Abstract
The objective of the present study was to find very early viral kinetic markers to predict nonresponse to HCV therapy in a group of HIV/HCV-coinfected patients. Twenty-six patients (15 HCV genotype-1 and 11 genotype-3) were treated with a 48-week regimen of peginterferon-alfa-2a (PEG-IFN) (180 μg/week) and weight based ribavirin (11mg/kg/day). Samples were collected at baseline; 4, 8, 12, 18, 24, 30, 36 and 42 hours; days 2, 3, 4, 7, 8, 15, 22, 29, 43 and 57 then weekly and monthly. Five patients discontinued treatment. Seven patients (27%) achieved a sustained virological response (SVR). Nadir HCV RNA levels were observed 1.6±0.3 days after initiation of therapy, followed by a 0.3- to 12.9-fold viral rebound until the administration of the second dose of PEG-IFN, which were not associated with SVR or HCV genotype. A viral decline <1.19 log for genotype-1 and <0.97 log for genotype-3, 2 days after starting therapy, had a negative predictive value (NPV) of 100% for SVR. The day 2 virologic response had a similar positive predictive value (PPV) for SVR as a rapid virologic response at week 4. In addition, a second-phase-viral-decline slope (i.e., measured from day 2 to 29) less than 0.3 log/wk had a NPV=100% for SVR.
CONCLUSIONS
First phase viral decline at day 2 and second-phase-viral-decline slope (<0.3 log/wk) are excellent predictors of nonresponse. Further studies are needed to validate these viral kinetic parameters as early on-treatment prognosticators of nonresponse in patients with HCV and HIV.
Keywords: HCV, HIV, Pegylated Interferon and Viral Kinetics
Introduction
Coinfection with human immunodeficiency virus (HIV) and HCV affects approximately 10 million people worldwide [1] and up to 100,000 persons in Brazil [2]. The prevalence of coinfection is as high as 90% among injection drug users and HIV-infected hemophiliacs [3]. Hepatitis C is considered an opportunistic infection in persons with HIV based on prevalence and high rates of HCV-related morbidity and mortality [4, 5]. Progression to cirrhosis and hepatocellular carcinoma occurs more rapidly in HIV/HCV-coinfected patients than in HCV-monoinfected individuals [6, 7]. Moreover, sustained virological response (SVR) rates to pegylated-interferon (PEG-IFN) and ribavirin are lower in HIV/HCV coinfected [8-12] than in HCV monoinfected patients [13, 14]. Therefore, treatment of chronic HCV infection has become a priority in HIV-coinfected patients.
HCV therapy for 48 weeks was recommended for all HCV genotypes in most guidelines designed for HIV/HCV co-infected patients [15, 16]. Efforts have been made to identify early predictors of nonresponse in order to limit side effects and to reduce cost in patients who are unlikely to respond with continued therapy [17-19]. Detailed viral kinetic studies, which allow analysis of viral decline before week 4 [20-25], provide a means to assess the predictive value of on treatment response at early time points.
The objective of the present study was to identify very early viral kinetic markers to predict nonresponse to HCV therapy in HIV/HCV-coinfected patients.
Materials and Methods
Patients
The study was approved by the ethics committee at the University of São Paulo Hospital das Clínicas and subjects provided written informed consent to participate. Twenty-six HIV-HCV coinfected patients were included. Subjects were HBsAg negative and had no evidence of other causes of liver disease or comorbid illnesses. All patients had well-controlled HIV infection for at least six months on stable highly active antiretroviral therapy (HAART) or without HAART. Standard indications and contraindications for the use of peginterferon and ribavirin were followed [26]. Baseline data are summarized in Table 1 and are further detailed in a recent publication [27].
Table 1.
Demographic and basal data.
|
Gender, n (%) Male / Female |
23 (88%) / 3 (12%) |
| Mean age [yr] | 41 (range, 32–56) |
|
Race, n (%) White / Black or mixed |
14 (54%) / 12 (46%) |
| Mean weight [kg] | 69 (range, 52 – 92) |
| Mean CD4 [cells/mm3] | 570 (range, 327–956) |
|
HAART@ n (%) on / off |
18 (69%) / 8 (31%) |
|
Mean HIV viral load [copies/mL] |
2215 (range, 170–9930) |
|
Mean HCV viral load [IU/mL] |
2,039,492 (range, 1380–10,500,000) |
|
HCV genotype n (%) 1 / 3 |
15 (58%) / 11 (42%) |
| Cirrhosis (metavir F4) | 4 (15%) |
Treatment procedure
Patients received peginterferon alfa-2a (180 μg/week) and weight based ribavirin (11 mg/kg/day) for 48 weeks. At the outset of the treatment, patients were hospitalized for 48 hours in order to facilitate blood sample collection. After discharge, they were monitored as outpatients. To standardize the treatment regimen, each dose of peginterferon was given on a Monday morning at 08:00. For the initial 4 weeks of treatment, patients took peginterferon under direct observation. At each visit, ribavirin was dispensed in a quantity sufficient to last until the subsequent visit. Each patient received oral instructions and a written schedule for medication and sample collection.
