Abstract
The effective use of targeted therapy is highly dependent upon the identification of responder patient populations. Loss of the Fbw7 tumor suppressor is frequently found in various types of human cancers including breast cancer, colon cancer 1 and T-cell acute lymphoblastic leukemia (T-ALL)2. In line with these genomic data, engineered deletion of Fbw7 in mouse T cells results in T-ALL3–5, validating Fbw7 as a T-ALL tumor suppressor. The precise molecular mechanisms by which Fbw7 exerts anti-tumor activity remain areas of intensive investigation and are thought to relate in part to Fbw7-mediated destruction of key cancer relevant proteins including c-Jun6, c-Myc 7, Cyclin E 8 and Notch-19, all of which possess oncogenic activity and are overexpressed in various human cancers including leukemia. Besides accelerating cell growth 10, overexpression of either c-Jun, c-Myc or Notch-1 can also provoke programmed cell death 11. Thus, considerable uncertainty surrounds how Fbw7-deficient cells evade cell death in the setting of upregulated c-Jun, c-Myc and/or Notch-1. Here we report that SCFFbw7 governs cellular apoptosis by targeting the pro-survival Bcl-2 family member, Mcl-1, for ubiquitination and destruction in a GSK3 phosphorylation-dependent manner. Human T-ALL cell lines showed a close relationship between Fbw7 loss and Mcl-1 overexpression. Correspondingly, T-ALL cell lines with defective Fbw7 are particularly sensitive to the multi-kinase inhibitor, sorafenib, but resistant to the Bcl-2 antagonist, ABT-737. On the genetic level, Fbw7 reconstitution or Mcl-1 depletion restores ABT-737 sensitivity, establishing Mcl-1 as a therapeutically relevant bypass survival mechanism for Fbw7-deficient cells to evade apoptosis. Therefore, our work provides novel molecular insight into Fbw7-direct tumor suppression with direct implications for the targeted treatment of Fbw7-deficient T-ALL patients.
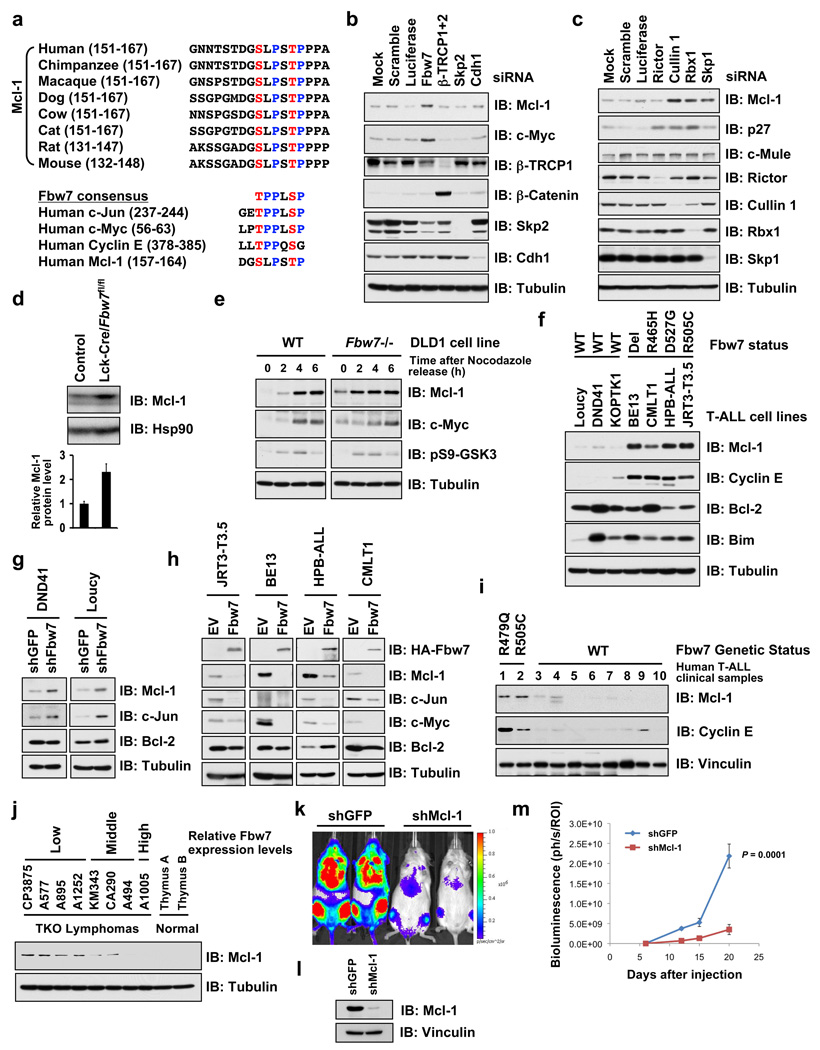
Mcl-1 is frequently overexpressed in various leukemias via mechanisms that are not fully understood 12. Mcl-1 is distinct from other Bcl-2 family members in its extremely unstable nature 13, which provides a mechanism for cells to switch into either survival or apoptotic mode in response to various stresses 14. While GSK3 phosphorylation regulates Mcl-1 stability 13, little is known about the identity of the E3 ubiquitin ligase that targets phosphorylated Mcl-1 for destruction. Upon examination of the GSK3 sites on Mcl-1, we surmised that they resemble a possible degron sequence that can be recognized by Fbw7 (Fig. 1a), prompting us to test the possibility that GSK3 phosphorylation of Mcl-1 triggers its degradation by Fbw7. Depletion of Fbw7 (Fig. 1b) or SCF components Cullin-1, Rbx1 and Skp1 (Fig. 1c), but not other F-box proteins we examined (Fig. 1b), resulted in a significant increase in Mcl-1. T-cell lineage-specific depletion of Fbw7 in Lck-Cre/Fbw7fl/fl mice 3 exhibited elevated Mcl-1 levels in their thymi (Fig. 1d) as well as resulting thymic lymphoma (Supplementary Fig. 1a) and acute leukemia cells (Supplementary Fig. 1b). Consistent with Wertz et al.15, Fbw7−/− DLD1 (Fig. 1e) and siFbw7 treated HeLa cells (Supplementary Fig. 1c) have elevated Mcl-1 expression mainly in the M and early G1 cell cycle phases. The clinical relevance of this finding is further demonstrated by the fact that T-ALL cell lines harboring Fbw7 mutations/deletions have a significant increase in Mcl-1 (Fig. 1f). Additionally, depletion of Fbw7 in DND41 or Loucy cells (with wild-type Fbw7) leads to increased Mcl-1 expression (Fig. 1g) while re-introduction of wild-type Fbw7 dramatically reduced Mcl-1 expression in Fbw7-deficient T-ALL cells (Fig. 1h), supporting a causal relationship between loss of Fbw7 activity and elevated Mcl-1 expression in the T-ALL cells examined. More importantly, elevated Mcl-1 expression is also observed in both primary human and murine T-ALL samples with deficient Fbw7 activity (Fig. 1i, Fig. 1j 2 and Supplementary Fig. 1a–b 3, 4), and depletion of Mcl-1 impaired T-ALL disease progression in vivo (Fig. 1k–m). Consistent with a post-translational mode of regulation, no changes in Mcl-1 mRNA levels were observed after depletion of Fbw7 in DLD1 cells (Supplementary Fig. 2d), and no positive relationship was observed between Mcl-1 mRNA levels and loss of Fbw7 in T-ALL cells (Supplementary Fig. 2e). The half-life of Mcl-1 was significantly extended in the thymi of Fbw7−/− mice and Fbw7-deficient human T-ALL cells (Supplementary Fig. 3a–c) and experimental manipulation of Fbw7 levels changed Mcl-1 stability accordingly (Supplementary Fig. 3d–e). Together, these results suggest that Mcl-1 is a downstream ubiquitination target for SCFFbw7.
