Abstract
Objective:
Urothelial carcinoma (UC) of the kidney is a relatively rare but aggressive form of kidney cancer. Differential diagnosis of renal UC from renal cell carcinoma (RCC) can be difficult, but is critical for correct patient management. We aimed to use global gene expression profiling to identify genes specifically expressed in urothelial carcinoma (UC) of the kidney, with purpose of finding new biomarkers for differential diagnosis of UC of both upper and lower tract from normal tissues.
Materials and methods:
Microarray gene expression profiling was performed on a variety of human kidney tumor samples, including clear cell, papillary, chromophobe, oncocytoma, renal UC and normal kidney controls. Differentially expressed mRNAs in various kidney tumor subtypes were thus identified. Protein expression in human UC tumor samples from both upper and lower urinary tract was evaluated by immunohistochemistry.
Results:
FXYD3 (MAT-8) mRNA was specifically expressed in UC of the kidney and not in normal kidney tissue or in any RCC tumor subtypes. FXYD3 mRNA levels displayed equal or better prediction rate for the detection of renal UC than the mRNA levels of selected known UC markers as p63, vimentin, S100P, KRT20 and KRT7. Finally, immunohistochemical staining of clinical UC samples showed that FXYD3 protein is overexpressed in majority of UC of the upper genitourinary tract (encompassing the kidney, ∼90%) and in majority of high grade bladder UC (∼84%, it’s <40% in low grade tumors, P < 0.001) compared to normal kidney and bladder tissues.
Conclusion:
FXYD3 may be a promising novel biomarker for the differential diagnosis of renal UC and a promising prognosis marker of UC from bladder. Because it was identified genome-widely, FXYD3 may have important biological ramifications for the genetic study of UC.
Keywords: FXYD3, FXYD, urothelial carcinoma, kidney, marker, microarray
Introduction
Urothelial carcinoma (UC) of the kidney, also known as transitional cell carcinoma, or urothelial cell carcinoma of upper urinary tract (UCC-UUT), is a relatively uncommon form of kidney cancer arising from the urothelium lining of the renal pelvis and calyces. UC accounts for the majority of bladder cancer; however, it only accounts for about 7% of renal neoplasms.1 UC of the renal pelvis is an aggressive tumor, which may invade the renal parenchyma, mimicking primary renal cell carcinoma (RCC).2 Similarly, advanced RCC can invade the pelvicalceal system. This can make differential diagnosis of RCCs and UC of the renal pelvis difficult. Correct diagnosis is critical for determining appropriate surgery and post- surgical treatments. For instance, UC of the upper urinary tract including renal pelvis, calyces and ureters will require radical nephrectomy with ureterectomy and bladder cuff resection. However, RCC will require only partial or radical nephrectomy without extensive ureter resection. Therefore, identifying markers that can distinguish UC from RCC is of great importance.3 A recent study shows that UC behaves identically in the upper and lower urinary tracts when stage and grade are considered. 4 We recently compared the gene expression profiles between upper and lower urinary tract UC, concluded that no significant profile differences exist between them.5 Therefore, any biomarkers for lower tract UC then will most likely be a biomarker for the upper tract UC.
Known diagnosis biomarkers for UCs are p63,6 vimentin7 and S100P.1 Phe et al studied the methylated genes as biomarkers in urothelial cell carcinomas of the urinary tract.8 p63 and S100P are UC specific biomarkers that are expressed in UC of the kidney, but not in other renal tumors. Vimentin, on the other hand, is down regulated in UC but up regulated in most RCCs. Other studies have shown that various cytokeratins can be used as effective biomarkers to distinguish between UC and RCC. Han and Duszak stated that coexpression of CK7 and CK20 is a feature of UC that can be used to properly identify and diagnose UC with extensive renal involvement.9 However, as far as the authors know, identifying the differential biomarkers at genome-wide level has not been conducted in the literature.
We aimed to identify genes that are highly specific in one type or subtype of kidney cancer and thus can serve as molecular biomarkers and diagnostic tools. In this study, we examined the gene expression profiles of 123 tumors of different subtypes and/ or normal kidney tissue samples with the goal of finding genes that are specific for UC of the kidney. The identified genes could have important biological ramifications, because of the ways it was identified genome-widely.