Viral Load Monitoring
Blood samples were collected at the following time points: at baseline; at 4, 8, 12, 18, 24, 30, 36 and 42 hours; on days 2, 3, 4, 7, 8, 15, 22, 29, 43, 57 and 84. . The end-of-treatment response and SVR were evaluated at weeks 48 and 72, respectively. Rapid virologic response (RVR) was defined as HCV RNA <10 IU/ml at day 29 of therapy, complete early virologic response (cEVR) was defined as undetectable HCV-RNA (<10 IU/ml) at week 12, and partial EVR (pEVR) as HCV RNA load decreased ≥2 log10, but detectable at week 12. Based on patient and physician discretion, continued therapy was permitted at week 12 in the absence of cEVR.
Specimens collected for viral quantification were prepared immediately, and the aliquots were frozen at −80°C until processing. The COBAS TaqMan HCV test (Roche Molecular Systems Inc., Branchburg, NJ), with a detection range of 10 to 10,000,000 HCV IU/mL was used for HCV quantification until week 12. The Amplicor HCV test (version 2.0; Roche Diagnostics Corp., Indianapolis, IN) was used for qualitative detection of HCV (limit of detection of 50 HCV IU/ml) at the end of therapy (EOT) (week 48) and follow up at week 72, and genotyping was performed with VERSANT HCV genotype assay (LiPA; Bayer Corp., Tarrytown, NY). The COBAS Amplicor HIV-1 Monitor test (version 1.5; Roche Molecular Systems Inc.), with a detection range of 50 to 100,000 HIV copies/mL, was used for quantification of HIV. All virologic assays were performed at TriCore Reference Laboratories (Albuquerque, NM). The remaining tests were performed in the Hepatitis Laboratory (LIM 47) of the Hospital das Clínicas.
Statistical Analysis
We used nonparametric methods in intention-to-treat (ITT) and per protocol (PP) analyses to compare baseline characteristics and viral response parameters of SVR vs. non SVR cases. To compare categorical variables, we used the two-tailed Fisher exact test. Multiple unrelated groups were compared using the Kruskal Wallis test (e.g., see viral decline patterns in HCV genotype-1 subjects; Fig. 1) and when indicated, subgroups analysis was performed using the Mann-Whitney test. To determine whether HCV RNA (log10) levels differ between SVR vs. non-SVR subjects in each time point, we used the two-way analysis of variance (ANOVA).
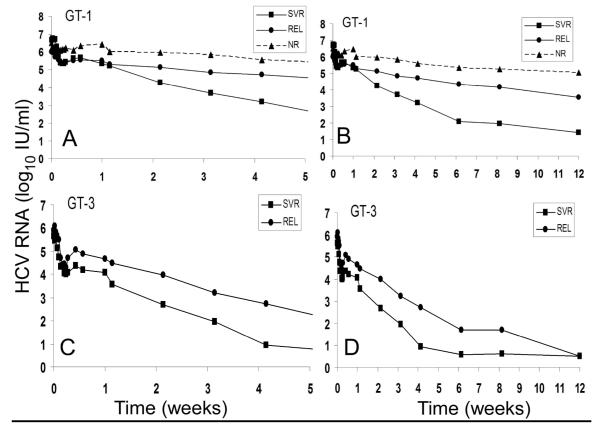
Figure 1.
HCV RNA kinetics per viral response group per genotype. (A) and (C) early viral kinetics from initiation of therapy until day 15, in patients infected with HCV genotype-1 (GT-1) or genotype-3 (GT-3), respectively. (B) and (D) viral kinetics from initiation of therapy until week 12, in patients infected with HCV genotype-1 (GT-1) or genotype-3 (GT-3), respectively. * For difference between XXX and XXX (p<xxx). ** For difference between XXX and XXX (p<xxx). Vertical lines represent standard error of the mean. Already at day 15, patients who achieved SVR and were infected with HCV genotype 1 had significantly lower viral load than patients who were relapsers (REL) or nonresponders (NR) (p=0.012 and p=0.008, respectively). Among Gen 3 patients, viral load in SVR patients was significantly (p=0.014) lower than REL patients at day 29.
To determine the discriminatory ability of log10 HCV RNA decline from baseline to identify likely SVR cases, we computed the area under the receiver operator characteristic curve (AUROC) at selected time points during therapy. The area under the receiver operator characteristic curve is a measure of the probability that in a randomly selected pair of SVR and non SVR patients, the marker (in this case, log10 HCV RNA decline from baseline) permits correct identification. We also report the positive and negative predictive values, which quantify the clinical value of a marker. The level of statistical significance was set at (p≤ 0.05). All tests were performed by SPSS v.17 Chicago, IL.
Results
Baseline characteristics and viral response
The baseline characteristics of the 26 subjects who were included in the study are presented in Table 1. Five patients discontinued therapy by week 12 (one was incarcerated, one died due to a complication not related to liver disease, two asked to stop treatment, and one was lost to follow-up). At the end of therapy, i.e., week 48, 18 (out of 21) achieved an end-of-treatment response (HCV RNA <50 IU/ml), while only seven patients had an SVR (Tables 2 and 3). Considering baseline characteristics (Table 1), HCV genotype was the only parameter associated with SVR based on intention to treat (ITT) p=0.09 and per protocol (PP) p=0.05 analysis.
Table 2.