Figure 1. Mcl-1 stability is controlled by Fbw7.
a, Sequence alignment of Mcl-1 with the c-Jun, c-Myc and Cyclin E Fbw7 phosphodegrons. The putative Fbw7 phosphodegron sequence present in Mcl-1 is conserved across different species.
b–c, Immunoblot analysis of HeLa cells transfected with the indicated siRNA oligonucleotides.
d, Immunoblot analysis of thymus cells derived from control mice or Fbw7 conditional knockout (Lck-Cre/Fbw7fl/fl) mice. Mcl-1 band intensity was normalized to Hsp90, then normalized to the control lane. Data was shown as mean ± SEM from three independent experiments.
e, Immunoblot analysis of wild-type (WT) or Fbw7−/− DLD1 cells after synchronization with nocodazole and release at the indicated time points.
f, Immunoblot analysis of the indicated human T-ALL cell lines.
g, DND41 and Loucy cells, which contain wild-type Fbw7, were infected with the indicated lentiviral shRNA constructs and selected with 1 µg/ml puromycin to eliminate the non-infected cells. Cell lysates were collected for immunoblot analysis with the indicated antibodies.
h, T-ALL cell lines with deficient Fbw7 were infected with Fbw7-expressing retrovirus construct (with empty vector as a negative control), and selected with 1 µg/ml puromycin to eliminate the non-infected cells. Cell lysates were collected for immunoblot analysis with the indicated antibodies.
i, Immunoblot analysis of the indicated primary human T-ALL clinical samples.
j. Immunoblot analysis of the indicated murine T-ALL cell lines derived from the Terc−/−Atm−/−p53−/− (TKO) mice.
k–m. In vivo effects of Mcl-1 depletion in Fbw7-deficient T-ALL cells. (k) An in vivo model of Fbw7-deficient T-ALL was created by orthotopic engraftment of CMLT1-luciferase cells in NOD-SCID-IL2Rγnull (NSG) mice (left; CMLT1-shGFP, right; CMLT1-shMcl-1). (l) immunoblot analysis of the engineered CMLT1 cell lines. (m) Mice were injected with 1×107 cells (n=7/group) via the lateral tail vein. Tumor burden was determined by quantification of total body luminescence, and are expressed as photons/second/standardized region of interest (ph/s/ROI). Data was represented as mean ± SEM with statistical significance determined by Student’s t-test.
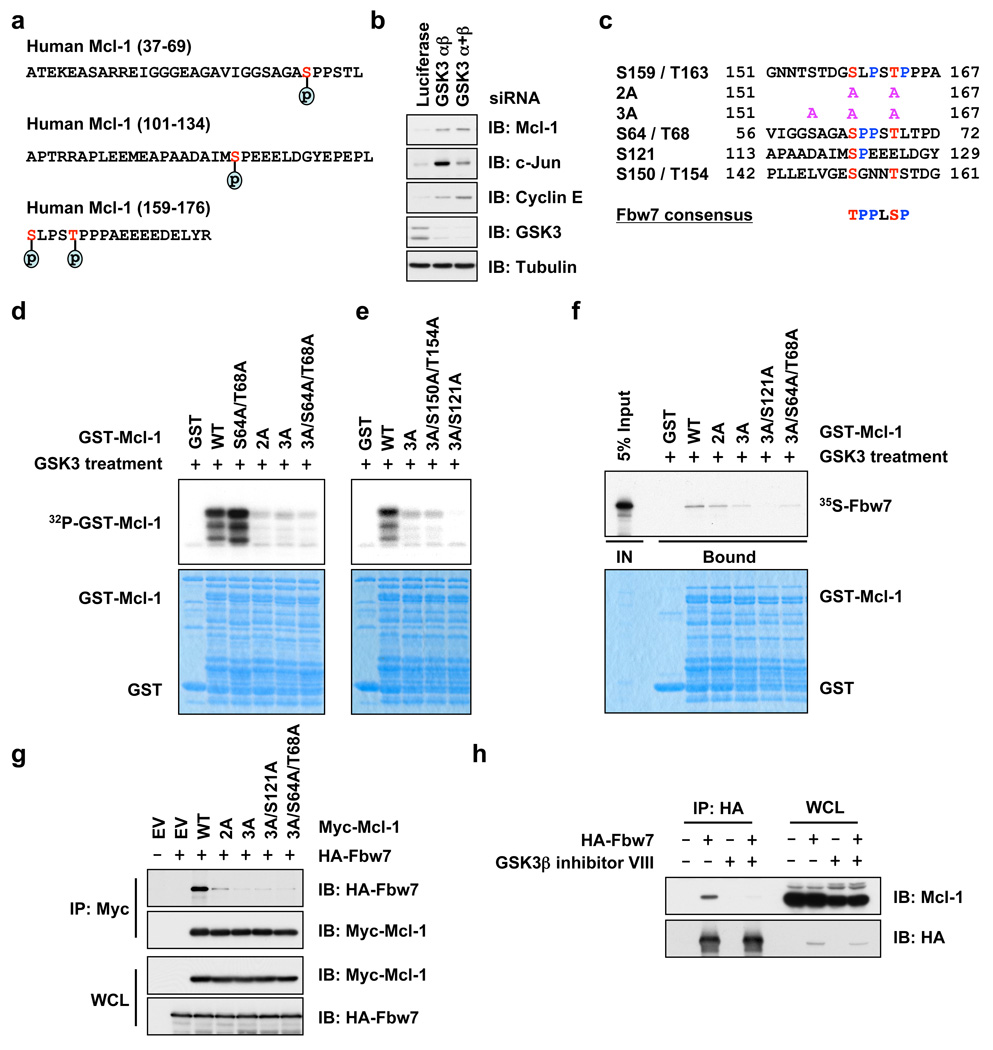
As proper substrate phosphorylation events are required for Fbw7 to recognize and target its substrates for ubiquitination 16, we next investigated which phosphorylation events that trigger Mcl-1 destruction by Fbw7. Mass spectrometry analysis revealed that Mcl-1 is phosphorylated at multiple sites in vivo (Fig. 2a and Supplementary Fig. 5a–c). In addition to Ser159 and Thr163 13, 17, Ser64 and Ser121 were also phosphorylated in vivo. Consistent with previous reports 13, 17, Mcl-1 destruction is promoted by GSK3 (Fig. 2b), but not ERK1/2 (Supplementary Fig. 5d–f). To further investigate the significance of each individual phosphorylation site, we created a panel of Mcl-1 mutants (Fig. 2c). Using in vitro kinase assays, we identified Ser159 and Thr163 as the major GSK3 phosphorylation sites17 and Ser121 as a minor GSK3 phosphorylation site (Fig. 2d–e and Supplementary Fig. 5g). Inactivation of these GSK3 phosphorylation sites impairs the interaction between Mcl-1 and Fbw7 both in vitro (Fig. 2f and Supplementary Fig. 5h) and in vivo (Fig. 2g and Supplementary Fig. 5i). Furthermore, pharmacological inhibition of GSK3 activity blocked the interaction between HA-Fbw7 and endogenous Mcl-1 (Fig. 2h) and inhibited the localization of Fbw7 to the mitochondria where Mcl-1 resides (Supplementary Fig. 5 j–k). These results indicated that GSK3-dependent phosphorylation of Mcl-1 is necessary for its interaction with Fbw7. Consistent with this Fbw7-Mcl-1 regulatory axis, Mcl-1 specifically interacts with Fbw7 (Supplementary Fig. 6a–b and 6j–l) and Cullin-1 (Supplementary Fig. 6c–d) and depletion of endogenous Cullin-1 increases Mcl-1 abundance (Supplementary Fig. 11a).