Materials and Methods
Case selection for gene expression profiling study
We have previously established gene expression data from 45 clear cell, 15 papillary and l9 chromophobe RCCs; 14 UCs of the kidney; 15 oncocytoma and 15 normal kidney tissue specimens as described previously. 10 In total, 123 samples of different kidney tumor subtypes and normal tissues were studied (Table 1). These specimens were obtained from Spectrum Health Hospital of Grand Rapids, MI, the Cooperative Human Tissue Network (CHTN), Baylor University Medical Center at Dallas. TX, the University of California, Los Angeles (UCLA), the French Kidney Cancer Study Group, and at Johns Hopkins University in Baltimore. MD. Patients’ clinicopathological information for the 14 UCs from renal pelvis was listed in Table 2. Among them, 8 samples with missing Grade information were due to the fact that they came from the same cancer center (French) that we were unable to trace the sample information back at this time other than that the patients was diagnosed with upper tract urothelial carcinoma at treatment time. Apart from them, all the rest samples showed high to modest grade information.
Table 1.
Summary of tumor samples analyzed in the gene expression experiments and IHC study.
|
Tissue samples in gene expression experiments |
Tissue samples used in IHC study |
||||
|---|---|---|---|---|---|
| Subtypes | Labels | Number of samples | Organ | Grade | #Samples |
| Clear cell RCC | CC | 45 | Bladder | Low | 33 |
| Chromophobe RCC | CH | 19 | High | 37 | |
| Normal kidney | NO | 15 | Total | 70 | |
| Oncocytoma | ON | 15 | Upper GU | Low | 18 |
| Papillary RCC | PA | 15 | High | 11 | |
| Urothelial carcinoma from kidney pelvis | UC | 14 | Total | 29 | |
| Total | 123 | Total | 99 | ||
| Total in all studies: | 222 | ||||
Abbreviation: Upper GU, upper genitourinary tract.
Table 2.
Patients’ clinicopathological information for the 14 UCs in the microarray study.
| PID | Age | Gender | Ethnicity | Diagnosis | Grade | Tumor size(cm) | Remarks |
|---|---|---|---|---|---|---|---|
| rUC007 | rUC | # | |||||
| rUC011 | rUC | # | |||||
| rUC014 | rUC | # | |||||
| rUC009 | rUC | # | |||||
| rUC012 | rUC | # | |||||
| rUC016 | rUC | # | |||||
| rUC010 | rUC | # | |||||
| rUC013 | rUC | # | |||||
| rUC023 | rUC | 2–3 | 4.5 | ||||
| rUC409 | 63 | M | W | Papillary rUC | 3 | 5.3 | * |
| rUC226 | 76 | F | rUC | high | 3.5 | ||
| rUC173 | 71 | M | W | rUC | high | 3 | * |
| rUC238 | 76 | F | rUC | high | 4.5 | ||
| rUC214 | 52 | M | W | rUC | high | 10 |
Note:
Indicating the samples being misclassified consistently in our study; rUC, Urothelial carcinoma from renal pelvis;
Indicating the patient’s information was missing and cannot be traced back at this time.
Examination of gene expression profiles in kidney tumors
All tissue was snap frozen in liquid nitrogen immediately following surgery and stored at −80 until extraction. The samples were homogenized in TRIZOL Reagent (Invitrogen, Carlsbad, CA). Ten micrograms of total RNA was processed for the expression microarrays using the Affymetrix GeneChip one- cycle target labeling kit (Affymetrix, Santa Clara, CA) according to the manufacturer’s recommended protocols. The resultant biotinylated cDNA was fragmented and then hybridized to the GeneChip human genome (54,675 probe sets in total, including more than 35,000 human genes; Affymetrix). The arrays were washed, stained, and scanned using the Affymetrix Model 450 Fluidics Station and Affymetrix Model 3000 scanner using the manufacturer’s recommended protocols. The scanned data was obtained by the Affymetrix GeneChip Operating Software (GCOS) version 1.4.
Evaluation and normalization of Affymetrix GeneChip data
Expression values were generated by using Microarray Suite v5.0 software (Affymetrix). The probes were filtered according to a new study.11 The hybridizations were normalized by using the robust multichip averaging (rma) algorithm to obtain a summary expression level for each retained gene. This resulted in more than 17,000 gene expression levels per sample, each of which then had one numeric value to represent its relative expression intensity across the study cohort.