Early Viral Kinetic parameters results
| GROUP | Patient No. | Genotype | Baseline HCV RNA (V0) log(IU/ml) |
First-phase viral decline Log10(V0/Vmin) |
Vmin (day) |
Viral rebound (V7 / Vmin) [min - max] |
Viral slope decline from day 2 to day 29 [log /wk] |
Viral slope decline from day 7 to day 15 [log /wk] |
Viral slope decline from day 7 to day 29 [log /wk] |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 3 | 6.2 | 1.88 | 2 | 0.3 | 1.12 | 0.82 | 1.11 |
| 1 | 2 | 3 | 5.8 | 1.57 | 1.5 | 1.7 | 0.98 | 1.18 | 1.01 |
| 1 | 3 | 3 | 5.2 | 1.67 | 1.5 | 2.2 | 0.48 | 0.41 | 0.62 |
| 1 | 6 | 3 | 5.0 | 1.47 | 1.75 | 7.3 | 0.72 | 1.77 | 0.73 |
| 1 | 7 | 3 | 6.0 | 1.49 | 1 | 1.0 | 1.25 | 1.21 | 1.40 |
| 1 | 4 | 1 | 7.0 | 2.26 | 1.75 | 2.3 | 0.36 | 0.65 | 0.38 |
| 1 | 5 | 1 | 6.3 | 1.35 | 1.5 | 0.9 | 0.98 | 1.28 | 1.00 |
| 1: | Mean±SD | 5.9±0.7 | 1.67±0.31 a | 1.6±0.3 | 0.3 - 7.3 | 0.84±0.33 g | 1.05±0.45 c | 0.89±0.34 e | |
| 2 | 8 | 3 | 5.6 | 1.36 | 1.25 | 2.9 | 0.52 | 0.40 | 0.58 |
| 2 | 9 | 3 | 6.1 | 2.09 | 2 | 2.0 | 1.38 | 1.53 | 1.54 |
| 2 | 13 | 3 | 6.1 | 0.54 | 1.75 | 7.8 | 0.08 | (0.28) | (0.02) |
| 2 | 10 | 1 | 5.3 | 1.03 | 1 | 1.2 | 0.29 | 0.35 | 0.30 |
| 2 | 11 | 1 | 5.7 | 0.64 | 1.75 | 1.6 | 0.54 | 0.26 | 0.40 |
| 2 | 12 | 1 | 5.3 | 2.04 | 1.75 | 1.8 | 0.28 | (0.03) | 0.34 |
| 2 | 14 | 1 | 6.5 | 0.43 | 1.75 | 1.4 | 0.08 | (0.06) | 0.12 |
| 2 | 15 | 1 | 5.7 | 0.76 | 1.5 | 1.3 | 0.24 | 0.57 | 0.13 |
| 2 | 16 | 1 | 6.4 | 1.36 | 1.75 | 1.5 | 0.11 | 0.20 | 0.18 |
| 2 | 17 | 1 | 6.7 | 1.35 | 1.5 | 9.5 | 0.12 | 0.42 | 0.30 |
| 2 | 18 | 1 | 6.6 | 0.00 | 2 | 1.0 | 0.06 | 0.29 | 0.07 |
| 2: | Mean±SD | 6.0±0.5 | 1.05±0.66 b | 1.6±0.3 | 1.0 - 9.5 | 0.34±0.39 h | 0.33±0.47 d | 0.36±0.43 f | |
| 3 | 19 | 1 | 6.5 | 0.17 | 2 | 1.3 | 0.26 | 0.16 | 0.35 |
| 3 | 20 | 1 | 6.8 | 0.92 | 1.5 | 6.2 | 0.20 | 0.67 | 0.24 |
| 3 | 21 | 1 | 6.1 | 0.59 | 1.25 | 4.0 | 0.04 | (0.02) | 0.03 |
| 3: | Mean±SD | 6.5±0.3 | 0.56±0.37 b | 1.6±0.4 | 1.3 - 6.2 | 0.17±0.11 h | 0.27±0.36d* | 0.21±0.16 f | |
| 4 | 24 | 3 | 3.1 | 1.67 | 1.25 | 0.5 | 1.68 | 2.31 | 2.31$ |
| 4 | 25 | 3 | 6.0 | 2.26 | 1.5 | 0.7 | 0.41 | 0.47 | 0.30 |
| 4 | 26 | 3 | 6.1 | 0.63 | 1.25 | 2.9 | 0.67 | 1.19 | 0.70 |
| 4 | 22 | 1 | 6.4 | 1.65 | 1.5 | 1.1 | 0.54 | 1.13 | 0.39 |
| 4 | 23 | 1 | 6.5 | 1.46 | 1.5 | 12.9 | 0.58 | 0.90 | 0.58 |
| 4: | Mean±SD | 5.6±1.4 | 1.53±0.59 | 1.4±0.1 | 0.5 - 12.9 | 0.56±0.03 | 1.01±0.16 | 0.49±0.14 | |
| All | Mean±SD | All | 6.0±0.8 | 1.26±0.64 | 1.6±0.3 | 0.3 – 12.9 | 0.54±0.45 | 0.68±0.62 | 0.51±0.42 |
Groups’ definition: 1) SVR, 2) REL, 3) NR as defined in Results and 4) discontinued therapy group; SD, one standard deviation; Vmin, viral nadir during the first week of treatment; V7, viral load at day 7;
calculated till day 15 due to negative HCV RNA from day 15 onwards;
() represent viral increase;
, groups marked with the same superscript letter were not significantly different from each other (P>0.05), but they were significantly different (P< 0.02; Kruskal Wallis and Mann-Whitney Tests) from all the other groups in the same column.
, groups marked with the same superscript letter were not significantly different from each other (P>0.05), but they were significantly different (P< 0.02; Kruskal Wallis and Mann-Whitney Tests) from all the other groups in the same column.