Figure 2. Phosphorylation of Mcl-1 by GSK3 triggers its interaction with Fbw7.
a, In vivo Mcl-1 phosphorylation sites detected by mass spectrum analysis.
b, Immunoblot analysis of HeLa cells transfected with the indicated siRNA oligonucleotides.
c, Illustration of the various Mcl-1 mutants generated for this study.
d–e, GSK3 phosphorylates Mcl-1 in vitro at multiple sites. Purified GSK3 protein (from New England Biolabs) was incubated with 5 µg of the indicated GST-Mcl-1 proteins in the presence of γ-32P-ATP. The kinase reaction products were resolved by SDS-PAGE and phosphorylation was detected by autoradiography.
f, Phosphorylation of Mcl-1 at multiple sites by GSK3 triggers its interaction with Fbw7 in vitro. Autoradiograms showing recovery of 35S-labeled Fbw7 protein bound to the indicated GST-Mcl-1 fusion proteins (GST protein as a negative control) incubated with GSK3 prior to the pull-down assays. IN, input (5% as indicated).
g, Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with HA-Fbw7 together with the indicated Myc-Mcl-1 constructs. Thirty hours post-transfection, cells were pretreated with 10 µM MG132 for 10 hours to block the proteasome pathway before harvesting.
h, Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with HA-Fbw7. Thirty hours post-transfection, cells were pretreated with 20 µM MG132 for 8 hours to block the proteasome pathway before harvesting. Where indicated, 25 µM of the GSK3β inhibitor VIII (with DMSO as a negative control) was added for 8 hours before harvesting.
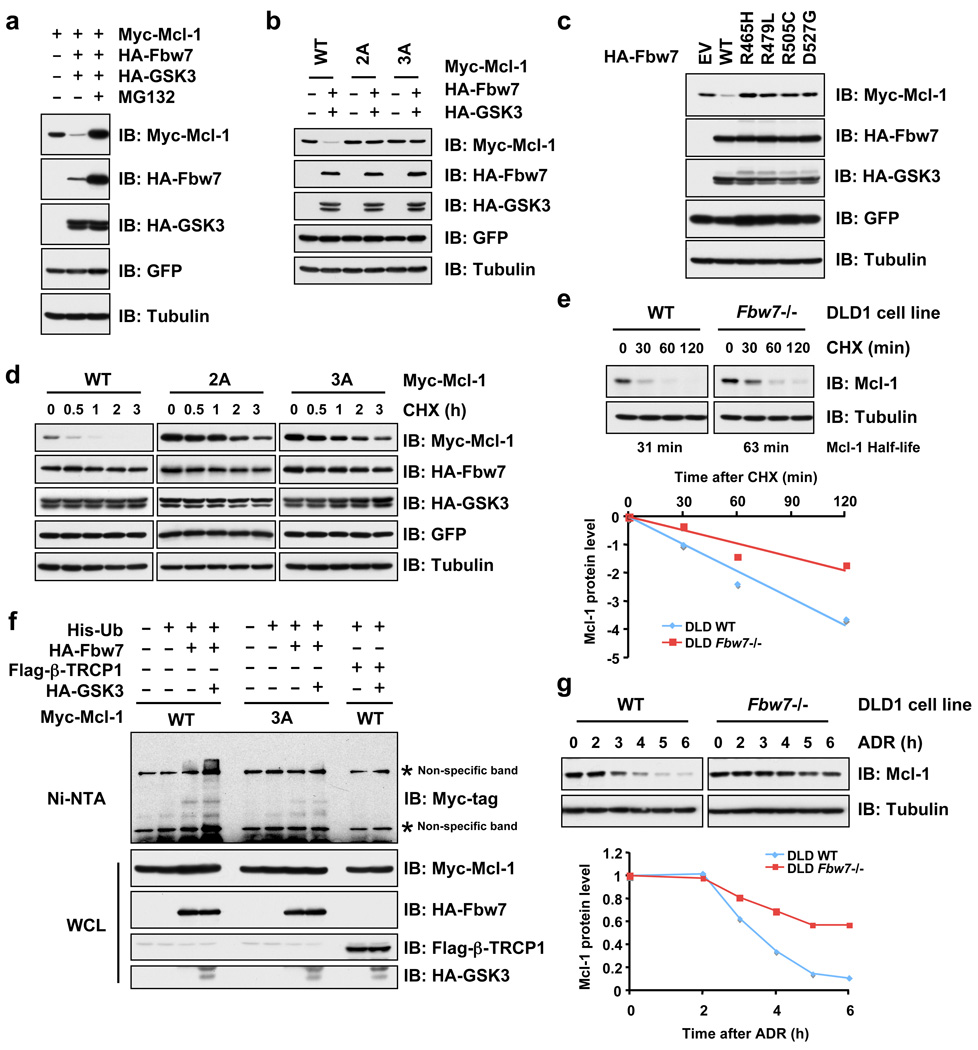
We next explored the mechanism by which Fbw7 alters Mcl-1 stability. Overexpression of Fbw7 and GSK3 significantly decreased Mcl-1 abundance (Fig. 3a and Supplementary Fig. 6h), while inactivation of the major GSK3 phosphorylation sites impaired Fbw7-mediated destruction (Fig. 3b and Supplementary Fig. 6e–g). All Fbw7 isoforms (particularly α and γ) participate in Mcl-1 stability control and Fbw7 dimerization is not required to degrade Mcl-1 (Supplementary Fig. 7a–e). Mutant Fbw7 constructs derived from T-ALL patients displayed reduced ability to interact with Mcl-l (Supplementary Fig. 6i), and were therefore unable to degrade Mcl-1 (Fig. 3c). Moreover, Fbw7/GSK3-mediated Mcl-1 destruction was blocked by MG132, indicating the involvement of the ubiquitin/proteasome pathway in this process (Fig. 3a). In support of this idea, co-expression of GSK3 and Fbw7 resulted in a marked reduction in the half-life of wild-type Mcl-1, but not the 2A or 3A Mcl-1 mutants (Fig. 3d) with reduced interaction with Fbw7 (Fig. 2g). Furthermore, loss of Fbw7 extends the half-life of endogenous Mcl-1 (Fig. 3e), and Fbw7 promotes Mcl-1 ubiquitination in a GSK3-dependent manner (Fig. 3f and Supplementary Fig. 8a–b and 8e). The decrease of Mcl-1 expression is also impaired in response to various DNA-damaging agents 18 in Fbw7−/− DLD1 cells (Fig. 3g and Supplementary Fig. 8f). These data together suggested a physiological role for Fbw7 in promoting Mcl-1 destruction in vivo in a GSK3 phosphorylation-dependent manner.
Figure 3. Fbw7 promotes Mcl-1 ubiquitination and destruction in a GSK3 phosphorylation-dependent manner.
a–c, GSK3 phosphorylation-dependent degradation of Mcl-1 by Fbw7. Immunoblot analysis of 293T cells transfected with the indicated Myc-Mcl-1 and HA-Fbw7 plasmids in the presence or absence of HA-GSK3. A plasmid encoding GFP was used as a negative control for transfection efficiency. Where indicated, the proteasome inhibitor MG132 was added.
d, 293T cells were transfected with the indicated Myc-Mcl-1 constructs together with the HA-Fbw7 and HA-GSK3 plasmids. Twenty hours post-transfection, cells were split into 60 mm dishes, and after another 20 hours, treated with 20 µg/ml cycloheximide (CHX). At the indicated time points, whole cell lysates were prepared and immunoblots were probed with the indicated antibodies.