Immunohistochemical staining
Paraffin-embedded tissue sections of 99 UCs were collected for this study from three participating institutions (Geisinger Health System, Tian-Jin Cancer Hospital and Northwestern Memorial Hospital) with Institutional Review Board approvals from each. These tissue samples represented a different cohort of samples from those used for the microarray study. Among them, 29 were from upper genitourinary tract (GU) and 70 were from the bladder. We graded them using the 2004 WHO classification.12 18 (upper tract) and 33 (bladder) were diagnosed as low-grade UC and the remaining were classified as high-grade UC (Table 1). Immunohistochemistry was carried out on all the cases as previously described.10,13–15 Briefly, 4 μm tissue sections were subjected to immunohistochemistry using a rabbit polyclonal antibody specific for FXYD3 (Sigma Aldrich, St. Louis, MO) at 1:50 dilution. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. Antigen retrieval was carried out in citrate buffer (10 mM, pH = 9) for 20 minutes at 100 °C in a microwave oven. The primary antibody was applied for 1 hour at room temperature. A subsequent reaction was performed with secondary mouse anti-rabbit antibody and biotin-free HRP enzyme labeled polymer of the EnVision-plus detection system (DAKO, Carpinteria, CA). A positive reaction was visualized with diaminobenzydine solution followed by counterstaining with hematoxylin. The staining was interpreted as either positive or negative by urologic pathologists.
Statistical methods
Fold change of gene expression levels between two conditions was defined to be the ratio of the mean values of expression levels of one condition over the other, if the ratio was greater than 1; or the negative reverse of the ratio if otherwise. Fisher’s exact test was used to test the independence between variables representing the column and row of contingency tables. The two tailed studentized t-test was used to test the mean differences between two groups. Linear models for microarray data (limma) were used to find differential genes between the tumor types. If there were more than two tumor types to be compared, all pairs were included in the fitting model (specified in the contrast matrix for the model). The genes were sorted by their significance of F-test. Coefficient variation (CV) of a numeric vector was defined to be its standard deviation (SD) divided by its mean. Supervised clustering was based on the limma selected genes. Unsupervised clustering was based on genes where the interquartile range (IQR) was greater than a specific value (0.5 in our case) and the CV was greater than a specific value (0.06 in our case). In all clustering studies, a hierarchical algorithm was used with the Euclidean distance for dissimilarities between the data samples. The slightly modified “plot.phylo” function from R package analysis of phylogenetics and evolution (ape) was used to plot the clustering results. The numbers of genes used were arbitrarily chosen to be 500. The Pearson’s method was used to calculate the correlations. All data analyses were programmed using R platform of version 2.6.2 or higher. A gene for a tumor sample was considered over- or under-expressed only if its expression intensity was at least three SDs away from the mean value of gene intensities from the normal references. A P-value of 0.05 or less was considered to be significant throughout the paper.
Results
Supervised and unsupervised clustering based on gene expression profiling
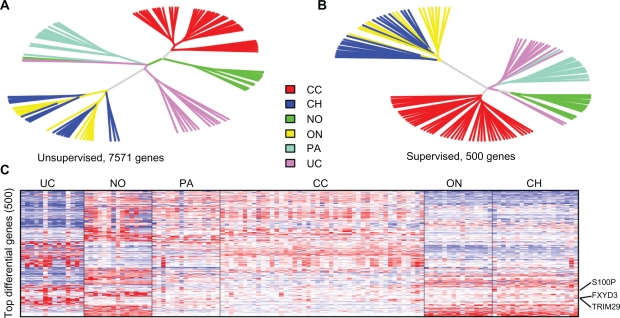
The filtered genes by IQR and CV were used to perform unsupervised clustering (–7500 genes, Fig. 1A), and the top 500 genes identified by the limma method were used to do a supervised clustering study of the cohort (Fig. 1B). Both approaches showed that 12 out of 14 UCs were tightly clustered together, distinct from the rest of the samples of other tumor subtypes or normal samples, indicating that UC of the kidney has a very unique gene expression profile from other renal cell neoplasm subtypes. They also indicated the potential usefulness of gene expression profiling in identifying biomarkers that are specific to renal UC. The two misclassified samples were both identified to be white male with high grade tumors, but with one of them having papillary elements (rUC 173 and rUC 409, see Table 2). We do not yet know exactly why their gene expression profiles were so different from the rest of the UCs. These 14 UC samples have been intensively studied in5 with the two misclassified samples consistently being misclassified in the study. The heatmap for the top 100 genes identified by limma method was shown in Figure 1C with FXYD3 being one of them. More details of its gene expression patterns will be discussed below.
Figure 1.
Unsupervised A) and supervised B) clustering on selected genes. The supervised clustering was based on selected genes most differentiable between the tumor types, while the unsupervised clustering was based on the filtered genes with maximal values of IQR and CV values among the study cohort. Most of the samples were clustered together correctly. UC is shown to have a particularly unique gene expression profile. C) Heatmap of the top differential genes between UC samples and normal references. FXYD3 is one of the top 3 genes shown in the heatmap. The other two are S100P and TRIM29, also shown in the plot.
