-buffered saline plus Tween-20, and secondary HRP-conjugated anti-rabbit antibody was applied for 0.5 hours at room temperature. Membranes were then washed and blots were incubated in ECL Western Blotting Detection reagents (GE Healthcare) and exposed to Hyperfilm (GE Healthcare).
Construction of EBV glycoproteins with YFP fragments
The constructs of EBV glycoproteins with the YFP amino or carboxyl terminal fragments were derived from the previously described HSV-1 split YFP glycoproteins that were generously provided by Gary Cohen and Roselyn Eisenberg (Atanasiu, Whitbeck et al. 2007). The amino and carboxyl terminal fragments of a variant enhanced yellow fluorescent protein (EYFP), called Venus, were PCR-amplified and ligated into the PCAGGS expression vector between the sites KpnI and XhoI. The N-terminal portion of EYFP (YFPN) contains residues 1-173 and the C-terminal portion of EYFP (YFPC) contains residues 173-239. EBV glycoproteins gB and gH and Rh-LCV glycoprotein gH were PCR-amplified using primers that excluded the stop codon. The glycoproteins were ligated in frame with the N or C-terminal portions of the EYFP open reading frame. EBV gB and Rh gH were ligated in frame into the PCAGGS-YFPN and PCAGGS-YFPC plasmids between the sites EcoRI and KpnI. EBV gH was ligated in frame into the PCAGGS-YFPN and PCAGGS-YFPC plasmids at the ClaI site. All glycoprotein contructs were sequenced by the Northwestern Biotech core sequencing facility to confirm the appropriate sequences were present and no mutations were introduced. DNA was isolated using Qiagen Maxi Kits (Qiagen).
Construction of EBV/Rh chimeric gB glycoproteins with YFP N-terminal fragment
EBV/Rh gB chimeras selected for further analysis were digested with their corresponding restriction enzyme pairs: ERh gB (456-628) PasI/PstI; ERh gB (628-807) PstI/AfeI; ERh gB (456-807) PasI/AfeI, and ERh gB (1-807) EcoRI/AfeI. The Rh gB inserts were ligated into the predigested EBV gB-YFPN glycoprotein construct in the pSG5 expression vector. All chimeric YFPN glycoprotein contructs were sequenced by the Northwestern Biotech core sequencing facility and DNA isolated using Qiagen Maxi Kits (Qiagen).
Bimolecular fluorescence complementation (BiFC) analysis
CHO-KI cells were seeded on glass coverslips in 6-well dishes and transfected using Lipofectamine 2000 as described above. For each glycoprotein split YFP construct, 200 ng of DNA was transfected, with the PCAGGS expression vector used to equalize DNA levels between samples. Media was replaced after 6 hours of transfection, and cells washed with PBS after 20 hours. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, followed by staining with either the rabbit polyclonal gB antibody (1:500 dilution) or the monoclonal gH/gL complex specific E1D1 antibody (1:50 dilution). Anti-rabbit and anti-mouse secondary antibodies carrying either a cy3 or cy5 fluorescent tag (Jackson Immuno) were used at a 1:500 dilution. Coverslips were mounted with ProLong Gold Antifade (Invitrogen) and imaged on a Zeiss LSM 510 META laser scanning confocal microscope. A UV 405 nm laser, Argon 488/543 laser, and HeNe 633 laser were used to excite the fluorescence of DAPI, YFP, cy3, and cy5, respectively. Images were collected using a 40X oil immersion objective, at 1,024 × 1,024 pixel resolution and a 16-bit intensity scale. The settings were adjusted while imaging the control samples and not modified between the remaining samples.