e, Wild-type (WT) or Fbw7−/− DLD1 cells were treated with 20 µg/ml cycloheximide (CHX). At the indicated time points, whole cell lysates were prepared and immunoblots were probed with the indicated antibodies. Mcl-1 band intensity was normalized to tubulin, then normalized to the t=0 controls.
f, Immunoblot analysis (IB) of whole cell lysates (WCL) and His-tag pull-down of HeLa cells transfected with the indicated plasmids. Twenty hours post-transfection, cells were treated with the proteasome inhibitor MG132 overnight before harvesting. His-tag pull-down was performed in the presence of 8 M urea to eliminate any possible contamination from Mcl-1-associated proteins.
g, Immunoblot analysis of wild-type (WT) or Fbw7−/− DLD1 cells treated with 10 µM adriamycin (ADR) for the indicated durations of time. Mcl-1 band intensity was normalized to tubulin, then normalized to the t=0 controls.
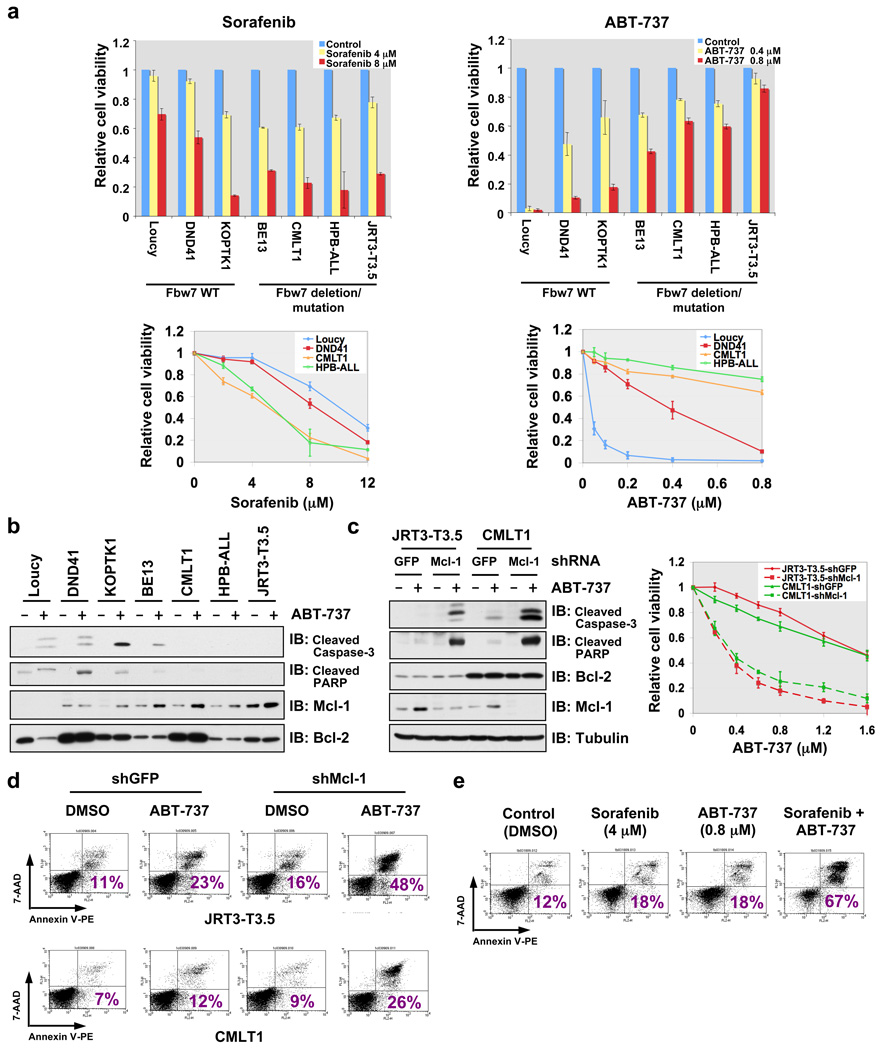
Next we explored how Fbw7 affects the cellular apoptotic response by modulating Mcl-1 abundance. As predicted, Fbw7−/− mouse thymocytes and Fbw7-deficient human T-ALL cells with increased Mcl-1 levels were less sensitive to apoptotic stimuli (Supplementary Fig. 9 a–f). More interestingly, compared with T-ALL cell lines with wild-type Fbw7, Fbw7-deficient T-ALL cells with elevated Mcl-1 expression (Fig. 1f and Supplementary Fig. 9h) were more sensitive to sorafenib, which can effectively reduce Mcl-1 expression (Fig. 4a, Supplementary Fig. 9g–i) 19, 20. Although sorafenib’s ability to repress Mcl-1 has been attributed to inactivating MAPK kinase and/or activating GSK-3 activity 19, the exact mechanism remains unclear. Nonetheless, this data suggests that Fbw7-deficient T-ALL cell lines might require elevated levels of Mcl-1 to evade apoptosis, a phenotype known as “oncogene addiction” 21. On the other hand, Fbw7-deficient T-ALL cell lines were more resistant to ABT-737 (Fig. 4a, Supplementary Fig. 9g and 9j). The BH3 mimetic ABT-737 is a pan-inhibitor of the Bcl-2 family of anti-apoptotic proteins, which is reported to effectively kill leukemia cells 22. However, leukemia cells with elevated Mcl-1 are refractory to ABT-737 23, 24 primarily because ABT-737 fails to inactivate Mcl-1 22. Experimental evidence from both 7-AAD/Annexin V double staining (Supplementary Fig. 9j) and immunoblots against apoptotic biomarkers (Fig. 4b) suggest that ABT-737-induced apoptosis is impaired in Fbw7-deficient T-ALL cells. Moreover, specific depletion of Mcl-1 in multiple Fbw7-deficient T-ALL cell lines restored their sensitivity to ABT-737 (Fig.4c–d), supporting the notion that increased Mcl-1 expression is the primary cause of desensitization to ABT-737 in vivo 23, 24. It also suggests that Fbw7-deficient T-ALL patients will not respond well to ABT-737 treatment. We further demonstrated that manipulation of Fbw7 activity or ectopic expression of a non-degradable Mcl-1 in human T-ALL cells affects their ABT-737 sensitivity (Supplementary Fig. 10a–b) and response to other apoptotic stimuli (Supplementary Fig. 10c–f).
Figure 4. Elevated Mcl-1 expression protects Fbw7-deficient T-ALL cell lines from ABT-737-induced apoptosis.
a, Cell viability assays showing that Fbw7-deficient T-ALL cell lines were more sensitive to sorafenib, but resistant to ABT-737 treatment. T-ALL cells were cultured in 10% FBS-containing medium with the indicated concentrations of sorafenib or ABT-737 for 48 hours before performing the cell viability assays. Data was shown as mean ± SD for three independent experiments.
b, Immunoblot analysis of the indicated human T-ALL cell lines with or without ABT-737 (0.8 µM) treatment.