Abbreviations: No, normal; UC, urothelial; PA, papillary; ON, oncocytoma; CH, chromophobe; CC, clear cell RCC.
Patterns of FXYD gene expressions in different renal tumors
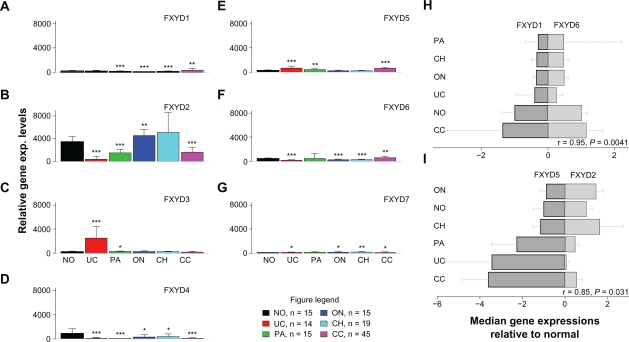
Microarray gene expression profiling identified the FXYD3 gene (also known as MAT-8) as one of the top three genes most differentially expressed between UC of the kidney and normal kidney tissue (average fold change = 10.5, P = 0.0006, Table 3), along other genes with similar gene expression patterns to that of FXYD3; these genes include S100P and TRIM29, as were marked in Figure 1C. S100P in UC has been studied in.1 TRIM29 also known as ATDC gene, was studied in lung cancer in.16 Among all kidney tumor subtypes and normal kidneys, FXYD3 is expressed in UC only (see below). It is a member of the FXYD family of proteins, which have been implicated in regulation of the Na, K+ ATPase and are characterized by a conserved FXYD motif.17–19 Based upon FXYD3’s up regulated expression in renal UC and its possible roles in UC tumors, we carefully examined the gene expression patterns of all FXYD gene members in different kidney tumors (Fig. 2). We found that FXYD3 expression was highly specific to UC of the kidney; it was not expressed in all other types of renal tumors and normal renal tissues (Fig. 2C). This UC specificity was not seen for the other FXYD family members. Interestingly, FXYD1 was specifically up regulated in clear cell renal cell carcinoma (Fig. 2A), while the other FXYD family members had variable gene expression patterns in the different kidney tumor subtypes (Fig. 2).
Table 3.
Comparison of FXYD3 mRNA with other mRNAs identified as potential biomarkers for urothelial carcinoma of the renal pelvis.
|
Fold changes to normal |
Significant cases1 | UC vs. normal2 | Significance3 | UC vs. RCC2 | Significance3 | |||
|---|---|---|---|---|---|---|---|---|
| Mean | Min | Max | ||||||
| FXYD3 | 10.5 | 1.07 | 30.4 | 13/14 (93%) | 0.00062 | *** | 0.00058 | *** |
| p63 | 4.81 | 1.37 | 13.3 | 12/14 (86%) | 0.0036 | ** | 0.0034 | ** |
| KRT20 | 37 | –1.36 | 181 | 7/14 (50%) | 0.046 | * | 0.046 | * |
| KRT7 | 11.2 | –4.92 | 28.4 | 10/14 (71%) | 0.00088 | *** | 0.00048 | *** |
| S100P | 107 | 3.9 | 275 | 12/14 (85.7%) | 0.0011 | ** | 0.0011 | ** |
| Vimentin | –3.61 | –10.2 | 2.07 | 0/14 (0%) | 0.032 | * | <0.0001 | *** |
Notes:
Significant cases were defined to be at least 3 SDs away from normal mean expressions;
P-values were obtained using two tailed student t-test;
Significance codes: 0–0.001 = ***, 0.001–0.01 = **, 0.01–0.05 = *, 0.05–0.1 = ., 0.1–1 = “ ”.
Figure 2.
Gene expression profiles of genes in the FXYD family and correlations. (A–G): gene expression profiling of normal and kidney tumors for genes FXYD1-FXYD7. Mean + SD in each group was shown. FXYD3 is the only gene that is UC specific. (H, I): Correlation of FXYD gene expression patterns in kidney tumors. The median values were used in each group. The correlation coefficients (r) and P-values were based on the Pearson method. Standard deviations were added in the plot. H) Significant positive correlation of gene expression patterns was seen between FXYD1 and FXYD6. I) Significant negative correlation of gene expression patterns was seen between FXYD2 and FXYD5. No other pairs in the family were found to be significantly correlated.
Notes: Significance codes: 0–0.001 = ***, 0.001–0.01 = **, 0.01–0.05 = *, 0.05–0.1 = ., 0.1–1 = “ ”.
