Abstract
Despite advances in the prevention and management of cardiovascular disease (CVD), this group of multifactorial disorders remains a leading cause of mortality worldwide. CVD is associated with multiple genetic and modifiable risk factors; however, known environmental and genetic influences can only explain a small part of the variability in CVD risk, which is a major obstacle for its prevention and treatment. A more thorough understanding of the factors that contribute to CVD is, therefore, needed to develop more efficacious and cost-effective therapy. Application of the ‘omics’ technologies will hopefully make these advances a reality. Epigenomics has emerged as one of the most promising areas that will address some of the gaps in our current knowledge of the interaction between nature and nurture in the development of CVD. Epigenetic mechanisms include DNA methylation, histone modification, and microRNA alterations, which collectively enable the cell to respond quickly to environmental changes. A number of CVD risk factors, such as nutrition, smoking, pollution, stress, and the circadian rhythm, have been associated with modification of epigenetic marks. Further examination of these mechanisms may lead to earlier prevention and novel therapy for CVD.
Introduction
Cardiovascular disease (CVD) is one of the leading causes of death worldwide. The prevalence of CVD has increased dramatically over the past 100 years, and this disease will remain a major health concern for the next two decades, even under the most optimistic scenarios.1–3 The emergence of CVD as a major public health problem during the 1940s initiated research to understand the factors associated with this disease. Projects such as the Framingham Heart Study were launched to find common biochemical, environmental, and behavioral factors or characteristics that contribute to CVD.4 As a result, several conditions, such as hypertension, obesity and diabetes, were identified and are commonly used today to detect and treat individuals at risk. This knowledge has contributed to the substantial decline in CVD mortality observed in Western Europe and in the USA over the past 30 years; however, this trend is primarily due to improvements in quality of care and treatment, and much less to the prevention of the disease itself. Lifestyle changes in low-income and middle-income countries have resulted in an increased prevalence of obesity and diabetes and, therefore, higher rates of CVD.5 Current recommendations for the treatment of CVD aim to reduce modifiable risk factors, such as high levels of LDL cholesterol.6 However, environmental and genetic effects can only explain a small part of the variability in CVD risk, even for well-established risk factors.7 New knowledge is needed, therefore, to find more efficacious and cost-effective preventive and therapeutic approaches.8
The ‘omics’ technologies, in particular genomics, transcriptomics, proteomics, and metabolomics, are revolutionizing our understanding of mammalian biology. Over 400 different areas of ‘omics’ have been catalogued, and are being used to advance the concept of systems biology.9 Epigenomics has emerged as one of the most promising approaches that will help to address gaps in our knowledge concerning inheritance of common traits. Epigenetics describes the mechanisms that enable cells to respond quickly to environmental changes and provide a link between genes and the environment. Individual variation in the epigenetic modification of genes can explain a larger part of the phenotypic variation observed in humans than differences in genotype alone.10 Several comprehensive reviews have focused on the relationship between epigenetics and CVD risk factors, including homocysteine—the biomarker most directly implicated in epigenetic mechanisms relating to CVD risk.11–23 Homocysteine-induced alterations in DNA methylation of vascular smooth muscle cells are involved in atherogenesis.24 However, the cardiovascular field is not as advanced as other areas in terms of epigenetic-related research. A search of the PubMed database using the terms “epigenetics and cancer” produced a total of 10,867 publications, of which 1,994 were reviews, whereas a search for “epigenetics and cardiovascular” returned only one-tenth of the manuscripts (1,031 in total and 110 reviews). The lack of understanding of the contribution of epigenetics to CVD is summarized in a review by Turunen and colleagues who highlight a number of important points.12 First, genomic DNA isolated from human atherosclerotic lesions is hypomethylated. However, data from peripheral blood lymphocytes in patients with coronary heart disease have been inconsistent, with reports of increases25 or decreases26,27 in methylation status. Likewise, no consistent data are available regarding histone modification in these lesions. Second, specific methylation changes, usually hypermethylation, have been found in the promoter region of genes associated with atherosclerosis, such as extracellular superoxide dismutase, estrogen receptor α, endothelial nitric oxide synthase, and 15-lipoxygenase. Third, inflammation—a driver of the formation, progression, and rupture of atherosclerotic plaques—has also been associated with hypermethylation.28 Whether epigenetic changes are causally related to the pathogenetic features in the lesions, or whether they merely represent a consequence of the ongoing pathological process remains unclear (Figure 1).12 Epigenetic changes could also partly explain the poorly understood environmental and dietary effects on atherogenesis, and the differences in the incidence of coronary heart disease observed in various populations. The emphasis of ongoing work is on environmental factors that cause epigenetic changes, and how these processes relate to CVD risk. Investigating the plasticity of the epigenome may also enable the development of much-needed novel CVD therapeutic approaches.29–33
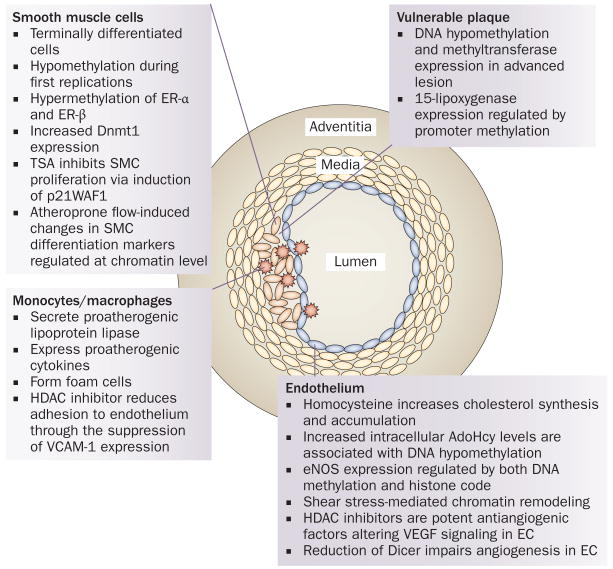
Figure 1.
Epigenetics and atherosclerosis. Epigenetic changes, such as methylation and deacetylation, are associated with atherosclerosis and have been identified in specific tissues in the arterial wall; however, causal relationships have not been established. Enzymatic regulators of epigenetic processes, such as HDAC inhibitors, inhibit atherosclerosis. Altered methylation patterns occur in conjunction with SMC proliferation, which is suppressed by the HDAC inhibitor TSA. HDAC inhibitors also reduce monocyte adhesion to the endothelium. Abnormal DNA methylation is associated with plasma homocysteine levels, and methylation also regulates eNOS expression. HDAC inhibitors reduce angiogenesis through VEGF signaling in endothelial cells. in vulnerable plaques, DNA and the 15-lipoxygenase promoter are hypomethylated. Abbreviations: AdoHcy, adenosylhomocysteine; Dicer, endoribonuclease Dicer; Dnmt1, DNA (cytosine-5)-methyltransferase 1; EC, endothelial cell; eNOS, endothelial nitric oxide synthase; ER, estrogen receptor; HDAC, histone deacetylase; p21WAF1, leucine carboxyl methyltransferase 1; SMC, smooth muscle cell; TSA, trichostatin A; VCAM, vascular cell adhesion molecule; VEGF, vascular endothelial growth factor. Reprinted from Biochim. Biophys. Acta 1790(9), Turunen, M. P., Aavik, E. & Ylä-Herttuala, S. Epigenetics and atherosclerosis. 886–891 © 2009, with permission from Elsevier.