, groups marked with the same superscript letter were not significantly different from each other (P>0.05), but they were significantly different (P< 0.02; Kruskal Wallis and Mann-Whitney Tests) from all the other groups in the same column.
, groups marked with the same superscript letter were not significantly different from each other (P>0.05), but they were significantly different (P< 0.02; Kruskal Wallis and Mann-Whitney Tests) from all the other groups in the same column.
, groups marked with the same superscript letter were not significantly different from each other (P>0.05), but they were significantly different (P< 0.02; Kruskal Wallis and Mann-Whitney Tests) from all the other groups in the same column.
, groups marked with the same superscript letter were not significantly different from each other (P>0.05), but they were significantly different (P< 0.02; Kruskal Wallis and Mann-Whitney Tests) from all the other groups in the same column.
, groups marked with the same superscript letter were not significantly different from each other (P>0.05), but they were significantly different (P< 0.02; Kruskal Wallis and Mann-Whitney Tests) from all the other groups in the same column.
, groups marked with the same superscript letter were not significantly different from each other (P>0.05), but they were significantly different (P< 0.02; Kruskal Wallis and Mann-Whitney Tests) from all the other groups in the same column.
d*, stands for a trend (p=0.065) with SVR group.
Table 3.
Virological response and HCV genotype
| Viral response parameter | All (n = 26) |
Genotype-1 (n=15) |
Genotype-3 (n=11) |
P value* |
|---|---|---|---|---|
| 1st phase viral decline, median [ Log10(V0/Vmin) ] and (IQR) |
0.92 (0.93) | 1.65 (0.68) | 0.009 | |
| slower phase slope decline (d7-d15), median [log/wk] and (IQR) |
0.35 (0.51) | 1.0 (0.88) | 0.031 | |
| slower phase slope decline (d7-d29), median [log/wk] and (IQR) |
0.30 (0.26) | 0.72 (0.67) | 0.008 | |
| RVR, <10 IU/ml at W4, n (%) | ||||
| Yes | 5 (19) | 0 (0) | 5 (45) | |
| No | 21 (81) | 15 (100) | 6 (55) | 0.007 |
| cEVR@, <10 IU/ml at W12, n(%) | ||||
| Yes | 13 (52) | 3 (20) | 10 (100) | |
| No | 12 (48) | 12 (80) | 0 (0) | <0.001 |
| pEVR@, ≥2log decline at W12, n (%) | ||||
| Yes | 20 (80) | 10 (67) | 10 (100) | |
| No | 5 (20) | 5 (33) | 0 (0) | 0.06 |
| EOT @, <100 IU/ml at W48, n (%) | ||||
| Yes | 18 (85) | 10 (77) | 8 (100) | 0.3 |
| No | 3 (15) | 3 (23) | 0 (0) | |
| REL@, >50 IU/ml at W24 follow up , n (%) | ||||
| Yes | 11 (61) | 8 (80) | 3 (37) | 0.1 |
| No | 7 (39) | 2 (20) | 5 (63) | |
| SVR @, <50 IU/ml at W24 follow up, n (%) | ||||
| Yes | 7 (33) | 2 (15) | 5 (62) | 0.05 |
| No | 14 (67) | 11 (85) | 3 (38) |
RVR, rapid viral response ; cEVR, complete early viral response; EOT, end of treatment response; REL, relapse during follow up; SVR, sustained viral response; Vmin, viral nadir during the first week of treatment (see Table 2); V0, viral load at baseline;
One patient discontinued treatment from week 11 and four patients after week 12.
p≤0.05 for a difference between Gen 1 and Gen 3. IQR, Interquartile range
Viral kinetic profiles and response
Viral response by genotype is presented in detail at 0-28 days and 0-84 days of therapy for the 21 patients with complete follow-up (Fig. 1). Patients were stratified into three groups based on their viral response (i.e., NR – nonresponders [i.e., detectable HCV RNA at the end of therapy (week 48); REL – relapsers [i.e., negative HCV RNA at week 48 but positive after follow up], SVR [i.e., negative HCV RNA at follow up] (Fig. 1). It is clear that regardless of the genotype, the viral load decay pattern at early on treatment time points distinguished among the SVR, REL and NR groups. There were distinct differences in the response pattern between genotype 1 (Figures 1A & 1B) and genotype 3 (Figures 1C & 1D). Patients infected with HCV genotype 3, including those in the REL group, showed a more rapid and intense initial reduction in viral load than that seen in patients infected with HCV genotype 1. Genotype 1 subjects who achieved an SVR had a fast and marked viral decline, which distinguished them from REL or NR. In genotype 1 patients, a 1.5 log drop at day 15 differentiated SVR from REL (p=0.012) and 1.9 log drop at day 15 distinguished SVR from NR (p=0.008). For genotype 3, a 1.5 log drop by week 4 distinguished between SVR and REL (p=0.014) even though all genotype 3 patients reached cEVR.
Viral kinetic parameters during the first 29 days of therapy in all 26 patients are presented in Table 2. Following the first dose of PEG-IFN, viral load dropped to a nadir, Vmin, of 1.3 ± 0.3 log10 from baseline within 1.6 ± 0.3 days (Table 2). Thereafter, a 0.3-to 12.9-fold viral rebound was observed before the second dose of PEG-IFN was given at day 7. Following the second dose of PEG-IFN, a slower phase of viral decline was observed (Fig. 1). While baseline viral load, day of Vmin, and viral rebound parameters were similar among the SVR, REL and NR groups, the first phase of viral decline from baseline and the viral slope decline (i.e., following the second dose of PEG-IFN) were significantly (p<0.02) higher in SVR compared to REL and NR (Table 2).