Abbreviations: No, normal; UC, urothelial; PA, papillary; ON, oncocytoma; CH, chromophobe; CC, clear cell RCC.
Correlation between gene expression patterns of the FXYD family genes in renal tumors
FXYD1 and FXYD6 displayed a positive correlation (r = 0.91, P = 0.015, Fig. 2H) while FXYD2 and FXYD5 displayed an inverse correlation (r = −0.866, P = 0.024, Fig. 2I) between the medians of gene expression levels among the studied RCC tumor types. No genes in the FXYD family were found to be significantly correlated with FXYD3, indicating the unique role it plays in UC. The negative correlation between FXYD2 and FXYD5 could possibly reflect negative feedback regulation between these two genes. Further study will be needed in this regard.
Comparison of FXYD3 with other genes as biomarkers of UC
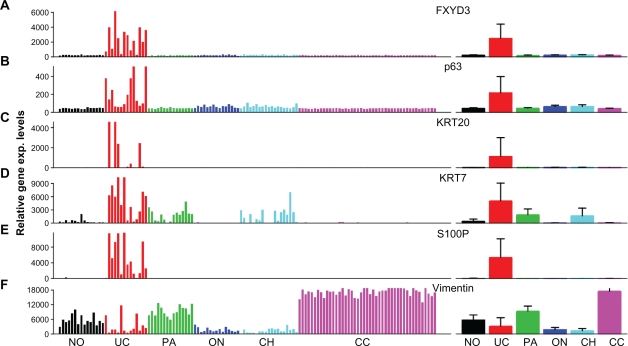
The high specificity of FXYD3 expression to renal UC, but not to normal kidney tissue or other kidney tumor subtypes, suggests possible application of FXYD3 as a biomarker for the differential diagnosis of UC of the kidney from other forms of kidney cancer. To investigate this possibility, we selected five other potential biomarkers for comparison at the mRNA level: p63, vimentin, KRT7, KRT20 and S100P. Apart from vimentin, which was down regulated in UC, the other genes were all up regulated in UC of the kidney and showed some degree of UC specificity (Fig. 3). However, we noted that these biomarkers displayed more variation in expression among the cohort of UC samples than did FXYD3. 93% of renal UC samples overexpressed FXYD3, while 86%, 50%, 71% and 85.7% of samples overexpressed p63, KRT20, KRT7 and S100P, respectively (Table 3, see section Statistical Methods for calculating over or under expressed). KRT7 was also less specific to UC, as it was overexpressed in some papillary and chromophobe RCCs (Fig. 3D). We also note that p63 showed a much smaller average fold change in mRNA expression between UCs and normal tissue (4.81 for p63 vs. 10.5 for FXYD3, Table 3. A formal student’s t-test for the ability of these genes to differentiate between normal tissue and renal UC, and between renal UC and other renal carcinomas showed that FXYD3 performs at least as well as the other genes (Table 3). In summary, FXYD3 appeared to be more specific for UC of the kidney than the other tested biomarkers.
Figure 3.
Gene expression profiling of genes identified to be potential biomarkers. Left: individual gene expression intensity plot. Right: mean intensity + SD plot. A) FXYD3 over expression is highly specific to UC, with at least a 3-fold increase over normal kidney in 13 out of 14 (92.9%) cases tested. B) Overexpression of p63 was detected in 7 of 14 (50%) UC samples. Overexpression is specific to UC, but not as consistent as FXYD3 overexpression. C) Overexpression of keratin 20 (KRT20) in UC. D) Overexpression of keratin 7 (KRT7) in UC. E) Overexpression of S100P was detected in 12 of 14 (85.7%) UC samples. Overexpression is highly specific to UC, comparable to FXYD3 overexpression. F) Vimentin was down regulated in the majority of UC cases, whereas it was up regulated in most clear cell and papillary RC cases. However, down-regulation was also observed in most chromophobe RCC and oncocytoma cases, limiting its use as a UC specific biomarker.
Abbreviations: CC, clear cell; PA, papillary; ON, oncocytoma; Ch, chromophobe; No, normal; UC, urothelial carcinoma.