In this Review, we propose that the identification and understanding of epigenetic factors will reduce longstanding gaps in our current knowledge of CVD risk. External influences and inherited traits that lead to epigenetic modifications are discussed, as well as their role in CVD risk. Studies evaluating potential therapeutic agents are also highlighted, including those focused on nutritional compounds that might act epigenetically in the prevention or treatment of CVD.
Epigenetics—then and now
Following the publication of the 3 billion base-pair sequence that makes up the human genome, the knowledge that we have accumulated through the use of increasingly powerful genomic approaches cannot fully explain most of our inheritance, the functionality of our cells, or their disruptions and accompanying disease states.34 Epigenetics will help us to advance this knowledge. The term epigenetics describes the concept itself, as ‘epi’ means above or over in Greek, but it was Conrad Waddington who, in 1940, provided the first broad and operational definition of epigenetics as “the causal interactions between genes and their products, which bring the phenotype into being”.35 Waddington also developed the related notion of genetic canalization—the process whereby a trait becomes buffered against allelic heterogeneity.36 Mechanistic evidence to support the concept was provided three decades later when Holliday and Pugh identified DNA methylation as the first mechanism involved in epigenetics.37 Moreover, methylation provided some explanation to puzzles such as X-chromosome inactivation and gene imprinting.38 The next breakthrough came with the identification of histone modifications in the mid-1990s, and the DNA world expanded from one dimension (linear sequence of base pairs) to three dimensions (nuclear topology), with the realization of the importance of chromatin structure in the regulation of the genome.39 At that time, methylation and histone modifications became known as epigenetic marks. The discovery of the noncoding RNAs known as microRNAs (miRNAs), provided further evidence of post-translational mechanisms that were classified as epigenetic marks. miRNAs are small RNAs, approximately 22 nucleotides in length, that repress the expression of mRNAs with which they are entirely or partially complementary (Figure 2).40 Although the specific mechanisms of mRNA repression remain unclear, binding of miRNA to mRNA triggers the activity of Argonaute proteins, which are critical for gene silencing in plants and animals.41 The study of miRNAs and their effects on translation is one of the most novel and active areas of epigenetic research; current knowledge supports the premise that miRNAs regulate key genetic programs in cardiovascular biology, and are critical for cardiac development, endothelial function, lipid metabolism, ventricular hypertrophy, and post-infarction dysrhythmias.42 Rayner et al. identified miR-33 as a regulator of cholesterol homeostasis that modulates both HDL biogenesis in the liver and cellular cholesterol efflux.43 Relationships between hydroxymethylcytosine residues and CVD have not yet been reported, but hydroxymethyl-2′-deoxycytidine is abundant in the brain and might participate in epigenetic regulation of neuronal function.44
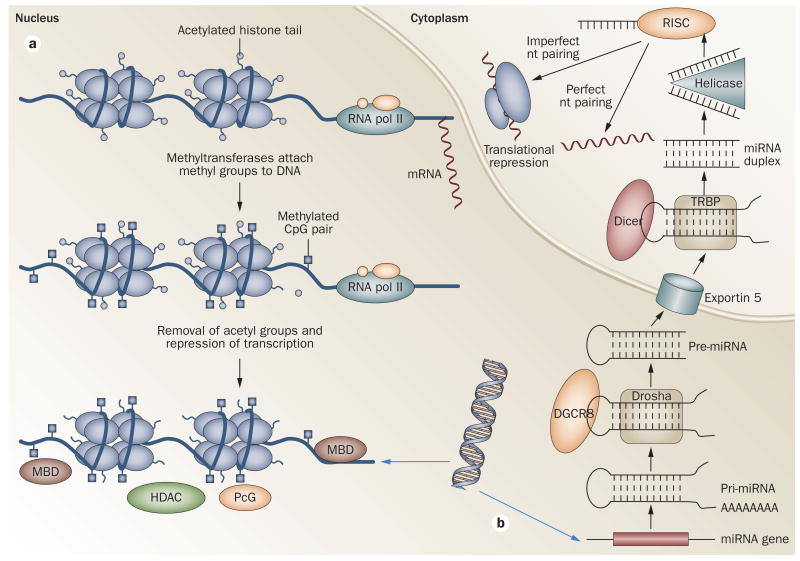
Figure 2.
Epigenetic modifications that regulate transcription or translation. a. CpG dinucleotides are methylated by METs at the 5′ position of the cytosine residue. MBDs bind to methylated CpG DNA and repress transcription. Histone deacetylation also results in transcription repression. PcGs bind to the DNA and block access of transcription factors to DNA. in addition, they interact with HDACs and/or METs to mediate gene silencing, and contribute to histone methylation. Conversely, HATs and methyltransferases catalyze histone acetylation and methylation, which increases accessibility to DNA and gene activation. b. in the nucleus, miRNAs are transcribed by RNA polymerase ii to generate pri-miRNAs. These transcripts are processed by the Drosha/DGCR8 complex, which results in the formation of pre-miRNAs. Exportin 5 transports the pre-miRNAs into the cytoplasm, where Dicer processes them into ∼22-nucleotide duplexes that contain the mature miRNA duplex. Single-stranded miRNA is catalyzed through the activity of a helicase and binds to RISC, which directs the miRNA to target mRNAs. Two situations may occur: if the miRNA and target are complementary, translational degradation occurs; alternatively, if the complementarity is partial, then translation repression takes place. Abbreviations: DGCR8, microprocessor complex subunit DGCR8; Dicer, endoribonuclease Dicer; HAT, histone acetyltransferase; HDAC, histone deacetyltransferase; MBD, methyl CpG-binding protein; MET, methyltransferase; miRNA, microRNA; nt, nucleotide; PcG, polycomb group; pol II, polymerase II; pri-miRNA, primary transcripts; RiSC, RNA-induced silencing complex; TRBP, TAR RNA-binding protein.