Viral kinetic profiles, HCV genotype, race and response
We further analyzed viral response in conjunction with genotype as shown in Table 3. First-phase viral decline was significantly higher in genotype 3 compared to genotype 1 patients (median 0.92 vs 1.65 log10, respectively, p=0.009). In addition, the slower-phase viral decline slope measured from day 7 until day 15 (median 1.0 vs 0.35 log/wk, respectively, p=0.031) or from day 7 until day 29 (0.72 vs 0.30 log/wk, respectively, p=0.008), were significantly higher in genotype 3 compared to genotype 1. All rapid viral responder (RVR) subjects were genotype 3 (p=0.007). At week 12, all genotype 3 subjects had a cEVR, while only 20% of genotype 1 (p<0.001) subjects achieved a cEVR. End-of-therapy response and relapse rate were similar by genotype (p=0.3 and p=0.1, respectively), but SVR rate was significantly (p=0.05) greater in genotype 3 patients. Interestingly, viral kinetic parameters and treatment response did not vary by race/ethnicity (White or Black) (not shown).
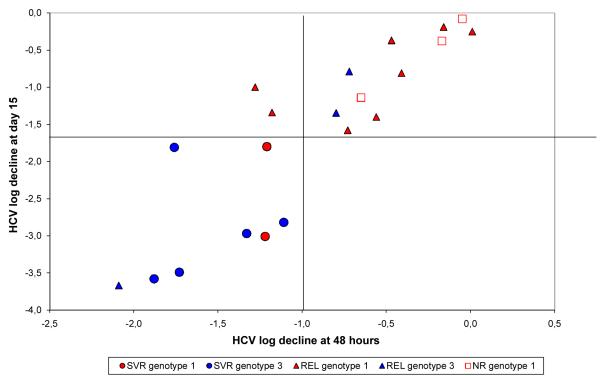
Finally, the relationship between log10 viral decline at days 2 and 15 per genotype and outcome of therapy is shown in Fig. 2. Patients who achieved an SVR had a profound viral decline (see left lower quadrant of Fig. 2). In contrast, relapsers (REL) and nonresponders (NR), scattered on the two upper quadrants, which corresponds to a slower pattern of viral decline slope (see also Table 2).
Figure 2.
A relationship between viral decline at days 2 and 15 of therapy per genotype and outcome (see also Table 2).
Very early predictors of successful treatment response
Change in HCV RNA level at very early on treatment time points was a strong predictor of SVR. In genotype 1 patients, an HCV RNA decline of 0.75 log10 from baseline at day 1, had good discriminatory ability (AUROC 0.95), a positive predictive value (PPV) of 67% and a negative predictive value (NPV) of 100% for SVR (Table 4). In the genotype 3 group, a day 2 HCV RNA decline cutoff value of 0.97 log10, had a good discriminatory ability (AUROC 0.82), with a PPV of 83% and NPV of 100% for the development of SVR (Table 4). In both groups, viral decline at day 2 had similar discriminatory ability with the same PPV and NPV to identify likely SVRs as RVR at week 4 (Table 4). According to these criteria, 2 out of 3 (67%) genotype-3 subjects and 10 out of 11 (91%) genotype-1 subjects, could have stopped (or extended) therapy based on day 2 HCV RNA decline.
Table 4.
Summary of ROC curve cut-off for selected early points and prediction of therapy outcome by HCV genotype.
| Genotype | Day/Week | Viral decline cut-off (log10 decay) |
Area Under the Curve |
Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|
| 1 | d1 | 0.75 | 0.95 | 100% | 91% | 67% | 100% |
| d2 | 1.19 | 0.95 | 100% | 91% | 67% | 100% | |
| w4 | 1.90 | 0,95 | 100% | 91% | 67% | 100% | |
|
| |||||||
| 3 | d1 | 1.30 | 0.80 | 60% | 91% | 100% | 60% |
| d2 | 0.97 | 0.83 | 100% | 67% | 83% | 100% | |
| w4 | 2.00 | 0.750 | 100% | 50% | 83% | 100% | |
PPV, positive predictive value; NPV, negative predictive value.
Coinfected patients with a slow second phase viral decline slope less than 0.3 log/wk (measured here from day 2 to day 29 using linear regression; Table 2) did not achieve SVR (NPV=100%), in agreement with publications in both HCV genotype 1-monoinfection and HIV/HCV coinfection patients [21, 22, 28, 29]. However, the positive predictive value for the second phase slope was low in both genotypes (PPV=67% in genotype 1 and PPV=71% in genotype 3), consistent with a recent study in coinfected patients [22]. According to these criteria, 1 out of 3 (33%) genotype-3 subjects and 10 out of 11 (91%) genotype-1 subjects, could have stopped (or extended) therapy at day 2.
Discussion
In this detailed viral kinetic study, we found that HIV/HCV coinfected patients who were treated with PEG-IFN-α-2a and ribavirin had nadir HCV RNA, Vmin, 1.6 ± 0.3 days which was followed by 0.3- to 12.9-fold viral rebound (until the administration of the second dose of PEG-IFN at day 7) regardless of genotype or outcome of therapy. This viral rebound is likely related to the PEG-IFN pharmacokinetic and pharmacodynamic features as recently shown using mathematical modeling [23, 24, 27]. These viral kinetic features were not associated with SVR and HCV genotype and may suggest that first phase viral decline (i.e., viral nadir) under PEG-IFN-α-2a can be estimated in future studies between 1 to 2 days post initiation of therapy.