FXYD3 immunohistochemistry in UC from bladder and from kidney pelvis
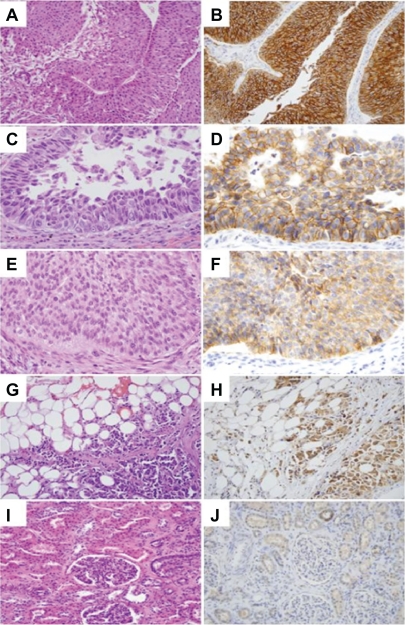
Immunohistochemical analysis was performed to evaluate the levels of FXYD3 in UC tissue sections from both the upper genitourinary tract and the bladder. The majority of UCs, regardless of origin, stained positive for FXYD3 (70/99, >70%). About 90% of UCs from renal pelvis stained positive for FXYD3, while this proportion decreased to about 63% of stained UCs from bladder. In contrast, normal kidney and bladder stromal tissues showed no staining (Figs. 4 and 5; Table 4). The benign urothelium showed mild to moderate staining. When the samples were divided into either high or low grade groups, there was a significant correlation between staining patterns (positive/negative) and histological grade (high/low) for the samples from bladder UC only (P < 0.0001, Table 4). Higher FXYD3 activities are strongly associated with higher tumor grade, suggesting a role for FXYD3 in tumor development and progression of bladder UC (Fig. 5). We concluded from this study that FXYD3 could play different roles in UCs from kidney pelvis and from bladder. It can be used to be an effective gene marker to distinguish between UC of kidney and normal tissues from kidney. It may have prognostic values in UC tumors from bladder.
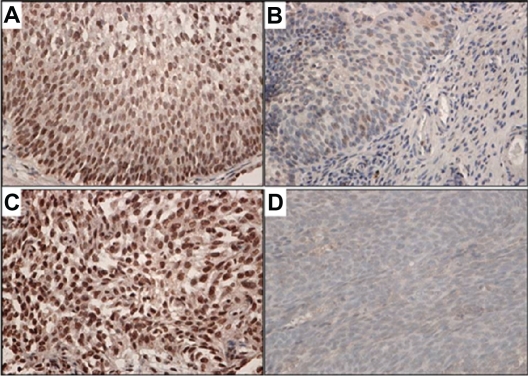
Figure 4.
FXYD3 immunohistostaining for UC of the renal pelvis. A, C, E, G, and I: H and E staining; B, D, F, H and J: FXYD3 staining. A, B) Low grade UC showing strong membranous and cytoplasmic staining. C, D) Area of high grade UC component showing strong membranous staining. E, F) Area of low grade UC component showing weak cytoplasmic staining. G, H) High grade UC cells invading perinephric fat showing strong FXYD3 staining. I, J) Normal appearing renal cortex showing no staining in kidney parenchyma including glomeruli and proximal tubules, while focal FXYD3 staining is occasionally seen in normal distal tubules.
Figure 5.
FXYD3 immunohisto staining for UC from bladder in A) low grade and C) high grade bladder UC in comparison with paired benign urothelium B) and D).
Table 4.
FXYD3 immunohistostaining results.
| Locations |
Positive/Total (%) |
Stat. | P1 | Signif.2 | |||
|---|---|---|---|---|---|---|---|
| Low grade | High grade | Total | |||||
| Upper GU | TMA3 | 13/14(92.9) | 6/6 (100) | 19/20 (95) | 0.451 | 0.502 | |
| Slides | 4/4 (100) | 3/5 (60) | 7/9 (77.8) | 2.06 | 0.151 | ||
| Sum | 17/18 (94.4) | 9/11 (81.8) | 26/29 (89.7) | 1.17 | 0.279 | ||
| Bladder | 13/33 (39.4) | 31/37 (83.8) | 44/70 (62.9) | 14.7 | 0.00012 | *** | |
| Sum in all locations | 30/51 (58.8) | 40/48 (83.3) | 70/99 (70.7) | 7.17 | 0.0074 | ** | |
Notes:
P-value was obtained using Fisher’s exact test on the contingency table of high/low grade and positive/negative staining;
Significance code: 0–0.001 = ***, 0.001–0.01 = **;
TMA = Tissue microarray.