Although current work focuses on epigenetic mechanisms of disease, epigenetic processes are essential to normal growth and development, and homeostasis. Some authors in the field propose a definition of epigenetics as the “molecular factors and processes around DNA that are mitotically stable and regulate genome activity independently of DNA sequence”.45 Hypermethylation of DNA and hypoacetylation of histones alter chromatin structure and regulate gene expression during vertebrate embryogenesis in mammalian and nonmammalian species.46 Cellular differentiation and neuronal plasticity are well-described examples of the critical role of epigenetic regulation in a physiologic system, but analogous examples are undoubtedly present in all systems.47 Evidence suggests, for instance, a role for miRNAs in the regulation of apoptosis, but further research is needed to provide a more comprehensive overview of these mechanisms.48
Epigenetics—adaptation to the environment
Positive natural selection describes the increase in the prevalence of beneficial traits in a population, and is one of the factors driving evolution. A trait under positive selection must be beneficial, thus increasing survival and reproduction, and the trait must also be heritable. Beneficial traits differ widely and may include anything from resistance to infections such as malaria,49 to lactose tolerance, which allows the utilization of a food source.50 This positive selection has been attributed to particular DNA variants that become more common because of the beneficial effects on those who carry them.51 However, a trait that may be advantageous at one point in human history may be detrimental under different environmental conditions. The classic example of this is the ‘thrifty’ genotype, a concept coined by the geneticist James Neel to explain the prevalence of obesity and diabetes.52 Neel hypothesized that people with diabetes often have allelic variations in certain genes that enable them to efficiently collect and utilize food.52 Carriers of these variations are, therefore, at a high risk of developing obesity and diabetes when exposed to the dietary and behavioral conditions of the developed world, which are characterized by an abundant and cheap supply of energy, and a dramatic reduction in energy expenditure. This thrifty genotype would have been beneficial during periods of history characterized by food instability.53
Genetic selection results from long-term exposure to specific environmental changes, and involves a response that is both slow and long-term. Whether selective forces will eventually eliminate or substantially reduce the thrifty genotype remains a matter of speculation. Although the thrifty genotype may provide an advantage in the context of an unstable food supply, metabolic diseases that are associated with thriftiness in the presence of abundant food are unlikely to affect survival before reproductive age. Hypothetically, the reduced fitness of thrifty genotype carriers, which would lead to decline in its frequency, may be offset by advances in medical care that ensure longer-term survival of at-risk genotypes. In contrast to the long-term nature of the selective forces that are hypothesized to have positively selected the thrifty genotype, epigenetic changes occur rapidly to alter metabolism. The ways in which epigenetic changes increase disease risk, perhaps in combination with evolutionarily mediated phenotypes, are described below.
Nutrition, epigenetics, and CVD risk
Throughout history, humans have had to cope with acute adverse environmental changes, such as periods of plague or famine. Famines provide an ideal setting to examine the biological adaptation and long-term consequences of unfavorable conditions, especially during human development. One of the most well documented events is the Dutch Hunger during the winter of 1944–1945.54 The trigger of this famine was a call by the Dutch government for a national railway strike to hinder German military initiatives. In retaliation, German authorities blocked all food supplies to the occupied west of the country in October 1944. Official rations consisted mostly of bread and potatoes, and by April 1945, people were surviving on as little as 500 calories per day. Widespread starvation was seen, especially in the cities of the western Netherlands.55 This event has permitted researchers to examine the long-term consequences of famine. The outcomes of these studies strongly support that in utero exposure to famine may result in adverse metabolic or mental health phenotypes, which depend on the sex of the exposed individual, and the timing of exposure during gestation as the embryo is more vulnerable around the time of conception. A sex-specific increased risk of obesity, dyslipidemia, and cerebrocardiovascular-related deaths has been observed six decades later among those whose mothers were exposed to famine early in gestation.56 For example, increased fat deposition with greater BMI and waist circumference was observed only in women, but detrimental changes in HDL cholesterol concentrations were observed only in men.57,58 Similar observations inspired the Barker's hypothesis almost three decades ago, which was developed following studies of birth and death records; a substantial correlation between birth weight and rates of adult death from ischemic heart disease was revealed, which related to the prenatal environment.59,60
Persistent epigenetic changes induced during early exposure to harsh conditions may explain the adverse phenotypes expressed later in life.61,62 This notion is supported by studies in experimental models that have shown the dramatic influence of maternal diet during pregnancy on the pattern of DNA methylation of specific genes, which resulted in permanent phenotypic changes such as coat color, body weight, and blood pressure.63,64 Proof that such mechanisms may be present in humans came from the work of Heijmans and colleagues, who studied individuals exposed to the Dutch Hunger.65 These investigators demonstrated that those who were exposed to famine in utero had very different methylation patterns in genes involved in growth and metabolic disease compared with controls (same-sex siblings who were not affected, or individuals exposed late in gestation). Methylation of IGF2 was lower, whereas methylation of the IL-10, LEP, ABCA1, GNASAS and MEG3 genes was higher.66 These results, combined with evidence of sex-specific methylation patterns,66,67 strongly suggest that early prenatal famine exposure results in persistent changes in DNA methylation.
Prenatal epigenetic contributions to adult CVD risk in humans are often inferred, but are difficult to confirm in observational studies. Animal models designed to evaluate the epigenetic mechanisms of intrauterine programming provide more concrete data in the context of controlled dietary interventions. Alkemade et al. demonstrated that the ApoE+/− offspring of ApoE−/− hypercholesterolemic mice were more susceptible to postnatal dietary-induced atherosclerotic changes than genetically similar offspring born to wild-type mothers.68 Neointima formation in susceptible offspring was associated with altered histone methylation and lysine methyltransferase expression in carotid endothelial and vascular smooth muscle cells.68 Although physiologic mechanisms for the differential response to diet cannot be concluded from this study, its examination of the combined effects of prenatal and postnatal influences raises intriguing possibilities for explaining differing CVD vulnerabilities, even in individuals with similar genetic backgrounds and dietary habits. In a second study examining epigenetic effects of intrauterine protein restriction, epigenetic changes were localized to specific genes that may contribute to CVD risk.69 Well-established metabolic consequences of reduced protein availability include insulin resistance, hypertension, and obesity. In this study, prenatal protein restriction was demonstrated to alter DNA methylation patterns in 204 genes in the fetal liver, including liver X receptor α (LXRA, also known as NR1H3), a nuclear receptor that targets downstream genetic regulators of lipid and fatty acid metabolism. Hypermethylation of the LXRA promoter was associated with reduced expression of LXR-α, together with reduced expression of genes encoding the established lipid regulators ABCG5, ABCG8 and ABCA1.69 The investigators were limited in their ability to detect differences in lipid metabolism in the fetal mice, and the potential consequences of additional methylation alterations (106 hypermethylated and 101 hypomethylated regions) are unknown. In spite of these limitations, the study's candidate gene approach demonstrates the potential usefulness of investigating known genes and their established genetic targets.