c, Specific depletion of endogenous Mcl-1 expression restored ABT-737 sensitivity in the indicated Fbw7-deficient T-ALL cell lines. Various T-ALL cells were cultured in 10% FBS-containing medium with the indicated concentrations of ABT-737 for 48 hours before performing the cell viability assays, or with or without ABT-737 (0.8 µM) treatment for 24 hours before collecting whole cell lysates for immunoblot analysis with the indicated antibodies. For cell viability assays, data was shown as mean ± SD for three independent experiments.
d, 7-Amino-Actinomycin D (7-AAD)/Annexin V double-staining FACS analysis to detect the percentage of ABT-737-induced apoptosis in the indicated Fbw7-deficient T-ALL cell lines where the endogenous Mcl-1 was depleted by lentiviral shRNA treatment (lentiviral shGFP was used as a negative control). Various T-ALL cells were cultured in 10% FBS-containing medium with or without ABT-737 (0.8 µM) treatment for 48 hours before the FACS analysis. Numbers indicate the percentage of apoptotic cells.
e, 7-AAD/Annexin V double-staining FACS analysis to demonstrate that sorafenib treatment restores ABT-737 sensitivity to Fbw7-deficient HPB-ALL cells. HPB-ALL cells were cultured in 10% FBS-containing medium with the indicated concentrations of sorafenib and/or ABT-737 for 48 hours before the FACS analysis. Numbers indicate the percentage of apoptotic cells.
Our results imply that inhibition of Mcl-1 could be used to restore ABT-737 sensitivity in Fbw7-deficient T-ALL cells. Given that the clinical application of siRNA- or shRNA-mediated target extinction is not yet mature due to delivery challenges, we instead exploited small molecule strategies to reduce Mcl-1 expression, specifically with the use of sorafenib (Supplementary Fig. 9h). To this end, combined use of sorafenib and ABT-737 produced a dose-dependent increase of ABT-737 sensitivity for HPB-ALL (Supplementary Fig. 10g), which correlated with a significant increase in the induction of apoptosis (Fig. 4e). Similar results were obtained with other Fbw7-deficient T-ALL cell lines (Supplementary Fig. 10h).
Our studies provide experimental evidence for a role of Fbw7 in governing the apoptotic pathway by controlling Mcl-1 destruction. Mcl-1 plays a key role in regulating the cellular apoptosis of T cells 14, but not other tissue types such as liver cells. Therefore, our studies also provide a possible mechanism for why loss of Fbw7 is frequently seen in T-ALL patients. Although other E3 ubiquitin ligases including c-Mule 25 and β-TRCP 17 have been implicated in Mcl-1 stability control, c-Mule activity was not implicated in GSK3-dependent regulation of Mcl-1 (Supplementary Fig. 11a–e) 17, 25. Additionally, no correlation was found between c-Mule and Mcl-1 expression in various T-ALL cells (Supplementary Fig. 11f), excluding a physiological role for c-Mule in regulating Mcl-1 abundance in T-ALL. We further found that depletion of Fbw7, but not β-TRCP, leads to a significant induction of Mcl-1 (Fig. 1b and Supplementary Fig. 11a–c). Array CGH analysis demonstrated a high frequency of Fbw7 loss 2, but not simultaneous loss of β-TRCP1 and β-TRCP2 in T-ALL (data not shown). Altogether, these data support the hypothesis that Fbw7 is a physiological E3 ubiquitin ligase for Mcl-1 with USP9X as the nominated deubiquitinase 26, and loss of Fbw7 contributes to T-ALL development via Mcl-1 upregulation. More importantly, our studies suggest that there is a correlation between Fbw7 genetic status and ABT-737 sensitivity and further provide insight into the usage of Mcl-1 inhibitors as a practical method to specifically kill Fbw7-deficient T-ALL cells. This work provides a basis for the rational treatment of T-ALL patients and motivates the development of specific Mcl-1 antagonists, or agents that significantly reduce Mcl-1 expression for the improved management of T-ALL patients.