Current therapeutic guidelines suggest the discontinuation of therapy based on failure to achieve a pEVR at week 12 [30]. Recently, the RVR at week 4 was found to be highly associated with SVR in coinfected patients [17-19]. Early prediction of nonresponse to pegylated-interferon/RBV is particularly important in HIV/HCV coinfected patients to permit early treatment discontinuation in those who are unlikely to respond. Here we found that the first phase viral decline from baseline and a slower phase decline slope (between day 7 and day 29) were associated (p<0.05) with SVR and were significantly (p<0.01) higher in genotype 3 than in genotype 1 subjects (Tables 2 and 3), in agreement with previous viral kinetic studies in patients mono-infected with HCV genotype 1 or genotype 3 [21, 31-34]. In addition, a second phase viral decline slope (i.e., measured from day 2 till day 29) less than 0.3 log/wk has a NPV of 100% for SVR. Perhaps even more importantly, day 2 viral response had similar predictive values of nonresponse as week 4 (Table 4). These results suggest that HCV RNA at day 2 of therapy could potentially identify patients who likely will not achieve SVR. Our results are in line with a recent publication [35] in which a similar very early predictor of noresponse at day 2 was identified in HCV monoinfected patients.
We did not find viral kinetic parameters or viral response patterns that differed by race/ethnicity in our analysis of patients in Brazil. Interestingly, HIV/HCV-coinfected African American patients exhibit significantly smaller declines in HCV RNA than non-Hispainc Whites [23, 24]. The same pattern was also found in African American vs. non-Hispanic Whites who were mono-infected with HCV [36, 37]. Recently, a genetic polymorphism near the IL28B gene, was associated with an approximately twofold change in response to treatment, with substantially greater frequency in European than African populations [38]. Very recently, Howell et al. [39] showed that compared to Caucasians, African Americans had a less vigorous decline of serum HCV RNA from day 0-2 (phase 1) and day 7-28 (phase 2) of early HCV kinetics (p < 0.01). Since in the current study there was no significant (p=0.5) difference in IL28B genotype frequencies between Black and white race/ethneicty (manuscript in preparation), it may partly explain the lack of association between race/ethnicity and viral kinetic parameters or viral response patterns. Larger studies are needed to provide a comprehensive viral kinetic comparison by race/ethnicity in South America, where patient ancestry may differ from that in the United States.
In a recent publication, we identified several PEG-IFN pharmacodynamic parameters that might identify likely SVR patients early in treatment via mathematical modeling [27]. Here, in order to identify other promising early predictors of successful treatment outcome, that can be easily addressed in future clinical settings, we evaluated the predictive value of early changes in serum HCV RNA levels relative to baseline, for its potential to identify likely nonresponders. As previously suggested in HCV monoinfection [34, 40], we found that a viral decline <1.19 log for genotype 1 and <0.97 log for genotype 3, respectively, had NPV of 100% for SVR as early as day 2 post initiation of therapy. The high negative predictive value at day 2 minimizes the likelihood of premature treatment discontinuation in patients who might achieve a successful therapeutic outcome. Notably, as we are approaching a new era of therapy with direct antiviral agents, an early prediction of nonresponse to PEG-IFN based therapy may help in designing more effective therapeutic strategies such as the inclusion of direct antiviral agents against HCV [41, 42]. Larger studies are needed to verify that these parameters can consistently identify nonresponders at very early time points.
Acknowledgments
The authors thank Fernando F.Tatsch and Lori Brisbin from Roche by the support on drug and tests supplies; Steve Young from Tricore Reference Laboratories for performing HIV and HCV load tests; Claudia Courtouke for helping with the original clinical data;Dr.José Luiz Boechat Paione and Hospital Ana Costa to allow patients hospitalization and blood drew along the clinical phase; LIM-47 team for excellent technical assistance, and the patients and their family members for the trust and participation.
Grant support: Study carried out with the assistance of Roche Laboratories of Brazil, which provided financial support in the form of the pegylated interferon α-2a, kits for molecular assays and the shipping of samples. H.D is supported by the University of Illinois Walter Payton Liver Center GUILD and by NIH grant P20-RR018754.
List of Abbreviations
- NPV
negative predictive value
- PPV
positive predictive value
- ANOVA
analysis of variance
- HAART
highly active antiretroviral therapy
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- ROC
receiver operating characteristic
- SVR
sustained virological response
- RVR
Rapid Virologic Response
- EVR
Early Virologic Response
- NR
Non Responders
- EOT
End of Therapy Response
- REL
Relapser
- ITT
intention to treat
- PP
per protocol
- Gen
Genotype
- NS
not significant
- IFN
interferon
- STAT-C
specifically target HCV therapy
Footnotes
Disclosure Financial Support
Conflict of Interest ESAA has received support for travel to international conferences, honoraria for scientific activities and research support from Roche Brazil.