Discussion
We have previously demonstrated a difference in gene expression profiles between renal pelvic UC and RCC.20 Here with more cases in our study, we report here that UC of the renal pelvis shows a very distinct genetic signature from that of other types of renal epithelial tumors. This difference can be exploited to identify biomarkers specific to the disease to aid in diagnosis and distinguish between RCC and UC of the kidney. Using this approach, we identified the FXYD3 gene as a marker that is specifically overexpressed at both the mRNA and protein levels in renal UC compared to normal kidney tissue. Moreover, comparison of FXYD3 with other known renal UC markers showed that FXYD3 mRNA levels displayed equal or better sensitivity and specificity for the detection of renal UC than did the mRNA levels of such known UC markers as p63, vimentin, KRT20 KRT7 and S100P. As a comparison study, we found that FXYD3 may serve as a prognostic marker for UC from bladder. This suggests different roles that FXYD3 plays in UCs from the two locations.
FXYD3 is a member of the FXYD family of ion transport regulatory proteins. These transmembrane proteins are characterized by a conserved FXYD motif and have been shown to associate with and modulate the function of Na,K ATPase. FXYD3 has also been suggested to act as a chloride channel or chloride channel regulator.21 Interestingly, FXYD3 has been found to be overexpressed in primary human breast cancer tumors,21 cell lines,22 androgen-dependent prostate cancer,23 and pancreatic cancers.19 Furthermore, siRNA-mediated knockdown of FXYD3 in separate experiments showed reduction in the proliferative activity of both PC-3 and LNCaP prostate cancer cells24 and T3M4 pancreatic cancer cells25 in vitro. These findings suggest a potential role for FXYD3 in the development and progression of certain cancers. Our finding that FXYD3 is overexpressed in the majority of UCs of bladder and kidney is consistent with a role for this protein in cancer development and is consistent to a recent study in,26 in which the authors studied gene expression patterns of a group of selected genes with FXYD3 included, on 86 upper tract UCs. They did not find prognostic significance in the study cohort. Interestingly, we note that FXYD3 protein expression is correlated with increased tumor grade in bladder UC, but not in UC of the renal pelvis/ calices. In view of the conclusion that gene expression profiles between the two locations are highly similar,5 our study here points to a subtle molecular differences in UC of different origins. The strong correlation of FXYD3 protein expression with high grade may suggest a potential prognostic role for FXYD3 in bladder UC, though further studies using patient survival data are required to establish this. The functional significance of FXYD3 expression in UC will also require further study.
Conclusions
Our results indicate that FXYD3 is a promising new biomarker for UC, particularly useful for the differential diagnosis of UC of the upper urinary tract from renal cell carcinoma. Overexpression of FXYD3 in UC also suggests that this protein may have a promoting role in the development and progression of UC in bladder.
Acknowledgments
We thank Vanessa Fogg and Sabrina Noyes for technical editing and proofreading during the writing of this manuscript.
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Higgins JP, et al. Placental S100 (S100P) and GATA3: markers for transitional epithelium and urothelial carcinoma discovered by complementary DNA microarray. Am J Surg Pathol. 2007;31(5):673–80. doi: 10.1097/01.pas.0000213438.01278.5f. [DOI] [PubMed] [Google Scholar]
- 2.Phatak S, Kolwadkar P. Renal and ureteral transitional cell carcinoma. A case report. 2006;16(4):907–9. [Google Scholar]
- 3.Kirkali Z, Tuzel E. Transitional cell carcinoma of the ureter and renal pelvis. Crit Rev Oncol Hematol. 2003;47(2):155–69. doi: 10.1016/s1040-8428(03)00079-9. [DOI] [PubMed] [Google Scholar]
- 4.Catto JW, et al. Behavior of urothelial carcinoma with respect to anatomical location. J Urol. 2007;177(5):1715–20. doi: 10.1016/j.juro.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, et al. Comparative gene expression profiling analysis of urothelial carcinoma of renal pelvis and bladder. BMC Medical Genomics. 2010 doi: 10.1186/1755-8794-3-58. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langner C, et al. P63 immunoreactivity distinguishes upper urinary tract transitional-cell carcinoma and renal-cell carcinoma even in poorly differentiated tumors. J Histochem Cytochem. 2003;51(8):1097–9. doi: 10.1177/002215540305100813. [DOI] [PubMed] [Google Scholar]