METHODS SUMMARY
Expression plasmid constructs, proteins, antibodies and cell lines are described in the Methods. The sequences of various siRNA oligos used in this study are also listed in the Methods section. Mcl-1 in vivo phosphorylation was detected by mass spectrometry analysis, and the identified major GSK3 phosphorylation sites were examined by in vitro kinase assays. All mutants were generated using PCR and the sequences were verified. Fbw7-mediated Mcl-1 destruction and ubiquitination were examined by cell-based ubiquitination and degradation assays. Cell viability assays were used to detect the response of various T-ALL cell lines to sorafenib and ABT-737. Annexin V/7-AAD double staining was used to detect the percentage of cellular apoptosis. A detailed description of the experimental procedures is provided in the Methods section.
METHODS
Plasmids
HA-Fbw7 and HA-GSK3 constructs were described previously 6. Fbw7 cDNA was subcloned using the Pfu polymerase (Stratagene) into the pBabe-Puro-HA retrovirus vector. Myc-Mcl-1 WT, Myc-Mcl-1 3A, and GST-Mcl-1 WT constructs were kind gifts from Dr. Mien-Chie Hung. Fbw7 and Mcl-1 mutants were generated with the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. HA-ERK1, shERK1 and shERK2 constructs were kind gifts from Dr. John Blenis. Flag-β-TRCP1, Flag-Ubiquitin, shTRCP1 and shTRCP1+2 retroviral constructs were kind gifts from Dr. Wade Harper. shFbw7 retroviral vector was purchased from Addgene, which has been validated and described previously 27. To generate the lentiviral shFbw7 and sh-c-Mule vectors, DNA oligos for shRNA against Fbw7 and c-Mule were annealed and subcloned into AgeI and EcoRI sites of the pLKO lentiviral plasmid. The following are DNA oligo sequences for the Fbw7 shRNA (sense; 5’-CCGGAACCTTCTCTGGAGAGAGAAACTCGAGTTTCTCTCTCCAGAGAAGGTTTTTTTG-3’, anti-sense; 5’-AATTCAAAAAAACCTTCTCTGGAGAGAGAAACTCGAGTTTCTCTCTCCAGAGAAGGTT-3’), and for c-Mule shRNA (sense; 5’-CCGGAATTGCTATGTCTCTGGGACACTCGAGTGTCCCAGAGACATAGCAATTTTTTTG-3’, antisense; 5’-AATTCAAAAAAATTGCTATGTCTCTGGGACACTCGAGTGTCCCAGAGACATAGCAATT-3’). Lentiviral shRNA constructs against GFP and Mcl-1 were obtained from Dr. William Hahn. WT-Mcl-1 and 3A-Mcl-1 cDNAs were amplified with PCR and subcloned into the BamH I and Sal I sites of the pLenti-GFP-Puro construct (Addgene, Cat. No.: 658-5).
Antibodies and Reagents
Anti-c-Myc antibody (sc-40), polyclonal anti-HA antibody (SC-805), anti-Cyclin A antibody (SC-751), anti-Plk1 antibody (SC-17783), anti-Cullin-1 antibody (sc-70895), anti-Rictor antibody (sc-81538), anti-p27 antibody (sc-528), anti-Skp1 antibody (sc-7163), anti-Mcl-1 antibody (sc-819) and anti-Cyclin E antibody (SC-247) were purchased from Santa Cruz. Anti-tubulin antibody (T-5168), polyclonal anti-FLAG antibody (F2425), monoclonal anti-FLAG antibody (F-3165), anti-β-Catenin antibody (C7207), anti-Vinculin antibody (V9131), peroxidase-conjugated anti-mouse secondary antibody (A4416) and peroxidase-conjugated anti-rabbit secondary antibody (A4914) were purchased from Sigma. Anti Mcl-1 antibody (4572), anti-Bcl-2 antibody (2872), anti-COX IV antibody (4850), anti-cleaved Caspase-3 (Asp175) antibody (9661), anti-cleaved PARP (Asp214) antibody (9541), anti-ERK1/2 antibody (4695), anti-c-Jun antibody (9162), anti-phospho-GSK3β (Ser-9) antibody (9336) and anti-Bim antibody (4582) were purchased from Cell Signaling. Anti-c-Mule antibody (A300–486A) was purchased from Bethyl. Monoclonal anti-HA antibody (MMS-101P) was purchased from Convace. Anti-Rbx1 antibody (RB-069P1) was purchased from Neomarker. Anti-Mcl-1 antibody (559027) was purchased from BD Pharmingen. Anti-GFP antibody (632380) and anti-Cullin-1 antibody (32–2400) were purchased from Invitrogen. Anti-Cdh1 antibody (CC43) was purchased from Oncogene. Oligofectamine, Lipofectamine and Plus reagents were purchased from Invitrogen. GSK3β inhibitor VIII was purchased from Calbiochem.
siRNAs
Human siRNA oligos against Fbw7, Skp2, Cdh1 and Cullin-1 have been described previously 6, 28, 29. A human siRNA oligo which can deplete both β-TRCP1 and β-TRCP2 (sense, 5’-AAGUGGAAUUUGUGGAACAUC-3’) was purchased from Dharmacon. Human siRNA oligos against c-Mule (sense, 5’-CAUGCCGCAAUCCAGACAUAU-3’) 25 and (sense, 5’-AAUUGCUAUGUCUCUGGGACA-3’) 30 have been validated previously and were purchased from Dharmacon. Luciferase GL2 siRNA oligo was purchased from Dharmacon. siRNA oligos to deplete endogenous Rbx1 (AACUGUGCCAUCUGCAGGAACAA), Cullin1 (GGUCGCUUCAUAAACAACAUU), and Rictor (AAACUUGUGAAGAAUCGUAUCUU) were synthesized by Dharmacon. Cocktailed siRNAs targeting Skp1 were purchased from Invitrogen (1299003). A GSK3α siRNA oligo (6312) and a GSK3α/β siRNA oligo (6301) were purchased from Cell Signaling. The GSK3β siRNA oligo (51012) was purchased from Ambion. As described previously, siRNA oligos were transfected into subconfluent cells with Oligofectamine or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions 6.