References
- 1.Arends JE, Boucher CA, Hoepelman AI. Hepatitis C virus and human immunodeficiency virus coinfection: where do we stand? Neth J Med. 2005;63:156–163. [PubMed] [Google Scholar]
- 2.Ferreira P, Navarro R, Araujo E, Barone A. In: Hepatite C. Ed M, editor. 2009. pp. 280–308. [Google Scholar]
- 3.Soriano V, Martin-Carbonero L, Maida I, Garcia-Samaniego J, Nunez M. New paradigms in the management of HIV and hepatitis C virus coinfection. Curr Opin Infect Dis. 2005;18:550–560. doi: 10.1097/01.qco.0000191509.56104.ec. [DOI] [PubMed] [Google Scholar]
- 4.Rockstroh JK. Management of hepatitis C/HIV coinfection. Curr Opin Infect Dis. 2006;19:8–13. doi: 10.1097/01.qco.0000200294.22661.e0. [DOI] [PubMed] [Google Scholar]
- 5.Sulkowski MS, Mast EE, Seeff LB, Thomas DL. Hepatitis C virus infection as an opportunistic disease in persons infected with human immunodeficiency virus. Clin Infect Dis. 2000;30:S77–84. doi: 10.1086/313842. [DOI] [PubMed] [Google Scholar]
- 6.Bica I, McGovern B, Dhar R, Stone D, McGowan K, Scheib R, Snydman DR. Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis. 2001;32:492–497. doi: 10.1086/318501. Epub 2001 Jan 2023. [DOI] [PubMed] [Google Scholar]
- 7.Chen TY, Ding EL, Seage GR, Iii, Kim AY. Meta-analysis: increased mortality associated with hepatitis C in HIV-infected persons is unrelated to HIV disease progression. Clin Infect Dis. 2009;49:1605–1615. doi: 10.1086/644771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung RT, Andersen J, Volberding P, Robbins GK, Liu T, Sherman KE, et al. Peginterferon Alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351:451–459. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, Lissen E, Gonzalez-Garcia J, Lazzarin A, et al. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–450. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 10.Carrat F, Bani-Sadr F, Pol S, Rosenthal E, Lunel-Fabiani F, Benzekri A, et al. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. Jama. 2004;292:2839–2848. doi: 10.1001/jama.292.23.2839. [DOI] [PubMed] [Google Scholar]
- 11.Ballesteros AL, Franco S, Fuster D, Planas R, Martinez MA, Acosta L, et al. Early HCV dynamics on Peg-interferon and ribavirin in HIV/HCV co-infection: indications for the investigation of new treatment approaches. Aids. 2004;18:59–66. doi: 10.1097/00002030-200401020-00007. [DOI] [PubMed] [Google Scholar]
- 12.Thomas DL. Options for treatment of hepatitis C in HIV-infected persons. J Hepatol. 2006;44:S40–43. doi: 10.1016/j.jhep.2005.11.011. Epub 2005 Nov 2028. [DOI] [PubMed] [Google Scholar]
- 13.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 14.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr., et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 15.Soriano V, Puoti M, Sulkowski M, Mauss S, Cacoub P, Cargnel A, et al. Care of patients with hepatitis C and HIV co-infection. Aids. 2004;18:1–12. doi: 10.1097/00002030-200401020-00001. [DOI] [PubMed] [Google Scholar]
- 16.Alberti A, Clumeck N, Collins S, Gerlich W, Lundgren J, Palu G, et al. Short statement of the first European Consensus Conference on the treatment of chronic hepatitis B and C in HIV co-infected patients. J Hepatol. 2005;42:615–624. doi: 10.1016/j.jhep.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Nunez M, Marino A, Miralles C, Berdun MA, Sola J, Hernandez-Burruezo JJ, et al. Baseline serum hepatitis C virus (HCV) RNA level and response at week 4 are the best predictors of relapse after treatment with pegylated interferon plus ribavirin in HIV/HCV-coinfected patients. J Acquir Immune Defic Syndr. 2007;45:439–444. doi: 10.1097/QAI.0b013e318061b5d9. [DOI] [PubMed] [Google Scholar]
- 18.Laguno M, Larrousse M, Murillas J, Blanco JL, Leon A, Milinkovic A, et al. Predictive value of early virologic response in HIV/hepatitis C virus-coinfected patients treated with an interferon-based regimen plus ribavirin. J Acquir Immune Defic Syndr. 2007;44:174–178. doi: 10.1097/QAI.0b013e31802b812d. [DOI] [PubMed] [Google Scholar]
- 19.Martin-Carbonero L, Nunez M, Marino A, Alcocer F, Bonet L, Garcia-Samaniego J, et al. Undetectable hepatitis C virus RNA at week 4 as predictor of sustained virological response in HIV patients with chronic hepatitis C. Aids. 2008;22:15–21. doi: 10.1097/QAD.0b013e3282f1da99. [DOI] [PubMed] [Google Scholar]