Smoking, epigenetics, and CVD risk
Prenatal exposure to tobacco smoke increases the risk of disease later in the child's life;70 these effects could in part be mediated by epigenetic changes. Breton et al. studied DNA methylation patterns in children exposed prenatally to tobacco smoke.71 DNA methylation patterns in the long interspersed element 1 (LINE-1) and in Alu elements, two classes of repetitive sequences that are normally hypermethylated, were examined. Moreover, the researchers investigated gene-specific CpG methylation. Exposed children had significantly lower levels of methylation of AluYb8 than controls (P = 0.03), whereas smoking-related effects on LINE-1 methylation were observed only in children who lacked functional glutathione S-transferase P (GSTP1), a detoxifying enzyme. Differential methylation of CpG loci in eight genes was identified through the screen, and these associations were modulated by specific alleles of other detoxifying genes. Life-long effects of in utero exposure to tobacco smoking may, therefore, be mediated through alterations in DNA methylation. Moreover, variants in detoxification genes may modulate these effects.
These observations support the notion that, at the personal level, we have a situation similar to that of the thrifty gene hypothesis; certain alleles have been selected through evolution that favor survival under nutritional hardship, but these alleles predispose to disease when calories are plentiful. The short-term epigenetic adaptations that take place shortly after conception under famine or other stress conditions, which are aimed at improving the survival of the newborn under such anticipated environmental conditions, may leave the individual ill-prepared to cope with excess calorie intake later in life. In the same way, the individual environmental stress of in utero tobacco exposure may produce epigenetic adaptations that augment evolutionarily selected responses to exacerbate metabolic dysregulation, leading to increased CVD risk. As in the case of maternal malnutrition, fetal exposure to smoking may produce short-term metabolic changes that compromise long-term survival.
Understanding the mechanisms that underlie the observed long latency periods between in utero tobacco exposure and CVD disease later in life is limited. Similar temporal patterns have been described in other common diseases, such as breast cancer, which is postulated to be modified in utero through epigenetic mechanisms.72 We are also not yet able to accurately quantify the relative contributions of in utero versus postnatal smoking or air pollution to CVD risk, or the contributions of epigenetic versus genetic interaction with the environment.
Environmentally induced epigenetic drift
Epigenetic changes are not restricted to the prenatal period. Elegant work by Fraga et al. demonstrated that monozygotic twins experience an epigenetic drift in relation to one another with advancing age.73 Differences in epigenetic modifications are accentuated by decreasing amounts of time shared together, and behavioral differences, such as smoking. Although most of this work has been carried out in the context of cancer, similar mechanisms may be relevant to CVD. Baccarelli et al. showed that pollution from traffic, an environmental challenge associated with increased CVD risk, affected DNA methylation in a cohort of individuals from Boston, MA, USA (n = 718).74,75 Their analysis focused on the repetitive elements LINE-1 and Alu. An association was found between exposure to black carbon and hypomethylation. Interestingly, the effect was only significant for the LINE-1 repetitive elements (P = 0.03). These data suggest that DNA methylation may be one of the mechanistic links between exposure to pollutants and the development of CVD. LINE-1 and Alu demethylation have also been detected in vascular smooth muscle cells that are exposed to homocysteine, another potential biomarker for CVD.24
In addition to the epigenetic changes in the newborn as a result of maternal smoking, Launay et al. demonstrated a direct effect of smoking on gene-specific methylation in adults.76 These researchers hypothesized that serotonin (5-hydroxytryptamine [5-HT]) released from activated platelets could be involved in vascular modifications induced by smoking. The metabolism of current smokers, never-smokers, and ex-smokers who had stopped smoking for a mean of 13 years, were studied. Both platelet 5-HT and plasma 5-hydroxyindoleacetic acid, the main metabolite of serotonin in humans, were associated with CVD risk. However, these associations lost statistical significance after adjustment for smoking status, which suggests that the influence of smoking had a key role in the study's results. These investigators also examined whether DNA methylation could explain some of their findings. Methylation frequency of the monoamine oxidase type B (MAOB) gene promoter was found to be significantly lower in smokers and ex-smokers than in never-smokers (P <0.0001), probably owing to an increase of demethylase activity caused by cigarette smoke.76 These results could have an impact beyond that of cardiovascular damage, considering that MAOB-dependent 5-HT catabolism is also involved in addiction to alcohol, predisposition to cancer, behavior, and mental health.
Ethnic and sex-related disparities
The contribution of nature versus nurture to the ethnic disparities observed in CVD risk is a matter of debate.77,78 Ethnic minorities in the USA suffer from both higher CVD rates and worse perinatal health than white individuals of non-Hispanic origin. The current knowledge provides a strong rationale for considering epigenetics as the link between the prenatal exposure to maternal stress and adult ethnicity-based health disparities in diseases such as hypertension, diabetes, stroke, and coronary heart disease.79 Whether some of these effects have transgenerational influences that could explain the persistence of health disparities, despite social improvements remains to be demonstrated.
CVD has traditionally been considered a male-related disease; however, similar numbers of men and women are affected by the disease, although CVD usually occurs a few years later in women.80 Several hypotheses have been put forward to explain these observations; for example, HDL levels are higher in women than in men.81 The molecular mechanisms for such sex-related differences are unclear, but could include the role of sex chromosomes and sex hormones. A substantial proportion of dimorphic gene expression might be under the control of sex-specific epigenetic marks. Sex-specific differences are apparent at a basic physiologic level, such as the differential methylation patterns and chromatin structure related to pulsatile (male) versus continuous (female) gonadotropin hormone secretion, which affects the expression of transcription factors, including STAT5b and hepatocyte nuclear factors (HNF) 3 gamma (HNF3γ), 4 alpha (HNF4α), and 6 (HNF6).82 Environmental factors, such as nutrition, chemicals and social behavior, can further influence epigenetic marks in a sex-specific manner, leading to the different CVD susceptibility between male and female individuals.82 Moreover, this epigenetic programming could be transmitted to subsequent generations in a sex-specific manner and lead to transgenerational effects. For example, maternal undernutrition may cause abnormalities in male but not female mice blastocysts that are linked to decreased expression of the noncoding H19 mRNA and postnatal hypertension.82
Chronobiology, epigenetics and CVD risk
Another intriguing risk factor for CVD is time, which in this case does not refer to the traditional process of aging, but rather to the time of day. The occurrence of ischemic heart disease is unevenly distributed over a 24 h period, peaking in the early morning and in the late afternoon or early evening.83 These temporal patterns are a result of circadian rhythms. In addition, both the pharmacokinetics and pharmacodynamics of cardiac medications are influenced by the time of their administration.84 Epidemiologic and pharmacologic evidence suggests that a chronobiological approach to ischemic heart disease can contribute new insight and opportunities to improve drug design and delivery to enhance therapeutic outcomes.85 However, before these developments are possible, a better understanding of the molecular mechanisms involved in circadian rhythms is needed.