Cell Culture
Cell culture including synchronization and transfection has been described 6, 28. Wild type and Fbw7−/− DLD1 cell lines were kind gifts from Dr. Bert Vogelstein. Murine T-ALL cell lines derived from Tal1 transgenic mice were kind gifts from Dr. Michele A. Kelliher. Human T-ALL cell lines were previously described 2. Loucy and CMLT1 T-ALL cell lines were obtained from Jon Aster. For various assays described below, as indicated in the figure legends, T-ALL cells were cultured in either 0.5% FBS or 10% FBS-containing medium for sorafenib (Alexis Biochemicals) or ABT-737 (Symansis) treatment. In the case of combined treatment with both sorafenib and ABT-737, T-ALL cells were maintained in 10% FBS-containing medium. Lentiviral shRNA virus packaging, retrovirus packaging, and subsequent infections were performed as described previously 28. For cell viability assays, cells were plated at 10,000 per well in 96-well plates, and incubated with the appropriate medium containing sorafenib, ABT-737 or DMSO for 48 h. Assays were performed with CellTiter-Glo Luminescent Cell Viability Assay kit according to the manufacturer’s instructions (Promega). For detection of apoptosis, cells treated with various drugs were stained with propidium iodide (Roche), or co-stained with Annexin V-PE and 7-amino-actinomycin D (Annexin V-PE Apoptosis Detection Kit I, BD Bioscience) according to the manufacturer’s instructions. Stained cells were sorted with Dako-Cytomation MoFlos sorter (Dako) at the Dana-Farber Cancer Institute FACS core facility.
Immunoblots and Immunoprecipitation
Cells were lysed in EBC (50 mM Tris pH 8.0, 120 mM NaCl, 0.5% NP-40) buffer supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). The protein concentrations of the lysates were measured using the Bio-Rad Bradford protein assay reagent on a Beckman Coulter DU-800 spectrophotometer. The lysates were then resolved by SDS-PAGE and immunoblotted with the indicated antibodies. For immunoprecipitation, 800 µg lysates were incubated with the appropriate antibody (1–2 µg) for 3–4 h at 4 °C followed by one-hour incubation with Protein-A Sepharose beads (GE Healthcare). Immuno-complexes were washed five times with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Quantification of the immunoblot band intensity was performed with Image J software.
Detection of Mcl-1 phosphorylation sites in vivo
To map Mcl-1 phosphorylation status in vivo, 293T cells were transfected with HA-Mcl-1 using the calcium phosphate method. Thirty hours post-transfection, 293T cells were treated with 10 µM MG132 for 16 hours to block the 26S proteasome pathway prior to collecting the whole cell lysates for HA-immunoprecipitation. After extensive washing with NETN buffer, the HA-immunoprecipitates were separated by SDS-PAGE and visualized by colloidal Coomassie Blue. The band containing Mcl-1 was excised and treated with DTT to reduce disulfide bonds and iodoacetamide to derivatize cysteine residues. In-gel digestion of the protein was done using trypsin or chymotrypsin. The resulting peptides were extracted from the gel and analyzed by nanoscale-microcapillary reversed phase liquid chromatography tandem mass spectrometry (LC-MS/MS). Peptides were separated across a 37-min gradient ranging from 4% to 27% (v/v) acetonitrile in 0.1% (v/v) formic acid in a microcapillary (125 µm × 18 cm) column packed with C18 reverse-phase material (Magic C18AQ, 5 µm particles, 200 Å pore size, Michrom Bioresources) and on-line analyzed on The LTQ Orbitrap XL™ hybrid FTMS (Thermo Scientific, Bremen, Germany). For each cycle, one full MS scan acquired on the Orbitrap at high mass resolution was followed by ten MS/MS spectra on the linear ion trap XL from the ten most abundant ions. MS/MS spectra were searched using the SEQUEST algorithm against a database created on the basis of a protein sequence database containing the sequence for Mcl-1, for common contaminants, such as human keratins protein with static modification of cysteine carboxymethylation, dynamic modification of methionine oxidation and serine, threonine and tyrosine phosphorylation. All peptide matches were filtered based on mass deviation, tryptic state, XCorr and dCn and confirmed by manual validation. The reliability of site-localization of phosphorylation events was evaluated using the Ascore algorithm.
Real-time RT-PCR Analysis
RNA was extracted using the Qiagen RNeasy mini kit, and the reverse transcription reaction was performed using the ABI Taqman Reverse Transcriptional Reagents (N808-0234). After mixing the resulting template with Mcl-1 (Hs00172036_m1) or GAPDH (Hs99999905_m1) primers and ABI Taqman Fast Universal PCR Master Mix (4352042), the real-time RT-PCR was performed with the ABI-7500 Fast Real-time PCR system. Fbw7 (Hs00217794_m1), Skp2 (Hs00180634_m1), β-TRCP1 (Hs00182707_m1), Mcl-1 (Hs00172036_m1) and GAPDH (Hs99999905_m1) primers were purchased from ABI.
Protein Degradation Analysis
Cells were transfected with Myc-Mcl-1 along with HA-Fbw7, or Flag-β-TRCP1, and GFP as a negative control, in the presence or absence of HA-GSK3 and/or HA-ERK1. For half-life studies, cycloheximide (20 µg/ml, Sigma) was added to the media 40 h post-transfection. At various time points thereafter, cells were lysed and protein abundances were measured by immunoblot analysis.
In vivo Ubiquitination Assay
Cells were transfected with a plasmid encoding Flag-Ubiquitin along with Myc-Mcl-1 and HA-Fbw7 in the presence or absence of HA-GSK3. Thirty-six hours after transfection, cells were treated with the proteasome inhibitor MG132 (30 µM, Calbiochem) for 6 hours, and then harvested. Anti-Myc immunoprecipitates were recovered and immunoblotted with the anti-Flag antibody. Alternatively, cells were transfected with His-Ubiquitin along with Myc-Mcl-1 and HA-Fbw7 in the presence or absence of HA-GSK3. Thirty-six hours after transfection, cells were harvested, and the lysates were incubated with Ni-NTA matrices (Qiagen) at 4 °C for 12 h in the presence of 8 M Urea pH 7.5. Immobilized proteins were washed five times with 8 M Urea pH 6.3 before being resolved by SDS-PAGE and immunoblotted with the anti-Myc antibody.
In vitro Ubiquitination Assay
The in vitro ubiquitination assays were performed as described previously 8. To purify the SCFFbw7 E3 ligase complex, 293T cells were transfected with vectors encoding GST-Fbw7, HA-Cullin-1, Myc-Skp1 and Flag-Rbx1. The SCFFbw7 E3 complexes were purified from the whole cell lysates using GST-agarose beads. Purified, recombinant GST-Mcl-1 proteins were incubated with purified SCFFbw7 complexes in the presence of purified, recombinant active E1, E2 (UbcH5a and UbcH3), ATP and ubiquitin. The reactions were stopped by the addition of 2X SDS-PAGE sample buffer and the reaction products were resolved by SDS-PAGE gel and probed with the indicated antibodies.