- 20.Dahari H, Markatou M, Zeremski M, Haller I, Ribeiro RM, Licholai T, et al. Early ribavirin pharmacokinetics, HCV RNA and alanine aminotransferase kinetics in HIV/HCV co-infected patients during treatment with pegylated interferon and ribavirin. J Hepatol. 2007;47:23–30. doi: 10.1016/j.jhep.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 22.Avidan NU, Goldstein D, Rozenberg L, McLaughlin M, Ferenci P, Masur H, et al. Hepatitis C viral kinetics during treatment with peg IFN-alpha-2b in HIV/HCV coinfected patients as a function of baseline CD4+ T-cell counts. J Acquir Immune Defic Syndr. 2009;52:452–458. doi: 10.1097/QAI.0b013e3181be7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talal AH, Ribeiro RM, Powers KA, Grace M, Cullen C, Hussain M, et al. Pharmacodynamics of PEG-IFN alpha differentiate HIV/HCV coinfected sustained virological responders from nonresponders. Hepatology. 2006;43:943–953. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 24.Rozenberg L, Haagmans BL, Neumann AU, Chen G, McLaughlin M, Levy-Drummer RS, et al. Therapeutic response to peg-IFN-alpha-2b and ribavirin in HIV/HCV co-infected African-American and Caucasian patients as a function of HCV viral kinetics and interferon pharmacodynamics. Aids. 2009;23:2439–2450. doi: 10.1097/QAD.0b013e32832ff1c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahari H, Layden-Almer JE, Perelson AS, Layden TJ. Hepatitis C Viral Kinetics in Special Populations. Curr Hepat Rep. 2008;7:97–105. doi: 10.1007/s11901-008-0022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tien PC. Management and treatment of hepatitis C virus infection in HIV-infected adults: recommendations from the Veterans Affairs Hepatitis C Resource Center Program and National Hepatitis C Program Office. Am J Gastroenterol. 2005;100:2338–2354. doi: 10.1111/j.1572-0241.2005.00222.x. [DOI] [PubMed] [Google Scholar]
- 27.Dahari H, Araujo E, Haagmans B, Layden TJ, Cotler SJ, Barone A, Neumann AU. Pharmacodynamics of PEG-IFN alpha-2a in HIV/HCV co-infected patients: Implications for treatment outcomes. Journal of Hepatology. 2010 doi: 10.1016/j.jhep.2010.03.019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferenci P. Predicting the therapeutic response in patients with chronic hepatitis C: the role of viral kinetic studies. J Antimicrob Chemother. 2004;53:15–18. doi: 10.1093/jac/dkh015. [DOI] [PubMed] [Google Scholar]
- 29.Perelson AS, Herrmann E, Micol F, Zeuzem S. New kinetic models for the hepatitis C virus. Hepatology. 2005;42:749–754. doi: 10.1002/hep.20882. [DOI] [PubMed] [Google Scholar]
- 30.NIH National Institutes of Health Consensus Development Conference: management of hepatitis C: 2002. Hepatology. 2002;36:S3–20. doi: 10.1053/jhep.2002.37117. [DOI] [PubMed] [Google Scholar]
- 31.Herrmann E, Lee JH, Marinos G, Modi M, Zeuzem S. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology. 2003;37:1351–1358. doi: 10.1053/jhep.2003.50218. [DOI] [PubMed] [Google Scholar]
- 32.Medeiros-Filho JE, de Carvalho Mello IM, Pinho JR, Neumann AU, de Mello Malta F, da Silva LC, Carrilho FJ. Differences in viral kinetics between genotypes 1 and 3 of hepatitis C virus and between cirrhotic and non-cirrhotic patients during antiviral therapy. World J Gastroenterol. 2006;12:7271–7277. doi: 10.3748/wjg.v12.i45.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawlotsky JM, Dahari H, Neumann AU, Hezode C, Germanidis G, Lonjon I, et al. Antiviral action of ribavirin in chronic hepatitis C. Gastroenterology. 2004;126:703–714. doi: 10.1053/j.gastro.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Layden JE, Layden TJ, Reddy KR, Levy-Drummer RS, Poulakos J, Neumann AU. First phase viral kinetic parameters as predictors of treatment response and their influence on the second phase viral decline. J Viral Hepat. 2002;9:340–345. doi: 10.1046/j.1365-2893.2002.00377.x. [DOI] [PubMed] [Google Scholar]
- 35.Durante-Mangoni E, Zampino R, Portella G, Adinolfi LE, Utili R, Ruggiero G. Correlates and prognostic value of the first-phase hepatitis C virus RNA kinetics during treatment. Clin Infect Dis. 2009;49:498–506. doi: 10.1086/600887. [DOI] [PubMed] [Google Scholar]
- 36.Layden-Almer JE, Ribeiro RM, Wiley T, Perelson AS, Layden TJ. Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin. Hepatology. 2003;37:1343–1350. doi: 10.1053/jhep.2003.50217. [DOI] [PubMed] [Google Scholar]
- 37.Howell CD, Dowling TC, Paul M, Wahed AS, Terrault NA, Taylor M, et al. Peginterferon pharmacokinetics in African American and Caucasian American patients with hepatitis C virus genotype 1 infection. Clin Gastroenterol Hepatol. 2008;6:575–583. doi: 10.1016/j.cgh.2008.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 39.Howell CD, Thompson AJ, Ryan K, Zambeeli S, Gorden A, Fried M, et al. IL28B genetic variation association with early viral kinetics and SVR in HCV henotype 1 the VIRAHEP-C study. J Hepatol. 2010;(Suppl) [Google Scholar]
- 40.Cotler SJ, Layden JE, Neumann AU, Jensen DM. First phase hepatitis c viral kinetics in previous nonresponders patients. J Viral Hepat. 2003;10:43–49. doi: 10.1046/j.1365-2893.2003.00401.x. [DOI] [PubMed] [Google Scholar]
- 41.Zeuzem S, Nelson DR, Marcellin P. Dynamic evolution of therapy for chronic hepatitis C: how will novel agents be incorporated into the standard of care? Antivir Ther. 2008;13:747–760. [PubMed] [Google Scholar]
- 42.Boehme RE, Cameron S. Key data from the 11th International Workshop on Adverse Drug Reactions and Co-Morbidities in HIV. Antivir Ther. 2009;14:1195–1208. doi: 10.3851/IMP1470. [DOI] [PubMed] [Google Scholar]