The circadian clock is regulated by transcriptional–translational feedback loops. This fine-tuned program of gene expression is possible through dynamic changes in chromatin transitions. Advances in this field have revealed intriguing links between circadian regulators, chromatin remodeling, and cellular metabolism.86 Specifically, the central clock protein CLOCK has histone acetyltransferase (HAT) enzymatic properties, and directs acetylation of histone H3 and its dimerization partner ARNT-like 1 at Lys537, an event essential for circadian function.87 In addition, some studies have suggested that miRNAs may be important regulators of circadian rhythm, which provides a new dimension to our understanding of biological clocks at the post-transcriptional level.88 Whereas no studies have yet linked circadian rhythms, epigenetics, and CVD, the evidence from other diseases, particularly cancer, suggests that epigenetic mechanisms may have a role in the circadian occurrence of acute cardiovascular events; this knowledge could help to identify novel preventive and therapeutic approaches to CVD.89,90
Epigenetics and CVD therapy
Despite the heritable nature of epigenetic modifications, these alterations are theoretically reversible through pharmacologic agents. In particular, enzymatic regulators of chromatin modification are well-characterized. Targeting these enzymes, which include HATs, histone deacetylases (HDACs), and histone methyltransferases, holds promise for modulating the transcriptional regulation of genes that are involved in atherogenesis, inflammation, smooth muscle cell proliferation, and matrix formation (Figure 2). RNA interference, which uses small RNAs to reduce gene expression post-transcriptionally, is a second approach that could be applied to CVD, possibly by targeting genes that are overexpressed in endothelial cells susceptible to atherosclerosis.91 Drug therapies that work through epigenetic mechanisms are currently limited to antineoplastic agents; phase II trials are underway for lymphoma treatment using the HDAC inhibitor romidepsin.92 Although therapy for CVD has not yet been designed with epigenetic actions in mind, existing therapies, such as statins, already seem to exploit some of these mechanisms.
HDAC inhibition
Recognition that the pleiotropic effects of statins may depend, in part, on epigenetic mechanisms has emerged from studies investigating cancer therapy and chemoprevention. Lin et al. demonstrated that trichostatin A and lovastatin inhibited HDAC activity and induced hyperacetylation of histone H3 in cancer cells, but not in normal human umbilical endothelial cells.93 These results raise questions about the potential differences in epigenetic modification of chromatin in normal cells compared with mutated tumor cells, but also suggest that tissue specificity will be an important consideration in the evaluation of studies of epigenetically active pharmaceutical agents. Evidence documenting HDAC inhibitors as an accessible and physiologically relevant target is not limited to cancer research, however, but has also accumulated from studies designed to investigate modulation of bile acid synthesis.94 In a hypercholesterolemic mouse model, bile acid-mediated repression of the gene encoding cholesterol 7-alpha-monooxygenase (CYP7A1) was shown to depend on increased nuclear concentration of HDAC7, which inhibited CYP7A1 expression. Furthermore, the HDAC inhibitors trichostatin A and valproic acid lowered plasma cholesterol levels in LDL-knockout mice by increasing CYP7A1 expression and bile acid synthesis.94 This study confirms previous evidence that statins act in part through epigenetic mechanisms, and also suggests the possibility of developing new classes of hypocholesterolemic agents for individuals for whom existing therapies are inadequate.
Although several studies of the mechanisms of action of statins have demonstrated inhibition of HDAC, which implies reduced deacetylation and potentially increased transcription, other studies have shown an opposite action for statins, where gene expression is reduced. Presumably, different genes are targeted in each of these cases. In cultured human endothelial cells, Dje N'Guessan et al. observed that histone modification seems to have a role not only in the inflammatory activation of endothelial cells by oxidized LDL, but also in the provision of some of the anti-inflammatory benefits of statins.95 First, the group showed that histone modification (acetylation of histone H4, and phosphorylation or acetylation of histone H3) contributed to oxidized LDL-mediated expression of proinflammatory interleukin (IL)-8 and chemoattractant C-C motif chemokine 2 by endothelial cells. Next, they showed that oxidized LDL-mediated cytokine release was reduced by pretreatment of the cells with either simvastatin or fluvastatin, and preincubation with both statins blocked modification of histone H3 and H4. Extending their investigations yet one step further, Dje N'Guessan et al. were also able to demonstrate that oxidized LDL reduced HDAC expression in coronary artery endothelium, an effect that could be partly reversed by statins.95 Taken together, this series of experiments suggests that control of gene expression by histone modification is maintained through a complex and dynamic balance. In spite of the unintended and possibly nonspecific epigenetically mediated benefits of statins, therapeutic agents designed to modify these processes are likely to require high selectivity and specificity.
HAT inhibition by dietary compounds
The therapeutic goal of avoiding harmful effects while reducing risk, combined with research supporting a chemopreventive role for certain plant-based foods, have stimulated investigation of the epigenetic properties of selected dietary compounds. Dietary polyphenols, including myricetin, quercetin, hydroxycinnamic acid, curcumin and garcinol, inhibited DNA methyltransferases in a KYSE 510 cancer cell line, supporting the possibility of modifying methylation status and altering disease risk.96
Cardiac hypertrophy is one condition for which dietary compounds have been examined. Early work demonstrated that the balance between histone acetylation and deacetylation regulates the hypertrophic response of cardiomyocytes to phenylephrine.97 In this study, inhibition of the coactivators nuclear cap-binding protein subunit 1 and p300 HAT reduced HAT activity and hypertrophy, whereas enhancing HAT activity promoted hypertrophy. Curcumin (diferuloylmethane), the most abundant polyphenol in the spice turmeric, inhibits HAT activity.98 In an animal model of heart failure, consumption of curcumin at a dose of 50 mg/kg/day inhibited p300 HAT activity, prevented ventricular hypertrophy, and preserved systolic function.99 The corresponding curcumin dose for a person weighing 70 kg is 3.5 g per day, which is less than the well-tolerated dose used in a clinical trial for treatment of pancreatic cancer (8 g), but is more than what is obtainable through a normal diet.100 In a second study linking p300 HAT inhibition to vascular function, pharmacologic concentrations of curcumin were used (80 mM) to inhibit p300 HAT activity and block shear-stress-induced endothelial nitric oxide (eNOS) transcription in human cells.101 Interestingly, the same study demonstrated that the proinflammatory cytokine nuclear factor kappa β (NFκβ) participated in shear stress-mediated endothelial activation to induce eNOS, and that curcumin inhibits acetyltransferase, which in turn modulates NFκβ, as well as eNOS. In light of the multifaceted nature of NFκβ (proinflammatory yet cytoprotective), the therapeutic implications of curcumin activities are unclear, and at high concentrations this compound may lead to a nonspecific inhibition of HAT.