In vitro Kinase Assay
GSK-3 was purchased from New England Biolabs. The in vitro kinase reaction was performed according to the manufacturer’s instructions. Briefly, 5 µg of the indicated GST fusion proteins were incubated with purified active GSK3 in the presence of 5 µCi [γ-32P] ATP and 20 µM cold ATP in the kinase reaction buffer for 20 min. The reaction was stopped by the addition of SDS-containing lysis buffer, resolved on SDS-PAGE, and detected by autoradiography.
Mcl-1 Binding Assays
Binding to immobilized GST proteins was performed as described previously 28. Where indicated, the GST-Mcl-1 proteins were incubated with GSK3 in the presence of ATP for 1 h prior to the binding assays.
Subcellular Fractionation
Mitochondrial and cytosolic (S100) fractions were prepared by resuspending HeLa cells in 0.8 ml ice-cold buffer A (250 mM sucrose, 20mM HEPES [pH 7.4], 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 17 µg/ml phenylmethylsulfonyl fluoride, 8 µg/ml aprotinin, 2 µg/ml leupeptin). Cells were then passed through an ice-cold cylinder cell homogenizer. Unlysed cells and nuclei were pelleted through a 10 min, 750 g spin. The recovered supernatant was spun at 10,000 g for 25 min. This pellet was resuspended in buffer A and represents the mitochondrial fraction. The supernatant was spun at 100,000 g for 1 hr. The supernatant from this final centrifugation represents the S100 (cytosolic) fraction.
Mice
Generation of conditional Fbw7 knockout mice (Lck-Cre/Fbw7fl/fl and Mx1-Cre/Fbw7fl/fl) was described previously 3, 4.
In vivo imaging
CMLT1 cells were infected with lentiviral vectors encoding a shRNA against Mcl-1 (shMcl-1) or an irrelevant control (shGFP). After selection in puromycin 1 µg/ml, cells were engineered for in vivo imaging by transduction with a retrovirus encoding a fusion of firefly luciferase fused to neomycin phosphotransferase, and then selected with G418 at 0.5 mg/ml. After selection, the luciferase activity of each engineered cell line was measured and found to have similar reading. Subsequently, equal numbers of viable cells (0.5–1 × 107 cells) were injected into NSG mice via the lateral tail vein. Tumor burden was determined using bioluminescence imaging (IVIS Spectrum, Caliper Life Sciences) after intraperitoneal injection of D-Luciferin 75 mg/kg. Total body luminescence was quantified using the Living Images software package (Caliper Life Sciences), and are expressed as photons/second/standardized region of interest (ph/s/ROI) encompassing the entire mouse. Data represented as mean ± SEM with statistical significance determined by Student’s t-test.
Supplementary Material
Acknowledgements
We thank Jack Lawler, Christoph Schorl, Qing Zhang, and Susan Glueck for critical reading of the manuscript, James DeCaprio, Mien-Chie Hung, Michelle A. Kelliher, Wade Harper and William Hahn for providing reagents, Lewis Cantley and Alex Toker for helpful suggestions, Ingrid Wertz and Vishva Dixit for sharing unpublished data, and members of the Wei and DePinho labs for useful discussions. W.W. is a Kimmel Scholar and V Scholar. This work was supported in part by the Emerald Foundation New Investigator award (W.W.) and by the Leukemia and Lymphoma Society Special Fellow award (W.W.). RAD is an American Cancer Society Research Professor and supported by the Robert A. and Renee E. Belfer Foundation Institute for Applied Cancer Science.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions:
H.I. performed most of the experiments with the critical assistance from S.S. and D.G.; A.T., L.W. and A.W.L. also helped in performing a portion of the experiments. I.O. performed the Fbw7 conditional knockout mice experiments and A.L.C. performed the orthotopic engraftment mice experiments. B.Z. performed the mass spectrometry analysis and Y.X. and R.S.M. helped to perform the experiments with the tumors derived from the TKO mice. A.G. helped to perform the experiments with the human T-ALL clinical samples. W.W., R.A.D. and K.I.N. designed the experiments with assistance from J.A., J. S., A.L.K., H.I., and T.L. W.W. supervised the study. W.W. wrote the manuscript with help from H.I. and S.S. All authors commented on the manuscript.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Reference
- 1.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 2.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onoyama I, et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuoka S, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008;22:986–991. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koepp DM, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 9.Gupta-Rossi N, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 10.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez I, Yuan J. A convoluted way to die. Neuron. 2001;29:563–566. doi: 10.1016/s0896-6273(01)00232-x. [DOI] [PubMed] [Google Scholar]
- 12.Akgul C. Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell Mol Life Sci. 2009;66:1326–1336. doi: 10.1007/s00018-008-8637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Opferman JT, et al. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 15.Wertz IE, et al. Regulation of antimitotic resistance by Mcl-1 and SCF/Fbw7. 2009 Submitted. [Google Scholar]
- 16.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 17.Ding Q, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijhawan D, et al. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panka DJ, Cho DC, Atkins MB, Mier JW. GSK-3beta inhibition enhances sorafenib-induced apoptosis in melanoma cell lines. J Biol Chem. 2008;283:726–732. doi: 10.1074/jbc.M705343200. [DOI] [PubMed] [Google Scholar]
- 20.Yu C, et al. The role of Mcl-1 downregulation in the proapoptotic activity of the multikinase inhibitor BAY 43–9006. Oncogene. 2005;24:6861–6869. doi: 10.1038/sj.onc.1208841. [DOI] [PubMed] [Google Scholar]
- 21.Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 22.Cragg MS, Harris C, Strasser A, Scott CL. Unleashing the power of inhibitors of oncogenic kinases through BH3 mimetics. Nat Rev Cancer. 2009;9:321–326. doi: 10.1038/nrc2615. [DOI] [PubMed] [Google Scholar]
- 23.Konopleva M, et al. Mechanisms of antileukemic activity of the novel Bcl-2 homology domain-3 mimetic GX15-070 (obatoclax) Cancer Res. 2008;68:3413–3420. doi: 10.1158/0008-5472.CAN-07-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Delft MF, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Schwickart M, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 27.Popov N, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 28.Gao D, et al. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol. 2009;11:397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benmaamar R, Pagano M. Involvement of the SCF complex in the control of Cdh1 degradation in S-phase. Cell Cycle. 2005;4:1230–1232. doi: 10.4161/cc.4.9.2048. [DOI] [PubMed] [Google Scholar]
- 30.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.