Acknowledgements
We thank Lindsey Hutt-Fletcher for her generous gift of the E1D1 and CL55 antibody cell line. Gary Cohen and Roz Eisenberg for kindly proving the constructs used for BiFC and Fred Wang for providing the DNA used fort he construction of the Rhesus glycoprotein expression vectors. Finally, we thank the members of the Longnecker and Jardetzky laboratories for help and support. This research was supported by AI076183 (R.L. and T.J.) and AI067048 (R.L.) from National Institute of Allergy and Infectious Diseases and CA117794 (R.L. and T.J.) from the National Cancer Institute. A.P. is supported by a pre-doctoral training award from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atanasiu D, Whitbeck JC, et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A. 2007;104(47):18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D, Whitbeck JC, et al. Bimolecular complementation defines functional regions of HSV gB that are involved with gH/gL as necessary steps leading to cell fusion. J Virol. 2010 doi: 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile E, Forghieri C, et al. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol. 2007;81(20):11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile E, Forghieri C, et al. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J Virol. 2009;83(20):10752–10760. doi: 10.1128/JVI.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock GJ, Decker LL, et al. EBV persistence in memory B cells in vivo. Immunity. 1998;9(3):395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- Backovic M, Jardetzky TS. Class III viral membrane fusion proteins. Curr Opin Struct Biol. 2009;19(2):189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M, Jardetzky TS, et al. Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity. J Virol. 2007;81(17):9596–9600. doi: 10.1128/JVI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M, Longnecker R, et al. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A. 2009;106(8):2880–2885. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran N, Oba DE, et al. Antigenic cross-reactions among herpes simplex virus types 1 and 2, Epstein-Barr virus, and cytomegalovirus. J Virol. 1987;61(4):1125–1135. doi: 10.1128/jvi.61.4.1125-1135.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova LS, Nishimura SL, et al. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc Natl Acad Sci U S A. 2009;106(48):20464–20469. doi: 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YG, Gordadze AV, et al. Evolution of two types of rhesus lymphocryptovirus similar to type 1 and type 2 Epstein-Barr virus. J Virol. 1999;73(11):9206–9212. doi: 10.1128/jvi.73.11.9206-9212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdary TK, Cairns TM, et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nature Structural and Molecular Biology. 2010;17(7):882–889. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson-Welsh L, Spear PG. Oligomerization of herpes simplex virus glycoprotein B. J Virol. 1986;60(2):803–806. doi: 10.1128/jvi.60.2.803-806.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini EA, Luka J, et al. Identification of an Epstein-Barr virus glycoprotein which is antigenically homologous to the varicella-zoster virus glycoprotein II and the herpes simplex virus glycoprotein B. Virology. 1987;157(2):552–555. doi: 10.1016/0042-6822(87)90300-x. [DOI] [PubMed] [Google Scholar]
- Fingeroth JD, Weis JJ, et al. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci U S A. 1984;81(14):4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M, Kieff E. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. J Virol. 1990;64(4):1507–1516. doi: 10.1128/jvi.64.4.1507-1516.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M, Ooka T, et al. Epstein-Barr virus glycoprotein homologous to herpes simplex virus gB. J Virol. 1987;61(2):499–508. doi: 10.1128/jvi.61.2.499-508.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald K, Desai P, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302(5649):1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Haan KM, Lee SK, et al. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology. 2001;290(1):106–114. doi: 10.1006/viro.2001.1141. [DOI] [PubMed] [Google Scholar]
- Hannah BP, Cairns TM, et al. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J Virol. 2009;83(13):6825–6836. doi: 10.1128/JVI.00301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah BP, Heldwein EE, et al. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J Virol. 2007;81(9):4858–4865. doi: 10.1128/JVI.02755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Lou H, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313(5784):217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- Hutchinson L, Browne H, et al. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66(4):2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Complementary methods for studies of protein interactions in living cells. Nat Methods. 2006;3(12):969–971. doi: 10.1038/nmeth1206-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1(3):1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7(6):449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner AN, Omerovic J, et al. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J Virol. 2006;80(19):9444–9454. doi: 10.1128/JVI.00572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK. Four consecutive arginine residues at positions 836-839 of EBV gp110 determine intracellular localization of gp110. Virology. 1999;264(2):350–358. doi: 10.1006/viro.1999.0012. [DOI] [PubMed] [Google Scholar]