By contrast, garcinol—a polyisoprenylated benozophenone derived from the Garcinia indica fruit—is less studied.102 Garcinol strongly inhibits p300 HAT, decreases global gene expression, and induces apoptosis in the HeLa cancer cell line.103 Garcinol was used to investigate the role of histone modification in the regulation of the EGR-1 gene, which encodes the zinc finger transcription factor early growth response 1 protein.104 EGR-1 has been implicated in a range of cardiovascular processes, including thrombus formation, endothelial cell migration, and proliferation.105,106 As a result of IL-1β exposure, EGR-1 transcription in aortic smooth muscle cells was stimulated through the acetylation of histone H3. This response was enhanced through histone deacetylase inhibition and prevented by garcinol, which reflects HAT inhibition.104
As these studies illustrate, research on the potential therapeutic use of epigenetically active dietary compounds is very preliminary. Although the epigenetic effects of some phenols are well established, particularly when the compounds are applied at high concentrations, many important questions remain unanswered. First, both dietary intake of food and bioavailability may be too low to achieve epigenetic activity, although intake of dietary supplements may produce sufficient plasma concentrations of these substances. Second, whether results from cell studies will translate to health benefits from dietary sources is unclear, particularly for compounds evaluated in cancer cell lines. Third, the apparent lack of enzyme or tissue specificity for the polyphenols represents another large area of uncertainty. Additional research is needed to clarify the mechanisms of action of these dietary compounds, and clinical trials should be carried out to establish their efficacy and tolerance.
Conclusions
Preclinical and clinical studies have demonstrated that exposure to common environmental challenges modifies epigenetic marks. Most research has focused on DNA methylation, and less is known about histone modifications and miRNA alterations. The reported epigenetic changes associated with factors such as diet, smoking, and pollution are similar to the epigenetic alterations found in patients with CVD. For example, methylation patterns differ in patients with coronary artery disease compared with controls, and methylation is reported to be altered in smokers.20,107 These patterns may support the hypothesis that epigenetic changes relate to increased CVD risk, but it is not possible to conclude causality from these associations. Prospective investigations are needed to determine whether individuals who are exposed to various environmental challenges and develop risk factors accumulate epigenetic alterations over time, and whether these alterations increase the incidence of CVD. An even greater challenge is to determine whether environmental epigenetic changes associated with CVD risk are heritable. The latter point is especially important considering that classical genetics can explain only a small part of CVD heritability—epigenetics could contribute to some of the observed genetic variance.108 However, although epigenetic modifications may contribute to risk and are, therefore, a potential solution to the problem of missing causality of CVD, they do not have as much influence on recurrent risk or heritability, so cannot explain the problem of missing heritability.109
An increased understanding of epigenetic mechanisms may lead to the development of novel and targeted CVD therapies that act at the risk-factor level, as well as directly on the vascular system. However, the difficulties of the task ahead must not be underestimated. Whereas the genomic information is the same in all our cells and during our entire lifespan, the epigenomic information varies from cell to cell and during the lifetime of the individual. Moreover, not all epigenetic marks and their combinations have been identified, and how they work in concert to regulate epigenetic mechanisms and gene expression is not yet known. A complete epigenomic map will require major advances in knowledge and computing power that greatly exceed those currently available for the study of classical DNA genetic variation.110
Acknowledgments
This work was supported by grants from the National Heart, Lung and Blood institute (HL-54776), and the National Institute of Diabetes and Digestive and Kidney Diseases (DK075030), and by contracts 53-K06-5-10 and 58-1950-9-001 from the USDA Department of Agricultural Research Service.
Footnotes
Author contributions: J. M. Ordovás and C. E. Smith contributed to discussion of content for the article, researched data to include in the manuscript, reviewed and edited the manuscript before submission, and revised the manuscript in response to the peer-reviewers' comments.
Competing interests: The authors declare no competing interests.
References
- 1.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacKinnon AU. The origin of the modern epidemic of coronary artery disease in England. J R Coll Gen Pract. 1987;37:174–176. [PMC free article] [PubMed] [Google Scholar]
- 3.Azambuja MI, Levins R. Coronary heart disease (CHD)—one or several diseases? Changes in the prevalence and features of CHD. Perspect Biol Med. 2007;50:228–242. doi: 10.1353/pbm.2007.0013. [DOI] [PubMed] [Google Scholar]
- 4.Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham. Study Am J Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gersh BJ, Sliwa K, Mayosi BM, Yusuf S. Novel therapeutic concepts: the epidemic of cardiovascular disease in the developing world: global implications. Eur Heart J. 2010;31:642–648. doi: 10.1093/eurheartj/ehq030. [DOI] [PubMed] [Google Scholar]
- 6.Grundy SM, et al. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 7.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sipido KR, et al. Identifying needs and opportunities for advancing translational research in cardiovascular disease. Cardiovasc Res. 2009;83:425–435. doi: 10.1093/cvr/cvp165. [DOI] [PubMed] [Google Scholar]
- 9.Omics.org. Alphabetically ordered list of omes and omics. 2009 [online], http://omics.org/index.php/Alphabetically_ordered_list_of_omes_and_omics.
- 10.Turan N, Katari S, Coutifaris C, Sapienza C. Explaining inter-individual variability in phenotype: is epigenetics up to the challenge? Epigenetics. 2010;5:16–19. doi: 10.4161/epi.5.1.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ. Epigenetics in atherosclerosis and inflammation. J Cell Mol Med. doi: 10.1111/j.1582-4934201001022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turunen MP, Aavik E, Ylä-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta. 2009;1790:886–891. doi: 10.1016/j.bbagen.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- 14.Krause B, Sobrevia L, Casanello P. Epigenetics: new concepts of old phenomena in vascular physiology. Curr Vasc Pharmacol. 2009;7:513–520. doi: 10.2174/157016109789043883. [DOI] [PubMed] [Google Scholar]
- 15.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mack CP. An epigenetic clue to diabetic vascular disease. Circ Res. 2008;103:568–570. doi: 10.1161/CIRCRESAHA.108.184358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stöger R. Epigenetics and obesity. Pharmacogenomics. 2008;9:1851–1860. doi: 10.2217/14622416.9.12.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gluckman PD, Hanson MA. Developmental and epigenetic pathways to obesity: an evolutionary-developmental perspective. Int J Obes. 2008;32(Suppl. 7):S62–S71. doi: 10.1038/ijo.2008.240. [DOI] [PubMed] [Google Scholar]