- 7.Skinnider BF, et al. Distribution of cytokeratins and vimentin in adult renal neoplasms and normal renal tissue: potential utility of a cytokeratin antibody panel in the differential diagnosis of renal tumors. Am J Surg Pathol. 2005;29(6):747–54. doi: 10.1097/01.pas.0000163362.78475.63. [DOI] [PubMed] [Google Scholar]
- 8.Phe V, Cussenot O, Roupret M. Interest of methylated genes as biomarkers in urothelial cell carcinomas of the urinary tract. BJU Int. 2009;104(7):896–901. doi: 10.1111/j.1464-410X.2009.08696.x. [DOI] [PubMed] [Google Scholar]
- 9.Han AC, Duszak R., Jr Coexpression of cytokeratins 7 and 20 confirms urothelial carcinoma presenting as an intrarenal tumor. Cancer. 1999;86(11):2327–30. [PubMed] [Google Scholar]
- 10.Yang XJ, et al. Classification of renal neoplasms based on molecular signatures. J Urol. 2006;175(6):2302–6. doi: 10.1016/S0022-5347(06)00255-2. [DOI] [PubMed] [Google Scholar]
- 11.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):el75. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauter G, et al. Noninvasive urothelial neoplasias: WHO classification of noninvasive papillary urothelial tumors. In: Eble JN, Epstein JI, Sesterhenn I, editors. World Health Organization classification of tumors. Pathology and genetics of tumors of the urinary system and male genital organs. Lyon: IARCC Press; 2004. p. 110. [Google Scholar]
- 13.Yang XJ, et al. A molecular classification of papillary renal cell carcinoma. Cancer Res. 2005;65(13):5628–37. doi: 10.1158/0008-5472.CAN-05-0533. [DOI] [PubMed] [Google Scholar]
- 14.Tretiakova MS, et al. Expression of alpha-methylacyl-CoA racemase in papillary renal cell carcinoma. Am J Surg Pathol. 2004;28(1):69–76. doi: 10.1097/00000478-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Chuang ST, et al. Over expression of insulin-like growth factor binding protein 3 in clear cell renal cell carcinoma. J Urol. 2008;179(2):445–9. doi: 10.1016/j.juro.2007.09.106. [DOI] [PubMed] [Google Scholar]
- 16.Ring BZ, et al. A novel five-antibody immunohistochemical test for subclassification of lung carcinoma. Mod Pathol. 2009;22(8):1032–43. doi: 10.1038/modpathol.2009.60. [DOI] [PubMed] [Google Scholar]
- 17.Crambert G, et al. FXYD3 (Mat-8), a new regulator of Na,K-ATPase. Mol Biol Cell. 2005;16(5):2363–71. doi: 10.1091/mbc.E04-10-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geering K. FXYD proteins: new regulators of Na-K-ATPase. Am J Physiol Renal Physiol. 2006;290(2):F241–50. doi: 10.1152/ajprenal.00126.2005. [DOI] [PubMed] [Google Scholar]
- 19.Sweadner KJ, Rael E. The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics. 2000;68:41. doi: 10.1006/geno.2000.6274. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi M, et al. Molecular subclassification of kidney tumors and the discovery of new diagnostic markers. Oncogene. 2003;22(43):6810–8. doi: 10.1038/sj.onc.1206869. [DOI] [PubMed] [Google Scholar]
- 21.Morrison BW, et al. Mat-8, a novel phospholemman-like protein expressed in human breast tumors, induces a chloride conductance in Xenopus oocytes. J Biol Chem. 1995;270:2176. doi: 10.1074/jbc.270.5.2176. [DOI] [PubMed] [Google Scholar]
- 22.Schiemann SSM, Brunner N, Weidle UH. Molecular analysis of two mammary carcinoma cell lines at the transcriptional level as a model system for progression of breast cancer. Clin Exp Metastasis. 1988;16:129–39. doi: 10.1023/a:1021941203905. [DOI] [PubMed] [Google Scholar]
- 23.Vaarala MH, et al. Differentially expressed genes in two LNCaP prostate cancer cell lines reflecting changes during prostate cancer progression. Lab Invest. 2000;80(8):1259–68. doi: 10.1038/labinvest.3780134. [DOI] [PubMed] [Google Scholar]
- 24.Grzmil M, et al. Up-regulated expression of the MAT-8 gene in prostate cancer and its siRNA-mediated inhibition of expression induces a decrease in proliferation of human prostate carcinoma cells. Int J Oncol. 2004;24(1):97–105. [PubMed] [Google Scholar]
- 25.Kayed H, et al. FXYD3 is overexpressed in pancreatic ductal adenocarcinoma and influences pancreatic cancer cell growth. Int J Cancer. 2006;118(1):43–54. doi: 10.1002/ijc.21257. [DOI] [PubMed] [Google Scholar]
- 26.Izquierdo L, et al. Molecular characterization of upper urinary tract tumours. BJU Int. 2010;106(6):868–72. doi: 10.1111/j.1464-410X.2009.09135.x. [DOI] [PubMed] [Google Scholar]