- Li Q, Spriggs MK, et al. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J Virol. 1997;71(6):4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Turk SM, et al. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69(7):3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane MP, Longnecker R. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc Natl Acad Sci U S A. 2004;101(50):17474–17479. doi: 10.1073/pnas.0404535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane MP, Longnecker R. Analysis of fusion using a virus-free cell fusion assay. Methods Mol Biol. 2005;292:187–196. doi: 10.1385/1-59259-848-x:187. [DOI] [PubMed] [Google Scholar]
- Moghaddam A, Koch J, et al. Infection of human B lymphocytes with lymphocryptoviruses related to Epstein-Barr virus. J Virol. 1998;72(4):3205–3212. doi: 10.1128/jvi.72.4.3205-3212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ibata K, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20(1):87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nemerow GR, Wolfert R, et al. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2) J Virol. 1985;55(2):347–351. doi: 10.1128/jvi.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhierl B, Feederle R, et al. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc Natl Acad Sci U S A. 2002;99(23):15036–15041. doi: 10.1073/pnas.232381299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma K, Nakamura M, et al. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology. 1999;254(2):235–244. doi: 10.1006/viro.1998.9530. [DOI] [PubMed] [Google Scholar]
- Omerovic J, Lev L, et al. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J Virol. 2005;79(19):12408–12415. doi: 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omerovic J, Longnecker R. Functional homology of gHs and gLs from EBV-related gamma-herpesviruses for EBV-induced membrane fusion. Virology. 2007;365(1):157–165. doi: 10.1016/j.virol.2007.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papworth MA, Van Dijk AA, et al. The processing, transport and heterologous expression of Epstein-Barr virus gp110. J Gen Virol. 1997;78(Pt 9):2179–2189. doi: 10.1099/0022-1317-78-9-2179. [DOI] [PubMed] [Google Scholar]
- Plate AE, Smajlovic J, et al. Functional analysis of glycoprotein L (gL) from rhesus lymphocryptovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J Virol. 2009;83(15):7678–7689. doi: 10.1128/JVI.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer JJ, Backovic M, et al. Analysis of Epstein-Barr virus glycoprotein B functional domains via linker insertion mutagenesis. J Virol. 2009;83(2):734–747. doi: 10.1128/JVI.01817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson AB, K. E, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. Epstein-Barr virus. [Google Scholar]
- Rivailler P, Carville A, et al. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood. 2004;104(5):1482–1489. doi: 10.1182/blood-2004-01-0342. [DOI] [PubMed] [Google Scholar]
- Rivailler P, Cho YG, et al. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J Virol. 2002;76(23):12055–12068. doi: 10.1128/JVI.76.23.12055-12068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivailler P, Jiang H, et al. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J Virol. 2002;76(1):421–426. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S, Bressanelli S, et al. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313(5784):187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- Silva AL, Omerovic J, et al. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J Virol. 2004;78(11):5946–5956. doi: 10.1128/JVI.78.11.5946-5956.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorem J, Longnecker R. Cleavage of Epstein-Barr virus glycoprotein B is required for full function in cell-cell fusion with both epithelial and B cells. J Gen Virol. 2009;90(Pt 3):591–595. doi: 10.1099/vir.0.007237-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PG, Longnecker R. Herpesvirus entry: an update. J Virol. 2003;77(19):10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck P, Haan KM, et al. Epstein-Barr virus entry into cells. Virology. 2000;277(1):1–5. doi: 10.1006/viro.2000.0624. [DOI] [PubMed] [Google Scholar]
- Tanner J, Weis J, et al. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50(2):203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- Thorley-Lawson DA, Babcock GJ. A model for persistent infection with Epstein-Barr virus: the stealth virus of human B cells. Life Sci. 1999;65(14):1433–1453. doi: 10.1016/s0024-3205(99)00214-3. [DOI] [PubMed] [Google Scholar]
- Wang F, Rivailler P, et al. Simian homologues of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci. 2001;356(1408):489–497. doi: 10.1098/rstb.2000.0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hutt-Fletcher LM. Compatibility of the gH homologues of Epstein-Barr virus and related lymphocryptoviruses. J Gen Virol. 2007;88(Pt 8):2129–2136. doi: 10.1099/vir.0.82949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]