- 19.Campión J, Milagro FI, Martínez JA. Individuality and epigenetics in obesity. Obes Rev. 2009;10:383–392. doi: 10.1111/j.1467-789X.2009.00595.x. [DOI] [PubMed] [Google Scholar]
- 20.Dong C, Yoon W, Goldschmidt-Clermont PJ. DNA methylation and atherosclerosis. J Nutr. 2002;132:2406S–2409S. doi: 10.1093/jn/132.8.2406S. [DOI] [PubMed] [Google Scholar]
- 21.Sharma P, et al. Mining literature for a comprehensive pathway analysis: a case study for retrieval of homocysteine related genes for genetic and epigenetic studies. Lipids Health Dis. 2006;5:1. doi: 10.1186/1476-511X-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenvinkel P, et al. Impact of inflammation on epigenetic DNA methylation—a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–499. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 23.Lund G, Zaina S. Atherosclerosis risk factors can impose aberrant DNA methylation patterns: a tale of traffic and homocysteine. Curr Opin Lipidol. 2009;20:448–449. doi: 10.1097/MOL.0b013e3283309928. [DOI] [PubMed] [Google Scholar]
- 24.Yideng J, et al. Homocysteine-mediated expression of SAHH, DNMTs, MBD2, and DNA hypomethylation potential pathogenic mechanism in VSMCs. DNA Cell Biol. 2007;26:603–611. doi: 10.1089/dna.2007.0584. [DOI] [PubMed] [Google Scholar]
- 25.Sharma P, et al. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008;27:357–365. doi: 10.1089/dna.2007.0694. [DOI] [PubMed] [Google Scholar]
- 26.Lee ME, Wang H. Homocysteine and hypomethylation. A novel link to vascular disease. Trends Cardiovasc Med. 1999;9:49–54. doi: 10.1016/s1050-1738(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 27.Castro R, Rivera I, Blom HJ, Jakobs C, Tavares de Almeida I. Homocysteine metabolism, hyperhomocysteinaemia and vascular disease: an overview. J Inherit Metab Dis. 2006;29:3–20. doi: 10.1007/s10545-006-0106-5. [DOI] [PubMed] [Google Scholar]
- 28.Libby P, Okamoto Y, Rocha VZ, Folco E. Inflammation in atherosclerosis: transition from theory to practice. Circ J. 2010;74:213–220. doi: 10.1253/circj.cj-09-0706. [DOI] [PubMed] [Google Scholar]
- 29.Pons D, et al. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;30:266–277. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- 30.Goyal BR, Patel MM, Soni MK, Bhadada SV. Therapeutic opportunities of small interfering RNA. Fundam Clin Pharmacol. 2009;23:367–386. doi: 10.1111/j.1472-8206.2009.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silvestri P, et al. MicroRNAs and ischemic heart disease: towards a better comprehension of pathogenesis, new diagnostic tools and new therapeutic targets. Recent Pat Cardiovasc Drug Discov. 2009;4:109–118. doi: 10.2174/157489009788452977. [DOI] [PubMed] [Google Scholar]
- 32.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buysschaert I, Schmidt T, Roncal C, Carmeliet P, Lambrechts D. Genetics, epigenetics and pharmaco-(epi)genomics in angiogenesis. J Cell Mol Med. 2008;12:2533–2551. doi: 10.1111/j.1582-4934.2008.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esteller M. Epigenetics in evolution and disease. Lancet. 2008;372:S90–S96. [Google Scholar]
- 35.Waddington CH. Organisers and Genes. Cambridge University Press; Cambridge: 1940. [Google Scholar]
- 36.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 37.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 38.Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochem Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- 39.Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 42.Barringhaus KG, Zamore PD. MicroRNAs: regulating a change of heart. Circulation. 2009;119:2217–2224. doi: 10.1161/CIRCULATIONAHA.107.715839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katey J, et al. miR-33 coordinates genes regulating cholesterol homeostasis. Science. in press. [Google Scholar]
- 44.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21:214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meehan RR. DNA methylation in animal development. Semin Cell Dev Biol. 2003;14:53–65. doi: 10.1016/s1084-9521(02)00137-4. [DOI] [PubMed] [Google Scholar]
- 47.Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86:305–341. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol. 2010;223:289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- 49.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krebs JR. The gourmet ape: evolution and human food preferences. Am J Clin Nutr. 2009;90:707S–711S. doi: 10.3945/ajcn.2009.27462B. [DOI] [PubMed] [Google Scholar]
- 51.Kelley JL, Swanson WJ. Positive selection in the human genome: from genome scans to biological significance. Annu Rev Genomics Hum Genet. 2008;9:143–160. doi: 10.1146/annurev.genom.9.081307.164411. [DOI] [PubMed] [Google Scholar]
- 52.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 53.Stöger R. The thrifty epigenotype: an acquired and heritable predisposition for obesity and diabetes? Bioessays. 2008;30:156–166. doi: 10.1002/bies.20700. [DOI] [PubMed] [Google Scholar]
- 54.Roseboom T, de Rooij S, Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum Dev. 2006;82:485–491. doi: 10.1016/j.earlhumdev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 55.Lumey LH, et al. Cohort profile: the Dutch Hunger Winter families study. Int J Epidemiol. 2007;36:1196–1204. doi: 10.1093/ije/dym126. [DOI] [PubMed] [Google Scholar]
- 56.Kyle UG, Pichard C. The Dutch Famine of 1944–1945: a pathophysiological model of long-term consequences of wasting disease. Curr Opin Clin Nutr Metab Care. 2006;9:388–394. doi: 10.1097/01.mco.0000232898.74415.42. [DOI] [PubMed] [Google Scholar]
- 57.Stein AD, et al. Anthropometric measures in middle age after exposure to famine during gestation: evidence from the Dutch famine. Am J Clin Nutr. 2007;85:869–876. doi: 10.1093/ajcn/85.3.869. [DOI] [PubMed] [Google Scholar]
- 58.de Rooij SR, Painter RC, Holleman F, Bossuyt PM, Roseboom TJ. The metabolic syndrome in adults prenatally exposed to the Dutch famine. Am J Clin Nutr. 2007;86:1219–1224. doi: 10.1093/ajcn/86.4.1219. [DOI] [PubMed] [Google Scholar]
- 59.Dover GJ. The Barker hypothesis: how pediatricans will diagnose and prevent common adult-onset diseases. Trans Am Clin Climatol Assoc. 2009;120:199–207. [PMC free article] [PubMed] [Google Scholar]
- 60.Wadhwa PD, Buss C, Entringer S, Swanson JM. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms. Semin Reprod Med. 2009;27:358–368. doi: 10.1055/s-0029-1237424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 63.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bogdarina I, Welham S, King PJ, Burns SP, Clark AJ. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ Res. 2007;100:520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tobi EW, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El-Maarri O, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 68.Alkemade FE, et al. Prenatal exposure to apoE deficiency and postnatal hypercholesterolemia are associated with altered cell-specific lysine methyltransferase and histone methylation patterns in the vasculature. Am J Pathol. 2010;176:542–548. doi: 10.2353/ajpath.2010.090031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Straten EM, et al. The liver X-receptor gene promoter is hypermethylated in a mouse model of prenatal protein restriction. Am J Physiol Regul Integr Comp Physiol. 2010;298:R275–R282. doi: 10.1152/ajpregu.00413.2009. [DOI] [PubMed] [Google Scholar]
- 70.Anonymous. Active and passive tobacco exposure: a serious pediatric health problem. A statement from the Committee on Atherosclerosis and Hypertension in Children, Council on Cardiovascular Disease in the Young, American Heart Association. Circulation. 1994;90:2581–2590. doi: 10.1161/01.cir.90.5.2581. [DOI] [PubMed] [Google Scholar]
- 71.Breton CV, et al. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xue F, Michels KB. Intrauterine factors and risk of breast cancer: a systematic review and meta-analysis of current evidence. Lancet Oncol. 2007;8:1088–1100. doi: 10.1016/S1470-2045(07)70377-7. [DOI] [PubMed] [Google Scholar]
- 73.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 74.Baccarelli A, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoffmann B, et al. Residential traffic exposure and coronary heart disease: results from the Heinz Nixdorf Recall Study. Biomarkers. 2009;14(Suppl. 1):74–78. doi: 10.1080/13547500902965096. [DOI] [PubMed] [Google Scholar]
- 76.Launay JM, et al. Smoking induces long-lasting effects through a monoamine-oxidase epigenetic regulation. PLoS ONE. 2009;4:e7959. doi: 10.1371/journal.pone.0007959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lai CQ, et al. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum Genet. 2009;125:199–209. doi: 10.1007/s00439-008-0612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dodge KA. Practice and public policy in the era of gene-environment interactions. Novartis Found Symp. 2008;293:87–97. doi: 10.1002/9780470696781.ch7. [DOI] [PubMed] [Google Scholar]
- 79.Kuzawa CW, Sweet E. Epigenetics and the embodiment of race: developmental origins of US racial disparities in cardiovascular health. Am J Hum Biol. 2009;21:2–15. doi: 10.1002/ajhb.20822. [DOI] [PubMed] [Google Scholar]
- 80.Mosca L, et al. Cardiovascular disease in women: a statement for healthcare professionals from the American Heart Association. Writing Group Circulation. 1997;96:2468–2482. doi: 10.1161/01.cir.96.7.2468. [DOI] [PubMed] [Google Scholar]
- 81.Abbott RD, et al. Joint distribution of lipoprotein cholesterol classes. The Framingham study Arteriosclerosis. 1983;3:260–272. doi: 10.1161/01.atv.3.3.260. [DOI] [PubMed] [Google Scholar]
- 82.Gabory A, Attig L, Junien C. Sexual dimorphism in environmental epigenetic programming. Mol Cell Endocrinol. 2009;304:8–18. doi: 10.1016/j.mce.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 83.Martino TA, Sole MJ. Molecular time: an often overlooked dimension to cardiovascular disease. Circ Res. 2009;105:1047–1061. doi: 10.1161/CIRCRESAHA.109.206201. [DOI] [PubMed] [Google Scholar]
- 84.Hermida RC, Ayala DE, Fernández JR, Calvo C. Chronotherapy improves blood pressure control and reverts the nondipper pattern in patients with resistant hypertension. Hypertension. 2008;51:69–76. doi: 10.1161/HYPERTENSIONAHA.107.096933. [DOI] [PubMed] [Google Scholar]
- 85.Portaluppi F, Lemmer B. Chronobiology and chronotherapy of ischemic heart disease. Adv Drug Deliv Rev. 2007;59:952–965. doi: 10.1016/j.addr.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 86.Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr Opin Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 87.Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SiRT1. Int J Biochem Cell Biol. 2009;41:81–86. doi: 10.1016/j.biocel.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 88.Pegoraro M, Tauber E. The role of microRNAs (miRNA) in circadian rhythmicity. J Genet. 2008;87:505–511. doi: 10.1007/s12041-008-0073-8. [DOI] [PubMed] [Google Scholar]
- 89.Rudic RD, Fulton DJ. Pressed for time: the circadian clock and hypertension. J Appl Physiol. 2009;107:1328–1338. doi: 10.1152/japplphysiol.00661.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoffman AE, et al. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res. 2010;70:1459–1468. doi: 10.1158/0008-5472.CAN-09-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang J, Burridge KA, Friedman MH. In vivo differences between endothelial transcriptional profiles of coronary and iliac arteries revealed by microarray analysis. Am J Physiol Heart Circ Physiol. 2008;295:H1556–H1561. doi: 10.1152/ajpheart.00540.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Piekarz RL, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin YC, et al. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 2008;68:2375–2383. doi: 10.1158/0008-5472.CAN-07-5807. [DOI] [PubMed] [Google Scholar]
- 94.Mitro N, et al. Insights in the regulation of cholesterol 7alpha-hydroxylase gene reveal a target for modulating bile acid synthesis. Hepatology. 2007;46:885–897. doi: 10.1002/hep.21819. [DOI] [PubMed] [Google Scholar]
- 95.Dje N'Guessan P, et al. Statins control oxidized LDL-mediated histone modifications and gene expression in cultured human endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:380–386. doi: 10.1161/ATVBAHA.108.178319. [DOI] [PubMed] [Google Scholar]
- 96.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 97.Gusterson RJ, Jazrawi E, Adcock IM, Latchman DS. The transcriptional co-activators CREB-binding protein (CBP) and p300 play a critical role in cardiac hypertrophy that is dependent on their histone acetyltransferase activity. J Biol Chem. 2003;278:6838–6847. doi: 10.1074/jbc.M211762200. [DOI] [PubMed] [Google Scholar]
- 98.Balasubramanyam K, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 99.Morimoto T, et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest. 2008;118:868–878. doi: 10.1172/JCI33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dhillon N, et al. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14:4491–4499. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 101.Chen W, Bacanamwo M, Harrison DG. Activation of p300 histone acetyltransferase activity is an early endothelial response to laminar shear stress and is essential for stimulation of endothelial nitric-oxide synthase mRNA transcription. J Biol Chem. 2008;283:16293–16298. doi: 10.1074/jbc.M801803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choi KC, et al. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69:583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 103.Balasubramanyam K, et al. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004;279:33716–33726. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 104.Wang B, et al. Phosphorylation and acetylation of histone H3 and autoregulation by early growth response 1 mediate interleukin 1beta induction of early growth response 1 transcription. Arterioscler Thromb Vasc Biol. 2010;30:536–545. doi: 10.1161/ATVBAHA.109.193821. [DOI] [PubMed] [Google Scholar]
- 105.Shin IS, et al. Early growth response factor-1 is associated with intraluminal thrombus formation in human abdominal aortic aneurysm. J Am Coll Cardiol. 2009;53:792–799. doi: 10.1016/j.jacc.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 106.Abdel-Malak NA, Mofarrahi M, Mayaki D, Khachigian LM, Hussain SN. Early growth response-1 regulates angiopoietin-1-induced endothelial cell proliferation, migration, and differentiation. Arterioscler Thromb Vasc Biol. 2009;29:209–216. doi: 10.1161/ATVBAHA.108.181073. [DOI] [PubMed] [Google Scholar]
- 107.Oka D, et al. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115:3412–3426. doi: 10.1002/cncr.24394. [DOI] [PubMed] [Google Scholar]
- 108.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anonymous. Moving AHEAD with an international human epigenome project. Nature. 2008;454:711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]